Abstract
Because of its wide range of substrates, caspase-3, a main executioner among apoptosis-related caspases, is thought to have many unknown substrates that have remained unidentified. This report describes our predictive method to facilitate the discovery of novel caspase-3 substrates. To develop a more reliable prediction method, we specifically examined improvement of the data quantity and quality of caspase-3 cleavage sites. The ScreenCap3 method is based on machine learning and on information not only of experimentally verified positive examples but also of negative examples, which were not cleaved by caspase-3. Using information of experimentally verified noncleavage sites, we elucidate novel patterns of amino acids around “actual” cleavage sites. Results show that ScreenCap3 provides substantial improvement in terms of precision, compared with existing methods. Therefore, ScreenCap3 is anticipated for use with proteomic screening and identification of novel caspase-3 substrates and their cleavage sites. ScreenCap3 is available at http://scap.cbrc.jp/ScreenCap3/.
Keywords: Apoptosis, Bioinformatics, Caspase-3, High-throughput proteomics
Caspases are a family of cysteine proteases that are crucially important to the initiation and progression of apoptosis. Depending on the role caspases play during apoptosis, they are classifiable further into initiator or executioner caspases 1,2. Among all executioner caspases, caspase-3 is probably a main executioner caspase because of its wide range of substrates. In addition, immunodepletion of caspase-3 has been found to abolish most proteolytic events, causing more severe defects than other executioner caspases do 2. In addition to apoptosis, caspase-3 is involved in important processes such as cell differentiation, cell–cell adhesion, neurodevelopment, and neuronal signaling 3–5. Because of its functional importance, the experimental identification of novel caspase-3 substrates has remained an active field of research 6. However, only one currently available computational method is designed specifically for the same purpose: CAT3 7. Although many multi-caspase and multi-protease prediction methods have been used widely in attempts to discover new caspase-3 substrates, their prediction results are degraded by their numerous false-positive results 7. A possible cause is the mixed data of different caspase substrates used in multi-protease predictors. To resolve this shortcoming, CAT3 was built exclusively using caspase-3 substrates. When tested on an independent set of 17 caspase-3 substrates, CAT3 demonstrated not only a comparable level of sensitivity, but also a considerably lower false-positive rate over multi-caspase predictors. Instead of different meta-features used in other methods, CAT3 applied a simple but effective approach using only primary sequence information to generate position-specific scoring matrices for prediction. For this reason, the authors of CAT3 inferred that the improvement can be attributed mainly to their caspase-3 specific dataset, thereby avoiding overgeneralization and lowering the false-positive rate 7.
We propose ScreenCap3, a web-based prediction method, to facilitate the discovery of novel caspase-3 substrates in human proteome. To develop a more reliable predictor, we specifically emphasized the improvement of both data quantity and quality of caspase-3 cleavage sites. Based on 267 human caspase-3 cleavage sites readily collected in CAT3, we compiled an additional list of 206 caspase-3 cleavage sites from the literature and the MEROPS database 8: a 77% increase over the number used in CAT3. To achieve the highest data quality, we applied three filtering procedures based on (i) experimental evidence at both the site and protein level presented in the original paper, (ii) cleavage sites classified as type “P” in MEROPS, indicating that peptide and protein substrates are thought to be physiological, and (iii) protein sequences with less than 80% sequence identity using CD-HIT software 9. Consequently, our updated dataset includes 473 experimentally verified cleavage sites from 301 substrates of caspase-3.
After increasing the quantity of caspase-3 cleavage sites, our next emphasis was the quality of negative examples. Through a literature review, we compiled local information related to 1291 aspartic acids from 48 proteins, including those tested in a recent study with a cell-free system 10, which cannot be cleaved by caspase-3 in vitro (hereinafter denoted as noncleavage D-sites). It is noteworthy that “plausible” noncleavage sites are commonly used in existing prediction methods: for each caspase-3 substrate, every aspartic acid without experimental evidence for caspase-3-mediated cleavage will be used as negative examples. Although generating negative examples in this manner is common in practice, one must be extremely cautious because some can be yet undiscovered cleavage sites. Moreover, this strategy often produces numerous negative examples, necessitating random selection procedures to ease the resulting highly skewed ratio between positive and negative sets. In fact, it has been proposed that experimentally verified negative data can be useful to improve the predictive performance 11. However, ScreenCap3 is the first predictor to turn this concept into practice for caspase-3 substrates.
The cleavage signature of caspase is commonly regarded as a tetrapeptide motif comprising four residues immediately upstream from the cleavage site (hereinafter, P4–P1). However, results of previous research indicate that some amino acid positions located close to the cleavage site are important for the discrimination of caspase-7 and caspase-3 for their specific substrates such as P6, P5, P2′, and P3′ (second and the third positions downstream from the cleavage site) 12. A series of window sizes was evaluated in the original paper presenting CAT3, demonstrating that the best predictive performance in terms of the Matthews correlation coefficient (MCC) was retrieved at the window size of P6–P2′. Based on these observations, we performed sequence motif analysis and made a cleavage signature of P6–P2′ window using iceLogo 13. Compared with existing logo-based tools such as WebLogo 14, the salient feature of iceLogo is that it accepts a user-defined background set in search of amino acids that differ significantly (p < 0.05) from the positive set. In this study, the updated set of cleavage sites served as the positive set. However, we generated two iceLogos using two background sets: (1) our experimentally verified noncleavage D-sites and (2) the “plausible” noncleavage D-sites, as most previous methods used. Figure 1 shows that several differences are apparent between those amino acids enriched (upper part of the iceLogo) or depleted (lower part of the iceLogo) at each position within the P6–P2′ peptide. This result justified our choice for a more reliable negative dataset used to develop ScreenCap3.
Figure 1.
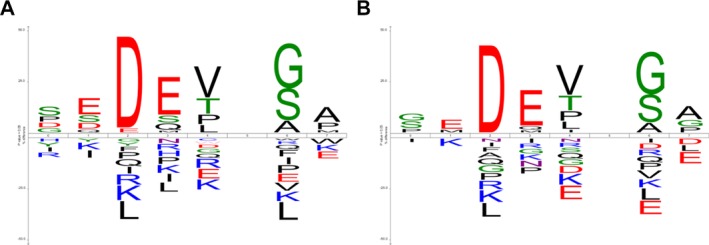
Two iceLogos generated using the same positive examples (experimentally verified cleavage sites) but different negative examples: (A) our experimentally verified noncleavage D-sites and (B) the “plausible” noncleavage D-sites, as most previous methods used.
The prediction of ScreenCap3 is performed from the primary sequence. It is based on a support vector machine (SVM) implemented using libsvm 3.17 15. We simply transformed the amino acid type at each position within each P6–P2′ peptide into a 20-dimensional binary vector with one element set to one and the rest to zero, generating 8 × 20 features for each example. The highly skewed ratio between positive and negative data is eased greatly to 1:3 when our noncleavage D-sites are used as negative examples. Therefore, we did not conduct random selection for negative examples. Instead, we used AUC rather than accuracy as a criterion for evaluation when conducting cross-validation of our unbalanced training dataset. The final model is optimized using a built-in grid search tool and a feature selection tool provided on the libsvm website (http://www.csie.ntu.edu.tw/∼cjlin/libsvmtools/). We also use an optional parameter (-b 1) provided in the libsvm package to produce probability estimates from 0 to 1 as the standard output of ScreenCap3 16. These probability estimates were further used to select appropriate cut-off values under fivefold cross-validation scheme. Users can find the full assessment in our online documentation: http://scap.cbrc.jp/ScreenCap3/documentations.php. As a result, we set the default cut-off value at a probability of 0.7 attributable to its corresponding false discovery rate (= 0.1) and the good balance among different performance metrics. However, it is noteworthy that a tradeoff exists between recall (also known as sensitivity), the percentage of known cleavage sites correctly predicted and precision (also known as positive predictive value (PPV)), the percentage of predicted cleavage sites that are experimentally verified. This cut-off value is adjustable to match different needs. For example, using probabilities greater than 0.7 can generate more precise predictions at a cost of detecting less real cleavage sites. For high-throughput screening, these cut-off values will be useful for selecting a manageable number of candidate sites to be tested experimentally.
In addition to CAT3, we compared ScreenCap3 with other existing methods designed for multi-caspase and multi-protease cleavage site predictions: Pripper 17 and SitePrediction 18, respectively. The evaluation was conducted using an independent test dataset containing 53 cleavage sites from 45 caspase-3 substrates compiled from a recent proteome profiling study 19. Among these tools, Pripper is a binary classifier, whereas CAT3 and SitePrediction make score-based prediction with different predefined cut-off values. While making predictions, we submitted the independent test data to SitePrediction web server and applied them to stand-alone versions of Pripper and CAT3. All predictions were made using default parameters and cut-off values of each tool. The performance is assessed using true positives, false positives, recall, precision, and MCC. Table1 shows that Pripper and SitePrediction (cut-off at 99%) can detect more than 70% cleavage sites, but at a cost of quadrupling the number of false positives. SitePrediction (cut-off at 99.9%) achieves the highest precision (0.49) among all but the lowest recall (0.34). Two predictors specific for caspase-3, ScreenCap3, and CAT3 achieve recall and precision in between Pripper and SitePedictions (two cut-off values). However, ScreenCap3 (cut-off at 0.7) achieves the best MCC performance of 0.41 compared to other methods. In fact, when we changed the cut-off value from 0.5 to 0.9 by 0.1, ScreenCap3 still consistently outperformed existing methods in MCC, with performance of 0.39–0.44.
Table 1.
Performance comparison with existing methods
| Method (cut-off) | #TPs (recall) | #FPs (precision) | MCC |
|---|---|---|---|
| SitePrediction (99%) | 42 (0.79) | 188 (0.18) | 0.32 |
| Prippera) | 38 (0.72) | 128 (0.23) | 0.35 |
| SitePrediction (99.9%) | 18 (0.34) | 19 (0.49) | 0.37 |
| CAT3 (30) | 23 (0.43) | 38 (0.38) | 0.37 |
| ScreenCap3 (0.7) | 26 (0.49) | 38 (0.41) | 0.41 |
Pripper is not a score-based predictor.
We conducted further assessment of the number of false positives reported by ScreenCap3 at the cut-off values attaining the same number of true-positive results predicted using each existing tool. Figure 2A presents the superiority of ScreenCap3 in reducing false positives. While attaining an equal number of false-positive results, ScreenCap3 also reports more true-positive results than each existing tool (Fig. 2B). In addition to specific cut-off values defined by each tool, we used a precision–recall curve (PR curve) to assess the overall predictive performance for three score-based predictors: ScreenCap3, CAT3, and SitePrediction. As Fig. 2C shows, ScreenCap3 performs uniformly better than CAT3 with higher precision values at every recall level. Comparison with SitePrediction demonstrates that ScreenCap3 attains higher precision values at almost all recall levels, except for a slightly lower precision value by 4% at the recall level of 60%. Moreover, we observed that D-sites with high scores predicted by ScreenCap3 have a much higher probability of being recognized and cleaved by caspase-3. For example, at recall of 20%, ScreenCap3 yields a precision higher than 90%, although the respective precisions of CAT3 and SitePrediction are 71 and 55%. This much-improved precision (PPV) at a low recall level reflects that ScreenCap3 is 20–35% more precise than existing tools. In other words, when applying strict cut-off values in ScreenCap3, experimenters can identify the same number of cleavage sites with less trial and error. In this regard, ScreenCap3 is the most appropriate large-scale screening tool in searching of potential capase-3 substrates. It is noteworthy that CAT3 and SitePrediction are unable to detect all verified cleavage sites even when the smallest score was used as the cut-off value. Consequently, the PR curves of CAT3 and SitePrediction are truncated at the recall level of 90%. These results show that CAT3 and SitePrediction do not make a prediction for every D-site, which causes some unavoidable false negatives.
Figure 2.
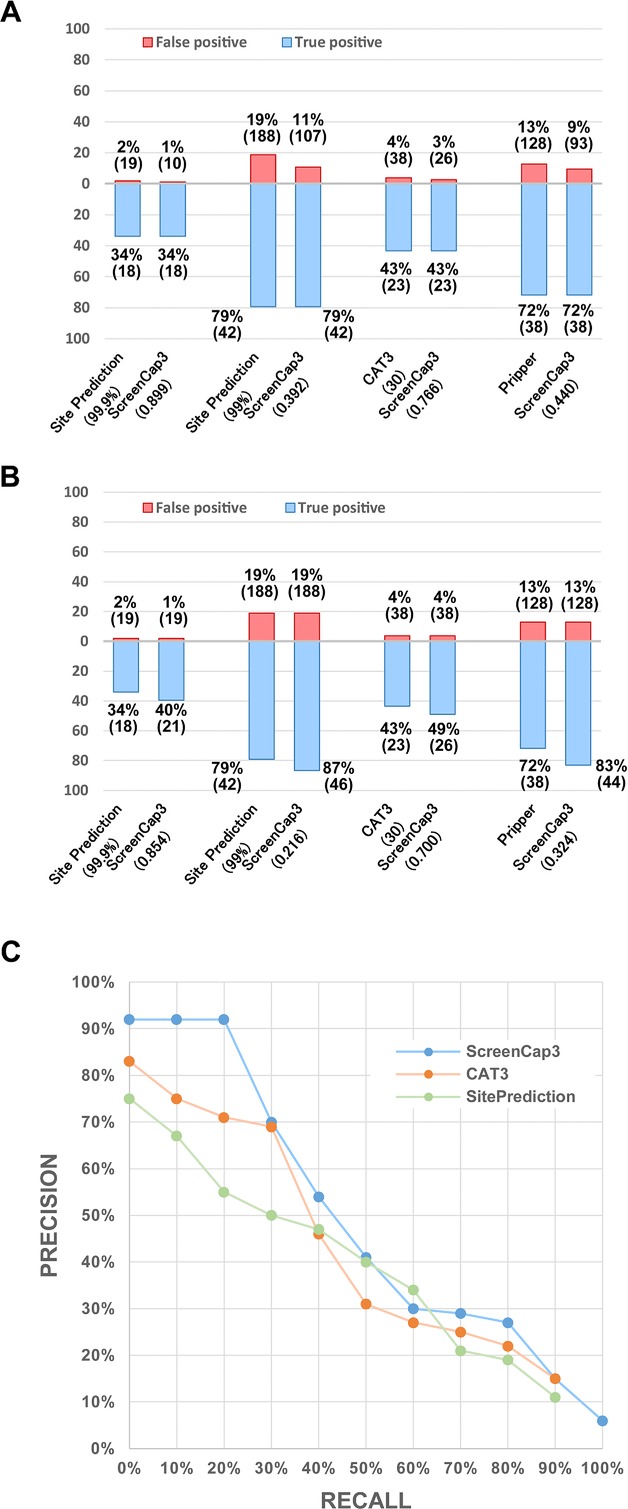
(A) Comparison of the number of false positives while attaining an equal number of true-positive results. Corresponding cut-off values are shown in parentheses. (B) Comparison of the number of true positives while attaining an equal number of false-positive results. Corresponding cut-off values are shown in parentheses. (C) PR curve of three tools at the site level. Recall is defined as TP/(TP + FN). Precision is defined as TP/(TP + FP), here TP = number of true positives, FN = number of false negatives, and FP = number of false positives.
As an extended test, we made predictions using two recently discovered caspase-3 substrates known to contain multiple cleavage sites. The dual specificity phosphatase Cdc25A, for instance, includes nine verified cleavage sites 20. ScreenCap3 achieves a recall of 44% and a precision of 100% while identifying all of them. SitePrediction (cut-off at 99.9%) also achieves a precision of 100% but recall drops to 33%. Regarding CAT3, it achieves only 22 and 50% in recall and precision, respectively. In the case of human PKC-interacting cousin of thioredoxin (PICOT) containing two cleavage sites 21, all three tools achieve 100% precision, but only ScreenCap3 attains a recall of 100%. Both CAT3 and SitePrediction (cut-off at 99.9%) miss one cleavage site verified in PICOT. Although these examples might be special cases, they suggest that ScreenCap3 can reduce the number of false-negative results because ScreenCap3 can make predictions for each single D-site.
We developed the ScreenCap3 web server with an intuitive user interface. Step-by-step instructions for novice users are available on our online documentations: http://scap.cbrc.jp/ScreenCap3/documentations.php. Using our intuitive submission interface, users can input either the protein sequence of interest in single FASTA format or UniProt 22 protein name (UniProt ID) such as XKR8_HUMAN, which has been identified recently as a substrate of caspase-3 23. After successful submission, users can retrieve the prediction results within seconds, with selected different cut-off values for filtering out those predicted cleavage sites with low probability.
We proposed ScreenCap3, a more reliable predictor of caspase-3 cleavage sites. Compared to the state-of-the-art caspase-3 predictor and other multi-protease tools, ScreenCap3 shows better overall performance and substantial improvement in precision when applying strict cut-off values. This feature makes ScreenCap3 a useful high-throughput screening tool for yet undiscovered capase-3 substrates and cleavage sites. Furthermore, ScreenCap3 is the first reported caspase predictor to use experimentally verified noncleavage sites as negative examples. This feature improves the performance, justifies our choice, and suggests potential for expanding this idea to prediction of other caspase families. ScreenCap3 is now available on: http://scap.cbrc.jp/ScreenCap3/.
Acknowledgments
We thank Dr. Kana Shimizu for helpful discussions. This work was supported by the Platform for Drug Discovery, Informatics, and Structural Life Science from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
The authors have declared no conflict of interest.
Glossary
- MCC
Matthews correlation coefficient
- PPV
positive predictive value
- PR
precision–recall
- SVM
support vector machine
References
- Riedl SJ, Shi Y. Molecular mechanisms of caspase regulation during apoptosis. Nat. Rev. Mol. Cell Biol. 2004;5:897–907. doi: 10.1038/nrm1496. [DOI] [PubMed] [Google Scholar]
- Walsh JG, Cullen SP, Sheridan C, Luthi AU. Executioner caspase-3 and caspase-7 are functionally distinct proteases. Proc. Natl. Acad. Sci. USA. 2008;105:12815–12819. doi: 10.1073/pnas.0707715105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amelio M, Cavallucci V, Cecconi F. Neuronal caspase-3 signaling: not only cell death. Cell Death Differ. 2010;17:1104–1114. doi: 10.1038/cdd.2009.180. [DOI] [PubMed] [Google Scholar]
- Nakamoto K, Kuratsu J, Ozawa M. β-catenin cleavage in non-apoptotic cells with reduced cell adhesion activity. Int. J. Mol. Med. 2005;15:973–979. [PubMed] [Google Scholar]
- Puga I, Rao A, Macian F. Targeted cleavage of signaling proteins by caspase 3 inhibits T cell receptor signaling in anergic T cells. Immunity. 2008;29:193–204. doi: 10.1016/j.immuni.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford ED, Wells JA. Caspase substrates and cellular remodeling. Annu. Rev. Biochem. 2011;80:1055–1087. doi: 10.1146/annurev-biochem-061809-121639. [DOI] [PubMed] [Google Scholar]
- Ayyash M, Tamimi H, Ashhab Y. Developing a powerful in silico tool for the discovery of novel caspase-3 substrates: a preliminary screening of the human proteome. BMC Bioinformatics. 2012;13:14. doi: 10.1186/1471-2105-13-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ, Bateman A. MEROPS: The database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2012;40:D343–D350. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Niu B, Gao Y, Fu L, Li W. CDHIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics. 2010;26:680–682. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadokoro D, Takahama S, Shimizu K, Hayashi S. Characterization of a caspase-3-substrate kinome using an N- and C-terminally tagged protein kinase library produced by a cell-free system. Cell Death Dis. 2010;1:e89. doi: 10.1038/cddis.2010.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Tan H, Perry AJ, Akutsu T. PROSPER: an integrated feature-based tool for predicting protease substrate cleavage sites. PLoS One. 2012;7:e50300. doi: 10.1371/journal.pone.0050300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demon D, Van Damme P, Vanden Berghe T, Deceuninck A. Proteome-wide substrate analysis indicates substrate exclusion as a mechanism to generate caspase-7 versus caspase-3 specificity. Mol. Cell Proteomics. 2009;8:2700–2714. doi: 10.1074/mcp.M900310-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaert N, Helsens K, Martens L, Vandekerckhove J, Gevaert K. Improved visualization of protein consensus sequences by iceLogo. Nat. Methods. 2009;6:786–787. doi: 10.1038/nmeth1109-786. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C-C, Lin C-J. LIBSVM: a library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011;2:27. [Google Scholar]
- Chen Y-W, Lin C-J. In: Feature Extraction. Guyon I, Nikravesh M, Gunn S, Zadeh LA, editors. Springer, Berlin Heidelberg; 2006. pp. 315–324. [Google Scholar]
- Piippo M, Lietzen N, Nevalainen OS, Salmi J, Nyman TA. Pripper: prediction of caspase cleavage sites from whole proteomes. BMC Bioinformatics. 2010;11:320. doi: 10.1186/1471-2105-11-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verspurten J, Gevaert K, Declercq W, Vandenabeele P. SitePredicting the cleavage of proteinase substrates. Trends Biochem. Sci. 2009;34:319–323. doi: 10.1016/j.tibs.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Timmer JC, Zhu W, Pop C, Regan T. Structural and kinetic determinants of protease substrates. Nat. Struct. Mol. Biol. 2009;16:1101–1108. doi: 10.1038/nsmb.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou ST, Yen YC, Lee CM, Chen MS. Pro-apoptotic role of Cdc25A: activation of cyclin B1/Cdc2 by the Cdc25A C-terminal domain. J. Biol. Chem. 2010;285:17833–17845. doi: 10.1074/jbc.M109.078386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun N, Kim C, Cha H, Park WJ. Caspase-3-mediated cleavage of PICOT in apoptosis. Biochem. Biophys. Res. Commun. 2013;432:533–538. doi: 10.1016/j.bbrc.2013.02.017. [DOI] [PubMed] [Google Scholar]
- The UniProt Consortium. Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 2011;39:D214–D219. doi: 10.1093/nar/gkq1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki J, Denning DP, Imanishi E, Horvitz HR, Nagata S. Xk-related protein 8 and CED-8 promote phosphatidylserine exposure in apoptotic cells. Science. 2013;341:403–406. doi: 10.1126/science.1236758. [DOI] [PubMed] [Google Scholar]


