Abstract
Purpose
We explored the impact of obesity, body composition, and genetic polymorphisms on the pharmacokinetics (PK) of daunorubicin in children with cancer.
Patients and methods
Patients ≤21 years receiving daunorubicin as an infusion of any duration <24 h for any type of cancer were eligible. Plasma drug concentrations were measured by high-performance liquid chromatography. Body composition was measured by dual-energy X-ray absorptiometry. Obesity was defined as a BMI >95 % for age or as body fat >30 %. NONMEM was used to perform PK model fitting. The Affymetrix DMET chip was used for genotyping. The impact of genetic polymorphisms was investigated using SNP/haplotype association analysis with estimated individual PK parameters.
Results
A total of 107 subjects were enrolled, 98 patients had PK sampling, and 50 patients underwent DNA analysis. Population estimates for daunorubicin clearance and volume of distribution were 116 L/m2/h ± 14 % and 68.1 L/m2 ± 24 %, respectively. Apparent daunorubicinol clearance and volume of distribution were 26.8 L/m2/h ± 5.6 % and 232 L/m2 ± 10 %, respectively. No effect of body composition or obesity was observed on PK. Forty-four genes with variant haplotypes were tested for association with PK. FMO3-H1/H3 genotype was associated with lower daunorubicin clearance than FMO3-H1/ H1, p = 0.00829. GSTP1*B/*B genotype was also associated with lower daunorubicin clearance compared to GSTP1*A/*A, p = 0.0347. However, neither of these associations was significant after adjusting for multiple testing by either Bonferroni or false discovery rate correction.
Conclusions
We did not detect an effect of body composition or obesity on daunorubicin PK. We found suggestive associations between FMO3 and GSTP1 haplotypes with daunorubicin PK that could potentially affect efficacy and toxicity.
Keywords: Daunorubicin, Daunorubicinol, Pharmacokinetics, Pharmacogenetics, Pediatrics, Body composition, Obesity
Introduction
Daunorubicin is an anthracycline antineoplastic drug widely used in childhood cancer. Despite its frequent use, however, daunorubicin’s pharmacokinetics (PK) and pharmacodynamics (PD) have not been studied widely in children and very little is known about the relationship between PK parameters and pharmacogenetic markers or covariates like obesity, body composition, age, gender, or ethnicity.
Daunorubicin is administered intravenously over a wide range of doses and schedules. As with other anthracyclines, daunorubicin is extensively metabolized through a complex process involving reduction to the C-13 alcohol metabolite daunorubicinol as well as conversion of both daunorubicin and daunorubicinol to various aglycones, with daunorubicinol the primary metabolite reported in human plasma (reviewed in Robert [1]). Daunorubicinol is more than a log less cytotoxic in vitro than the parent drug daunorubicin [2]. However, alcohol metabolites of anthracyclines may contribute significantly to cardiac toxicity, an important late effect of treatment [3–10].
Recent reports have begun to elucidate the genetic contribution to variability in anthracycline PK/PD, predominately focusing on doxorubicin (reviewed in Lau 2010 [11]). Among the genes involved in anthracycline uptake and efflux in cells are MDR1 [12]; the glutathione conjugate transporter RLIP76 [13]; and the organic cation transporter SLC22A16 [14]. Genes implicated in anthracycline metabolism include the carbonyl reductases CBR1 and CBR3 [15], and the pregnane X receptor PXR [16]. Anthracyclines and their metabolites may also act as substrates for the glutathione S-transferases (GSTM1, GSTP1, and GSTT1) [17, 18]. To our knowledge, studies of the impact of these genes on daunorubicin PK in children have not been performed. Furthermore, the influence of body size on anthracycline PK and treatment outcomes is unclear [19–22]. We therefore performed a study to describe daunorubicin PK in children and to explore the impact of obesity, body composition, and genetic polymorphisms on daunorubicin PK.
Patients and methods
Subjects and clinical methods
IRB approval and written informed consent were obtained according to federal and institutional guidelines. Patients ≤21 years of age, receiving daunorubicin as an infusion of any duration <24 h on either a 1-day or 2-day schedule, were eligible. There were no restrictions on diagnosis or concomitant chemotherapy, and there were no organ function requirements as the decision to administer daunorubicin was not part of the study. Pregnant and breast feeding patients and those with a large prosthesis that might interfere with body composition testing were excluded.
Height and weight were measured and used to calculate body surface area (BSA) and body mass index (BMI). Body composition was measured by dual-energy X-ray absorptiometry (DEXA) in subjects enrolling at institutions with this capability. A CBC, serum creatinine, ALT, AST, bilirubin, total protein, albumin, alkaline phosphatase, and gamma glutamyl transferase were obtained at baseline. Patients were on numerous concomitant medications, and it was not possible to analyze their effect.
Pharmacokinetic samples and assay
Blood samples were obtained in sodium heparin tubes stored on wet ice until centrifugation at 4 °C within 15 min of collection to separate the plasma, which was then frozen at −20 to −80 °C until analysis. For single dose regimens, samples were obtained prior to the drug infusion, at the midpoint of the infusion for infusions ≥30 min in duration, and at 0, 0.5, 1, 1.5, 2, 3, 4, 6, 8, 12, 24, 48, and 72 h after the end of the infusion. For 2 day dosing, samples were obtained on day 1 prior to the infusion, at 0, 0.5, 1, 2, 4, and 6 h after the end of the day 1 infusion, immediately prior to the day 2 infusion, and at 0, 0.5, 1, 1.5, 2, 4, 6, 8, 12, 24, 48, and 72 h after the end of the day 2 infusion. Samples were initially collected only from a site different from the daunorubicin infusion site. Subsequently, a catheter- clearing procedure was validated to permit samples to be obtained from a central venous catheter that had been used to administer drug. Plasma concentrations of daunorubicin and its metabolite daunorubicinol were measured using a validated high-performance liquid chromatography assay with fluorescence detection [23, 24]. The recovery of daunorubicin and daunorubicinol was >90 %. The limit of quantitation was 17 nM for daunorubicin and 10 nM for daunorubicinol. The intraday coefficient of variation was <3 %, and the interday variation was <7 % for both daunorubicin and daunorubicinol.
Pharmacokinetic model
We modeled the PK of daunorubicin and daunorubicinol using NONMEM VI (ICON Development Solutions, Elliott City, MD) on a personal computer using a DIGITAL Visual FORTRAN compiler (version 6.1). Final model selection was based on visual inspection of the fit of the model to the data. A stepwise modeling approach similar to that described Hempel et al. [25] was used.
For daunorubicin, a two-compartment model was built using the ADVAN5 subroutine. Exponential error models were used to describe the interindividual variance in each PK parameter, with a proportional residual error model. To analyze the pharmacokinetics of the metabolite daunorubicinol, the pharmacokinetic parameters for daunorubicin were fixed to the estimated parameters and used as inputs to estimate the parameters of daunorubicinol. A one-compartment model was used for daunorubicinol with the NONMEM subroutine ADVAN7. Exponential error models were used to describe the interindividual variance in the apparent volume of distribution and apparent clearance for daunorubicinol. The residual error model included both proportional and additive terms. The final model was used to generate estimates of the individual PK parameters for each patient. These estimates were then used in post hoc statistical analyses to explore the impact of obesity, body composition, and genetic polymorphisms on the PK parameters. PK parameters were linearly scaled based on BSA.
Analysis of the effect of body composition
Two approaches were used to evaluate potential body size or composition effects. First, we incorporated BMI, BMI percentile, z-BMI, and body fat content as covariates in the NONMEM base models, screening each PK parameter for significant covariate effects graphically using S-PLUS and LOESS curve-fitting. Promising covariates were then incorporated into the model, and we used the likelihood ratio test to evaluate the improvement relative to the base model. A change in the objective function of at least 3.84 for each degree of freedom (corresponding to a p < 0.05) was required to consider the improvement statistically significant.
Second, we conducted a post hoc analysis comparing the individual PK parameters of obese versus non-obese subjects. The analysis was repeated using two different definitions of obesity: (1) BMI greater than the 95th percentile and (2) for the subset of patients (n = 48) who had DEXA measurement of body composition, body fat >30 %. To make these comparisons, we used an unpaired t test. All p values are two-sided, and we considered a p < 0.05 to be statistically significant. To screen for the effect of other factors on PK, we used S-PLUS to examine plots of each parameter against the available clinical variables (age, gender, race/ethnicity, and baseline laboratory values).
Other analyses of potential variables effecting PK
We investigated the potential effect of other clinical variables (age, gender, race/ethnicity, and baseline laboratory data) on each PK parameter graphically using S-PLUS and LOESS curve-fitting.
Covariates that showed potential relationships with PK parameters were then evaluated by direct assessment within NONMEM. With the incorporation of each covariate, a likelihood ratio test was used to evaluate the improvement relative to the base model. A change in the objective function of at least 3.84 for each degree of freedom (corresponding to a p < 0.05) was required to consider the improvement statistically significant.
Analysis of the effect of genetic polymorphisms
Approximately 5 mL of whole blood was collected in K2-EDTA-coated vacutainers. DNA was isolated using the QIAamp DNA Blood Maxi Kit (Qiagen Inc, Valencia, CA) according to manufacturer’s protocol. DNA was analyzed via Nanodrop 8000 (Thermo Scientific, Wilmington, DE) spectrophotometric readings (260/280) for quantity and quality and diluted to 60 ng/µL. Samples were evaluated by PicoGreen after dilution, and those that did not contain enough DNA were amplified using the Repli-g method (Qiagen Inc). Sufficient high-quality DNA was available for 50 individuals to be genotyped on Affymetrix DMET Plus arrays. Of the 1,834 successfully genotyped SNPs, 595 had a minor allele frequency >0.05 and were in Hardy–Weinberg equilibrium (p > 0.001). These 595 SNPs were tested for association with daunorubicin and daunorubicinol clearance (L/m2/h) using a linear additive model in the software PLINK [26]. The clearances were log2-transformed prior to association testing, which created approximately normal distributions. The DMET Plus software also provides haplotypes (i.e., *1/*2 alleles) for 57 of the genes on the array. Forty-four genes had at least one individual with a variant haplotype, and these haplotypes were tested for association with daunorubicin and daunorubicinol clearance. Only haplotypes classified as unique (“UNIQ” by the DMET Plus software) were used in the association tests. Linear regression was performed using R statistical software.
Results
One hundred and seven subjects enrolled; seven withdrew prior to the completion of PK sampling. PK data were analyzable for 98 patients. Subject characteristics are shown in Table 1. The median age was 12 years (range 0.5–20.4), median weight was 51.4 kg (range 7.5–126), and median BMI was 20.2 (range 13.2–39.9). Five patients were underweight (BMI <10th percentile for age), 65 were normal weight (BMI 10th–85th percentile), twelve were overweight (BMI 85th–95th percentile), and 16 were obese (BMI >95th percentile). Forty-eight patients had body composition measured by DEXA scan. Median body fat was 26 % (range 12–46), and 15 patients had body fat >30 %.
Table 1.
Patient characteristics (n = 98)
| Age (years) | |
| Median | 12 |
| Range | 0.5–20.4 |
| Gender | |
| Male | 67 |
| Female | 31 |
| Race | |
| White | 70 |
| Black | 12 |
| Other/unknown | 16 |
| Ethnicity | |
| Non-Hispanic | 75 |
| Hispanic | 15 |
| Other/unknown | 8 |
| Diagnoses | |
| ALL | 60 |
| AML | 32 |
| Lymphoma | 5 |
| Other | 1 |
| Weight (kg) | |
| Median | 51.4 |
| Range | 7.5–126 |
| BMI (kg/m2) | |
| Median | 20.2 |
| Range | 13.2–39.9 |
| Body fat (%; n = 48) | |
| Median | 26.4 |
| Range | 11.9–45.7 |
| Infusion time (min) | |
| ≤15 | 33 |
| 15–30 | 31 |
| 60 | 1 |
| 3 h | 1 |
| 6 h | 32 |
The PK model consisted of two compartments for daunorubicin and one for daunorubicinol. The population PK parameters are shown in Table 2. Daunorubicin clearance and volume of distribution were 116 L/m2/h ± 14 % and 68.1 L/m2 ± 24 %. The apparent daunorubicinol clearance and volume of distribution were 26.8 L/m2/h ± 5.6 % and 232 L/m2 ± 10 %. Interindividual variability was large, particularly for the volume of distribution parameters for both daunorubicin and daunorubicinol.
Table 2.
Population pharmacokinetic model parameters for patients receiving short and long daunorubicin infusions
| Parameter | Population mean |
Interindividual variability |
|---|---|---|
| CL—Daunorubicin (L/h/m2) | 116 (±14 %) | 63.6 % ± 24.1 % |
| V1—Daunorubicin (L/m2) | 68.1 (±24 %) | 151 % ± 23 % |
| V2—Daunorubicin (L/m2) | 128 (±25 %) | 80.1 % ± 26 % |
| Q—Daunorubicin (L/hr/m2) | 312 (±15 %) | 99.0 % ± 31 % |
| CLm/F—Daunorubicinol (L/h/m2) | 26.8 (±5.6 %) | 63.6 % ± 22.6 % |
| Vm/F—Daunorubicinol (L/m2) | 232 (±10 %) | 108.6 % ± 27.9 % |
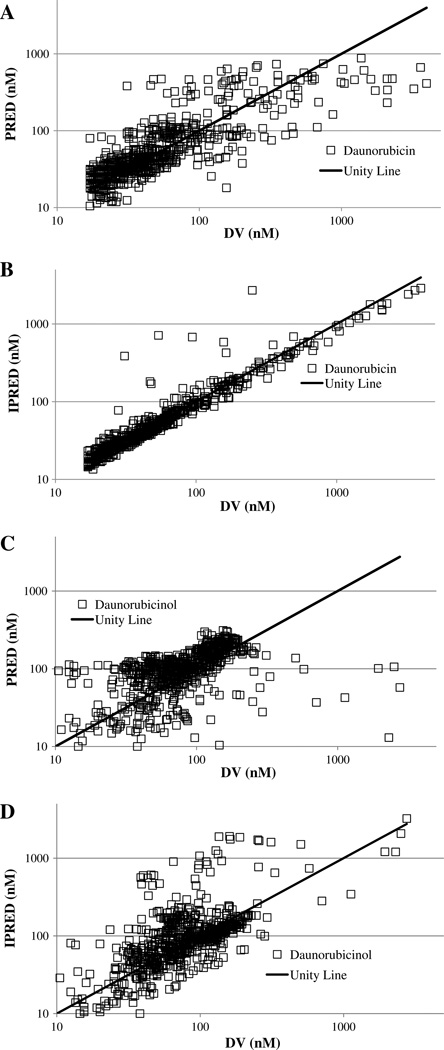
The diagnostic plots for the model are shown in Fig. 1. Figure 1a and b shows the population (PRED) and individual estimated (IPRED) model concentrations plotted against the measured plasma concentrations (DV) for daunorubicin. Figure 1c and d shows the population (PRED) and individual estimated (IPRED) model concentrations plotted against the measured plasma concentrations (DV) for daunorubicinol.
Fig. 1.
a Comparison of population predicted (PRED) and measured daunorubicin concentrations (DV). b Comparison of individual predicted (IPRED) and measured daunorubicin concentrations (DV). c Comparison of population predicted (PRED) and measured daunorubicinol concentrations (DV). d Comparison of individual predicted (IPRED) and measured daunorubicinol concentrations (DV)
No significant effects of body composition covariates on PK parameters were identified. Table 3 shows the PK parameters in the patients defined as obese based on BMI criteria (BMI greater or equal to 95 %) compared with nonobese patients. Table 4 shows the PK parameters in patients defined as obese on the basis of body fat measurements (body fat >30 %) compared with non-obese patients. No statistically significant differences were observed in either comparison.
Table 3.
Effect of BMI on daunorubicin and daunorubicinol pharmacokinetic parameters
| Parameter (units) | BMI <95 % (median ± SD) N = 82 |
BMI ≥95 % (median ± SD) N = 16 |
p value |
|---|---|---|---|
| CL—Daunorubicin (L/h/m2) | 125 ± 73.6 | 121 ± 54.3 | 0.80 |
| V1—Daunorubicin (L/m2) | 54.8 ± 214 | 76.5 ± 258 | 0.92 |
| V2—Daunorubicin (L/m2) | 317 ± 266 | 370 ± 154 | 0.65 |
| Q—Daunorubicin (L/h/m2) | 137 ± 214 | 198 ± 102 | 0.66 |
| CLm/F—Daunorubicinol (L/h/m2) | 31.9 ± 20.7 | 27.9 ± 10.5 | 0.083 |
| Vm/F—Daunorubicinol (L/m2) | 329 ± 186 | 258 ± 252 | 0.058 |
Table 4.
Effect of percentage body fat on daunorubicin and daunorubicinol pharmacokinetic parameters
| Parameter (units) | FAT % <30 (median ± SD) N = 33 |
FAT % >30 (median ± SD) N = 15 |
p value |
|---|---|---|---|
| CL—Daunorubicin (L/h/m2) | 123 ± 83.6 | 112 ± 54.3 | 0.39 |
| V1—Daunorubicin (L/m2) | 62.7 ± 180 | 37.5 ± 142 | 0.13 |
| V2—Daunorubicin (L/m2) | 309 ± 227 | 261 ± 178 | 0.14 |
| Q—Daunorubicin (L/h/m2) | 149 ± 130 | 106 ± 73.1 | 0.053 |
| CLm/F—Daunorubicinol (L/h/m2) | 36.0 ± 26.1 | 30.6 ± 8.8 | 0.23 |
| Vm/F—Daunorubicinol (L/m2) | 387 ± 211 | 424 ± 174 | 0.18 |
No significant relationships between age, gender, race/ethnicity, or baseline laboratory values and PK were observed. For laboratory values, this was an expected result since the overwhelming majority of patients enrolled in the study had no significant abnormalities in organ function or baseline laboratory values. Patients were on numerous concomitant medications, and it was not possible to analyze their effect on PK.
PK–PG results
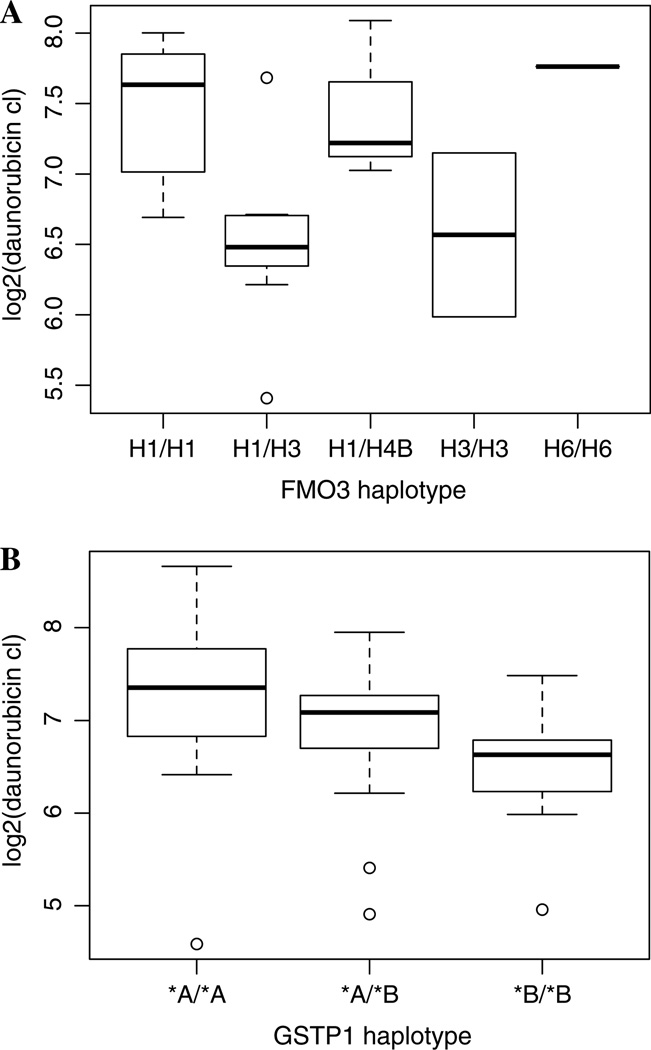
Of the 1,834 genotyped SNPs from 50 patients, 595 had a minor allele frequency (MAF) >0.05 and were in Hardy– Weinberg equilibrium (p > 0.001). These 595 SNPs were tested for association with daunorubicin and daunorubicinol clearance using a linear additive model. Eighteen SNPs associated with daunorubicin clearance (Supplemental Table S1, p < 0.05) and 37 associated with daunorubicinol clearance (Supplemental Table S2, p < 0.05). None of the associations remained significant after adjusting for multiple comparisons. Of 44 genes with variant haplotypes tested for association with the two phenotypes, the best results (p < 0.05) were for FMO3 and GSTP1. In univariate analyses, individuals with FMO3-H1/H3 (n = 7) have significantly lower daunorubicin clearance than individuals with FMO3-H1/H1 (n = 8), p = 0.00829. Individuals with GSTP1*B/*B (n = 7) have significantly lower daunorubicin clearance than individuals with GSTP1*A/*A (n = 19), p = 0.0347. Figure 2 illustrates the effect on clearance. However, neither of these associations was significant after adjusting for multiple testing by either Bonferroni or false discovery rate correction.
Fig. 2.
a Boxplot of FMO3 genotype versus daunorubicin clearance. Individuals with FMO3-H1/H3 (n = 7) have significantly lower daunorubicin clearance than individuals with FMO3-H1/H1 (n = 8), p = 0.00829. b Boxplot of GSTP1 genotype versus daunorubicin clearance. Individuals with GSTP1*B/*B (n = 7) have significantly lower daunorubicin clearance than individuals with GSTP1*A/*A (n = 19), p = 0.0347
Discussion
In this study, we described daunorubicin PK in children with cancer and examined the influence of obesity, body composition, and genetic polymorphisms on daunorubicin PK. There were no significant differences in daunorubicin clearance and daunorubicinol clearance between obese and non-obese patients. Associations between FMO3-H1/H3 and GSTP1*B/*B genotypes and lower daunorubicin clearance were suggested, though neither of these associations was significant after adjusting for multiple testing.
Despite daunorubicin’s use in the treatment of a wide range of malignancies, there are limited data available in the literature on its PK. Our population model has several similarities to what has been previously reported for daunorubicin PK in other studies. Hempel et al. [25] developed a population model for daunorubicin and daunorubicinol in infants with leukemia. Their model was similar structurally (two compartments for daunorubicin and one for daunorubicinol), and they estimated daunorubicin clearance to be 43.9 L/m2/h ± 65 %. The apparent clearance of daunorubicinol was 19.1 L/m2/h ± 9 %, similar to what we report. In a study in adults with sixteen patients receiving daunorubicin 50 mg/m2 as a 10-min infusion, Callies et al. estimated somewhat higher clearances of 168 L/h ± 36 % for daunorubicin and 54.3 L/h ± 27 % for daunorubicinol [27]. Assuming an average BSA of 1.73 m2, the normalized estimates from the Callies study are 97.1 and 31.3 L/m2/h. In our study, large interindividual variability was observed in the volume of distribution parameter estimates. This probably reflects the wide variation of drug doses and schedules administered. Estimates of volume of distribution, particularly for daunorubicin, are influenced by the early concentration time points and may be influenced by the infusion time and the sampling schedule.
Glutathione S-transferases (GSTs) are a polygenic group of enzymes involved in the phase 2 detoxification of a wide range of xenobiotics and chemotherapeutic agents by catalyzing the conjugation of reactive electrophiles to glutathione [28]. Anthracyclines can be substrates for the glutathione S-transferases (GSTM1, GSTP1, and GSTT1) [17, 18], and GSTs are also involved in the detoxification of reactive oxygen species, which can be formed from anthracyclines [29, 30]. GSTP1 is polymorphic with two single-nucleotide substitutions in the coding sequence (1578A>G and 2293C>T), giving rise to Ile105Val and Ala114Val amino acid substitutions [30]. Both amino acid residues lie within the substrate-binding site of GSTP1 [31]. Three functional haplotypes have been identified: GSTP1*A (105Ile;114Ala), GSTP1*B (105Val;114Ala), and GSTP1*C (105Val;114Val) [32]. Therefore, GSTP1*B is a haplotype containing the variant at 105, a polymorphism close to the substrate-binding site that changes the enzymatic activity for particular substrates. In our study, those carrying the variant had lower adjusted daunorubicin clearance than individuals with GSTP1*A/*A or wild type.
There is a paucity of published data on the role of GSTP1 and daunorubicin; however, data from studies with doxorubicin indicate that GSTs have an important role in anthracycline pharmacology. In support of our hypothesis, previous work has shown that breast cancer patients carrying the homozygous GSTP 105Val variant had a lower risk of chemoresistance when treated with doxorubicin [33]. Our results are also compatible with other studies in multiple myeloma, breast and colon cancer, and Hodgkin’s lymphoma that showed better outcomes (e.g., progression-free survival or overall survival) for patients with GSTP 105Val following chemotherapy with drugs known to be GSTP1 substrates [34–37]. Similarly, a study of 68 children with intermediate- and high-risk ALL found that patients who were homozygous for the valine variant were at lower risk for CNS relapse [38]. The authors suggested that impaired detoxification ability in patients with the homozygous valine mutation led to increased active drug exposure and reduced the risk of CNS relapse.
Our work supports the hypothesis that the significance of GSTP1 polymorphisms may be explained at least in part by pharmacokinetic effects. Lower clearance of parent drug in individuals homozygous for the valine allele, as seen in our study, could result in increased exposure to active drug in these patients, which in turn could improve outcomes. In addition, previous work has shown that polymorphisms in GSTs have important pharmacodynamic effects. In studies with cultured human lymphocytes, the genetic damage induced in vitro by doxorubicin was strongly influenced by GSTP1 genotype. Lymphocytes from individuals homozygous for the valine variant had a significant increase in chromosomal damage compared with individuals with at least one wild-type allele [39].
Compared to the GSTs, significantly less is known about the flavin-containing monooxygenase (FMO) enzymes. FMO3 is a hepatic microsomal enzyme that oxidizes a host of drugs, xenobiotics, and other chemicals. Numerous variants in the gene encoding FMO3 have been identified. Some of the variants alter enzymatic activity and, consequently, substrate metabolism. [40] Most notably, FMO3 plays an important role in the N-oxidation of the antipsychotic drug olanzapine. [41].
Although GSTP1 and FMO3 genotype may have an impact on anthracycline pharmacology, it is clearly not the only important genetic factor. Doxorubicin clearance has shown to be lower and AUC higher in patients with genetic variations such as at least one ABCB1 c.1236C>T allele [42], or homozygosity for c.146A>G and c.312T>C polymorphisms in the influx transporter SLC22A16 [43]. Accordingly, doxorubicin clearance is higher and AUC lower in patients with a CC-GG-CC genotype at the ABCB1 1236-2677-3435 loci [42]. In our study, ABCB-1genotype did not affect daunorubicin clearance; however, the C allele of another SNP in ABCB1 (rs2235033), which is linked to the C allele of ABCB1 c.1236C>T, was associated with increased daunorubicinol clearance in the univariate analysis (Table S2). The clinical impact of this change is unknown, but it is possible that patients with faster clearance and lower daunorubicinol exposure could have less risk of cardiotoxicity.
In summary, our current work is the largest pharmacokinetic study of daunorubicin and its metabolite daunorubicinol in children. The study explored the effect of obesity, body composition, and drug metabolizing genotypes on daunorubicin and daunorubicinol PK. Our study design did not permit analysis of the toxicity or efficacy of daunorubicin, so it is still possible that obesity, body composition, and other drug metabolizing genotypes may have important clinical effects. Future studies should be conducted to further evaluate these factors and to evaluate the influence of GSTP1 and FMO3 haplotype on daunorubicin PK. Understanding the impact of these clinical and genetic factors on pharmacokinetics and pharmacodynamics may lead to safer and more effective therapy with anthracyclines in children.
Supplementary Material
Acknowledgments
We would like to acknowledge the contributions of Dolly Yang. This study was supported in part by the Better Pharmaceuticals for Children Act through a supplement to National Cancer Institute grant U10 CA098543 (COG), the Pharmacogenomics of Anticancer Agent Research Grant NIH/NIGMS GM61393 (MED), a Leukemia Lymphoma SCOR grant (MED), University of Chicago Comprehensive Cancer Center (MED), and the NIH/NCI National Research Service Award F32CA165823 (HEW).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00280-014-2535-4) contains supplementary material, which is available to authorized users.
Contributor Information
Patrick Thompson, Texas Children’s Cancer Center, Baylor College of Medicine, 1102 Bates Ave., Suite 1570, Houston, TX 77030, USA.
Heather E. Wheeler, Section of Hematology-Oncology, Department of Medicine, University of Chicago, 900 East 57th Street, KCBD, Room 7100, Chicago, IL 60637, USA
Shannon M. Delaney, Section of Hematology-Oncology, Department of Medicine, University of Chicago, 900 East 57th Street, KCBD, Room 7100, Chicago, IL 60637, USA
Rachel Lorier, Section of Genomic Pediatrics, Department of Pediatrics, Children’s Research Institute and Human and Molecular Genetics Center, Medical College of Wisconsin, Milwaukee, WI, USA.
Ulrich Broeckel, Section of Genomic Pediatrics, Department of Pediatrics, Children’s Research Institute and Human and Molecular Genetics Center, Medical College of Wisconsin, Milwaukee, WI, USA.
Meenakshi Devidas, Texas Children’s Cancer Center, Baylor College of Medicine, 1102 Bates Ave., Suite 1570, Houston, TX 77030, USA.
Gregory H. Reaman, US Food and Drug Administration, Silver Spring, MD, USA
Kathleen Scorsone, Texas Children’s Cancer Center, Baylor College of Medicine, 1102 Bates Ave., Suite 1570, Houston, TX 77030, USA.
Lillian Sung, Hospital for Sick Children, Toronto, ON, Canada.
M. Eileen Dolan, Section of Hematology-Oncology, Department of Medicine, University of Chicago, 900 East 57th Street, KCBD, Room 7100, Chicago, IL 60637, USA.
Stacey L. Berg, Email: sberg@txch.org, Texas Children’s Cancer Center, Baylor College of Medicine, 1102 Bates Ave., Suite 1570, Houston, TX 77030, USA.
References
- 1.Robert J. Anthracyclines. In: Grochow LB, Ames MM, editors. A clinician’s guide to chemotherapy pharmacokinetics and pharmacodynamics. Baltimore: Williams and Wilkins; 1998. pp. 93–173. [Google Scholar]
- 2.Kuffel MJ, Ames MM. Comparative resistance of idarubicin, doxorubicin and their C-13 alcohol metabolites in human MDR1 transfected NIH-3T3 Cells. Cancer Chemother Pharmacol. 1995;36:223–226. doi: 10.1007/BF00685850. [DOI] [PubMed] [Google Scholar]
- 3.Boucek RJ, Jr, Olson RD, Boucek DE, et al. The major metabolite of doxorubicin is a potent inhibitor of membrane-associated ion pumps. A correlative study of cardiac muscle with isolated membrane fractions. J Biol Chem. 1987;262:15851–15856. [PubMed] [Google Scholar]
- 4.Olson RD, Mushlin PS, Brenner DE, et al. Doxorubicin cardiotoxicity may be caused by its metabolite, doxorubicinol. Proc Natl Acad Sci U S A. 1988;85:3585–3589. doi: 10.1073/pnas.85.10.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cusack BJ, Young SP, Driskell J, et al. Doxorubicin and doxorubicinol pharmacokinetics and tissue concentrations following bolus injection and continuous infusion of doxorubicin in the rabbit. Cancer Chemother Pharmacol. 1993;32:53–58. doi: 10.1007/BF00685876. [DOI] [PubMed] [Google Scholar]
- 6.Mushlin PS, Cusack BJ, Boucek RJ, Jr, et al. Time-related increases in cardiac concentrations of doxorubicinol could interact with doxorubicin to depress myocardial contractile function. Br J Pharmacol. 1993;110:975–982. doi: 10.1111/j.1476-5381.1993.tb13909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stewart DJ, Grewaal D, Green RM, et al. Concentrations of doxorubicin and its metabolites in human autopsy heart and other tissues. Anticancer Res. 1993;13:1945–1952. [PubMed] [Google Scholar]
- 8.Forrest GL, Gonzalez B, Tseng W, et al. Human carbonyl reductase overexpression in the heart advances the development of doxorubicin-induced cardiotoxicity in transgenic mice. Cancer Res. 2000;60:5158–5164. [PubMed] [Google Scholar]
- 9.Wang GX, Wang YX, Zhou XB, et al. Effects of doxorubicinol on excitation–contraction coupling in guinea pig ventricular myocytes. Eur J Pharmacol. 2001;423:99–107. doi: 10.1016/s0014-2999(01)01096-2. [DOI] [PubMed] [Google Scholar]
- 10.Olson LE, Bedja D, Alvey SJ, et al. Protection from doxorubicin-induced cardiac toxicity in mice with a null allele of carbonyl reductase 1. Cancer Res. 2003;63:6602–6606. [PubMed] [Google Scholar]
- 11.Lal S, Mahajan A, Chen WN, et al. Pharmacogenetics of target genes across doxorubicin disposition pathway: a review. Curr Drug Metab. 2010;11:115–128. doi: 10.2174/138920010791110890. [DOI] [PubMed] [Google Scholar]
- 12.Ueda K, Cardarelli C, Gottesman MM, et al. Expression of a full-length cDNA for the human “MDR1” gene confers resistance to colchicine, doxorubicin, and vinblastine. Proc Natl Acad Sci U S A. 1987;84:3004–3008. doi: 10.1073/pnas.84.9.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma R, Singhal SS, Cheng J, et al. RLIP76 is the major AT P-dependent transporter of glutathione-conjugates and doxorubicin in human erythrocytes. Arch Biochem Biophys. 2001;391:171–179. doi: 10.1006/abbi.2001.2395. [DOI] [PubMed] [Google Scholar]
- 14.Okabe M, Unno M, Harigae H, et al. Characterization of the organic cation transporter SLC22A16: a doxorubicin importer. Biochem Biophys Res Commun. 2005;333:754–762. doi: 10.1016/j.bbrc.2005.05.174. [DOI] [PubMed] [Google Scholar]
- 15.Lal S, Sandanaraj E, Wong ZW, et al. CBR1 and CBR3 pharmacogenetics and their influence on doxorubicin disposition in Asian breast cancer patients. Cancer Sci. 2008;99:2045–2054. doi: 10.1111/j.1349-7006.2008.00903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sandanaraj E, Lal S, Selvarajan V, et al. PXR pharmacogenetics: association of haplotypes with hepatic CYP3A4 and ABCB1 messenger RNA expression and doxorubicin clearance in Asian breast cancer patients. Clin Cancer Res. 2008;14:7116–7126. doi: 10.1158/1078-0432.CCR-08-0411. [DOI] [PubMed] [Google Scholar]
- 17.Rocha JC, Cheng C, Liu W, et al. Pharmacogenetics of outcome in children with acute lymphoblastic leukemia. Blood. 2005;105:4752–4758. doi: 10.1182/blood-2004-11-4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pander J, Gelderblom H, Guchelaar HJ. Insights into the role of heritable genetic variation in the pharmacokinetics and pharmacodynamics of anticancer drugs. Expert Opin Pharmacother. 2007;8:1197–1210. doi: 10.1517/14656566.8.9.1197. [DOI] [PubMed] [Google Scholar]
- 19.Lange BJ, Gerbing RB, Feusner J, et al. Mortality in overweight and underweight children with acute myeloid leukemia. JAMA. 2005;293:203–211. doi: 10.1001/jama.293.2.203. [DOI] [PubMed] [Google Scholar]
- 20.Hijiya N, Panetta JC, Zhou Y, et al. Body mass index does not influence pharmacokinetics or outcome of treatment in children with acute lymphoblastic leukemia. Blood. 2006;108:3997–4002. doi: 10.1182/blood-2006-05-024414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palle J, Frost BM, Peterson C, et al. Doxorubicin pharmacokinetics is correlated to the effect of induction therapy in children with acute myeloid leukemia. Anticancer Drugs. 2006;17:385–392. doi: 10.1097/01.cad.0000198911.98442.16. [DOI] [PubMed] [Google Scholar]
- 22.Thompson PA, Rosner GL, Matthay KK, et al. Impact of body composition on pharmacokinetics of doxorubicin in children: a Glaser pediatric research network study. Cancer Chemother Pharmacol. 2008 doi: 10.1007/s00280-008-0854-z. [DOI] [PubMed] [Google Scholar]
- 23.Berg S, Cowan K, Balis F, et al. Pharmacokinetics of Taxol and Doxorubicin administered alone and in combination by continuous 72-hour infusion. J Natl Cancer Inst. 1994;86:143–145. doi: 10.1093/jnci/86.2.143. [DOI] [PubMed] [Google Scholar]
- 24.Dobbs N, James C. Estimation of doxorubicin and doxorubicinol by high performance liquid chromatography and advanced automated sample processor. J Chromatogr Biomed Appl. 1987;420:184–188. doi: 10.1016/0378-4347(87)80171-8. [DOI] [PubMed] [Google Scholar]
- 25.Hempel G, Relling MV, de Rossi G, et al. Pharmacokinetics of daunorubicin and daunorubicinol in infants with leukemia treated in the interfant 99 protocol. Pediatr Blood Cancer. 2010;54:355–360. doi: 10.1002/pbc.22266. [DOI] [PubMed] [Google Scholar]
- 26.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Callies S, de Alwis DP, Mehta A, et al. Population pharmacokinetic model for daunorubicin and daunorubicinol co-administered with zosuquidar.3HCl (LY335979) Cancer Chemother Pharmacol. 2004;54:39–48. doi: 10.1007/s00280-004-0775-4. [DOI] [PubMed] [Google Scholar]
- 28.Mannervik B, Alin P, Guthenberg C, et al. Identification of three classes of cytosolic glutathione transferase common to several mammalian species: correlation between structural data and enzymatic properties. Proc Natl Acad Sci U S A. 1985;82:7202–7206. doi: 10.1073/pnas.82.21.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Brien M, Kruh GD, Tew KD. The influence of coordinate overexpression of glutathione phase II detoxification gene products on drug resistance. J Pharmacol Exp Ther. 2000;294:480–487. [PubMed] [Google Scholar]
- 30.Hayes JD, Pulford DJ. The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit Rev Biochem Mol Biol. 1995;30:445–600. doi: 10.3109/10409239509083491. [DOI] [PubMed] [Google Scholar]
- 31.Marciniak P, Drobnik K, Ziemnicka K, et al. Transferase S-glutathione class pi gene (GSTP1) polymorphism in thyroid cancer patients. Endokrynol Pol. 2006;57:509–515. [PubMed] [Google Scholar]
- 32.Ali-Osman F, Akande O, Antoun G, et al. Molecular cloning, characterization, and expression in Escherichia coli of full-length cDNA s of three human glutathione S-transferase Pi gene variants. Evidence for differential catalytic activity of the encoded proteins. J Biol Chem. 1997;272:10004–10012. doi: 10.1074/jbc.272.15.10004. [DOI] [PubMed] [Google Scholar]
- 33.Romero A, Martin M, Oliva B, et al. Glutathione S-transferase P1 c.313A>G polymorphism could be useful in the prediction of doxorubicin response in breast cancer patients. Ann Oncol. 2011 doi: 10.1093/annonc/mdr483. [DOI] [PubMed] [Google Scholar]
- 34.Dasgupta RK, Adamson PJ, Davies FE, et al. Polymorphic variation in GSTP1 modulates outcome following therapy for multiple myeloma. Blood. 2003;102:2345–2350. doi: 10.1182/blood-2003-02-0444. [DOI] [PubMed] [Google Scholar]
- 35.Sweeney C, McClure GY, Fares MY, et al. Association between survival after treatment for breast cancer and glutathione S-transferase P1 Ile105Val polymorphism. Cancer Res. 2000;60:5621–5624. [PubMed] [Google Scholar]
- 36.Stoehlmacher J, Park DJ, Zhang W, et al. Association between glutathione S-transferase P1, T1, and M1 genetic polymorphism and survival of patients with metastatic colorectal cancer. J Natl Cancer Inst. 2002;94:936–942. doi: 10.1093/jnci/94.12.936. [DOI] [PubMed] [Google Scholar]
- 37.Hohaus S, Di Ruscio A, Di Febo A, et al. Glutathione S-transferase P1 genotype and prognosis in Hodgkin’s lymphoma. Clin Cancer Res. 2005;11:2175–2179. doi: 10.1158/1078-0432.CCR-04-1250. [DOI] [PubMed] [Google Scholar]
- 38.Stanulla M, Schaffeler E, Arens S, et al. GSTP1 and MDR1 genotypes and central nervous system relapse in childhood acute lymphoblastic leukemia. Int J Hematol. 2005;81:39–44. doi: 10.1532/ijh97.e0418. [DOI] [PubMed] [Google Scholar]
- 39.Ramos DL, Gaspar JF, Pingarilho M, et al. Genotoxic effects of doxorubicin in cultured human lymphocytes with different glutathione S-transferase genotypes. Mutat Res. 2011;724:28–34. doi: 10.1016/j.mrgentox.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 40.Hisamuddin IM, Yang VW. Genetic polymorphisms of human flavin-containing monooxygenase 3: implications for drug metabolism and clinical perspectives. Pharmacogenomics. 2007;8:635–643. doi: 10.2217/14622416.8.6.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soderberg MM, Haslemo T, Molden E, et al. Influence of FMO1 and 3 polymorphisms on serum olanzapine and its N-oxide metabolite in psychiatric patients. Pharmacogenomics J. 2012 doi: 10.1038/tpj.2012.47. [DOI] [PubMed] [Google Scholar]
- 42.Lal S, Wong ZW, Sandanaraj E, et al. Influence of ABCB1 and ABCG2 polymorphisms on doxorubicin disposition in Asian breast cancer patients. Cancer Sci. 2008;99:816–823. doi: 10.1111/j.1349-7006.2008.00744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lal S, Wong ZW, Jada SR, et al. Novel SLC22A16 polymorphisms and influence on doxorubicin pharmacokinetics in Asian breast cancer patients. Pharmacogenomics. 2007;8:567–575. doi: 10.2217/14622416.8.6.567. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.