Abstract
The Hippo pathway regulates tissue growth and organ size, and inactivation contributes to cancer. Signals flow through Mst/Lats kinases, which phosphorylate and promote cytoplasmic localization of the transcriptional regulators Yap and Taz to inhibit transcription. Here, we identify the multidomain-containing guanine nucleotide exchange factor (GEF) Arhgef7, or βPix, as a positive Hippo pathway regulator. We show that βPix, which localizes to the cytoplasm, binds both Lats and Yap/Taz and thereby promotes Lats-mediated phosphorylation of Yap/Taz in a GEF-independent manner. βPix is required downstream of both cell density sensing and actin cytoskeletal rearrangements, and we demonstrate that loss of βPix expression in normal mammary epithelial cells strongly reduces Yap/Taz phosphorylation, promotes nuclear localization and increases target gene expression. Conversely, increased expression of βPIX in breast cancer cell lines re-couples the Hippo kinase cassette to Yap/Taz, promoting localization of Yap/Taz to the cytoplasm and inhibiting cell migration and proliferation. These studies thus define βPix as a key component that links the Hippo kinase cassette to Yap/Taz in response to multiple upstream Hippo pathway activators.
Keywords: Hippo, Arhgef7, Lats, mechanotransduction, Yap/Taz
Introduction
The Hippo signalling pathway is a major regulator of cell proliferation and tissue growth control. First uncovered using genetic screens in Drosophila, the conservation of this pathway in mammals has been firmly established (Halder & Johnson, 2011; Irvine, 2012; Ramos & Camargo, 2012; Harvey et al, 2013; Yu & Guan, 2013). At the core of the mammalian Hippo pathway is a kinase cassette comprised of the Drosophila Hippo homologs, mammalian STE20-like protein kinase 1/2 (Mst1/2, gene name Stk4/3) and large tumour suppressors 1 and 2 (Lats1/2). Upon activation, Mst1/2 in association with the adaptor protein Salvador (Sav1) phosphorylates and activates Mob1A/B-bound Lats1/2 kinases that in turn phosphorylate the related transcriptional regulators, Yes-associated protein (Yap) and transcriptional co-activator with PDZ-binding motif (Taz, gene name Wwtr1). When Hippo is inactive, Yap/Taz are primarily localized in the nucleus and in association with diverse transcription factors such as Teads, Runx and Smads regulate the expression of target genes including connective tissue growth factor (Ctgf), Ankrd1 and Cyr61. However, phosphorylation of Yap/Taz by Lats1/2 kinases leads to their cytoplasmic accumulation and enhanced ubiquitin-dependent degradation that thereby prevents transcriptional activity (Halder & Johnson, 2011; Irvine, 2012; Harvey et al, 2013; Yu & Guan, 2013).
Disruption of Hippo signalling in mouse models promotes tumour formation, and overexpression and constitutive nuclear localization of Yap/Taz occurs in many human cancers (Harvey et al, 2013). Yap/Taz-regulated transcriptional programmes are associated with tumour initiation, progression and metastasis by promoting cell proliferation, migration, survival and epithelial–mesenchymal transition (Harvey et al, 2013). Functional interactions of Yap/Taz with many cancer-associated signalling networks also contribute to their tumour-promoting activities and to related physiological processes such as the regulation of stem cell maintenance and differentiation (Irvine, 2012; Ramos & Camargo, 2012; Attisano & Wrana, 2013).
Cell–cell contact was one of the first identified regulators of Hippo signalling (Zhao et al, 2007; Ota & Sasaki, 2008) and is sensed and transmitted to the pathway by proteins that are involved in maintenance of cell architecture, such as polarity complexes and junctional proteins (Genevet & Tapon, 2011; Boggiano & Fehon, 2012; Schroeder & Halder, 2012). The apically localized Crumbs complex, that includes angiomotin, activates the Hippo kinase cassette in flies and mammals, possibly in connection with the sub-apically localized Kibra/NF2/Willin complex (Chen et al, 2010; Grzeschik et al, 2010; Ling et al, 2010; Robinson et al, 2010; Varelas et al, 2010b; Genevet & Tapon, 2011; Zhao et al, 2011; Boggiano & Fehon, 2012; Schroeder & Halder, 2012). Other components of polarity complexes including Scribble and adherens junction proteins, such as α-catenin and Ajuba, and protocadherins, such as fat, also promote Hippo pathway activity (Sopko & McNeill, 2009; Das Thakur et al, 2010; Kim et al, 2011; Schlegelmilch et al, 2011; Boggiano & Fehon, 2012; Reddy & Irvine, 2013). Yap/Taz activity is also modulated by rearrangements of the actin cytoskeleton that can occur with changes in cell morphology, attachment to the extracellular matrix and in response to mechanical forces (Dupont et al, 2011; Wada et al, 2011; Halder et al, 2012; Zhao et al, 2012). Pathways emanating from G protein-coupled receptors can also positively or negatively regulate the Hippo pathway (Yu et al, 2012).
Despite extensive interest in understanding how the Hippo pathway is regulated, mechanistic details and the molecular mediators that connect upstream signals to the Hippo kinase cassette remain elusive. Using LUMIER, a high-throughput protein–protein interaction screen (Barrios-Rodiles et al, 2005; Miller et al, 2009; Varelas et al, 2010b), we identified Arhgef7, more commonly known as βPix (for PAK-interacting exchange factor beta) as a Taz interacting protein. βPix is a member of Dbl family of guanine nucleotide exchange factor (GEF) for the small GTPases Rac1 and Cdc42 (Rosenberger & Kutsche, 2006; Staruschenko & Sorokin, 2012). βPix contains several protein–protein interaction domains, including Src homology 3 (SH3), a tandem Dbl homology (DH) and pleckstrin homology (PH) domain that mediates GEF activity, and a carboxy-terminal leucine zipper (LZ) domain (Rosenberger & Kutsche, 2006; Staruschenko & Sorokin, 2012). As a multidomain-containing protein, βPix is thought to act as a signal organizer by scaffolding the formation of multiprotein complexes. In this study, we show that βPix is required for Hippo pathway activity in response to multiple upstream stimuli. Mechanistically, we demonstrate that βPix scaffolds Lats to Yap/Taz, thereby promoting Yap/Taz phosphorylation and cytoplasmic sequestration. Thus, we delineate βPix, as a novel regulator of the Hippo core kinase cassette.
Results
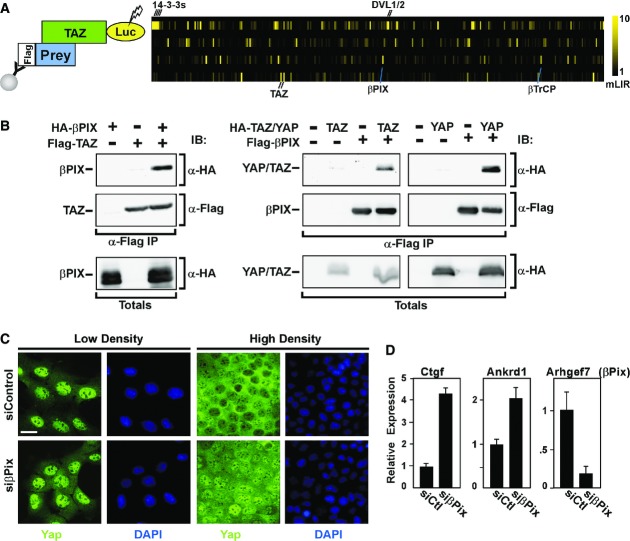
βPix interacts with Yap and Taz
Yap/Taz are the key effectors of the Hippo pathway. Thus, to identify putative pathway modulators, we undertook LUMIER, a mammalian cell-based protein–protein interaction screen (Barrios-Rodiles et al, 2005; Miller et al, 2009; Varelas et al, 2010a), to uncover novel TAZ binding partners. For this, TAZ, fused to Firefly luciferase, was used as a bait to screen a library of Flag-tagged proteins and interactions were detected by conducting a luciferase assay on anti-Flag immunopreciptates (Fig1A). Identified binding proteins included known partners, such as DVL1/2, 14-3-3 proteins, βTrCP and TAZ, which forms a dimer (Kanai et al, 2000; Tian et al, 2007; Varelas et al, 2010a), and also revealed a novel interaction with ARHGEF7, commonly known as βPix. βPix and the closely related αPix are proteins comprised of diverse binding domains including an SH3, a LZ and centrally localized tandem DH and PH domains, characteristic of GEFs (Rosenberger & Kutsche, 2006; Staruschenko & Sorokin, 2012). To confirm the association of TAZ with βPIX, we performed immunoprecipitations of either Flag-tagged TAZ or βPIX, followed by anti-HA immunoblotting, and observed an interaction between the two proteins regardless of whether TAZ or βPIX was immunoprecipitated (Fig1B). Similarly, YAP interacted with βPIX (Fig1B) and with the closely related αPIX protein (see Supplementary Fig S2).
Figure 1. βPix binds Yap/Taz and regulates Yap/Taz localization and transcriptional activity.
- Detection of TAZ binding partners using LUMIER assay. The interaction of luciferase-tagged TAZ with individual Flag-tagged proteins was assessed by luciferase assay. Each Flag-tagged protein tested in the screen is represented as a vertical bar, and the median luminescence intensity ratio (mLIR), which reflects the intensity of the interaction with TAZ, is represented by the yellow tone. Several interacting partners are marked.
- βPIX interacts with YAP and TAZ. Lysates from HEK293T cells co-transfected with Flag- or HA-tagged βPIX, YAP or TAZ as indicated were subjected to anti-Flag immunoprecipitation (α-Flag IP) and the presence of an associated protein detected by anti-HA immunoblotting. Equivalent protein expression levels were confirmed (Totals).
- βPix regulates Yap/Taz localization in polarized epithelial cells. EpH4 cells, transfected with control siRNA or siRNA targeting βPix, were plated at low or high cell densities. After 48 h, cells were fixed and Yap/Taz localization was visualized by confocal immunofluorescence microscopy. Scale bar, 25 μm.
- βPix regulates Yap/Taz transcriptional activity. EpH4 cells were transfected with siControl (siCtl) or siβPix, and RNA was extracted at 48 h. A representative experiment showing the relative expression of the Yap/Taz target genes Ctgf and Ankrd1, and the βPix knockdown efficiency as determined by qPCR is plotted as the mean ± the range.
Source data are available online for this figure.
βPix is required for cytoplasmic localization of Yap/Taz in response to diverse cues
High cell density and the formation of cell–cell junctions activate the Hippo pathway and result in cytoplasmic retention of the transcriptional regulators Yap/Taz (Genevet & Tapon, 2011; Boggiano & Fehon, 2012; Schroeder & Halder, 2012). Thus, to understand the role of βPix in regulating Yap/Taz activity, we sought to determine the effect of loss of βPix expression on Yap/Taz localization in response to cell density. For this, we first used mouse mammary EpH4 cells in which cytoplasmic localization of Yap/Taz occurs upon assembly of the Crumbs complex during epithelial cell polarization (Varelas et al, 2010b). Consistent with previous findings, Yap/Taz was primarily nuclear in sparse cultures, but was predominantly found in the cytoplasm at high density (Fig1C). Notably, abrogation of βPix expression using a pool of four siRNAs markedly attenuated the cytoplasmic sequestration of Yap/Taz (Fig1C). Analysis of expression of the well-characterized Yap/Taz target genes, Ctgf and Ankrd1, revealed a concomitant upregulation of expression of both genes upon loss of βPix (Fig1D). In mouse mammary NMuMG cells, high cell density also promotes the accumulation of cytoplasmic Yap/Taz (Supplementary Fig S1A, see also 3Fig C) and abrogation of βPix expression in these cells, similarly promoted nuclear accumulation of Yap/Taz in high-density cultures and increased expression of the Yap/Taz target genes Ctgf, Ankrd1 and Cyr61 (Fig2A). Deconvolving of the βPix siRNAs confirmed that all four individual siRNAs efficiently reduced βPix expression and concomitantly activated Yap/Taz target gene expression (Supplementary Fig S1B). While βPix is widely expressed, αPix displays a more limited distribution pattern (Rosenberger & Kutsche, 2006; Staruschenko & Sorokin, 2012), and consistent with this, αPix is not expressed in NMuMG cells nor did siRNA-mediated targeting of αPix alter Yap/Taz target gene expression (Fig2A). These results demonstrate that βPIX is important for cell density-dependent activation of the Hippo pathway in mammary epithelial cells.
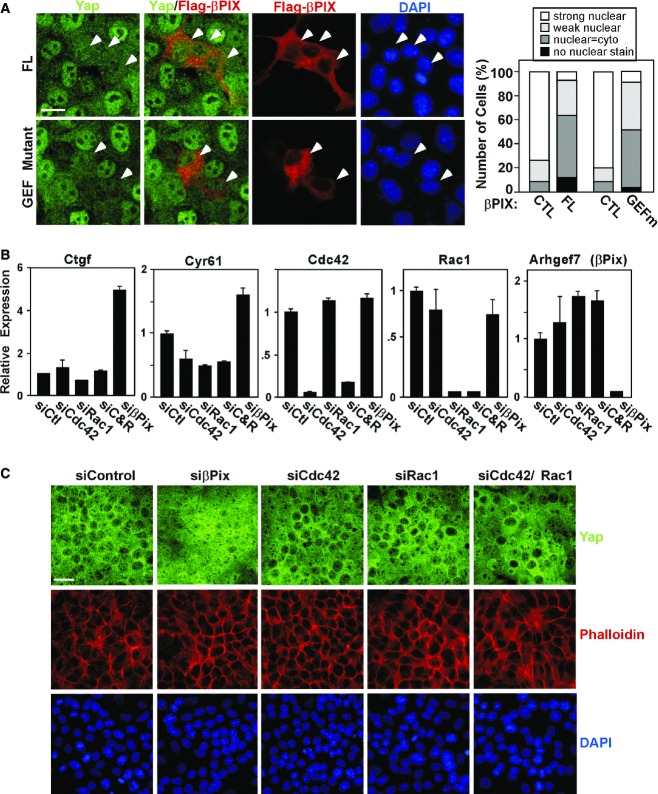
Figure 3. βPix functions independent of Rac1 and Cdc42 to regulate Yap/Taz localization.
- A βPix GEF activity is dispensable for regulation of Yap/Taz. NMuMG cells were transfected with Flag-tagged wild-type or L238R/L239S double-mutant version of βPIX (GEFm), which lacks GEF activity. βPix and Yap localization was analysed by immunofluorescence microscopy. A representative experiment showing Yap localization quantitated from n > 40 cells per condition is plotted. Scale bar, 20 μm.
- B, C NMuMG cells transfected with control siRNA or siRNA targeting βPix, Rac1 or Cdc42 were plated at high cell density. A representative experiment (B) showing the relative expression of Yap/Taz target genes, Ctgf and Cyr61, and the knockdown efficiency of βPix, Rac1 and Cdc42 was determined by qPCR and is plotted as the mean ± the range. Yap/Taz localization was visualized by immunofluorescence confocal microscopy (C). Scale bar, 25 μm.
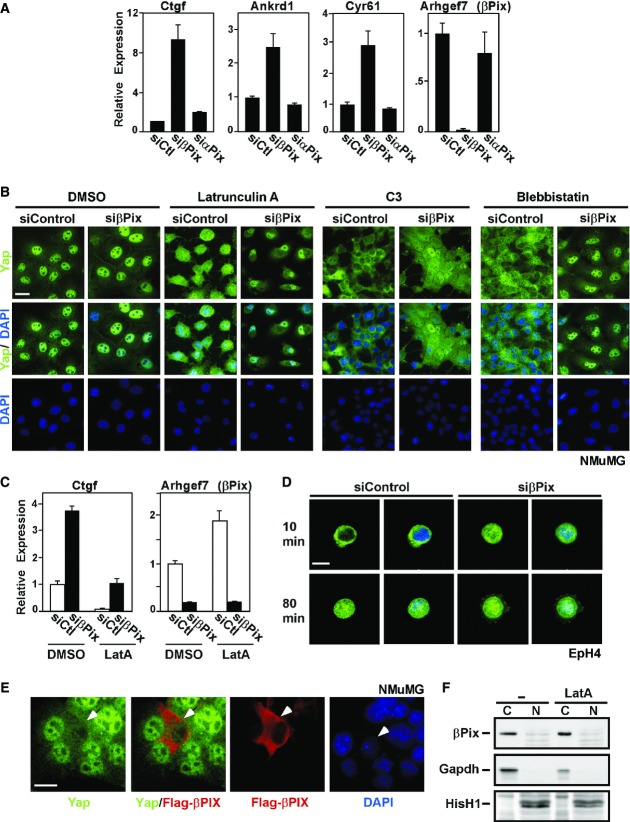
Figure 2. βPix regulates Yap/Taz localization and transcriptional activity during actin cytoskeleton reorganization.
- A representative experiment showing the relative expression of the Yap/Taz target genes Ctgf, Cyr61 and Ankrd1 in NMuMG cells transfected with control siRNA or siRNA targeting βPix or αPix was measured by qPCR and is plotted as the mean ± the range.
- NMuMG cells, transfected with control siRNA or βPix siRNA and cultured at low density for 48 h, were treated with the F-actin inhibitor latrunculin A (LatA, 0.5 μM), the Rho inhibitor C3 (3 μg/ml) or the non-muscle myosin inhibitor blebbistatin (Blebb, 50 μM) for 4 h. Cells were fixed and Yap/Taz localization was analysed by immunofluorescence confocal microscopy. Scale bar, 25 μm.
- A representative experiment showing the relative expression of Yap/Taz target genes in LatA-treated NMuMG cells transfected with control siRNA or siRNA targeting βPix was determined by qPCR and is plotted as the mean ± the range.
- EpH4 cells transfected with control siRNA or βPix siRNA cultured at low cell density for 48 h were trypsinized, kept in suspension for 1 h and then re-plated on fibronectin-coated chambers for either 10 or 80 min. Samples were fixed and Yap/Taz localization was analysed by immunofluorescence confocal microscopy. Scale bar, 15 μm.
- βPix is localized in the cytoplasm and promotes Yap/Taz cytoplasmic translocation. NMuMG cells were transfected with Flag-tagged βPIX, and endogenous Yap and βPIX localization was analysed by immunofluorescence microscopy using anti-YAP and anti-Flag antibodies, respectively. Scale bar, 15 μm.
- NMuMG cells were treated with DMSO or LatA, nuclear (N) and cytoplasmic (C) fractions were isolated and samples analysed by immunoblotting. Gapdh and histone H1 were used as cytoplasmic and nuclear markers, respectively.
Changes in the organization of the actin cytoskeleton can regulate the subcellular distribution of Yap/Taz though little is known of the mechanisms that connect dynamic actin cytoskeletal rearrangements to Yap/Taz (Genevet & Tapon, 2011; Boggiano & Fehon, 2012; Schroeder & Halder, 2012). Thus, to examine whether βPix is required for regulation of Yap/Taz in response to actin cytoskeleton dynamics, we abrogated the expression of βPix in NMuMG cells and treated them with various actin-disrupting agents, including latrunculin A, which disrupts F-actin, blebbistatin, which inhibits myosin-II-ATPase and C3, an inhibitor of Rho GTPase. As previously reported, cells plated at low density displayed primarily nuclear Yap/Taz that re-localized to the cytoplasm upon disruption of the actin cytoskeleton (Fig2B). However, in the absence of βPix, Yap/Taz cytoplasmic accumulation was markedly attenuated, and concordantly, Yap/Taz target gene expression was enhanced (Fig2B and C). Similar results were obtained in LatA-treated EpH4 cells (Supplementary Fig S1C and D). Cell detachment/attachment can also regulate the subcellular distribution of Yap/Taz (Zhao et al, 2012). Accordingly, detachment of EpH4 cells obtained from low-density cultures followed by a brief re-plating, caused pronounced cytoplasmic sequestration of Yap/Taz, which re-accumulated in the nucleus after 80 min of cell attachment (Fig2D). Abrogation of βPix expression attenuated the cytosolic sequestration and promoted nuclear retention of Yap/Taz upon cell detachment (Fig2D). These results suggest that βPix plays a key role in mediating the cytoplasmic localization of Yap/Taz during cytoskeletal remodelling and high cell density. We therefore next asked whether overexpression of βPix might enhance cytoplasmic localization of Yap/Taz under low-density conditions, where Yap is predominantly nuclear. Indeed, transient overexpression of βPIX in low-density NMuMG cells led to redistribution of Yap/Taz such that levels of Yap/Taz in nucleus and cytoplasm were roughly equivalent (Fig2E and quantitated in Figs3A and 6A). Collectively, these results show that βPix promotes increased cytoplasmic localization of Yap/Taz and concomitantly inhibits target gene activation in diverse contexts, including epithelial cell polarization, high cell density and disruption of the actin cytoskeleton.
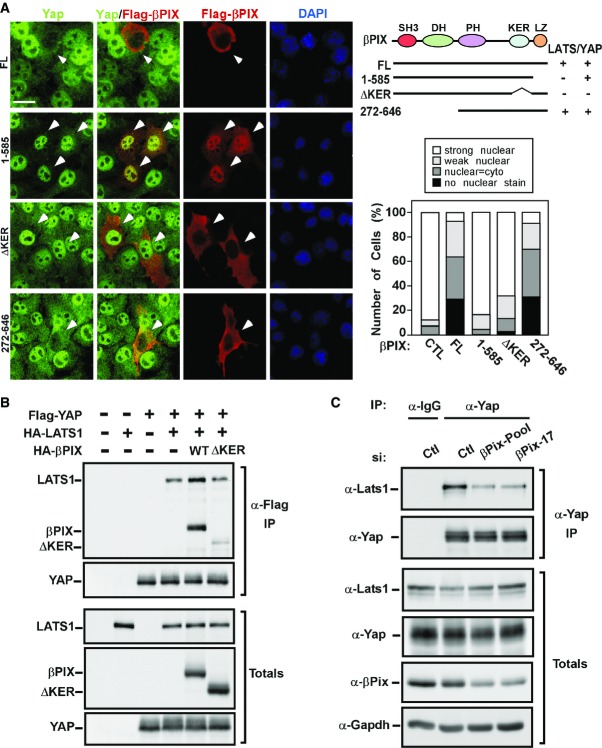
Figure 6. A C-terminal region of βPix is important for regulating Yap localization.
- NMuMG cells were transfected with the indicated Flag-βPIX WT or mutant constructs. Localization of endogenous Yap along with expressed Flag-βPIX constructs was determined by immunofluorescence confocal microscopy using anti-Yap and anti-Flag antibodies, respectively. Quantification of the percentage of cells with the indicated patterns of Yap localization from a representative experiment with n > 30 per condition is shown. Scale bar, 20 μm.
- βPix enhances Yap and Lats1 interaction. HEK293T cells co-transfected with Flag-tagged YAP and HA-tagged LATS1 in the presence or absence of HA-tagged WT or ΔKER mutant βPIX were lysed and subjected to α-Flag IP. The interaction between YAP and LATS1 was determined by immunoblotting using an anti-HA antibody.
- βPix knockdown reduces the strength of interaction between Yap and Lats1. EpH4 cells were transfected with either control siRNA or a pool and a single siβPix. After 48 h, cells were lysed and subjected to immunoprecipitation using either α-IgG or α-Yap antibodies. The strength of interaction between Yap and Lats1 was determined by immunoblotting using anti-Lats1 antibody. Equivalent protein expression levels and βPix knockdown efficiency were confirmed by immunoblotting (Totals).
Source data are available online for this figure.
βPix promotes Hippo-dependent Yap/Taz phosphorylation and interaction with Lats
To better understand how βPix might modulate Yap/Taz, we first examined the subcellular localization of endogenous βPix in NMuMG cells by subcellular fractionation. βPix was predominantly localized in the cytoplasm in both control cells and cells treated with the actin-disrupting agent latrunculin A (Fig2F). This is consistent with the strong cytoplasmic localization of transiently overexpressed Flag-βPIX observed by immunofluorescence microscopy (see Fig2E). These results suggest βPix acts in the cytoplasm rather than the nucleus to prevent Yap/Taz nuclear accumulation. Since βPix has GEF activity towards the monomeric RhoGTPases, Cdc42 and Rac1 (Rosenberger & Kutsche, 2006; Staruschenko & Sorokin, 2012), both of which are known modulators of the actin cytoskeleton, we considered whether βPix GEF activity was important for regulating Yap localization. However, overexpression of the GEF domain double-point mutant (L238R/L239S) that abrogates GEF activity (Manser et al, 1998) drove cytoplasmic localization of Yap/Taz in low-density NMuMG cells, similar to overexpressed WT βPIX (Fig3A). We also examined whether abrogation of Cdc42 and Rac1 expression affected Yap/Taz localization and activity. While loss of βPix promoted nuclear Yap/Taz localization and increased target gene expression as above, loss of Cdc42 or Rac1, either individually or together, had no effect (Fig3B and C). These findings are consistent with previous observations indicating that Cdc42/Rac1 are not involved in attachment-mediated control of Yap localization (Zhao et al, 2012) and together indicate that GEF activity is not the means through which βPix controls Yap/Taz.
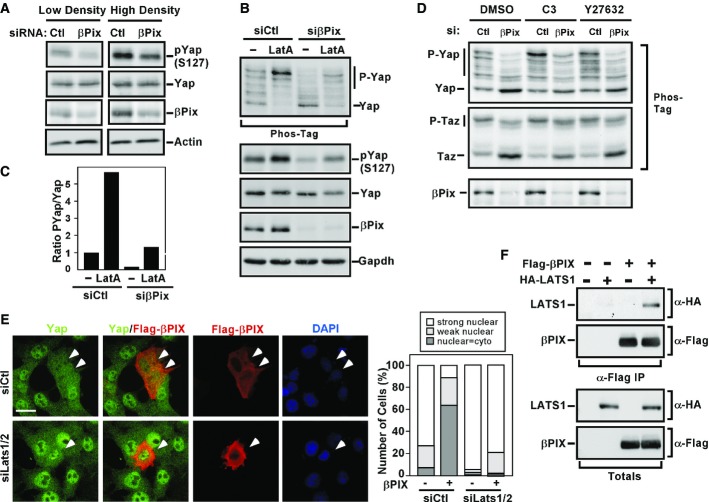
The Hippo pathway signals through a core kinase cassette comprised of Mst1/2 and Lats1/2 kinases that promote phosphorylation of Yap/Taz, thereby driving Yap/Taz localization to the cytoplasm (Halder & Johnson, 2011; Yu & Guan, 2013). Therefore, we explored the involvement of the Hippo pathway in βPix-mediated regulation of Yap localization by examining the phosphorylation status of Yap on Ser127, the Lats target site that mediates cytoplasmic sequestration. In EpH4 cells, immunoblotting with a phospho-Ser127 Yap antibody revealed that abrogation of βPix expression reduced the levels of phospho-Yap in cells cultured at either low or high density (Fig4A). In NMuMG cells treated with the actin-disrupting compound, LatA, we observed the expected robust increase in phosphorylation of Yap, as determined by Yap mobility shift in a Phos-Tag gel and by immunoblotting in a standard gel using the phospho-Ser127 Yap antibody (Fig4B and C). Of note, abrogation of βPix expression decreased phosphorylated Yap both in basal conditions and in the context of LatA. Similar results were obtained with C3 and the ROCK inhibitor, Y27632, for both Yap and Taz (Fig4D). Thus, βPix is required for efficient phosphorylation of Yap/Taz.
Figure 4. βPix regulates Yap phosphorylation.
- A EpH4 cells transfected with control siRNA or siRNA targeting βPix were cultured at low and high cell densities for 48 h. Cells lysates were subjected to immunoblotting to assess Yap phosphorylation using anti-phospho-YAP antibodies.
- B–D NMuMG cells transfected with control siRNA or βPix siRNA were treated with latrunculin A (B, C), C3 or Y27632 (D) for 4 h. Cell lysates, separated on Phos-Tag or regular gels, were analysed by immunoblotting using the indicated antibodies. Total levels of Yap, βPix and Gapdh or actin as loading controls were determined as indicated. The ratio of P-Yap to total Yap in the Phos-Tag gel (B) was quantitated by measuring the intensity of the upper band (P-Yap) over all bands (total Yap) in (C).
- E βPix acts upstream of Lats1/2 kinases to regulate Yap/Taz activity. NMuMG cells were transfected with control siRNA or siRNAs targeting Lats1 and Lats2 (siLats1/2) and 24 h later with Flag-tagged βPIX. Localization of βPIX and Yap/Taz was analysed by immunofluorescence microscopy. A representative experiment showing Yap localization quantitated from n > 20 cells per condition is plotted. Scale bar, 20 μm.
- F βPIX interacts with LATS1. Lysates from HEK293T cells co-transfected with Flag-tagged βPIX and HA-tagged LATS1 were subjected to anti-Flag immunoprecipitation (α-Flag IP), and the presence of LATS1 was detected by anti-HA immunoblotting. Equivalent protein expression levels were confirmed (Totals).
Our results thus far show that βPix promotes phosphorylation of Yap/Taz on the Lats kinase target site (Halder & Johnson, 2011; Yu & Guan, 2013). We therefore explored whether Lats is required for βPix function. Overexpression of βPIX induces cytoplasmic sequestration of Yap/Taz (Figs2E and 3A) and as expected, abrogation of Lats1/2 expression using siRNAs enhanced the nuclear accumulation of Yap/Taz (Fig4E). Importantly, loss of Lats1/2 expression prevented the βPIX-induced cytoplasmic accumulation of Yap/Taz, suggesting that βPix functions upstream of Lats kinases to regulate Yap/Taz activity (Fig4E). We next examined whether βPix might directly function at the level of Lats by first testing for physical interaction. Immunoprecipitation of Flag-βPIX revealed that LATS1 interacted with βPIX (Fig4F). Thus, Lats1 interacts with and is required for βPix function towards Yap/Taz.
βPix binds Lats1 and Yap via an internal domain in the carboxy-terminus
To map the determinants of the interactions between βPix and Lats1 or Yap, we constructed a series of βPIX deletion mutants and assessed interactions by immunoprecipitation and immunoblotting (Fig5). Deletion of the amino terminus comprising the SH3 and DH domains (βPIX 272–646), which bind PAK1 or are required for GEF activity, respectively, was dispensable for interaction with YAP or LATS1. This is consistent with our observation that GEF activity is not required to promote cytoplasmic Yap localization (Fig3A). We next explored the C-terminal region and observed that a truncation mutant (construct 1–495) lacking the last 151 amino acids, displayed reduced interaction with both YAP and LATS1. The C-terminal deletion encompasses two regions, a lysine (K)- and glutamate (E)-rich region (KER, amino acids 496–555, which has been reported to mediate interactions with other βPix partners such as Git1 (Flanders et al, 2003; Audebert et al, 2004; Jin et al, 2004; Hoefen & Berk, 2006; Chahdi & Sorokin, 2008) or Naa10p (Hua et al, 2011) and is also referred to as the GB (Git-binding) domain, as well as a C-terminal leucine zipper motif (LZ; amino acids 586–646) that is required for βPix dimerization. An internal deletion of the KER that retains an intact LZ (ΔKER; lacking amino acid 496–555) also failed to interact with YAP and LATS1. Mapping of the interaction between YAP- and the βPIX-related protein, αPIX, similarly showed that deletion of the corresponding KER (construct 1–625) abrogated interaction with YAP, while amino terminal deletions or a LZ point mutant did not (Supplementary Fig S2). Thus, the KER is required for interaction between α/βPIX and both YAP and LATS1. Analysis of the requirement for the LZ was more complex, since this region mediates βPix dimerization and is essential to maintain βPix in the cytoplasm (Kim et al, 2001). Interestingly, while the LZ deletion mutant (1–585) retained interaction with YAP, both of which are localized to the nucleus (see Fig6A), the interaction with LATS1, which is localized to the cytoplasm (Supplementary Fig S2D), was disrupted. Thus, for YAP interaction, neither the LZ or βPIX dimerization per se is required. In the case of LATS1, the βPIX KER domain-only deletion restored cytoplasmic βPIX, but still failed to interact with LATS1. These findings suggest that loss of interaction between the LZ mutant and LATS1 is secondary to their nuclear versus cytoplasmic compartmentalization. Thus, a region (KER) in α/βPIX, rich in charged residues including Lys and Glu, is required for interaction with both YAP and LATS. As βPix typically exists as either a dimer, or even a trimer in vivo (Schlenker & Rittinger, 2009), our findings are compatible with the notion that βPix can simultaneously recruit both Yap and Lats into a multimeric complex.
Figure 5. A C-terminal region of βPIX is required for binding to YAP and LATS.
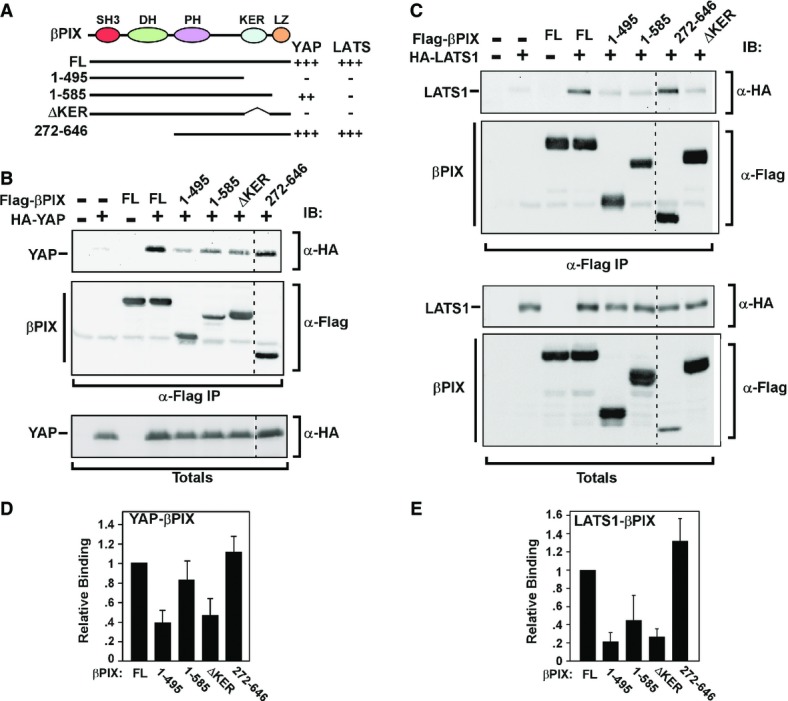
- A A schematic depicting the different βPIX cDNA constructs used for mapping interactions is shown. Positive or negative interactions with YAP and LATS1 are indicated on the right.
- B, C HEK293T cells were co-transfected with wild-type or mutant constructs of Flag-βPIX along with HA-YAP (B) or HA-LATS1 (C). Cell lysates were subjected to anti-Flag IP, and the presence of YAP or LATS1 was determined by anti-HA immunoblotting. A dashed line on blots indicates removal of a sample lane.
- D, E Quantitation of βPIX interaction mapping from replicate experiments YAP (D), n = 3, and LATS1 (E), n = 2, is plotted.
Source data are available online for this figure.
Disruption of Lats/Yap binding to βPix prevents cytoplasmic sequestration of Yap
We next examined the ability of βPIX mutants to regulate Taz/Yap localization. For this, NMuMG cells were transiently transfected with the Flag-tagged βPIX constructs and Yap/Taz localization was examined by immunofluorescence microscopy. As noted above, while the majority of control cells not expressing WT βPIX had nuclear Yap/Taz, cells overexpressing either WT βPIX, or the amino terminal deletion mutant lacking the PAK binding (SH3) and GEF (DH) domains displayed marked enrichment in cytoplasmic Yap/Taz (Fig6A). As previously reported (Kim et al, 2001), the LZ mutant is primarily localized to the nucleus and did not alter the nuclear localization of Yap. In contrast, the βPIX mutant lacking the KER (ΔKER, amino acids 496–555) was localized in the cytoplasm like the WT βPIX, but failed to promote Yap/Taz cytoplasmic sequestration. Since the KER deletion mutant fails to associate with either Yap or Lats, and the LZ mutant, which binds only Yap, both fail to drive cytoplasmic Yap sequestration, these results indicate that the interaction of βPix with both Yap and Lats is required for control of Taz/Yap localization.
The ability of βPix to bind both Yap and Lats1 suggests that βPix may act to scaffold to promote the association of Lats1 with its substrate Yap. To test this possibility, we examined the effect of βPix on the interaction between Yap and Lats1. We observed that overexpression of WT βPIX, but not a βPIX mutant lacking the KER domain, enhanced the interaction of LATS1 with YAP as determined by YAP immunoprecipitation followed by LATS1 immunoblotting (Fig6B). In line with a scaffolding function for βPix, abrogation of the expression of βPix using either a pool or a single siRNA dramatically reduced the interaction of endogenous Yap with Lats1 in EpH4 cells (Fig6C). Altogether, these results indicate that βPix can act as a scaffold to promote the interaction of Lats with its substrate, Yap/Taz.
βPIX attenuates the tumourigenic properties of MDA-MB-231 breast cancer cells
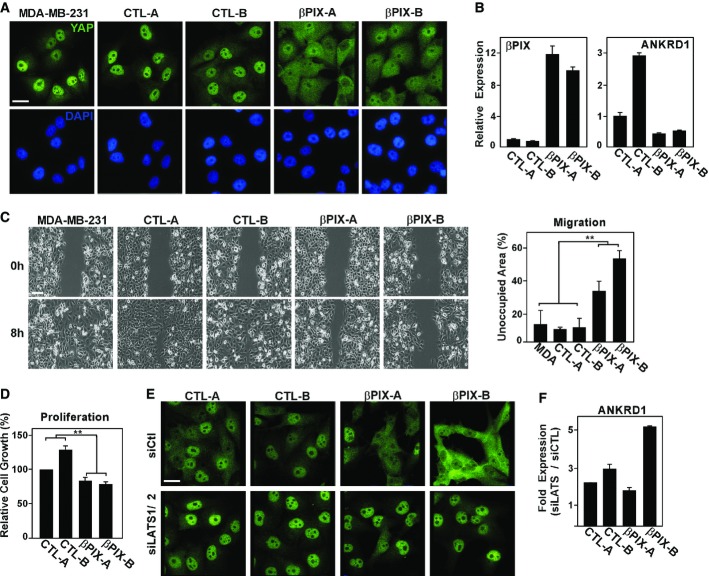
Studies in cells, mice and human tumour samples indicate that Yap/Taz display oncogenic activities, whereas the Hippo pathway, which restrains nuclear Yap/Taz, is tumour suppressive (Harvey et al, 2013). Tumour cells have thus acquired the ability to bypass the Hippo pathway, thereby permitting the emergence of the pro-tumourigenic Yap/Taz-mediated transcriptional programme. Consistent with this, in breast cancer cells, overexpression of Yap/Taz promotes proliferation, migration and tumour initiation, while loss of Taz inhibits tumour formation (Chan et al, 2008; Lei et al, 2008; Zhao et al, 2008; Cordenonsi et al, 2011; Lamar et al, 2012; Harvey et al, 2013; Serrano et al, 2013; Hiemer et al, 2014; Mi et al, 2014; Sorrentino et al, 2014). We therefore sought to determine whether βPix might re-couple the Hippo pathway to Yap/Taz in a cancer context. The triple-negative breast cancer cell line, MDA-MB-231, displays nuclear YAP/TAZ and constitutively expresses YAP/TAZ target genes; thus, we used these cells to generate clones stably overexpressing βPIX (Supplementary Fig S4A). Analysis of YAP/TAZ localization showed that in control MDA-MB-231 cell clones, YAP/TAZ were predominantly nuclear, while in two independently derived βPIX overexpressing clones, abundant cytoplasmic YAP/TAZ was detected, with some cells displaying nuclear exclusion (Fig7A). A concomitant reduction in YAP/TAZ target gene expression was also observed in the βPIX-expressing clones (Fig7B). Thus, increased expression of βPIX inhibits the nuclear localization and transcriptional activity of YAP/TAZ in these breast cancer cells.
Figure 7. Effect of βPix expression on tumourigenic properties of MDA-MB-231 breast cancer cells.
- A YAP is mainly cytoplasmic in MDA-MB-231 cells stably expressing βPIX. Parental MDA-MB-231 cells or MDA-MB-231 cells stably expressing Flag-βPIX or empty vector were fixed, and YAP localization was visualized by immunofluorescence microscopy. Scale bar, 25 μm.
- B A representative experiment showing the expression levels of βPIX and YAP/TAZ target gene ANKRD1, determined by qPCR and plotted as the mean ± the range.
- C βPIX regulates cell migration. A wound was introduced in a confluent monolayer of MDA-MB-231 cells stably expressing either control vector or βPIX, and the migration of the cells into the wound was assessed by live-cell phase-contrast microscopy. Quantitation of cell migration, determined by measuring the cell-free area within the wound, of three independent experiments, n = 4, is plotted as the mean ± SEM. Scale bar, 120 μm.
- D βPIX attenuates cell proliferation. An SRB assay was used to measure cell proliferation. Data are plotted as the mean ± SEM of six independent experiments, n = 5.
- E, F LATS1/2 kinases are required for βPIX-mediated YAP/TAZ inactivation. MDA-MB-231 cells stably expressing control vector or βPIX were transfected with siCTL or siLATS1/2, and YAP localization was analysed by immunofluorescence microscopy (E). Scale bar, 25 μm. Expression level of YAP target gene, ANKRD1, was determined by qPCR (F) and is plotted as the relative expression in siLATS1/2 over siControl (CTL).
We next determined how ectopic βPIX expression affected properties typically associated with tumourigenesis. MDA-MB-231 cells simultaneously depleted of both YAP and TAZ were difficult to culture; nevertheless, we confirmed that individual loss of YAP or TAZ decreased the rate of cell migration and proliferation (Supplementary Fig S3), consistent with the notion that YAP/TAZ contribute to these biological responses. Clones overexpressing βPIX displayed a marked attenuation of cell migration in an in vitro wound healing scratch assay (Fig7C) as well as a decreased rate of cell proliferation (Fig7D). Thus, expression of βPIX, which enhances the cytoplasmic localization of YAP/TAZ results in responses that are characteristic of tumour suppressive activity, in a manner that parallels that of loss of YAP/TAZ.
Although YAP/TAZ are predominantly nuclear, MDA-MB-231 cells nevertheless retain some LATS activity as siRNA-mediated depletion of LATS1/2 resulted in a more pronounced nuclear accumulation of YAP/TAZ and enhanced YAP/TAZ target gene expression (Figure7E and F). Thus, while upstream signals are disconnected from the core Hippo kinases, LATS1/2 are expressed and are able to limit YAP/TAZ activity. We thus examined the effect of loss of LATS1/2 on YAP/TAZ localization in the βPIX-expressing clones and observed that siLATS1/2 overcame the effects of βPIX and resulted in strong nuclear localization of YAP/TAZ (Figure7E and F), and enhanced expression of ANKRD1 (Figure7F). Thus, βPIX-mediated regulation of YAP/TAZ is dependent on the Hippo pathway kinase LATS. Taken together, our findings show that enhanced expression of βPIX can re-engage the Hippo core kinases, LATS1/2, and that βPIX thereby functions as a tumour suppressor to restrain the pro-oncogenic properties of YAP/TAZ in metastatic breast cancer cells.
Discussion
The Hippo pathway has been established as a key regulator of tissue growth and cell fate, and disruption of the pathway promotes tumourigenic processes such as cell migration and proliferation (Halder & Johnson, 2011; Ramos & Camargo, 2012; Harvey et al, 2013; Yu & Guan, 2013). Several upstream mediators, including determinants of apical–basal polarity, mechanical forces acting through the actin cytoskeleton and G protein-coupled receptors, have emerged as pathway activators (Genevet & Tapon, 2011; Boggiano & Fehon, 2012; Schroeder & Halder, 2012). How these signals are transduced to the core Mst/Lats kinase cassette is the subject of intense investigation. Ultimately, however, activation of the Hippo pathway results in Lats kinase-mediated phosphorylation of Yap/Taz, which then drives Yap/Taz localization to the cytoplasm. Here, we identify βPix as a key positive regulator of the Hippo kinase. Mechanistically, we demonstrate that βPix binds both Lats and its substrate target, Yap, and that loss of βPIX impairs the interaction between Lats and Yap/Taz, resulting in decreased Yap/Taz phosphorylation. Moreover, we show that increased expression of βPIX in cancer cells can restore cytoplasmic localization of Yap/Taz in a Lats-dependent manner. Altogether, our findings suggest a model in which βPix promotes Hippo pathway activity by scaffolding Lats to its Yap/Taz substrates to stimulate phosphorylation and localization to the cytoplasm (Fig8).
Figure 8. Model of the mechanism for βPix function in Hippo signalling.
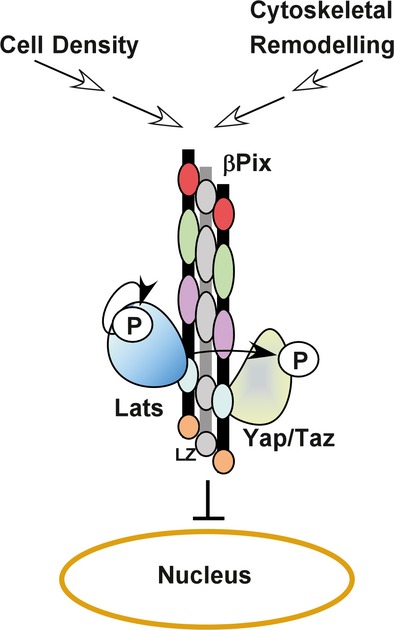
The Hippo pathway is activated by upstream signals such as high cell density and actin cytoskeleton remodelling. βPix, which exists as a dimer or a trimer in the cytoplasm, forms a multiprotein complex with both Lats and Yap, acting as a scaffold to promote phosphorylation of Yap/Taz by Lats and resulting in cytoplasmic accumulation of Yap/Taz.
Signals emanating from polarity determinants, such as the Crumbs/Amot complex, processes such as mechanotransduction that act through the actin cytoskeleton and mediators of cell density sensing, all can control Yap localization (Genevet & Tapon, 2011; Boggiano & Fehon, 2012; Schroeder & Halder, 2012). In general, these signals flow to the Mst/Lats kinase cassette, and while Mst1/2-independent Yap phosphorylation has been reported (Yu et al, 2012, 2013; Zhao et al, 2012; Kim et al, 2013), there appears to be a more ubiquitous requirement for Lats. Our data demonstrate that βPix functions at the level of Lats, consistent with the notion that βPix acts in the Hippo pathway downstream of multiple cues. Accordingly, we observed a requirement for βPix in regulating Yap activity in response to cell–cell contact and cell density, actin cytoskeleton disruption and in attachment/detachment events. In all the cases, loss of βPix expression resulted in retention of Yap/Taz in the nucleus and continued activation of a Yap/Taz transcriptional programme.
In solid tumours, YAP and/or TAZ are frequently overexpressed and unlike normal cells, most cancer cells have acquired the means to bypass Hippo-dependent regulation. YAP/TAZ become constitutively nuclear and act to promote the tumourigenic phenotype (Harvey et al, 2013). In breast cancer cells for example, overexpression of TAZ and YAP promotes proliferation, migration, invasion, epithelial–mesenchymal transition (EMT), acquisition of cancer stem cell (CSC) properties and sustains CSC self-renewal (Chan et al, 2008; Lei et al, 2008; Zhao et al, 2008; Cordenonsi et al, 2011; Lamar et al, 2012; Harvey et al, 2013; Hiemer et al, 2014). In MDA-MB-231 cells, the Hippo pathway is inactive, possibly through loss of the upstream component NF2 (Dupont et al, 2011), resulting in a pronounced nuclear accumulation of YAP/TAZ. Remarkably, we found that ectopic expression of βPIX alone in MDA-MB-231 cells was sufficient to restrain YAP/TAZ activity and yielded a concomitant suppression of cell proliferation and cell migration. This activity of βPIX was dependent on the presence of LATS1/2. Thus, we uncovered a tumour suppressor function for βPix that acts via re-coupling of the Hippo kinase cassette to its Yap/Taz substrates. Given that these cells lack NF2 (Dupont et al, 2011), these findings also suggest the intriguing possibility that βPix might function to link NF2 to the Hippo kinase cassette.
Our interaction mapping studies revealed that Yap and Lats both bind to an approximately 50 amino acid region located just upstream of the carboxy-terminus that we termed the KER and which is highly conserved in both αPix and βPix. This region does not contain any recognizable protein–protein interaction motifs, but rather is rich in charged amino acids including Lys and Glu. Nevertheless, this region also mediates interactions with other proteins such as Git1 (Hoefen & Berk, 2006; Rosenberger & Kutsche, 2006; Staruschenko & Sorokin, 2012). We showed that loss of βPix attenuated the interaction of Lats1 with Yap, while overexpression of βPIX enhanced association in a KER domain-dependent manner. βPix was first characterized as a dimer, although a more recent study suggests that trimers may be the more typical state (Schlenker & Rittinger, 2009). Taken together, we speculate that simultaneous binding of Yap and Lats to individual βPix proteins within the context of a multimerized complex provides the scaffolding function and allows for enhanced Yap phosphorylation (Fig8). In this study, we focused on βPix, since abrogation of βPix expression alone was sufficient to inhibit Hippo pathway activity in our cell models. This is consistent with a more ubiquitous expression pattern for βPix, as compared to αPix (Rosenberger & Kutsche, 2006; Staruschenko & Sorokin, 2012). However, the KER region is highly conserved and also mediates αPix binding to Yap/Taz and Lats. Interestingly, mutations in αPIX (ARHGEF6) have been associated with X-linked intellectual disability in humans (Kutsche et al, 2000) and αPix/Arhgef6-deficient mice display alterations in synaptic and immune system function (Missy et al, 2008; Ramakers et al, 2012), whereas βPix mutants display early embryonic lethality (Missy et al, 2008). Thus, it will be important to determine whether αPix might also modulate Hippo signalling in these distinct contexts.
αPix and βPix have been most studied for their function as guanine nucleotide exchange factors (GEFs) for the Rho GTPases, Cdc42 and Rac1, although the mechanisms whereby GEF activity is controlled remain unclear (Rosenberger & Kutsche, 2006; Staruschenko & Sorokin, 2012). We showed that loss of Cdc42, Rac1 or both has no effect on Yap/Taz subcellular localization or target gene activation, suggesting that GEF activity is not required for βPix function in the Hippo pathway. Consistent with this, a βPIX construct harbouring point mutations in the GEF domain that prevent guanine nucleotide exchange (Manser et al, 1998) still functioned to sequester Yap/Taz in the cytoplasm. Pix has also been studied as a binding partner for p21-activating kinase 1, PAK1 (Chan & Manser, 2012), and can stimulate PAK, via the GEF activity. Although we did not directly test whether PAK activity alters Yap/Taz function, deletion of the SH3 domain of βPIX, which mediates binding to PAK1, was not required to promote Yap localization to the cytoplasm. Thus, our findings suggest that the well-characterized role of βPix in complex with Cdc42/Rac1 and PAK1 is molecularly distinct from its promotion of Hippo pathway activity. Instead, our studies demonstrate that βPix functions in the Hippo pathway by scaffolding Yap and Lats. A general role for βPix as a scaffold is suggested by the ability of βPix to bind a diverse array of proteins, some of which form large macromolecular assemblies, to control cellular processes such as focal adhesion formation and function, cell migration or G protein-coupled receptor signalling. For instance, βPix can bind to 14-3-3 proteins, p66Shc, Scribble, Cbl and the multidomain-containing Git1 (Flanders et al, 2003; Audebert et al, 2004; Jin et al, 2004; Hoefen & Berk, 2006; Chahdi & Sorokin, 2008). Although, mechanistic understanding of how βPix functions in so many processes is not well understood, the ability of βPix to engage in diverse protein complexes indicates that scaffolding functions for βPix are likely to be widespread. Whether any of the known βPix interactors might cooperate in regulating the Hippo pathway is an interesting area for future investigations.
Materials and Methods
Cell culture and transfection
For cell culturing, NMuMG cells were grown in DMEM supplemented with 10% FBS and 10 μg/ml insulin, EpH4 and HEK293T cells in DMEM with 10% FBS, and MDA-MB-231 cells in RPMI with 5% FBS. Cells were transfected with Dharmacon siGENOME pools of four individual siRNAs (Thermo Scientific) using Lipofectamine RNAiMAX (Life Technologies), or with cDNAs using Lipofectamine LTX or Lipofectamine 3000 (Life Technologies) according to the manufacturer's instructions.
Plasmids and chemicals
The βPIX construct was generated by PCR using an isoform of human βPIX (NM_001113513.1) and was N-terminally tagged with Flag or HA in a pCMV5 vector. βPIX and αPIX deletion constructs were generated by PCR-mediated site-directed mutagenesis. Flag- or HA-tagged constructs for LATS1, TAZ and YAP in pCMV5 were previously described (Varelas et al, 2010a). For MDA-MB-231 cells stably expressing βPIX, Flag-tagged βPIX was subcloned into pBABE-puro vector (addgene #1764; Mani et al, 2007). pBABE-puro empty vector was used as a control. Chemicals used in this study were as follows: latrunculin A (Tocris Bioscience #3973), C3 (Cytoskeleton Inc #CT04), Y-27632 (Sigma-Aldrich #Y0503) and blebbistatin (Sigma-Aldrich #B0506).
Immunoblotting, immunoprecipitation and subcellular fractionation
Cells were lysed in lysis buffer (50 mM Tris–HCl, 150 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, 1 mM DTT containing phosphatase and protease inhibitors). Lysates were separated on SDS–PAGE gels, and immunoblotting was performed using standard protocols as previously described (Labbe et al, 2000). Phos-Tag gels, using reagents purchased from Waco Chemicals, were prepared according to manufacturer's instructions. Nuclear and cytoplasmic fractions were prepared using the NE-PER Nuclear and Cytoplasmic Extraction Reagent (Thermo Scientific #78833). For immunoprecipitations, cell lysates were subjected to anti-Flag or anti-Lats1 immunoprecipitation and proteins collected using protein G-Sepharose prior to analyses by immunoblotting. The antibodies used were as follows: pYAP (D9W2I; Cell Signalling #13008); YAP (Cell Signalling #4912), TAZ (Cell Signalling #2149), Lats1 (C66B5; Cell Signalling #3477), Cool1/βPix (Cell Signalling #4515), rat anti-HA (Roche #1867423) and anti-Flag M2 (Sigma-Aldrich #F1804).
Immunofluorescence microscopy
Cell were plated in 4-well Lab-Tek chambers (#154526) and fixed with 4% paraformaldehyde for 10 min at room temperature. After washing with PBS, cells were permeabilized with 0.5% Triton X-100 in PBS for 10 min at room temperature. Samples were washed three times with 0.01% PBS–Tween and then blocked in 2% BSA–PBS for 30 min before treatment with primary antibody. Samples were then incubated with primary antibodies (mouse anti-YAP 1:300; Santa Cruz sc-101199; or rabbit anti-Flag 1:500, Sigma F7425) in 2% BSA–PBS overnight at 4°C. After washing three times with 0.01% PBS–Tween, slides were incubated with the secondary antibodies, goat anti-rabbit Alexa Fluor 488 (Life Technologies #A11305, 1:1,000 in 2% BSA–PBS) or goat anti-mouse Alexa Fluor 546 (Invitrogen #A11029, 1:1,000 in 2% BSA–PBS) for 1–2 h at room temperature. Slides were washed three times with 0.01% PBS–Tween and once with PBS and mounted with ProLong Gold Antifade Reagent (Life Technologies #P36035). Cell nuclei were visualized by DAPI staining and Alexa Fluor 568-Phalloidin (Life Technologies #12380) was used for actin cytoskeleton staining. Images were captured using a spinning disc confocal scanner (CSU10, Yokogawa) on Leica DMI6000B microscope, and Volocity software was used for image acquisition and processing. For quantification of Yap localization transfected with different Flag-tagged βPIX cDNA constructs, a minimum of 30 transfected cells were counted and nuclear/cytoplasmic localization of Yap was evaluated in transfected cells compared to the surrounding non-transfected cells.
Quantitative Real-Time PCR
Total RNA was purified using PureLink RNA Mini Kit (Life Technologies). cDNA was synthesized using 1 μg of purified RNA using oligo-dT primers and M-MLV Reverse Transcriptase (Invitrogen #28025-013). Real-Time PCR was performed using the SYBR Green master mix (Applied Biosystems) on the ABI Prism 7900 HT system (Applied Biosystems). Relative gene expression was quantified by ΔΔCt method and normalized to Gapdh. The sequences of the primers used are listed in Supplementary Table S1.
Wound healing and cell growth assays
For wound healing migration assay, cells were seeded in a 6-well plate and were grown to confluency. The wound was introduced by scraping with a sterile 200-μl pipette tip, and the unfilled area was quantified by ImageJ at the 8 h time point. Cell growth was determined using the sulforhodamine B (SRB) assay. Cells were plated overnight in 96-well dishes and after 48 h were fixed with 10% (w/v) trichloroacetic acid and stained as previously described (Bao et al, 2012). The amount of SRB present in each well was determined by optical density reading at 490 nm.
Acknowledgments
We would like to thank Tania Christova for helpful insights and Jeff Wrana for critical comments on the manuscript. This work was supported by Grants #496138 and #246295 to L.A. from the Canadian Institute for Health Research (CIHR). LA is Canada Research Chair.
Author contributions
EH and LA conceived the project and designed experiments; EH, KS, SS and AS performed experiments; EH, LA and KS analysed the data; EH and LA wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary information for this article is available online: http://emboj.embopress.org
Supplementary Figures
Supplementary Table S1
Review Process File
Source Data for Figure 1B
Source Data for Figure 5B, C
Source Data for Figure 6B, C
References
- Attisano L, Wrana JL. Signal integration in TGF-beta, WNT, and Hippo pathways. F1000Prime Rep. 2013;5:17. doi: 10.12703/P5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audebert S, Navarro C, Nourry C, Chasserot-Golaz S, Lecine P, Bellaiche Y, Dupont JL, Premont RT, Sempere C, Strub JM, Van Dorsselaer A, Vitale N, Borg JP. Mammalian Scribble forms a tight complex with the betaPIX exchange factor. Curr Biol. 2004;14:987–995. doi: 10.1016/j.cub.2004.05.051. [DOI] [PubMed] [Google Scholar]
- Bao R, Christova T, Song S, Angers S, Yan X, Attisano L. Inhibition of tankyrases induces Axin stabilization and blocks Wnt signalling in breast cancer cells. PLoS ONE. 2012;7:e48670. doi: 10.1371/journal.pone.0048670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrios-Rodiles M, Brown KR, Ozdamar B, Bose R, Liu Z, Donovan RS, Shinjo F, Liu Y, Dembowy J, Taylor IW, Luga V, Przulj N, Robinson M, Suzuki H, Hayashizaki Y, Jurisica I, Wrana JL. High-throughput mapping of a dynamic signaling network in mammalian cells. Science. 2005;307:1621–1625. doi: 10.1126/science.1105776. [DOI] [PubMed] [Google Scholar]
- Boggiano JC, Fehon RG. Growth control by committee: intercellular junctions, cell polarity, and the cytoskeleton regulate Hippo signaling. Dev Cell. 2012;22:695–702. doi: 10.1016/j.devcel.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahdi A, Sorokin A. Endothelin-1 couples betaPix to p66Shc: role of betaPix in cell proliferation through FOXO3a phosphorylation and p27kip1 down-regulation independently of Akt. Mol Biol Cell. 2008;19:2609–2619. doi: 10.1091/mbc.E07-05-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SW, Lim CJ, Guo K, Ng CP, Lee I, Hunziker W, Zeng Q, Hong W. A role for TAZ in migration, invasion, and tumorigenesis of breast cancer cells. Cancer Res. 2008;68:2592–2598. doi: 10.1158/0008-5472.CAN-07-2696. [DOI] [PubMed] [Google Scholar]
- Chan PM, Manser E. PAKs in human disease. Prog Mol Biol Transl Sci. 2012;106:171–187. doi: 10.1016/B978-0-12-396456-4.00011-0. [DOI] [PubMed] [Google Scholar]
- Chen CL, Gajewski KM, Hamaratoglu F, Bossuyt W, Sansores-Garcia L, Tao C, Halder G. The apical-basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proc Natl Acad Sci U S A. 2010;107:15810–15815. doi: 10.1073/pnas.1004060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordenonsi M, Zanconato F, Azzolin L, Forcato M, Rosato A, Frasson C, Inui M, Montagner M, Parenti AR, Poletti A, Daidone MG, Dupont S, Basso G, Bicciato S, Piccolo S. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147:759–772. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- Das Thakur M, Feng Y, Jagannathan R, Seppa MJ, Skeath JB, Longmore GD. Ajuba LIM proteins are negative regulators of the Hippo signaling pathway. Curr Biol. 2010;20:657–662. doi: 10.1016/j.cub.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, Elvassore N, Piccolo S. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- Flanders JA, Feng Q, Bagrodia S, Laux MT, Singavarapu A, Cerione RA. The Cbl proteins are binding partners for the Cool/Pix family of p21-activated kinase-binding proteins. FEBS Lett. 2003;550:119–123. doi: 10.1016/s0014-5793(03)00853-6. [DOI] [PubMed] [Google Scholar]
- Genevet A, Tapon N. The Hippo pathway and apico-basal cell polarity. Biochem J. 2011;436:213–224. doi: 10.1042/BJ20110217. [DOI] [PubMed] [Google Scholar]
- Grzeschik NA, Parsons LM, Allott ML, Harvey KF, Richardson HE. Lgl, aPKC, and Crumbs regulate the Salvador/Warts/Hippo pathway through two distinct mechanisms. Curr Biol. 2010;20:573–581. doi: 10.1016/j.cub.2010.01.055. [DOI] [PubMed] [Google Scholar]
- Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder G, Dupont S, Piccolo S. Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nat Rev Mol Cell Biol. 2012;13:591–600. doi: 10.1038/nrm3416. [DOI] [PubMed] [Google Scholar]
- Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- Hiemer SE, Szymaniak AD, Varelas X. The transcriptional regulators TAZ and YAP direct transforming growth factor beta-induced tumorigenic phenotypes in breast cancer cells. J Biol Chem. 2014;289:13461–13474. doi: 10.1074/jbc.M113.529115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoefen RJ, Berk BC. The multifunctional GIT family of proteins. J Cell Sci. 2006;119:1469–1475. doi: 10.1242/jcs.02925. [DOI] [PubMed] [Google Scholar]
- Hua KT, Tan CT, Johansson G, Lee JM, Yang PW, Lu HY, Chen CK, Su JL, Chen PB, Wu YL, Chi CC, Kao HJ, Shih HJ, Chen MW, Chien MH, Chen PS, Lee WJ, Cheng TY, Rosenberger G, Chai CY, et al. N-alpha-acetyltransferase 10 protein suppresses cancer cell metastasis by binding PIX proteins and inhibiting Cdc42/Rac1 activity. Cancer Cell. 2011;19:218–231. doi: 10.1016/j.ccr.2010.11.010. [DOI] [PubMed] [Google Scholar]
- Irvine KD. Integration of intercellular signaling through the Hippo pathway. Semin Cell Dev Biol. 2012;23:812–817. doi: 10.1016/j.semcdb.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Smith FD, Stark C, Wells CD, Fawcett JP, Kulkarni S, Metalnikov P, O'Donnell P, Taylor P, Taylor L, Zougman A, Woodgett JR, Langeberg LK, Scott JD, Pawson T. Proteomic, functional, and domain-based analysis of in vivo 14-3-3 binding proteins involved in cytoskeletal regulation and cellular organization. Curr Biol. 2004;14:1436–1450. doi: 10.1016/j.cub.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Kanai F, Marignani PA, Sarbassova D, Yagi R, Hall RA, Donowitz M, Hisaminato A, Fujiwara T, Ito Y, Cantley LC, Yaffe MB. TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J. 2000;19:6778–6791. doi: 10.1093/emboj/19.24.6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Lee SH, Park D. Leucine zipper-mediated homodimerization of the p21-activated kinase-interacting factor, beta Pix. Implication for a role in cytoskeletal reorganization. J Biol Chem. 2001;276:10581–10584. doi: 10.1074/jbc.C000806200. [DOI] [PubMed] [Google Scholar]
- Kim NG, Koh E, Chen X, Gumbiner BM. E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proc Natl Acad Sci U S A. 2011;108:11930–11935. doi: 10.1073/pnas.1103345108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Kim M, Lee S, Kuninaka S, Saya H, Lee H, Lee S, Lim DS. cAMP/PKA signalling reinforces the LATS-YAP pathway to fully suppress YAP in response to actin cytoskeletal changes. EMBO J. 2013;32:1543–1555. doi: 10.1038/emboj.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsche K, Yntema H, Brandt A, Jantke I, Nothwang HG, Orth U, Boavida MG, David D, Chelly J, Fryns JP, Moraine C, Ropers HH, Hamel BC, van Bokhoven H, Gal A. Mutations in ARHGEF6, encoding a guanine nucleotide exchange factor for Rho GTPases, in patients with X-linked mental retardation. Nat Genet. 2000;26:247–250. doi: 10.1038/80002. [DOI] [PubMed] [Google Scholar]
- Labbe E, Letamendia A, Attisano L. Association of Smads with lymphoid enhancer binding factor 1/T cell-specific factor mediates cooperative signaling by the transforming growth factor-beta and wnt pathways. Proc Natl Acad Sci U S A. 2000;97:8358–8363. doi: 10.1073/pnas.150152697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamar JM, Stern P, Liu H, Schindler JW, Jiang ZG, Hynes RO. The Hippo pathway target, YAP, promotes metastasis through its TEAD-interaction domain. Proc Natl Acad Sci U S A. 2012;109:E2441–E2450. doi: 10.1073/pnas.1212021109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei QY, Zhang H, Zhao B, Zha ZY, Bai F, Pei XH, Zhao S, Xiong Y, Guan KL. TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol Cell Biol. 2008;28:2426–2436. doi: 10.1128/MCB.01874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling C, Zheng Y, Yin F, Yu J, Huang J, Hong Y, Wu S, Pan D. The apical transmembrane protein Crumbs functions as a tumor suppressor that regulates Hippo signaling by binding to Expanded. Proc Natl Acad Sci U S A. 2010;107:10532–10537. doi: 10.1073/pnas.1004279107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani SA, Yang J, Brooks M, Schwaninger G, Zhou A, Miura N, Kutok JL, Hartwell K, Richardson AL, Weinberg RA. Mesenchyme Forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-like breast cancers. Proc Natl Acad Sci U S A. 2007;104:10069–10074. doi: 10.1073/pnas.0703900104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manser E, Loo TH, Koh CG, Zhao ZS, Chen XQ, Tan L, Tan I, Leung T, Lim L. PAK kinases are directly coupled to the PIX family of nucleotide exchange factors. Mol Cell. 1998;1:183–192. doi: 10.1016/s1097-2765(00)80019-2. [DOI] [PubMed] [Google Scholar]
- Mi W, Lin Q, Childress C, Sudol M, Robishaw J, Berlot CH, Shabahang M, Yang W. Geranylgeranylation signals to the Hippo pathway for breast cancer cell proliferation and migration. Oncogene. 2014 doi: 10.1038/onc.2014.251. doi: 10.1038/onc.2014.251. [DOI] [PubMed] [Google Scholar]
- Miller BW, Lau G, Grouios C, Mollica E, Barrios-Rodiles M, Liu Y, Datti A, Morris Q, Wrana JL, Attisano L. Application of an integrated physical and functional screening approach to identify inhibitors of the Wnt pathway. Mol Syst Biol. 2009;5:315. doi: 10.1038/msb.2009.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missy K, Hu B, Schilling K, Harenberg A, Sakk V, Kuchenbecker K, Kutsche K, Fischer KD. AlphaPIX Rho GTPase guanine nucleotide exchange factor regulates lymphocyte functions and antigen receptor signaling. Mol Cell Biol. 2008;28:3776–3789. doi: 10.1128/MCB.00507-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota M, Sasaki H. Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. Development. 2008;135:4059–4069. doi: 10.1242/dev.027151. [DOI] [PubMed] [Google Scholar]
- Ramakers GJ, Wolfer D, Rosenberger G, Kuchenbecker K, Kreienkamp HJ, Prange-Kiel J, Rune G, Richter K, Langnaese K, Masneuf S, Bosl MR, Fischer KD, Krugers HJ, Lipp HP, van Galen E, Kutsche K. Dysregulation of Rho GTPases in the alphaPix/Arhgef6 mouse model of X-linked intellectual disability is paralleled by impaired structural and synaptic plasticity and cognitive deficits. Hum Mol Genet. 2012;21:268–286. doi: 10.1093/hmg/ddr457. [DOI] [PubMed] [Google Scholar]
- Ramos A, Camargo FD. The Hippo signaling pathway and stem cell biology. Trends Cell Biol. 2012;22:339–346. doi: 10.1016/j.tcb.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy BV, Irvine KD. Regulation of Hippo signaling by EGFR-MAPK signaling through Ajuba family proteins. Dev Cell. 2013;24:459–471. doi: 10.1016/j.devcel.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson BS, Huang J, Hong Y, Moberg KH. Crumbs regulates Salvador/Warts/Hippo signaling in Drosophila via the FERM-domain protein Expanded. Curr Biol. 2010;20:582–590. doi: 10.1016/j.cub.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberger G, Kutsche K. AlphaPIX and betaPIX and their role in focal adhesion formation. Eur J Cell Biol. 2006;85:265–274. doi: 10.1016/j.ejcb.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Schlegelmilch K, Mohseni M, Kirak O, Pruszak J, Rodriguez JR, Zhou D, Kreger BT, Vasioukhin V, Avruch J, Brummelkamp TR, Camargo FD. Yap1 acts downstream of alpha-catenin to control epidermal proliferation. Cell. 2011;144:782–795. doi: 10.1016/j.cell.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenker O, Rittinger K. Structures of dimeric GIT1 and trimeric beta-PIX and implications for GIT-PIX complex assembly. J Mol Biol. 2009;386:280–289. doi: 10.1016/j.jmb.2008.12.050. [DOI] [PubMed] [Google Scholar]
- Schroeder MC, Halder G. Regulation of the Hippo pathway by cell architecture and mechanical signals. Semin Cell Dev Biol. 2012;23:803–811. doi: 10.1016/j.semcdb.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Serrano I, McDonald PC, Lock F, Muller WJ, Dedhar S. Inactivation of the Hippo tumour suppressor pathway by integrin-linked kinase. Nat Commun. 2013;4:2976. doi: 10.1038/ncomms3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko R, McNeill H. The skinny on Fat: an enormous cadherin that regulates cell adhesion, tissue growth, and planar cell polarity. Curr Opin Cell Biol. 2009;21:717–723. doi: 10.1016/j.ceb.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Sorrentino G, Ruggeri N, Specchia V, Cordenonsi M, Mano M, Dupont S, Manfrin A, Ingallina E, Sommaggio R, Piazza S, Rosato A, Piccolo S, Del Sal G. Metabolic control of YAP and TAZ by the mevalonate pathway. Nat Cell Biol. 2014;16:357–366. doi: 10.1038/ncb2936. [DOI] [PubMed] [Google Scholar]
- Staruschenko A, Sorokin A. Role of betaPix in the kidney. Front Physiol. 2012;3:154. doi: 10.3389/fphys.2012.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Kolb R, Hong JH, Carroll J, Li D, You J, Bronson R, Yaffe MB, Zhou J, Benjamin T. TAZ promotes PC2 degradation through a SCFbeta-Trcp E3 ligase complex. Mol Cell Biol. 2007;27:6383–6395. doi: 10.1128/MCB.00254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varelas X, Miller BW, Sopko R, Song S, Gregorieff A, Fellouse FA, Sakuma R, Pawson T, Hunziker W, McNeill H, Wrana JL, Attisano L. The Hippo pathway regulates Wnt/beta-catenin signaling. Dev Cell. 2010a;18:579–591. doi: 10.1016/j.devcel.2010.03.007. [DOI] [PubMed] [Google Scholar]
- Varelas X, Samavarchi-Tehrani P, Narimatsu M, Weiss A, Cockburn K, Larsen BG, Rossant J, Wrana JL. The Crumbs complex couples cell density sensing to Hippo-dependent control of the TGF-beta-SMAD pathway. Dev Cell. 2010b;19:831–844. doi: 10.1016/j.devcel.2010.11.012. [DOI] [PubMed] [Google Scholar]
- Wada K, Itoga K, Okano T, Yonemura S, Sasaki H. Hippo pathway regulation by cell morphology and stress fibers. Development. 2011;138:3907–3914. doi: 10.1242/dev.070987. [DOI] [PubMed] [Google Scholar]
- Yu FX, Zhao B, Panupinthu N, Jewell JL, Lian I, Wang LH, Zhao J, Yuan H, Tumaneng K, Li H, Fu XD, Mills GB, Guan KL. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150:780–791. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Guan KL. The Hippo pathway: regulators and regulations. Genes Dev. 2013;27:355–371. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Zhang Y, Park HW, Jewell JL, Chen Q, Deng Y, Pan D, Taylor SS, Lai ZC, Guan KL. Protein kinase A activates the Hippo pathway to modulate cell proliferation and differentiation. Genes Dev. 2013;27:1223–1232. doi: 10.1101/gad.219402.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, Zheng P, Ye K, Chinnaiyan A, Halder G, Lai ZC, Guan KL. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinnaiyan AM, Lai ZC, Guan KL. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Lu Q, Wang LH, Liu CY, Lei Q, Guan KL. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 2011;25:51–63. doi: 10.1101/gad.2000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Wang L, Wang CY, Yu J, Guan KL. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev. 2012;26:54–68. doi: 10.1101/gad.173435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Table S1
Review Process File
Source Data for Figure 1B
Source Data for Figure 5B, C
Source Data for Figure 6B, C