Abstract
Caenorhabditis elegans mutants deleted for TDP-1, an ortholog of the neurodegeneration-associated RNA-binding protein TDP-43, display only mild phenotypes. Nevertheless, transcriptome sequencing revealed that many RNAs were altered in accumulation and/or processing in the mutant. Analysis of these transcriptional abnormalities demonstrates that a primary function of TDP-1 is to limit formation or stability of double-stranded RNA. Specifically, we found that deletion of tdp-1: (1) preferentially alters the accumulation of RNAs with inherent double-stranded structure (dsRNA); (2) increases the accumulation of nuclear dsRNA foci; (3) enhances the frequency of adenosine-to-inosine RNA editing; and (4) dramatically increases the amount of transcripts immunoprecipitable with a dsRNA-specific antibody, including intronic sequences, RNAs with antisense overlap to another transcript, and transposons. We also show that TDP-43 knockdown in human cells results in accumulation of dsRNA, indicating that suppression of dsRNA is a conserved function of TDP-43 in mammals. Altered accumulation of structured RNA may account for some of the previously described molecular phenotypes (e.g., altered splicing) resulting from reduction of TDP-43 function.
Keywords: neurodegeneration, RNA editing, RNA structure, splicing
Introduction
TAR-DNA binding protein 43 (TDP-43) was originally discovered as a negative regulator of HIV DNA transcription (Ou et al, 1995). Subsequently, TDP-43 was identified as a major component of aggregates in a variety of degenerative neurological conditions including amyotrophic lateral sclerosis (ALS) and frontotemporal lobar dementia (FTLD) (Chen-Plotkin et al, 2010). In affected tissues, TDP-43 is disrupted, with the formation of large, insoluble cytoplasmic aggregates and concurrent nuclear depletion. While mutations in TDP-43 have been identified in a small number of familial cases of ALS (Pesiridis et al, 2009), mislocalized wild-type TDP-43 appears in the vast majority of sporadic ALS and FTLD-affected tissue and in a significant percentage of other neurological disorders. These observations suggest that the wild-type function of TDP-43 is central to the disease cascade.
TDP-43 is a ubiquitously expressed RNA/DNA-binding protein containing two RNA Recognition Motifs (RRMs) that allow binding to single-stranded RNA as well as single- and double-stranded DNA (Ayala et al, 2005). TDP-43 proteins preferentially associate with (UG)n repeats in RNA, but this sequence is neither necessary nor sufficient for TDP-43 association in vivo (Buratti & Baralle, 2001; Polymenidou et al, 2011). TDP-43 is involved in many RNA-related processes including transcription, pre-mRNA splicing, mRNA stability, and micro-RNA biogenesis (Da Cruz & Cleveland, 2011). Several recent studies have characterized neuronal transcripts affected by TDP-43 (Polymenidou et al, 2011; Tollervey et al, 2011). These studies indicate that TDP-43 preferentially binds long intronic regions in pre-mRNA and maintains these transcripts by an unknown mechanism. Alternative splicing analysis in several TDP-43 knockdown models indicates TDP-43 functions to ensure correct alternative splicing of pre-mRNAs (Polymenidou et al, 2011; Hazelett et al, 2012). However, changes in splicing are not observed in the majority of transcripts with decreased levels. Therefore, the reason TDP-43 is required to maintain the levels of certain transcripts is unknown.
Knockout of TDP-43 in most model organisms is lethal or results in severe phenotypes (Feiguin et al, 2009; Sephton et al, 2010). Surprisingly, complete deletion of the C. elegans ortholog, TDP-1, causes minor defects (Zhang et al, 2012). Despite the lack of severe phenotypes in the tdp-1 mutant, TDP-1 has molecular properties similar to its mammalian homolog. TDP-1 binds the canonical TDP-43 binding sequence [(UG)n] with high affinity (Ayala et al, 2005), and we have shown that TDP-1 can substitute for human TDP-43 in in vivo splicing assays (Ash et al, 2010). These results imply that while tdp-1 loss of function may be less consequential in the worm, TDP-1's basic molecular roles are likely conserved.
In this study, we investigated the role of TDP-1 on the transcriptome. We discovered that TDP-1 functions to maintain the amount of mature RNA transcripts originating from potentially double-stranded precursor RNAs and to limit nuclear dsRNA accumulation in multiple tissues. Immunoprecipitation using a dsRNA-specific antibody revealed that tdp-1 mutant animals accumulate a variety of double-stranded transcripts indicating a global effect of TDP-1 on RNA structure or stability. Analysis of TDP-1 binding by deep sequencing of anti-TDP-1 chromatin immunoprecipitation (ChIP) indicated that TDP-1 associates with highly structured regions co-transcriptionally. The reduction of dsRNA accumulation is likely to be a conserved function of TDP-43 proteins because knockdown of mammalian TDP-43 in HeLa cells and M17 neuronal cells also causes dsRNA accumulation.
Results
RNA transcripts aberrantly represented in tdp-1(ok803)
To determine TDP-1's effect on the transcriptome, we deep-sequenced RNA from tdp-1 mutant animals to identify changes in RNA metabolism or abundance. While two deletion alleles exist for tdp-1, tdp-1(ok803) and tdp-1(ok781), only the tdp-1(ok803) allele appeared to be a clear null (Supplementary Fig S1B and C); therefore, our study is focused on this allele. We created and sequenced poly(A)-selected cDNA libraries from wild-type and tdp-1(ok803) animals. Mapping of sequenced reads showed over 50% of all amplifiable annotated genes were well represented (roughly 14,000 transcripts). A differential gene expression comparison (RPKMs) between tdp-1(ok803) mutants and wild-type revealed over 1,700 transcripts aberrantly represented with close to an equal number of transcripts underrepresented as overrepresented (Fig1A; Supplementary Table S1). qRT–PCR verification of a select set of these abundance changes is shown in Supplementary Fig S2. Gene ontology analysis of aberrantly represented transcripts in tdp-1 mutants indicated very few pathways were enriched among over- and underexpressed genes (Supplementary Table S2). Most enriched pathways were associated with developmental processes and stage-specific molting, which may be an artifact due to the mild growth delay reported in tdp-1 mutant animals (Zhang et al, 2012).
Figure 1. TDP-1 maintains transcripts with potential dsRNA structure.
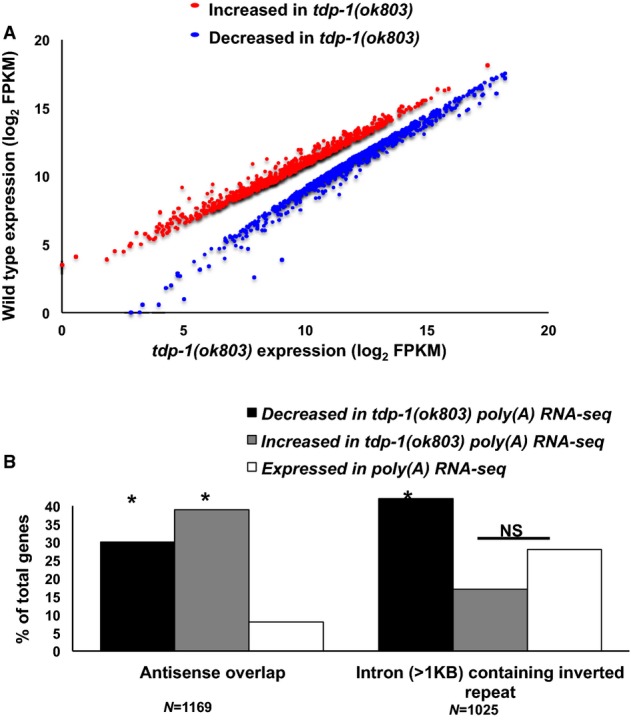
- Scatter plot comparing the log2 of transcript RPKMs (Reads Per Kilobase of transcript per Million mapped reads) between wild-type and tdp-1(ok803) poly(A)-selected RNA-seq experiments. Only transcripts significantly (corrected P < 0.05) increased (red) or decreased (blue) compared to wild-type are shown. Scatter plot represents significantly different genes calculated from two independent biological replicates of both wild-type and tdp-1(ok803) poly(A) RNA-seq prepared from L4 animals.
- Percentage increased and decreased transcripts in tdp-1(ok803) poly(A) RNA-seq that have antisense overlap with another spliced gene or have intronic sequences (> 1 kb) that contain inverted repeats. *P < 1 × 10−4 (hypergeometric distribution); NS, not significant relative to control gene sets.
Because TDP-43 functions in alternative splicing, we also asked whether transcripts with altered abundance in the mutant had splicing abnormalities. Splicing analysis identified ∼350 genes with significant (P < 0.001) changes in splice site representation (Supplementary Table S3, independent verification in Supplementary Fig S3); however, the majority of transcripts altered in splicing were not altered in abundance compared to wild-type. While splicing abnormalities may contribute to tdp-1 loss of function defects, splicing differences do not readily explain changes in transcript abundance in tdp-1(ok803) animals.
TDP-1 regulates genes with inherent double-stranded structure
Mammalian TDP-43 maintains the abundance of transcripts with long introns. One interpretation of this observation is that transcripts require TDP-43 because of an inherent characteristic of the RNA transcript itself and not due to its involvement in a common functional pathway. To look for common features in RNA transcripts affected by TDP-1, we examined the most dramatically altered transcripts in the tdp-1 deletion using the IGV genome browser and online databases (wormbase, aceview) for common characteristics within the RNA molecule. Interestingly, we noticed that a large proportion of altered transcripts had potential double-stranded structure. Specifically, many over/underexpressed transcripts contained either antisense overlap with another gene or multiple inverted repeats within intronic regions.
To quantify these effects, we first analyzed the percentage of altered genes with antisense overlap to another coding gene. While about 8% of the worm genome is arranged antisense to another gene (Thierry-Mieg & Thierry-Mieg, 2006), approximately 35% of increased and decreased transcripts were arranged in this manner, representing a highly significant enrichment (P = 5.3 × 10−181, hypergeometric distribution (hgd)) (Fig1B). We also determined the percentage of genes containing intronic inverted repeats that were aberrantly represented in tdp-1 deletion. We limited our analysis to inverted repeats contained within introns greater than 1 kb. While about 25% of all expressed genes contained an intron inverted repeat (2,641), 40% of underrepresented genes in tdp-1(ok803) RNA-seq contained inverted repeats, representing a significant enrichment (P = 4 × 10−5, hgd, P = 3.2 × 10−3, chi-square test) (Fig1B). Among overexpressed transcripts, inverted repeat containing introns were not significantly enriched, suggesting that intronic RNA structure in tdp-1 mutants results in reduction of mRNA abundance.
Transcripts with antisense overlap to another RNA and transcripts containing inverted repeats are capable of forming either inter- or intra-molecular dsRNA. Because deletion of tdp-1 perturbed the abundance of these transcripts, we hypothesized that TDP-1 has a fundamental function in the formation, or metabolism, of dsRNA.
TDP-1 limits the accumulation of double-stranded RNA
To address the hypothesis that TDP-1 affects dsRNA metabolism, we looked directly at the amount and localization of dsRNA in tdp-1(ok803) mutant worms by immunostaining with a dsRNA-specific antibody, J2. The J2 antibody is specific to dsRNA duplexes greater than 40 bp (Schonborn et al, 1991) and was used previously to visualize double-stranded RNA in vivo (Kaneko et al, 2011). We stained populations of wild-type and tdp-1(ok803) worms with the J2 antibody. As shown in Fig2, tdp-1(ok803) worms had a significant increase in nuclear inclusions reactive with the J2 antibody. This increase was observed in multiple tissues including: gut (Fig2A), muscle, and the anterior head region (Supplementary Fig S4). Importantly, detection of these inclusions was ablated by pretreatment with the dsRNA-specific RNAse V1, but not by treatment with the ssRNA-specific RNAse T1 (Fig2A, bottom two panels). To confirm the specificity of the J2 antibody in worms, we also immunostained a transgenic strain expressing a dsRNA-binding domain fused GFP (Supplementary Fig S5). J2 and GFP loci tightly overlapped, indicating they likely recognize the same substrates.
Figure 2. TDP-1 limits the amount of nuclear dsRNA.
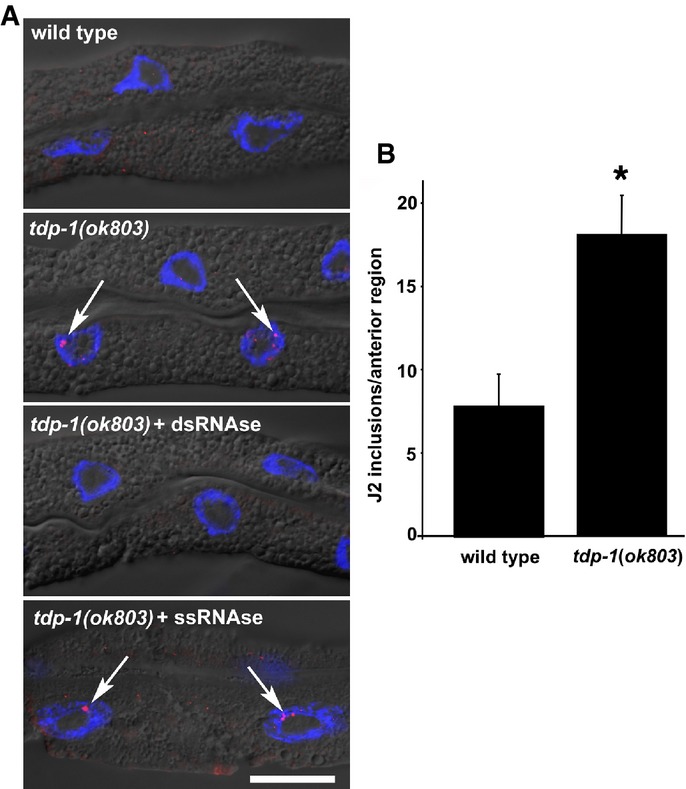
- Fixed, isolated intestinal tissue probed with anti-dsRNA antibody (J2). The J2 antibody recognizes dsRNA stretches of 40 bp or more in a sequence-independent manner. Intensely stained inclusions (red dots, indicated by arrows) were detected in intestinal nuclei (blue, DAPI counterstain) specifically in tdp-1(ok803) mutant worms (middle panel). J2-reactive inclusions were observed in 28% tdp-1(ok803) intestinal nuclei scored (30/107), but not detected in wild-type controls (0/122) or tdp-1(ok803) (0/105) fixed tissue pretreated with dsRNA nuclease (V1) before J2 staining. J2-reactive foci were still observed (arrows, bottom panel) in intestinal nuclei pretreated with the ssRNA-specific nuclease (T1) (29%, 12/41). Scale bar, 20 μm.
- Quantification of anterior J2 inclusions using ImageJ software. tdp-1(ok803) worms had significantly more J2 inclusions (*P < 0.01, Student's t-test, error bars = SEM). Representative projection images used to generate this data are shown in Supplementary Fig S4.
TDP-1 is required to maintain normally low levels of A-to-I RNA editing
Since nuclear dsRNA is limited by the activity of A-to-I dsRNA-specific deaminases (ADARs) (Bass, 1998), we hypothesized that TDP-1 could function in promoting A-to-I RNA editing. To address this possibility, we investigated the amount of A-to-I editing in tdp-1(ok803) mutants. In order to determine whether identified A-to-I edits represented true physiological targets, we sequenced RNA in parallel from a strain deleted for the only functional worm ADAR, adr-2(gv42). Using a compiled list of all nucleotide positions that showed a discrepancy between the RNA and DNA sequence (see Materials and Methods), we hand-annotated 210 regions containing convincing evidence of A-to-I RNA editing (Supplementary Table S4). Regions were only considered edited if A-to-I conversions were observed (by inspection of sequencing reads in IGV genome browser) in all three independent biological replicates and not observed in RNA-seq from adr-2(gv42) mutants (examples of representative regions scored as positive for editing are shown in Supplementary Fig S6). The majority (> 90%) of editing events occurred in introns and 3′ UTRs, and all observed editing was consistent with promiscuous editing, which occurs in long dsRNA (average of 123 positions targeted per region, see Supplementary Table S4). We found no reproducible evidence of exonic A-to-I editing. While our list is likely not an exhaustive list of all edited transcripts in C. elegans (only larval stage 4 was analyzed and the read depth of many intronic regions was too low to assess editing), our analysis did successfully identify eight of the ten published transcripts characterized as edited due to preferential digestion with T1 RNAse (Morse et al, 2002) as well as 83 of the regions identified by Wu et al (2011). To confirm that regions positive for A-to-I editing contained sequence motifs capable of dsRNA formation, we assayed for the presence of inverted repeat/loop sequences within identified editing targets (see Materials and Methods). We found that almost all (> 98%) of the regions identified in this study contained two or more inverted repeat sequences and/or overlapped with transposons/retrotransposons (Supplementary Table S5). Transcripts containing both inverted repeats and transposons can form intra-molecular dsRNA and are known targets of A-to-I editing in vivo (Nishikura, 2010). These results indicate that our analysis identified editing only within potentially structured RNA sequences, which are suitable targets of worm ADAR.
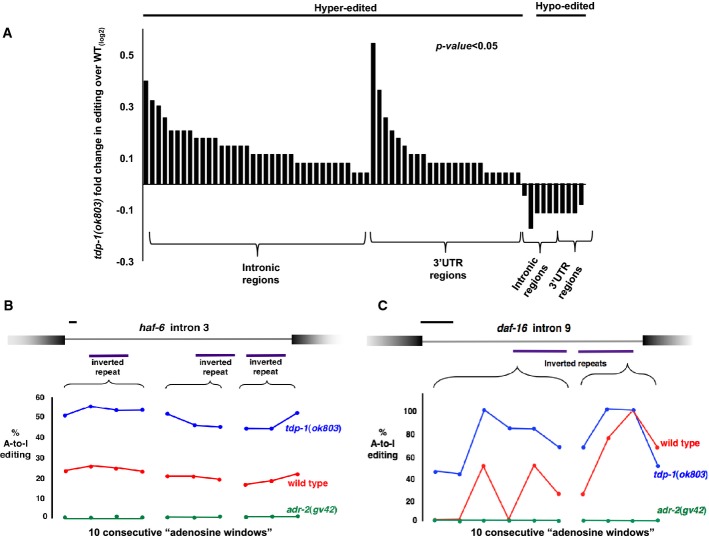
To determine whether tdp-1 maintains A-to-I RNA editing within these regions, we calculated the percent editing across all identified edited regions that were well represented (> 20 reads, 153/210 regions) in both wild-type and tdp-1 mutants. Roughly 50% of well-represented regions showed significantly (P < 0.05) altered editing (Fig3A), and in almost all (> 85%) cases, editing was increased in the tdp-1 deletion transcripts compared to wild-type. The most common hyper-editing occurred in intronic regions. Two examples of increased intronic editing in tdp-1(ok803) RNA-seq are shown (Fig3B). Examination of transcript abundance and splicing of the worm ADAR genes revealed no substantial change between wild-type and tdp-1 mutants (Supplementary Fig S7A). While there is no available antibody for worm ADR-2, we observed no change in ADR-1 protein levels between wild-type and mutant (Supplementary Fig S7B), suggesting that tdp-1 does not limit A-to-I RNA editing by controlling ADAR function. Although the majority of editing increases in tdp-1 mutants were mild (average fold increase ∼1.4-fold), increased amounts of A-to-I editing indicate that the accumulation of dsRNA in tdp-1(ok803) mutant worms is not due to a reduction in RNA editing. Rather, increases in the structure or stability of dsRNA could result in a slightly higher frequency of editing in tdp-1 mutants.
Figure 3. TDP-1 limits levels of A-I RNA editing.
- A Fold change in % A-to-I editing in tdp-1(ok803) compared to wild-type RNA-seq is shown. For all regions, the average percent editing for each potentially edited nucleotide was calculated. Bars in graph represent the average percent editing across individual edited regions altered in tdp-1(ok803) (73 altered/154 analyzed). Genomic location of edited regions, actual change in percent editing and actual P-values are listed in Supplementary Table S4.
- B, C Examples of hyper-edited intronic regions in tdp-1(ok803) mutant RNA-seq: intron 3 of ABC-transporter haf-6 (B) [ChrI: 1170900-1173858] and intron 9 of FOXO transcription factor daf-16 (C) [ChrI: 10774096-10774765]. Purple bars indicate annotated structural elements containing editing. Regions were divided into 10 equal windows and mean fraction edited for each window is plotted (y-axis). Connected points indicate editing in the same structural element. Percent editing in wild-type (blue), tdp-1(ok803) (red) and adr-2(gv42) (green) is shown. Scale bars (upper left, black), 100 bp.
Immunoprecipitation with an anti-dsRNA antibody recovers structured transcripts
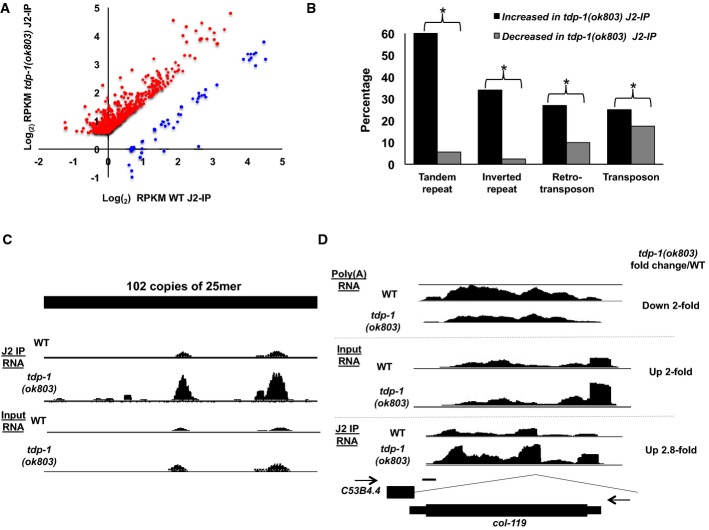
The J2 antibody can be used to immunoprecipitate dsRNA, as shown for Alu transcripts in mammalian cell lysates (Kaneko et al, 2011). Therefore, we analyzed J2 immunoprecipitates from both wild-type and mutant whole-worm lysates (in triplicate) to characterize the specific dsRNAs that accumulate in tdp-1(ok803) mutants. Because J2 immunoprecipitation was done in untreated lysate, entire transcripts containing 40 bp or more of dsRNA anywhere within the molecule will be precipitated. J2-immunoprecipitated RNA and total RNA were sequenced in a strand-specific manner. Pairwise correlation of J2-IP RNA-seq replicates indicated that the J2-IP replicates were highly similar (Supplementary Fig S8).
First, we analyzed the wild-type RNA-seq data to validate J2 immunoprecipitation (J2-IP) as an effective method to isolate dsRNA. Comparison between input and J2-immunoprecipitated RNA confirmed that J2-IP selected for a subset of transcripts (Fig4A). As a positive control, we calculated the fold enrichment (over input) of J2-IP RNA-seq for all expressed repetitive regions (Fig4B). Consistent with J2-IP selecting for dsRNA, we observed a fourfold to fivefold increase in expression level (RPKMs) of annotated transposon/retrotransposon sequences, as well as a twofold to fourfold increase in the expression level (RPKMs) of tandem/inverted repeats. We did not see an overall increase in the enrichment of exon sequences, confirming J2-IP is specific for transcripts originating from regions known to produce dsRNA. Examples of an enriched transposon and retrotransposon are depicted in Fig4C. We verified the specificity of J2-IP for dsRNA transposons as this enrichment was abolished in lysates pretreated with dsRNA-specific RNase (Supplementary Fig S9).
Figure 4. Immunoprecipitation with J2 antibody selects for dsRNA in wild-type worm extracts.
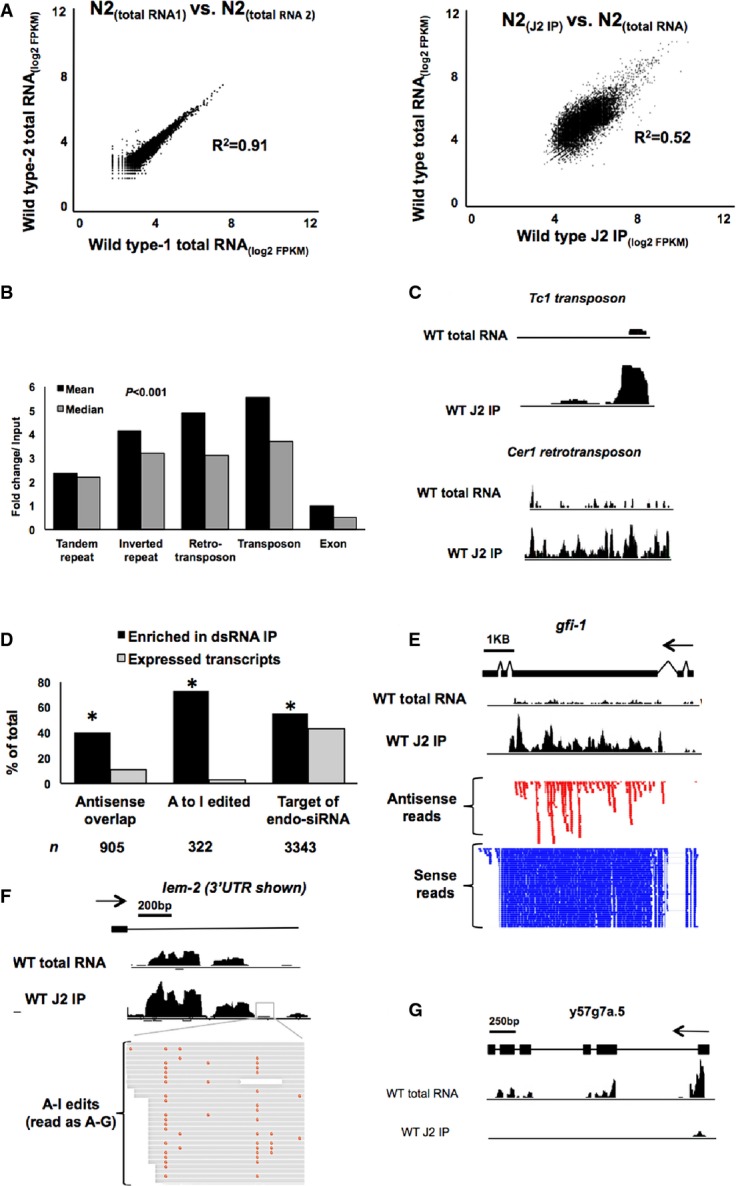
- A The J2 anti-dsRNA antibody immunoprecipitates a subset of RNAs from wild-type extracts. Plotted are RPKMs(log2) for two independent wild-type input RNA-seq datasets (left graph) and for a J2-IP RNA-seq dataset versus input RNA (right graph). 9,086 transcripts are shown. R2 values for each plot are displayed in the graph.
- B The fold change (in RPKM) over input (y-axis) of all significantly enriched repetitive transcripts (type indicated on x-axis) and all exons (corrected P < 0.001) in wild-type J2-IP RNA-seq. Graph shows both mean (black bars) and median (gray bars) fold enrichment/input.
- C Coverage tracks (black bumps) of alignments to consensus sequences of representative transposons enriched in J2-IP RNA compared to total RNA. Coverage track height was set to the same value for both input RNA and J2-immunoprecipitated RNA, and height is proportional to abundance of each region.
- D The percentage of total transcripts (gray bars) with potential dsRNA of three types (antisense overlap determined from Thierry-Mieg & Thierry-Mieg, 2006, A-to-I RNA editing taken from Supplementary Table S4 and siRNA targets taken from Warf et al, 2012) and the percentage of each of those transcript types enriched by J2-IP (in three independent biological replicates; black bars, *P-value < 0.001 (hypergeometric distribution); n = number of genes analyzed in each structural category).
- E–G Examples of transcripts enriched in the J2-IP RNA-seq. The gfi-1 locus (E) expresses an antisense transcript, as shown by both antisense (red) and sense (blue) reads. The 3′ UTR of lem-2 (F) undergoes dsRNA-specific A-to-I RNA editing. The editing pattern in the boxed region is shown. (G) Example of transcript not enriched by J2-IP (y57g7a.5). Black arrows indicate direction of transcription.
To ask which gene transcripts were enriched by J2-IP, we compared normalized read counts between input RNA-seq and J2-IP RNA-seq for each gene. Only genes that were significantly increased (P < 0.05, FDR < 0.1, > 1.5-fold) over input were considered “selected” in the IP. We identified 2,875 transcripts enriched in the wild-type J2-IP, representing 27% of all genes expressed in the experiment (10,630). Most (95%) of the enriched RNAs originated from protein-coding genes. Both antisense-overlap genes and A-to-I edited genes were significantly enriched (P < 1 × 10−15 hgd) by J2-IP (Fig4D). As expected, transcripts targeted by endogenous small interfering RNAs (siRNAs), which often trigger siRNA production due to the presences of double-stranded regions (Fischer, 2010), were also enriched by J2-IP (P < 1 × 10−143, hgd, endo-siRNA target genes taken from Warf et al, 2012). Representative examples of these classes of enriched transcripts are shown as well as an unenriched gene (Fig4E–G). Transcripts with the structural characteristics mentioned represented 73% of all coding genes enriched in the J2-IP (Supplementary Table S6).
TDP-1 functions to limit the structure or stability of dsRNA
To identity transcripts with more dsRNA structure/stability in tdp-1(ok803) mutants, we used DESeq software to compare normalized (to input) expression levels of all genes expressed in the J2-IP between tdp-1 mutants and wild-type (Supplementary Fig S10). After filtering out low expressed genes and genes not enriched by J2-IP over input in either sample (see Materials and Methods and Supplementary Methods), we identified 1,422 genes with increased J2-IP enrichment (P < 0.05, FDR < 0.1) in tdp-1 mutants compared to wild-type, representing about 40% of all transcripts selected by J2-IP. In contrast, only 166 genes (5% of all J2-IP selected transcripts) were identified as having less dsRNA structure/stability in tdp-1 mutants (Fig5A; Supplementary Table S7). This result indicates that TDP-1 limits the structure or stability of many dsRNA transcripts, resulting in accumulation of dsRNA when TDP-1 is absent. We confirmed the enrichment of several transcripts in the tdp-1(ok803) J2-IP by RT–PCR and showed this enrichment is abolished by pretreatment with dsRNA-specific RNAse (Supplementary Fig S9).
Figure 5. Double-stranded RNA transcripts are preferentially recovered in tdp-1(ok803) mutant extract.
- Graph comparing the log2 RPKM (normalized to input) of tdp-1(ok803) J2-IP (y-axis) versus wild-type J2-IP (x-axis) for all genes significantly increased (red dots) and decreased (blue dots) in representation (P < 0.05, FDR < 0.1 for all changes). Plot derived from data shown in Supplementary Fig S10 using three independent biological replicates of wild-type and tdp-1(ok803) J2-IP.
- The percentage (x-axis) of all expressed repetitive elements (type shown on y-axis) that are significantly increased (black bars) and decreased (gray bars) in tdp-1 mutant J2-IP compared to wild-type J2-IP (P < 0.05 for all changes). *P < 1 × 10−10 chi-square test. Plot derived from data shown in Supplementary Fig S12.
- A representative example of a tandem repeat region that is increased for dsRNA structure/stability in tdp-1(ok803) J2-IP. Coverage tracks from both the J2-IP RNA-seq and input RNA-seq are shown; region displayed: chromosome I: 10,130,500–10,133,500.
- Coverage tracks of col-119 gene (transcribed antisense with the intron of expressed gene C53B4.4) in poly(A) RNA-seq (top), input (total) RNA-seq (middle) and J2-IP RNA-seq (bottom) (note: scale of coverage tracks for J2-IP is normalized to input for display purposes). Gene models for both genes are shown below coverage tracks, the direction of transcription is indicated by black arrows). Scale bars (black line), 200 bp. See also Supplementary Fig S11.
Analysis of transcripts increased in tdp-1 mutant J2-IP did not indicate that any specific class of dsRNA (e.g. antisense to another gene or target of siRNA) was being affected in tdp-1 mutants. Rather, we found that most transcripts that form dsRNA structure simply have more dsRNA structure/stability in the absence of TDP-1 (examples of enriched antisense-overlap genes shown in Supplementary Fig S11A). However, the exception to this observation was A-to-I RNA edited transcripts, which were significantly under represented (P = 1 × 10−5, hgd, P = 9 × 10−260, chi-square) among genes increased in tdp-1(ok803) J2-IP. This result suggests that hyper-editing of these transcripts could counteract any potential increases in dsRNA structure in tdp-1 mutants.
To determine whether tdp-1 controls the structure of a specific functional class of transcripts, we performed a gene ontology analysis of genes whose transcripts have altered dsRNA structure/stability in tdp-1 mutants. Among genes with increased recovery in tdp-1(ok803) J2-IP, we found an enrichment for genes functioning in nucleosome assembly (mostly replicative histone transcripts, 59/66 increased in dsRNA structure) as well as genes involved in translation and protein modification/turnover (Supplementary Table S8). Examination of enriched replicative histone transcripts in tdp-1 mutants showed that both 9 and antisense transcripts originating from histone genes were observed in tdp-1(ok803) and wild-type total RNA-seq data. However, J2-IP recovered the antisense histone-region transcripts only in tdp-1(ok803) (Supplementary Fig S11B), suggesting that these transcripts form intermolecular hybrids whose structure or stability is preserved in the absence of TDP-1 function. Interestingly, genes with decreased recovery in tdp-1(ok803) J2-IP were enriched for neuronal-specific genes, many of which are highly edited (Supplementary Table S8, bottom). Since ADAR enzymes are prominently expressed in the nervous system (Savva et al, 2012), one interesting possibility is that an initial increase in dsRNA structure in tdp-1 mutants results in excessive editing that ultimately leads to decreased dsRNA structure in these transcripts.
A major fraction of transcripts with increased enrichment in tdp-1 mutant J2-IP also mapped to repetitive elements. Comparison of normalized expression levels (RPKMs) of annotated repeat regions (taken from wormbase) indicated that most repeat regions showed increased dsRNA structure/stability in tdp-1 mutants compared to wild-type (Fig5B; Supplementary Fig S12), with the most dramatically affected class being tandem repeats. 60% of all expressed tandem repeats showed increased recovery in tdp-1(ok803) J2-IP compared to only 5% in wild-type. An example of a tandem repeat increased for dsRNA structure/stability is shown in Fig5C.
Transcripts with excess secondary structure are aberrantly represented in mature RNA
If excess dsRNA structure/stability results in altered levels of mature transcripts, we would predict transcripts that are over/under-expressed in the poly(A)-selected pool would be preferentially recovered by J2-IP. Indeed, transcripts aberrantly represented in poly(A) RNA-seq from tdp-1(ok803) animals were significantly enriched (P = 1.7 × 10−17, hgd) for transcripts isolated by J2-IP from tdp-1(ok803) (Supplementary Table S7 and Supplementary Fig S13). This result is consistent with the hypothesis that increased secondary structure within the pre-mRNA interferes with normal mRNA maturation causing altered representation in poly(A)-selected mRNA. As an example, Fig5D depicts col-119 (encodes a collagen protein), which is transcribed antisense to another expressed transcript. In the mature poly(A) RNA pool, this transcript is decreased. However, col-119 transcript is overrepresented twofold in total RNA, suggesting that the reduction in poly(A) RNA results from defective maturation, as opposed to decreased expression/transcription. J2-IP enriched this transcript almost threefold in tdp-1(ok803) compared to wild-type controls, indicating this transcript has a high degree of aberrant dsRNA structure/stability that interferes with its maturation.
TDP-1 functions to limit the double-stranded structure of intronic regions
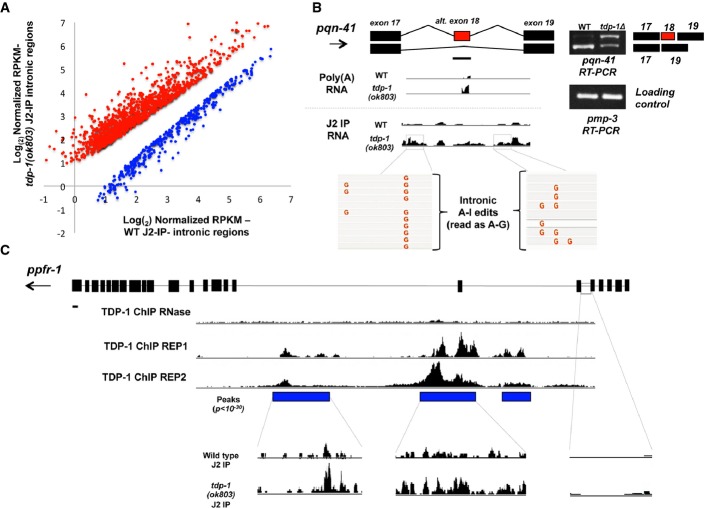
TDP-43 homologs sustain the expression of transcripts with long introns presumably by binding the intronic sequences and allowing expression or stabilization of the transcript (Polymenidou et al, 2011). We propose that TDP-1 suppresses double-stranded structure within introns and may thereby prevent transcript degradation. To ask whether intronic sequences contain excess structure in tdp-1 mutant animals, we compared normalized expression levels (RPKMs) for all intronic regions (> 250 bp) represented in the J2-IP RNA-seq. Among enriched regions, 1,287 introns were significantly different (P < 0.05, FDR < 0.1) between tdp-1(ok803) and wild-type, with 75% showing an increased abundance in tdp-1(ok803) J2-IP (Fig6A; Supplementary Fig S14). Visual inspection of introns altered for J2-IP enrichment in tdp-1(ok803) mutants indicated that ∼31% contained anecdotal evidence of A-to-I editing (scored by visual inspection of sequencing reads) and 80% overlapped with annotated complex and/or simple structural elements (see Supplementary Table S9). Both of these features are consistent with the formation of dsRNA. Many edited intronic regions observed in the J2-IP RNA-seq were not identified as edited in our initial analysis (due to low coverage of intronic regions), suggesting that immunoprecipitation with the J2 antibody is capable of enriching for low abundance or rapidly degraded regions. Interestingly, introns with decreased recovery in tdp-1(ok803) J2-IP were highly enriched for A-to-I editing (∼50%, P = 4 × 10−56, chi-square test) and conversely, regions with increased structure were unenriched for editing (∼25%, P = 5.8 × 10−10, chi-square test), suggesting that hyper-editing in tdp-1 mutants may result in a decrease in dsRNA in some intronic regions.
Figure 6. TDP-1 directly limits the dsRNA structure or stability of intronic RNA.
- Log2 fold change of tdp-1(ok803) J2-IP/wild-type J2-IP (x-axis) for all expressed intronic regions selected in the J2-IP graphed according to increasing number of reads that map to each region (x-axis). Abundance levels for introns were normalized to input (see Materials and Methods for details). All introns selected in the J2-IP that are significantly (P < 0.05, FDR < 0.1, three biologically independent replicates) increased (red dots) and decreased (blue dots) between tdp-1 mutant and wild-type J2-IP are shown. See Supplementary Table S9.
- Example of a tdp-1(ok803) J2-IP enriched intronic region that exhibits altered splicing in tdp-1(ok803) polyA RNA-seq. Coverage tracks from poly(A) RNA-seq and J2-IP RNA-seq are displayed for pqn-41 (ChrIII: 1,931,953–1,934,515). A-to-I RNA editing patterns in the boxed regions for each transcript are also depicted. Arrows depict the direction of transcription, and scale bars (solid black line below gene model) are set to 250 bp.
- ChIP-seq pattern for TDP-1 within the long, structured gene ppfr-1 (ChrI: 9,364,769-9,393,087). The location of significant TDP-1 peaks in both replicates as well as in the RNase control is shown by blue boxes below the ChIP-seq coverage tracks. The enrichment by anti-dsRNA immunoprecipitation (J2-IP) within regions bound by TDP-1 is displayed below significant peaks. Both wild-type (top) and tdp-1(ok803) (bottom) J2-IP tracks are shown to indicate excess RNA structure when TDP-1 is absent. The J2-IP enrichment for an intron not bound by TDP-1 is also shown. Arrows depict the direction of transcription, and scale bars (solid black line below gene model) are set to 250 bp.
Source data are available online for this figure.
Double-stranded RNA structure, as well as A-to-I RNA editing, is known to modulate alternative splicing (Nishikura, 2010), so we asked whether transcripts with altered intronic dsRNA in the mutant were the same transcripts with altered splicing in poly(A) RNA. Comparison of J2-IP enriched introns and genes with altered splicing in tdp-1(ok803) mutants revealed a significant enrichment (94/163 that could be assayed, P = 7.5 × 10−14, hgd) for intronic regions with differential recovery in tdp-1(ok803) J2-IP among transcripts altered in splicing. As a representative example, Fig6B shows the pqn-41 transcript, which displays enhanced inclusion of exon 18 in tdp-1(ok803) poly(A) RNA-seq and enrichment by J2-IP of the intronic regions bracketing that exon in tdp-1(ok803) mutants. A-to-I RNA editing was also observed in these introns consistent with the formation of dsRNA. Additional examples are provided (Supplementary Fig S15). These results suggest that some of the tdp-1(ok803) splicing abnormalities could be due to excess dsRNA formation and/or A-to-I editing within the intronic regions of transcripts altered in splicing.
TDP-1 associates co-transcriptionally with genes whose dsRNA stability is increased in tdp-1 mutants
To determine whether TDP-1 acts directly on transcripts with potential double-stranded structure, we asked whether TDP-1 protein associates with genes containing excessive dsRNA in tdp-1(ok803) mutants. As TDP-1 orthologs bind primarily intronic regions, we suspected that TDP-1 might associate with nascent pre-mRNAs co-transcriptionally in order to limit dsRNA structure or stability in these regions. Chromatin immunoprecipitation (ChIP) sequencing is widely used to determine the association site of RNA-binding/processing factors that co-transcriptionally process nascent RNA (Swinburne et al, 2006). Therefore, we chose to analyze TDP-1 binding via chromatin immunoprecipitation followed by deep sequencing. In parallel, we also performed TDP-1 ChIP-seq on extract pretreated with RNase to confirm the ChIP signal was dependent on association with nascent RNA and not an association with the DNA, as has been done previously (Bieberstein et al, 2014). TDP-1 ChIP-seq was normalized using the RNase-treated ChIP-seq. Approximately 3,500 genes contained significant (P < 0.01) TDP-1 binding sites (Supplementary Table S10). We were not able to find any significant TDP-1 peaks not dependent on RNA (RNase ChIP sample was normalized using total genomic sequencing, data not shown) verifying that TDP-1 ChIP peaks represent co-transcriptional association of TDP-1 with nascent RNA. 64% of TDP-1 peaks were contained in introns that were 32-fold longer than the average intron length (mean size of bound intron: 2,120 bp versus 65 bp genome average). As TDP-43 orthologs show a preferential association with UG-rich sequences (Ayala et al, 2005), we asked whether worm TDP-1 also associates with UG repeats. Analysis of UG repeat sequences indicated TDP-1 associated with ∼70% of all UG repeats of at least six repeats long (n = 201). Further, TDP-1 association increased with increasing UG-repeat size (Supplementary Fig S16). While this result indicates TDP-1 association is enriched in UG-rich regions, many TDP-1 bound regions did not contain UG repeats and not all UG repeats were bound by TDP-1, mirroring results reported for mammalian TDP-43 binding.
Analysis of TDP-1 binding in genes with increased dsRNA structure/stability in tdp-1 mutants (as assayed by J2-IP enrichment) showed about 40% of these genes contained clear TDP-1 binding sites (P = 1 × 10−21, hgd). For J2-IP enriched intronic regions, TDP-1 binding showed a more significant enrichment (51% bound, P = 8 × 10−141). A large fraction (37%) of TDP-1 ChIP peaks also overlapped with annotated repeat regions such as transposons and tandem repeats (2,061 peaks overlapped out of 5,587 peaks total), confirming that TDP-1 associates with potentially double-stranded transcripts and mirroring mammalian studies showing TDP-43 binds transposable elements (Li et al, 2012). An example of TDP-1 binding sites within intronic regions containing excess dsRNA structure/stability is shown along with the J2-IP enrichment of these regions (Fig6C). In order to provide independent support for our ChIP-seq results, we performed an RNA-immunoprecipitation (UV-CLIP) experiment using an antibody against TDP-1 followed by RT–PCR. As shown in Supplementary Fig S15, the RIP experiments confirmed TDP-1's association with a subset of introns that were bound by TDP-1 in the ChIP-seq. Taken together, these results support the model that TDP-1 associates co-transcriptionally with nascent pre-mRNAs to limit potential dsRNA structure, thereby allowing correct maturation of transcripts.
TDP-1 maintains normal olfactory response by limiting the action of pathways that act on dsRNA
Our results indicated that TDP-1 functions to control the structure or stability of double-stranded RNA, but it is not clear whether there is a neurological consequence of excess dsRNA. Although tdp-1 loss of function mutants are reported to have reduced mobility (Zhang et al, 2012), we were unable to detect movement deficits in tdp-1(ok803) animals under standard conditions (Supplementary Fig S17A and B). However, TDP-1 might function in other neuronally mediated processes, such as chemotaxis. We assayed the response of tdp-1 mutant animals to attractive volatile compounds (butanone and isoamyl alcohol) that are sensed by the AWCR and AWCL neurons (Bargmann, 2006). Determination of chemotaxis indices (CI) revealed significant (P < 0.001) olfactory deficits in tdp-1(ok803) mutant animals (Fig7A). Similar defects were also observed in the non-null allele, tdp-1(ok781) (Supplementary Fig S17C).
Figure 7. TDP-1 maintains chemotaxis by limiting RNA interference.
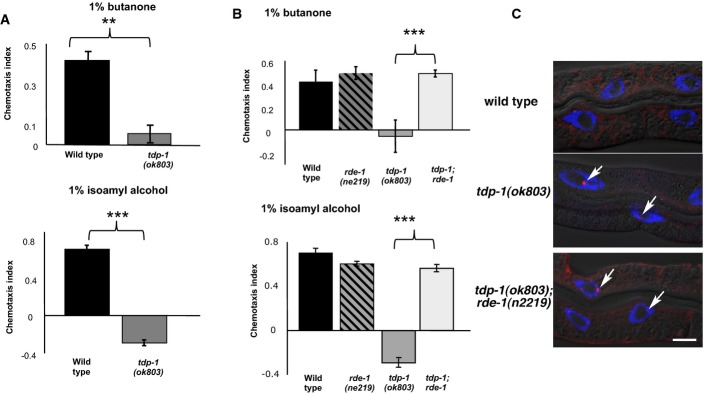
- The chemotaxis index (y-axis) for wild-type and tdp-1(ok803) animals toward 1% butanone and 1% isoamyl alcohol are shown. Negative values signify repulsion. Error bars represent SEM. **P < 0.01, ***P < 0.001 (Student's t-test).
- Rescue of chemotaxis toward 1% butanone (top two graphs) and 1% isoamyl alcohol (bottom two graphs) in tdp-1(ok803);rde-1(ne219) double-mutant animals. Assays were done in triplicates at least 3 independent times. Error bars represent SEM. ***P < 0.001 (Student's t-test).
- Immunostaining of isolated worm nuclei in wild-type, tdp-1(ok803) mutants and tdp-1(ok803); rde-1(n2219) double mutants. Single plane images distally overlaid with DIC image. The presence of J2 foci in tdp-1(ok803); rde-1(n2219) mutants indicates that deletion of rde-1 does not suppress the accumulation of dsRNA in tdp-1 mutants.
We were curious if the chemotaxis defects in tdp-1 mutants were due to increased dsRNA accumulation or an alternate function of tdp-1. If aberrant dsRNA results in chemotaxis defects, it could either be because dsRNA itself is intrinsically deleterious or because the dsRNA leads to the hyper-activation of downstream processes, such as inappropriate production of endogenous siRNAs. To ask whether the tdp-1(ok803) olfactory defects were due to the RNAi pathway, we created tdp-1(ok803) strains that also had a loss of function mutation for a gene essential to the RNAi pathway, rde-1(ne219). Chemotaxis assays on this strain showed that mutation of rde-1 completely suppressed the olfactory defect in tdp-1(ok803) animals (Fig7B). Staining with the J2 antibody detected similar amounts of dsRNA in the tdp-1(ok803), rde-1(ne219) double-mutant as compared to the tdp-1(ok803) single-mutant worms (Fig7C), confirming that the mutation in rde-1 did not suppress the primary accumulation of dsRNA. These results indicate that specific neuronal defects in tdp-1 mutants require the action of pathways downstream of dsRNA. Importantly, deletion of worm adr-2 also results in a chemotaxis defect that can be rescued by mutation in rde-1 (Tonkin & Bass, 2003), indicating that defective chemotaxis is a major phenotypic consequence of excess dsRNA in C. elegans and this defect is due to metabolism of double-stranded transcripts by the RNAi pathway.
Limiting dsRNA accumulation is a conserved function of human TDP-43
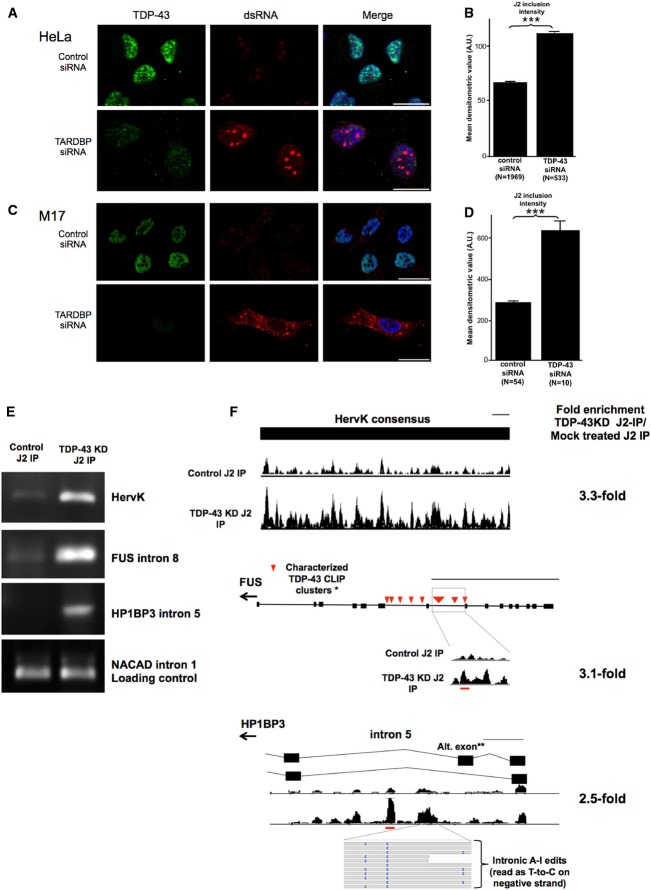
TDP-1 limited the accumulation of double-stranded RNA in worms. To determine whether this is a conserved function of mammalian TDP-43, we treated human-derived M17 neuroblastoma or HeLa cells with antisense TDP-43 siRNA and then immunostained the cells with J2 antibody. Consistent with the worm results, we observed a dramatic increase in dsRNA accumulation in both cell types. Within HeLa cells (Fig8A), the J2-immunopositive signal was concentrated in defined nuclear foci that were significantly brighter (P < 0.001) in TDP-43 siRNA-treated cells than in control cells (Fig8B). M17 neuronal cells also displayed increased J2 staining upon TDP-43 knockdown (Fig8C), but the staining of dsRNA was more diffuse in these cells and occurred in both the nucleus and the cytoplasm. Quantification of immunofluorescence indicated that dsRNA was also significantly increased in M17 cells upon TDP-43 knockdown (Fig8D). J2 staining specificity was confirmed by differential nuclease digestion (Supplementary Fig S18). The different subcellular localization of accumulating dsRNA in HeLa cells and M17 neuroblastoma cells suggests that TDP-43 could have distinct functions on dsRNA in these two cell types. Alternatively, different cell types may have distinctive mechanisms to metabolize or sequester dsRNA.
Figure 8. Mammalian TDP-43 also functions to limit the accumulation of dsRNA.
- Cultured HeLa cells depleted for TDP-43 (green) with TARDBP siRNA display increased intensity of J2 anti-dsRNA labeling (red) within nuclear foci. Scale bars, 20 μm.
- Densitometric quantification of the intensity of nuclear dsRNA foci. Graph shows the mean J2-dsRNA densitometric value, error bars = SEM. ***P < 0.0001 (Student's t-test). N = number of objects detected in each group.
- Cultured M17 cells display increased nuclear and cytoplasmic dsRNA staining (red) upon knockdown of TDP-43. Scale bars, 20 μm.
- Quantification of nuclear dsRNA in TDP-43 knockdown M17 cells. Graph shows the mean J2-dsRNA densitometric value, error bars = SEM. ***P < 0.0001 (Student's t-test). N = number of objects detected in each group.
- Semi-quantitative RT–PCR of RNA precipitated from J2-IP on lysate from mock-treated M17 cells (left panel) and TDP-43 knockdown M17 cells (right panel). RT–PCR was done in triplicates (only one replicate shown).
- Expression tracks from J2-IP RNA-seq in regions shown to have increased recovery in TDP-43 knockdown J2-IP compared to mock-treated controls. The gene model for each region is shown above the expression tracks. Evidence of A-to-I RNA editing in human sequencing read is also shown for HP1BP3. Herv-K expression tracks represent reads mapping to a consensus sequence of the Herv-K genome. All expression tracks are normalized to the number of reads in each sample, and the height of the track indicates reads depth. The fold enrichment by J2-IP in TDP-43 knockdown over control (normalized to total RNA-seq) for each region is shown to the right. The thin black line indicates scale, and scale is set to 500 bp. Red lines show the location of primer sequences relative to the gene. Expression tracks for NCAD1 (loading control) are shown in Supplementary Fig S20.
Source data are available online for this figure.
Increased dsRNA accumulation in TDP-43 knockdown cells suggests that human TDP-43 may also control dsRNA structure in the same transcript types observed in the tdp-1 deletion. We do not predict that human TDP-43 will affect orthologs of worm transcripts as the common feature among affected transcripts is the inherent structure of the transcript (i.e., inverted repeats) not necessarily the function of its resultant protein. To ask whether TDP-43 knockdown in human cells resulted in increased dsRNA in the same transcript types as worm tdp-1 deletion, we performed J2-IPs followed by RT–PCR of RNA isolated from TDP-43 knockdown cells and mock-treated controls. As our analysis in worms indicated that repetitive sequences and intronic regions were the most affected by tdp-1 loss of function, we focused on these transcript types. As a representative example of a repetitive sequence, we assayed the Herv-K retrotransposon, which is misregulated in ALS pathological samples (Douville et al, 2011). Comparison of the intensity of the RT–PCR product for mock-treated cells and TDP-43 siRNA-treated cells indicated that TDP-43 knockdown dramatically increased the dsRNA structure of the Herv-K transcript (Fig8E). We then asked whether intronic regions associated with human TDP-43 had increased J2-IP recovery in knock down cells by assaying Fus/TLS intron 8, which is bound by TDP-43 in three locations (Tollervey et al, 2011). Again, the J2-IP signal in TDP-43 knock down cells was more pronounced than in mock-treated samples. Finally, we asked whether transcripts dependent on TDP-43 for correct splicing contained increased dsRNA within intronic regions. We selected five transcripts with reproducible alternative splicing differences in TDP-43 pathological tissue compared to healthy tissue (Tollervey et al, 2011). Of these five candidates, four showed increased recovery in the J2-IP from TDP-43 knockdown cells compared to control (Fig8E; Supplementary Fig S19A). To confirm these results, we created RNA-seq libraries from an independent J2-IP on TDP-43 knockdown and mock-treated cells (along with input controls). After mapping and normalization to input control levels, we examined the J2-IP enrichment of regions tested by RT–PCR (Fig8F; Supplementary Fig S19B). In all cases, we were able to confirm increased J2-IP enrichment in TDP-43 knock down (over input expression levels) compared to control. These results indicate that mammalian TDP-43 limits dsRNA structure in some of the same transcript types as worm TDP-1.
To rule out the possibility that dsRNA accumulation in human cells results from decreased ADAR2 function, we asked whether knocking down TDP-43 in human M17 neuroblastoma cells led to reduced GluR2 Q607R editing, an ADAR2-specific editing site (Nishikura, 2010). We analyzed the editing by gene-specific RT–PCR followed by Sanger sequencing. We found no correlation between the amount of TDP-43 expression in M17 cells and the efficiency of editing in the GluR2 transcript (Supplementary Fig S20), indicating that dsRNA accumulation is not due to decreased ADAR2 function.
TDP-43 is an RNA chaperone with strand displacement activity
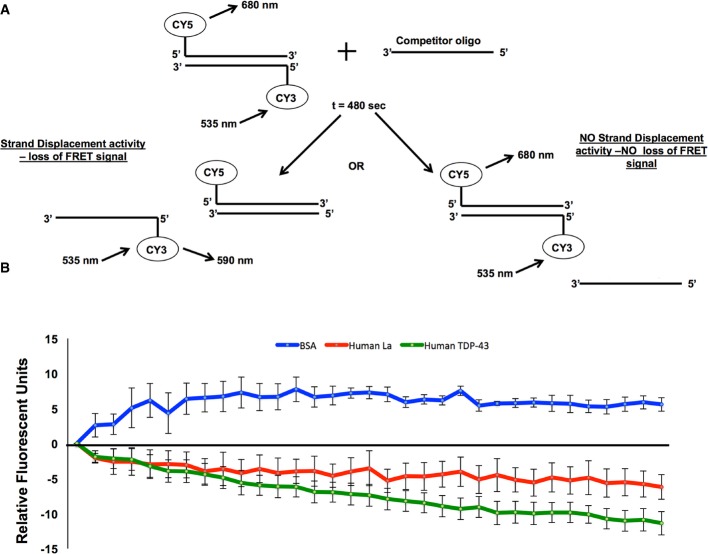
Our data indicated that TDP-43 does not limit double-stranded RNA accumulation by maintaining A-to-I RNA editing or RNA interference, as both of these processes appear hyper-active in tdp-1 deletion animals. Therefore, we postulated that TDP-43 may act directly on the RNA molecule as an RNA chaperone to limit dsRNA structure. In order to test this theory, we assayed recombinant human TDP-43 in vitro for strand displacement activity, an activity well characterized for many RNA chaperones, including human La protein (Naeeni et al, 2012). In brief, two complementary RNA oligos labeled with Cy5 or Cy3, respectively, were allowed to anneal at 37°C. Following annealing, an excess of unlabeled bottom strand was added, Cy3 was excited and the relative change in FRET (Cy5 excitation) was measured over time (see Fig9A). In the presence of our negative control (BSA), the relative FRET signal increased (Fig9B), which is expected as addition of competitor oligo results in an increase in the concentration of one of the partners in the annealing reaction (Rajkowitsch & Schroeder, 2007). However, in the presence of our positive control (human La), the relative FRET signal decreased over time due to the unlabeled bottom strand displacing the labeled strand, an activity that requires an RNA chaperone. Importantly, in the presence of recombinant human TDP-43, the relative FRET signal also significantly decreased (P = 5 × 10−31, paired t-test, BSA compared to TDP-43) at a level similar or greater than the decrease observed for human La (Fig9B). This result indicates that human TDP-43 is capable of RNA chaperone activity in vitro and suggests that TDP-43 could function as an RNA chaperone on nascent RNA in vivo in order to limit inappropriate dsRNA structure/folding during transcription.
Figure 9. Human TDP-43 displays strand dissociation activity in vitro.
- Assay for strand displacement activity (adapted from Naeeni et al, 2012).
- RNA strand dissociation activity was measured as a change in relative FRET signal (y-axis) over time (x-axis) of two complementary, fluorescently labeled (Cy5 and Cy3, respectively) RNA oligos (annealed for 480 s at 37°C) following the addition of tenfold molar excess of a competitor oligo complementary to the Cy5-labeled RNA molecule. RNA molecules were excited at 535 nm, and FRET signal was recorded at 680 nm. Initial FRET fluorescence was set to zero and then measured every 15 s for 480 s. Data represent results from six to seven independent assays. Error bars, SEM calculated between replicates at each 15 s time point. The assay was done in the presence of Bovine Serum Albumin (BSA) protein (blue line); recombinant human Lupus Antigen (Human La) (red line), and recombinant human Tar-DNA Binding protein 43 (TDP-43) (green line).
Discussion
TDP-43 is involved in many different RNA processes, making the identification of this protein's role in neurodegeneration complex. Because a large number of binding partners have been identified for TDP-43 (Freibaum et al, 2010), it is possible that TDP-43 affects many aspects of RNA metabolism by participating in multiple protein complexes. An alternative, although not mutually exclusive, possibility is that TDP-43 interacts with RNA in a way that is important for many different RNA-mediated processes. In this study, we uncovered a fundamental role for TDP-1 in directly controlling dsRNA accumulation and processing. Furthermore, we showed that knockdown of human TDP-43 also increases dsRNA accumulation, implying that functions described for worm TDP-1 have relevance to mammalian TDP-43.
The hypothesis that a fundamental function of TDP-1/TDP-43 is to control the accumulation of dsRNA is strongly supported by the studies of Polymenidou et al, who examined gene expression in mouse brain after TDP-43 knockdown. Strikingly, 29 of the top 50 transcripts overexpressed after TDP-43 knockdown are type 1 interferon-inducible genes (Samarajiwa et al, 2009) (Supplementary Table S12). Included in the up-regulated genes in TDP-43 knock-down mice were the mouse orthologs of PKR (up 3.9-fold), RIG-1 (up 4.1-fold) and MDA5 (up 3.4-fold), which specifically respond to cytoplasmic dsRNA (Li et al, 2011). Because interferon activation is a major consequence of excessive dsRNA mammals, it would be interesting to determine whether increased interferon activation contributes to neuronal toxicity in mammals.
While worms lack an interferon response, our data indicate that processes downstream of dsRNA (i.e., RNAi) result in the neuronal phenotypes of tdp-1 mutants. In addition, Vaccaro et al (2012) demonstrated that the tdp-1(ok803) allele renders worms hypersensitive to oxidative stress and interacts with the C. elegans insulin-like signaling (ILS) pathway. Interestingly, daf-16 (the key transcription factor controlling ILS in C. elegans) is hyper-edited in tdp-1(ok803) (Fig3C), suggesting increased dsRNA structure in tdp-1(ok803) worms could contribute to the altered stress sensitivity.
Identification of dsRNA by immunoprecipitation and deep sequencing
In this study, we utilized anti-dsRNA immunoprecipitation experiments to globally identify transcripts containing double-stranded structure. Isolation and sequencing of dsRNA from lysates have the advantage of identifying transcripts likely to contain double-stranded RNA in vivo, in contrast to protocols that rely on isolation and refolding of RNA in vitro (Underwood et al, 2010). Refolding after RNA isolation could potentially disrupt RNA interactions or introduce structure that normally does not exist in vivo. While immunoprecipitation of dsRNA from lysates could recover transcripts that are not themselves double-stranded, but are associated with dsRNA, almost 75% of J2-immunoprecipitated transcripts had dsRNA characteristics. This result indicates that this method does preferentially identify transcripts containing dsRNA. In addition, anti-dsRNA immunoprecipitation isolated a significant amount of pre-mRNA or excised intronic sequences allowing the analysis of structured transcripts that affect pre-mRNA processing, but are normally too transient to detect.
Characteristics of transcripts with increased double-stranded RNA structure in tdp-1 mutants
Immunoprecipitation of dsRNA in tdp-1 mutants indicates that many transcripts rely on TDP-1 to limit intra-strand and/or inter-strand structure or stability. The variety among these transcripts is quite striking, such that dsRNA immunoprecipitation in tdp-1 mutants enriched for both coding and intronic RNAs, as well as most replicative histone transcripts. We do not know which of these transcripts constitute the J2-immunopositive foci, or whether foci in different cell types contain different transcripts. The visible dsRNA foci in TDP-43 knockout/knockdown likely contain only a sub-population of accumulating dsRNA molecules that are retained (or persist) due to innate characteristics of those transcripts. Importantly, expression of tandem array transgenes in C. elegans, which is known to produce dsRNA (Knight & Bass, 2002), can also lead to the accumulation of J2-immunoreactive nuclear foci (C. D. Link, unpublished results). This observation suggests both that J2 foci can result from accumulation of a limited set of transcripts, and the detection of J2-immunoreactive foci does not require removal of endogenous TDP-1 (as would be expected, for example, if TDP-1/TDP-43 binding to transcripts obscured epitopes recognized by the J2 antibody).
Importantly, the transcripts enriched by J2-IP in tdp-1 mutants undergo distinctly different processing events. Some of the enriched transcripts undergo pre-mRNA splicing (most mRNAs), and some do not (replicative histone transcripts and ncRNAs). Similarly, ncRNAs and replicative histone transcripts have unique 3′-end formation pathways that differ from the canonical cleavage and polyadenylation machinery used by mRNAs (Wilusz & Spector, 2010). Given the varied maturation pathways of transcripts whose double-stranded structure is decreased by TDP-1, it is unlikely that TDP-1 reduces this structure by controlling steps in RNA processing (i.e., splicing or 3′-end formation). Rather, TDP-1 is likely to play a fundamental role in limiting RNA structure of many origins in a variety of transcript types.
Transcript abundance and splicing in tdp-1 mutants
In the absence of tdp-1, potentially structured transcripts are both increased and decreased in the mature (poly-A) RNA pool. These same transcripts are associated with TDP-1 and contain increased amounts of dsRNA structure/abundance in tdp-1 mutants. We do not know the exact mechanism by which increased dsRNA formation/stability alters the abundance of potentially structured mRNAs in tdp-1 mutants. However, increased structure could either increase or decrease transcript abundance depending on the individual regulatory pathways acting on each transcript. For example, increased structure could trigger siRNA production, which is known to down-regulate transcript abundance of antisense-overlap transcripts in multiple systems (Ghildiyal et al, 2008; Gu et al, 2009; Gullerova et al, 2011). Alternatively, increased intra- or inter-molecular structure in the RNA could preclude association of factors that promote degradation (such as micro-RNAs or RNA-binding proteins), increasing transcript abundance. Consistent with both of these possibilities, many aberrantly represented overlapping gene pairs in tdp-1 mutants are co-altered, such that if one gene in the pair is increased/decreased, the other gene in the pair is also increased/decreased.
Further, inappropriate formation of dsRNA in introns could result in instability and degradation of those transcripts through the action of dsRNA-binding proteins and dsRNA nucleases. Nuclear dsRNA-binding proteins are known to destabilize structured pre-mRNA (Danin-Kreiselman et al, 2003; Kadener et al, 2009). Our data also indicate that a large number of excessively structured introns display altered splicing patterns in tdp-1 mutants. The intronic A-to-I RNA editing could contribute to the splicing abnormalities as editing is known to alter splice site recognition (Laurencikiene et al, 2006). dsRNA structure itself also affects splicing through multiple mechanisms (Warf & Berglund, 2010). Formation of a dsRNA stem-loop is responsible for the skipping of exon 7 in the Smn2 transcripts. Interestingly, TDP-43 binds Smn2 pre-mRNA and stimulates inclusion of this same exon (Singh et al, 2007). Whether TDP-43 affects the structure of SMN2 RNA, however, is currently unknown.
How does TDP-1 limit dsRNA accumulation?
Data presented here indicate that TDP-43 can act as an RNA chaperone in vitro. TDP-43 may function in this capacity to maintain correct RNA folding during active transcription. In the absence of TDP-43, inappropriate RNA folding could result in abbarent dsRNA structure leading to defects in RNA processing and degradation. We cannot rule out the possibility that TDP-43 limits dsRNA accumulation through additional or alternative pathways. TDP-43 could also act as an RNA chaperone post-transcriptionally. TDP-43 has been shown to localize to a variety of RNA-containing cellular bodies in both the nucleus and cytoplasm (Da Cruz & Cleveland, 2011). Perhaps, TDP-43 maintains the function of these bodies by ensuring the correct structure of the transcripts contained within. Importantly, TDP-43 is a known component of nuclear paraspeckles, which are nuclear bodies that retain edited and structured RNAs (Fox & Lamond, 2010). TDP-43 could control the retention/release of transcripts in paraspeckles by altering RNA structure. Consistent with this possibility, mouse Slc7a2 RNA, which was the first transcript identified as retained in paraspeckles due to an edited hairpin in the 3′ UTR, is bound by mouse TDP-43 in four distinct regions within the 3′ UTR (Polymenidou et al, 2011). Interestingly, FUS/TLS and Matrin3, other RNA-binding proteins causative in ALS (Kwiatkowski et al, 2009; Johnson et al, 2014), are also components of paraspeckles (Naganuma et al, 2012).
Is dsRNA accumulation relevant to ALS/FTLD pathology?
TDP-43 is aberrantly localized to the cytoplasm in several pathological situations. Whether dsRNA is involved in this aberrant localization is an intriguing question. TDP-43 mislocalization is associated with CUG repeat expansion diseases (Schwab et al, 2008; Hart & Gitler, 2012), as well as with the recently identified C90orf72 ALS-FTD mutation, which consists of an expanded GGGGCC repeat (DeJesus-Hernandez et al, 2011). Transcripts containing either of these RNA repeats form extensive dsRNA structures. Furthermore, age-associated decreases in the mouse ADAR2 ortholog is concurrent with aberrant TDP-43 localization in motor neurons (Hideyama et al, 2012). Decreased ADAR2 function would be predicted to increase nuclear dsRNA. Finally, NEAT1 RNA, which nucleates paraspeckles, is induced in mouse brains by dsRNA viruses (Saha et al, 2006) and is highly up-regulated in human brains affected by FTLD (Tollervey et al, 2011), consistent with a potential increase of dsRNA accumulation in FTLD pathology. Therefore, future studies to determine whether aberrant dsRNA formation causes, or is caused by, the pathological mislocalization of TDP-43 are paramount.
Materials and Methods
Caenorhabditis elegans strains
Maintenance and growth of worms were performed as described in (Brenner, 1974), and all strains were raised at 20°C unless otherwise noted. All transgenic strains used in this study were created by gonad injection and integration of DNA array. Supplementary Table S12 lists all strains created or used in this work.
Chemotaxis assays
Chemotaxis assays were performed as described previously (Kauffman et al, 2011). 100-mm agar plates were spotted with 1 μl of attractant diluted 1:100 or 1 μl dilution buffer and 1 μl 1 mM sodium azide to paralyze worms arriving in either attractant or buffer-only spot. Worms were synchronized by alkaline hypochlorite treatment and hatched off overnight in S-basal buffer. Animals were then allowed to grow to 1-day-old adults at 20°C (about 72 h). Approximately 200 animals were added to each assay plate in a defined origin position in 50 μl of S-basal buffer, released simultaneously, and worms were allowed to crawl freely for 2 h. The total number of animals in the attractant spot, buffer-only spot and in undefined locations around the plate were counted. The chemotaxis assay was determined according to the equation: 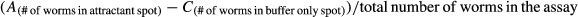 .
.
Thrashing assays
Liquid thrashing assays were performed using synchronized 1-day-old adults grown at 20°C. Thrashes were counted for 30 s under a dissecting microscope by hand. For automated thrash counting, C. elegans body bends were captured using the wrMTrck plugin developed by Jesper S Pedersen (http://www.phage.dk/plugins/wrmtrck.html). Body bends of 4 L4 worms thrashing in 1× S basal were captured for 30 s. Three independent biological replicates were analyzed for wild-type and tdp-1(ok803) animals, resulting in a total of 12 worms per group. All the movies were filtered as suggested in the manual prior to body bend analysis.
Caenorhabditis elegans immunoblotting and immunohistochemistry
Immunoblotting of worm and human proteins was done according to Ash et al, 2010. 20 μg of total protein was loaded per lane, and the resultant blots were probed with primary antibodies against anti-TDP-1 (1:2,000, in house), anti-tubulin (1:4,000, Sigma), anti-TDP-43 (1:2,000, Abnova) or anti-GAPDH (1:4,000, BioDesign). Blots were probed with secondary HRP-conjugated antibodies (1:5,000, Jackson) and developed in ECL Plus (Amersham). For immunohistochemistry, permeabilized whole-mount preparations were probed with J2 antibody (English & Scientific Consulting Lot: J2-1102 and J2-1103) at 2–5 μg/ml for 16 h at 4°C and Alexa dye-conjugated goat anti-mouse secondary antibody (Invitrogen) at 4–10 μg/ml for 2 h at room temperature. To assay the sensitivity of J2 immunoreactivity, permeabilized worms were incubated with either RNase V1 (0.003 U/μl, 30 min at 37°C) or RNase T1 (30 U/ml, 30 min, room temperature) in RNA structure buffer (all from Invitrogen, AM2275). RNase-treated worms were washed and probed with J2 as described above. Images were acquired with a Zeiss Axiophot microscope equipped with digital deconvolution optics (Intelligent Imaging Innovations), and image brightness and contrast were digitally adjusted in Photoshop.
Extracts
Extracts for J2-IP experiments were made from young adult animals grown on solid plates at 15°C. Worms used for TDP-1 immunoprecipitation followed by RT–PCR were cross-linked according to Zisoulis et al, 2010. Worms used for J2-IP were not cross-linked. Extract was prepared by bead beating as described in Saldi et al, 2007. Extracts were used immediately for immunoprecipitations or dissolved in TRIzol (for input RNA).
RNA immunoprecipitations
Immunoprecipitations were preformed as described (Saldi et al, 2007). 20 μl of Protein A magnetic beads (Dynabeads, Invitrogen) were washed, blocked, and bound by antibody at a ratio of 10 μl/IP for both anti-TDP-1 (made in house) and anti-J2. Protein concentration of each extract was determined by Bradford assay (Bio-Rad 500-0111), and 100 μg of protein was added to each IP. dsRNase-treated extract was incubated with 1 μl of RNase V1 for 30 min (room temperature) prior to addition to beads. Tubes were rocked at 15°C for 2 h (anti-J2 IPs) or 4°C overnight (anti-TDP-1 IPs). The supernatant was removed, beads were washed, and immunoprecipitated RNA was removed from beads by TRIzol extraction. DNA was removed from immunoprecipitated RNA using TURBO DNase (Invitrogen) and dissolved in nuclease-free water.
Chromatin immunoprecipitation (ChIP)
ChIP was performed as described (Garrido-Lecca & Blumenthal, 2010). 20 μl TDP-1 antibody was used. DNA was purified using a QIAGEN column (QIAquick, # 28104) and eluted twice with 30 μl of water. For RNase-treated extract, a combination of RNases (RNase A, T1 and V1) was added to the extract and rocked for 15 min at room temperature prior to addition to the ChIP. Completion of RNase was confirmed on an aliquot of treated and untreated extract on acrylamide gel (data not shown).
Cell culture and immunohistochemistry
M17 neuroblastoma and HeLa cells were cultured in OptiMEM (Invitrogen), and when confluent, 1.75 × 104 cells/well plated to 24-well plates on coverslips. TDP-43 was knocked down for 72 h with 20 nM TARDBP siRNA (QIAGEN) (Prudencio et al, 2012) or All Star control siRNA (QIAGEN) and siLentFect reagent (Bio-Rad) and fixed in 4% PFA in DEPC-treated PBS and permeabilized. Nuclease treatment of HeLa cells was performed at 37°C for 30 min with 100 U/ml of RNase-free DNase I and RNase A (QIAGEN), ShortCut RNase III (NEB) or ultrapure benzonase nuclease (Sigma) each in their recommended buffers. Protein Block Serum Free (Dako) was used for blocking. Primary antibody incubation was done overnight in Antibody Diluent (Dako) with 0.25 μl/ml RNase OUT and 1:1,000 anti-TDP-43-Cterm (ProteinTech) and 1:1,000 J2, and secondary antibodies were 1:1,000 (donkey) anti-rabbit-AF488 and anti-mouse-AF568 (Alexafluor). Coverslips were counterstained with 0.1 μg/ml Hoechst. Images were taken by Zeiss AxioImager Z1 with Apotome at 63× with fixed exposures times.
RNA isolation, cDNA library preparation, high-throughput sequencing
RNA for poly(A) and total RNA sequencing libraries was extracted from whole animals by TRIzol extraction. Genomic DNA was removed using TURBO DNase (Invitrogen). For total RNA libraries, 5 μg of RNA was run through a Ribo-Zero column (Epicenter, #R2C1046) to remove ribosomal RNA. For poly(A)-selected libraries, RNA was selected using sera-mag magnetic oligo dT beads (Therma Scientific). Libraries were created using Illumina TruSeq kits (RS-122-2001). RNA recovered by immunoprecipitation with the J2 antibody (three biologically independent lysates) of young adult worms as well as input material (as a loading control) was converted into strand-specific total RNA libraries using V2 Scriptseq (Epicenter #SSV21106) kits following manufacturer's instructions, except reverse transcription was done with SuperScript III (Invitrogen #18080-044) using incrementally increasing temperatures from 42 to 59°C to allow for transcription though structured RNAs. rRNA was not removed from J2-IP RNA samples. Immunoprecipitated DNA from ChIP samples was converted in sequencing libraries using ChIP-Seq DNA Sample Prep Kit (IP-102–1001). Libraries were sequenced on Illumina HiSeq 2000 platforms. Data were deposited under GEO accession number GSE61581.
Sequencing alignment, gene expression quantification, and J2-IP analysis
Detailed algorithms and specifications for sequencing analysis including read filtering, mapping, and differential expression calculations are provided in the Supplementary Methods and Supplementary Fig S14. In brief, reads were trimmed and filtered, and rRNA reads were removed. Reads were aligned to the WS220 C. elegans genome. Differential expression in poly(A) selected libraries between wild-type and tdp-1(ok803) mutant was calculated using the Bioconductor package edgeR (release 2.12) (Robinson et al, 2010). Identification of transcripts enriched by J2-IP in wild-type animals was done by comparing RPKMs of expressed genes/repeat regions in the J2-IP RNA-seq to input RNA-seq. Genes/repeat regions that were significantly (corrected P < 0.05) increased in abundance > 1.5-fold over input were considered enriched by J2-IP. To determine genes, repeat regions and introns differentially selected by J2-IP in wild-type versus tdp-1(ok803) extract (three biological replicates of each), read counts for well-expressed regions were normalized to input (see Supplementary Methods) and read into DESeq v1.14.0. A significance cut-off of P < 0.05 and FDR < 0.1 was used.
Identification of adr-2-dependent editing locations
For wild-type, adr-2(gv42), and tdp-1(ok803) total RNA samples, the alignments were piled up using an in-house program. Positions in wild-type and tdp-1(ok803) samples with a discrepancy between the genomic sequence and mRNA sequence consistent with A-to-I editing (A-G and C-T transitions) were identified. Potential edited positions were compared to the adr-2(gv42) sample and sites also showing A-to-G or C-to-T changes were discarded. To identify editing in introns enriched by J2 immunoprecipitation (not identified as edited in our initial analysis of total RNA due to insufficient read depth), reads mapping to all J2-IP enriched introns were inspected visually on the IGV genome browser and hand-annotated as edited if editing was well represented and was present in all three biological replicates. Additional details in Supplementary Methods.
Calculation of RNA editing frequency in tdp-1(ok803) mutants compared to wild-type
The Integrative Genome Viewer (IGV) tool, count, was used to transform alignment files into pileups for downstream analysis. Well-expressed (> 20 reads) edited regions were used to compare percent editing between wild-type and tdp-1(ok803). The total number of edited nucleotides was divided by the total number of potentially edited nucleotides (determined by evidence of editing in either sample) for each location. Significant changes were determined by performing a two-proportion z-test between wild-type and tdp-1(ok803) percent edited ratios; P < 0.05 for significant changes.
Gene ontology term enrichment
David (Huang da et al, 2009) was used to calculate enriched gene ontology (GO) terms. A list of unranked significantly up- and down-regulated genes was compared to a background list containing all C. elegans genes expressed in our RNA-seq samples.
Splicing analysis
To capture splicing changes between wild-type and tdp-1(ok803) mutants, poly(A)-selected RNA from each strain was resequenced 100 bp paired-end at a high read depth (∼280 million reads per sample) to achieve the depth and mapping specificity to analyze alternative splicing. The Cuffdiff (v1.30) differential splicing test was used for splicing analysis.
ChIP peak identification
Filtered and trimmed ChIP-seq samples were aligned to WS220 using Bowtie. Significant TDP-1 ChIP peaks were identified using MACS software (version 1.42) (Zhang et al, 2008). Two replicates of TDP-1 ChIP-seq were done, and a TDP-1 ChIP-seq pretreated with RNase was used as the control file. Bandwidth was set to 300, a 2-fold to 30-fold change over control was required, and the P-value cut-off was set at P < 0.01. Regions positive for TDP-1 peaks were counted only if an overlapping peak existed in both biological replicates, and there was no peak in the RNase-treated sample.
Strand displacement assay
Strand displacement assay was done as described in Rajkowitsch & Schroeder (2007) with the following modifications: Labeled and competitor RNA oligos were obtained from IDT, and recombinant TDP-43 and La protein were obtained from GenWay Biotech, Inc. RNA oligo sequences were taken from Naeeni et al, 2012. Initial annealing was done at 37°C for 480 s in 40 μl assay buffer. Labeled oligos were used at a final concentration of 1 μM, and recombinant protein was used at 10 μM. Competitor oligo was added in 10 μl for a final concentration of 100 μM in 50 μl. The mixture was shaken for 2 s, excited at 535 nm, and florescence was read at 690 nm every 15 s for 480 s at 37°C. The assay was done in a 384-well black microtiter plate (Greiner Bio-one). All measurements were taken in a SpectraMax M5 microplate reader machine, and SoftMax Pro software was used to calculate relative florescent units for each sample. The assay was done seven independent times, and replicates deviating more than two standard deviations from the mean were removed, such that data represent six replicates of BSA, six replicates of human La and seven replicates of human TDP-43.
Acknowledgments
We would like to thank Kyle Dack for assistance with the chemotaxis assays. Some nematode strains were provided by the Caenorhabditis Genetics Center, funded by the NIH National Center for Research Resources. This work was supported by NIH grants NS063964 (CDL), GM42432 (TB), AG026251, NS063964, ES20395 (LP), DOD grants W81XWH-10-1-0512-1 and W81XWH-09-1-0315AL093108 (LP), and the CIHR Frederick Banting and Charles Best Canada Graduate Scholarship (GW).
Author contributions
TS and CL conceived the project, designed the experiments, and wrote the manuscript. TS produced data in Figs1 and 3, 4, 5, 6, 7, 8 and 9 and most Supplementary Figures and contributed to bioinformatics analysis. CL produced data in Figs2 and 7, Supplementary Figs S4 and S5. PA produced data in Fig8, Supplementary Figs S18 and S20. GW and LS contributed to data in Fig3. PG did the bulk of the data processing and bioinformatics. AG contributed to data in Fig6. CR produced data in Supplementary Figs S1 and S2. VD contributed to data in Fig7. TG contributed to data in Supplementary Fig S20. TB and LP made intellectual contributions to the design and analysis of the experiments and helped with the drafting of the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary information for this article is available online: http://emboj.embopress.org
Supplementary Figures
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3
Supplementary Table S4
Supplementary Table S5
Supplementary Table S6
Supplementary Table S7
Supplementary Table S8
Supplementary Table S9
Supplementary Table S10
Supplementary Table S11
Supplementary Table S12
Supplementary Table S13
Supplementary Table S14
Supplementary Table S15
Supplementary Methods
Legends for Supplementary Figures
Source Data for Supplementary Figure S1
Source Data for Supplementary Figure S3
Source Data for Supplementary Figure S7
Source Data for Supplementary Figure S9
Source Data for Supplementary Figure S15
Source Data for Supplementary Figure S19
Source Data for Supplementary Figure S20
Review Process File
Source Data for Figure 6
Source Data for Figure 8
References
- Ash PE, Zhang YJ, Roberts CM, Saldi T, Hutter H, Buratti E, Petrucelli L, Link CD. Neurotoxic effects of TDP-43 overexpression in C. elegans. Hum Mol Genet. 2010;19:3206–3218. doi: 10.1093/hmg/ddq230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala YM, Pantano S, D'Ambrogio A, Buratti E, Brindisi A, Marchetti C, Romano M, Baralle FE. Human, Drosophila, and C.elegans TDP43: nucleic acid binding properties and splicing regulatory function. J Mol Biol. 2005;348:575–588. doi: 10.1016/j.jmb.2005.02.038. [DOI] [PubMed] [Google Scholar]
- Bargmann CI. 2006. Chemosensation in C. elegans (October 25, 2006), WormBook, ed C. elegans. The Research Community, WormBook. doi: 10.1895/wormbook.1.123.1, http://www.wormbook.org.
- Bass BLWH. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell. 1998;55:1089–1098. doi: 10.1016/0092-8674(88)90253-x. [DOI] [PubMed] [Google Scholar]
- Bieberstein NI, Straube K, Neugebauer KM. Chromatin immunoprecipitation approaches to determine co-transcriptional nature of splicing. Methods Mol Biol. 2014;1126:315–323. doi: 10.1007/978-1-62703-980-2_23. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E, Baralle FE. Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J Biol Chem. 2001;276:36337–36343. doi: 10.1074/jbc.M104236200. [DOI] [PubMed] [Google Scholar]
- Chen-Plotkin AS, Lee VM, Trojanowski JQ. TAR DNA-binding protein 43 in neurodegenerative disease. Nat Rev Neurol. 2010;6:211–220. doi: 10.1038/nrneurol.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Cruz S, Cleveland DW. Understanding the role of TDP-43 and FUS/TLS in ALS and beyond. Curr Opin Neurobiol. 2011;21:904–919. doi: 10.1016/j.conb.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danin-Kreiselman M, Lee CY, Chanfreau G. RNAse III-mediated degradation of unspliced pre-mRNAs and lariat introns. Mol Cell. 2003;11:1279–1289. doi: 10.1016/s1097-2765(03)00137-0. [DOI] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung GY, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douville R, Liu J, Rothstein J, Nath A. Identification of active loci of a human endogenous retrovirus in neurons of patients with amyotrophic lateral sclerosis. Ann Neurol. 2011;69:141–151. doi: 10.1002/ana.22149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiguin F, Godena VK, Romano G, D'Ambrogio A, Klima R, Baralle FE. Depletion of TDP-43 affects Drosophila motoneurons terminal synapsis and locomotive behavior. FEBS Lett. 2009;583:1586–1592. doi: 10.1016/j.febslet.2009.04.019. [DOI] [PubMed] [Google Scholar]
- Fischer SE. Small RNA-mediated gene silencing pathways in C. elegans. Int J Biochem Cell Biol. 2010;42:1306–1315. doi: 10.1016/j.biocel.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Fox AH, Lamond AI. Paraspeckles. Cold Spring Harb Perspect Biol. 2010;2:a000687. doi: 10.1101/cshperspect.a000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freibaum BD, Chitta RK, High AA, Taylor JP. Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J Proteome Res. 2010;9:1104–1120. doi: 10.1021/pr901076y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido-Lecca A, Blumenthal T. RNA polymerase II C-terminal domain phosphorylation patterns in Caenorhabditis elegans operons, polycistronic gene clusters with only one promoter. Mol Cell Biol. 2010;30:3887–3893. doi: 10.1128/MCB.00325-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, Zamore PD. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu R, Zhang Z, DeCerbo JN, Carmichael GG. Gene regulation by sense-antisense overlap of polyadenylation signals. RNA. 2009;15:1154–1163. doi: 10.1261/rna.1608909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullerova M, Moazed D, Proudfoot NJ. Autoregulation of convergent RNAi genes in fission yeast. Genes Dev. 2011;25:556–568. doi: 10.1101/gad.618611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart MP, Gitler AD. ALS-associated ataxin 2 polyQ expansions enhance stress-induced caspase 3 activation and increase TDP-43 pathological modifications. J Neurosci. 2012;32:9133–9142. doi: 10.1523/JNEUROSCI.0996-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelett DJ, Chang CJ, Lakeland DL, Morton DB. Comparison of parallel high-throughput RNA sequencing between knockout of TDP-43 and its overexpression reveals primarily nonreciprocal and nonoverlapping gene expression changes in the central nervous system of drosophila. G3 (Bethesda) 2012;2:789–802. doi: 10.1534/g3.112.002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hideyama T, Teramoto S, Hachiga K, Yamashita T, Kwak S. Co-occurrence of TDP-43 mislocalization with reduced activity of an RNA editing enzyme, ADAR2, in aged mouse motor neurons. PLoS One. 2012;7:e43469. doi: 10.1371/journal.pone.0043469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Johnson JO, Pioro EP, Boehringer A, Chia R, Feit H, Renton AE, Pliner HA, Abramzon Y, Marangi G, Winborn BJ, Gibbs JR, Nalls MA, Morgan S, Shoai M, Hardy J, Pittman A, Orrell RW, Malaspina A, Sidle KC, Fratta P, et al. Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat Neurosci. 2014;17:664–666. doi: 10.1038/nn.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S, Rodriguez J, Abruzzi KC, Khodor YL, Sugino K, Marr MT, 2nd, Nelson S, Rosbash M. Genome-wide identification of targets of the drosha–pasha/DGCR8 complex. RNA. 2009;15:537–545. doi: 10.1261/rna.1319309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko H, Dridi S, Tarallo V, Gelfand BD, Fowler BJ, Cho WG, Kleinman ME, Ponicsan SL, Hauswirth WW, Chiodo VA, Kariko K, Yoo JW, Lee DK, Hadziahmetovic M, Song Y, Misra S, Chaudhuri G, Buaas FW, Braun RE, Hinton DR, et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011;471:325–330. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman A, Parsons L, Stein G, Wills A, Kaletsky R, Murphy C. C. elegans positive butanone learning, short-term, and long-term associative memory assays. J Vis Exp. 2011:e2490. doi: 10.3791/2490. , doi:10.3791/2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SW, Bass BL. The role of RNA editing by ADARs in RNAi. Mol Cell. 2002;10:809–817. doi: 10.1016/s1097-2765(02)00649-4. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski TJ, Jr, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, Valdmanis P, Rouleau GA, Hosler BA, Cortelli P, de Jong PJ, Yoshinaga Y, Haines JL, Pericak-Vance MA, Yan J, Ticozzi N, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- Laurencikiene J, Kallman AM, Fong N, Bentley DL, Ohman M. RNA editing and alternative splicing: the importance of co-transcriptional coordination. EMBO Rep. 2006;7:303–307. doi: 10.1038/sj.embor.7400621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XL, Ezelle HJ, Hsi TY, Hassel BA. A central role for RNA in the induction and biological activities of type 1 interferons. Wiley Interdiscip Rev RNA. 2011;2:58–78. doi: 10.1002/wrna.32. [DOI] [PubMed] [Google Scholar]
- Li W, Jin Y, Prazak L, Hammell M, Dubnau J. Transposable elements in TDP-43-mediated neurodegenerative disorders. PLoS One. 2012;7:e44099. doi: 10.1371/journal.pone.0044099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse DP, Aruscavage PJ, Bass BL. RNA hairpins in noncoding regions of human brain and Caenorhabditis elegans mRNA are edited by adenosine deaminases that act on RNA. Proc Natl Acad Sci USA. 2002;99:7906–7911. doi: 10.1073/pnas.112704299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeeni AR, Conte MR, Bayfield MA. RNA chaperone activity of human La protein is mediated by variant RNA recognition motif. J Biol Chem. 2012;287:5472–5482. doi: 10.1074/jbc.M111.276071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naganuma T, Nakagawa S, Tanigawa A, Sasaki YF, Goshima N, Hirose T. Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 2012;31:4020–4034. doi: 10.1038/emboj.2012.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou SH, Wu F, Harrich D, Garcia-Martinez LF, Gaynor RB. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J Virol. 1995;69:3584–3596. doi: 10.1128/jvi.69.6.3584-3596.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesiridis GS, Lee VM, Trojanowski JQ. Mutations in TDP-43 link glycine-rich domain functions to amyotrophic lateral sclerosis. Hum Mol Genet. 2009;18:R156–R162. doi: 10.1093/hmg/ddp303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, Ling SC, Sun E, Wancewicz E, Mazur C, Kordasiewicz H, Sedaghat Y, Donohue JP, Shiue L, Bennett CF, Yeo GW, Cleveland DW. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci. 2011;14:459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudencio M, Jansen-West KR, Lee WC, Gendron TF, Zhang YJ, Xu YF, Gass J, Stuani C, Stetler C, Rademakers R, Dickson DW, Buratti E, Petrucelli L. Misregulation of human sortilin splicing leads to the generation of a nonfunctional progranulin receptor. Proc Natl Acad Sci USA. 2012;109:21510–21515. doi: 10.1073/pnas.1211577110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkowitsch L, Schroeder R. Dissecting RNA chaperone activity. RNA. 2007;13:2053–2060. doi: 10.1261/rna.671807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Murthy S, Rangarajan PN. Identification and characterization of a virus-inducible non-coding RNA in mouse brain. J Gen Virol. 2006;87:1991–1995. doi: 10.1099/vir.0.81768-0. [DOI] [PubMed] [Google Scholar]
- Saldi T, Wilusz C, MacMorris M, Blumenthal T. Functional redundancy of worm spliceosomal proteins U1A and U2B''. Proc Natl Acad Sci USA. 2007;104:9753–9757. doi: 10.1073/pnas.0701720104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarajiwa SA, Forster S, Auchettl K, Hertzog PJ. INTERFEROME: the database of interferon regulated genes. Nucleic Acids Res. 2009;37:D852–D857. doi: 10.1093/nar/gkn732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savva YA, Rieder LE, Reenan RA. The ADAR protein family. Genome Biol. 2012;13:252. doi: 10.1186/gb-2012-13-12-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonborn J, Oberstrass J, Breyel E, Tittgen J, Schumacher J, Lukacs N. Monoclonal antibodies to double-stranded RNA as probes of RNA structure in crude nucleic acid extracts. Nucleic Acids Res. 1991;19:2993–3000. doi: 10.1093/nar/19.11.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab C, Arai T, Hasegawa M, Yu S, McGeer PL. Colocalization of transactivation-responsive DNA-binding protein 43 and huntingtin in inclusions of Huntington disease. J Neuropathol Exp Neurol. 2008;67:1159–1165. doi: 10.1097/NEN.0b013e31818e8951. [DOI] [PubMed] [Google Scholar]
- Sephton CFGS, Atkin S, Dewey CM, Mayer P, 3rd, Herz J, Yu G. TDP-43 is a developmentally regulated protein essential for early embryonic development. J Biol Chem. 2010;285:6826–6834. doi: 10.1074/jbc.M109.061846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NN, Singh RN, Androphy EJ. Modulating role of RNA structure in alternative splicing of a critical exon in the spinal muscular atrophy genes. Nucleic Acids Res. 2007;35:371–389. doi: 10.1093/nar/gkl1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinburne IA, Meyer CA, Liu XS, Silver PA, Brodsky AS. Genomic localization of RNA binding proteins reveals links between pre-mRNA processing and transcription. Genome Res. 2006;16:912–921. doi: 10.1101/gr.5211806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierry-Mieg D, Thierry-Mieg J. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7:S12. 11–S12. 14. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, Konig J, Hortobagyi T, Nishimura AL, Zupunski V, Patani R, Chandran S, Rot G, Zupan B, Shaw CE, Ule J. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci. 2011;14:452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin LA, Bass BL. Mutations in RNAi rescue aberrant chemotaxis of ADAR mutants. Science. 2003;302:1725. doi: 10.1126/science.1091340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood JG, Uzilov AV, Katzman S, Onodera CS, Mainzer JE, Mathews DH, Lowe TM, Salama SR, Haussler D. FragSeq: transcriptome-wide RNA structure probing using high-throughput sequencing. Nat Methods. 2010;7:995–1001. doi: 10.1038/nmeth.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccaro A, Tauffenberger A, Ash PE, Carlomagno Y, Petrucelli L, Parker JA. TDP-1/TDP-43 regulates stress signaling and age-dependent proteotoxicity in Caenorhabditis elegans. PLoS Genet. 2012;8:e1002806. doi: 10.1371/journal.pgen.1002806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warf MB, Berglund JA. Role of RNA structure in regulating pre-mRNA splicing. Trends Biochem Sci. 2010;35:169–178. doi: 10.1016/j.tibs.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warf MB, Shepherd BA, Johnson WE, Bass BL. Effects of ADARs on small RNA processing pathways in C. elegans. Genome Res. 2012;22:1488–1498. doi: 10.1101/gr.134841.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Spector DL. An unexpected ending: noncanonical 3′ end processing mechanisms. RNA. 2010;16:259–266. doi: 10.1261/rna.1907510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Lamm AT, Fire AZ. Competition between ADAR and RNAi pathways for an extensive class of RNA targets. Nat Struct Mol Biol. 2011;18:1094–1101. doi: 10.1038/nsmb.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Hwang HY, Hao H, Talbot C, Jr, Wang J. Caenorhabditis elegans RNA-processing protein TDP-1 regulates protein homeostasis and life span. J Biol Chem. 2012;287:8371–8382. doi: 10.1074/jbc.M111.311977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zisoulis DG, Lovci MT, Wilbert ML, Hutt KR, Liang TY, Pasquinelli AE, Yeo GW. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat Struct Mol Biol. 2010;17:173–179. doi: 10.1038/nsmb.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3
Supplementary Table S4
Supplementary Table S5
Supplementary Table S6
Supplementary Table S7
Supplementary Table S8
Supplementary Table S9
Supplementary Table S10
Supplementary Table S11
Supplementary Table S12
Supplementary Table S13
Supplementary Table S14
Supplementary Table S15
Supplementary Methods
Legends for Supplementary Figures
Source Data for Supplementary Figure S1
Source Data for Supplementary Figure S3
Source Data for Supplementary Figure S7
Source Data for Supplementary Figure S9
Source Data for Supplementary Figure S15
Source Data for Supplementary Figure S19
Source Data for Supplementary Figure S20
Review Process File
Source Data for Figure 6
Source Data for Figure 8