Abstract
Purpose
Circadian genes may be involved in regulating cancer-related pathways, including cell proliferation, DNA damage response and apoptosis. We aimed to assess the role of genetic variation in core circadian rhythm genes with the risk of fatal prostate cancer and morning void urinary 6-sulfatoxymelatonin levels.
Methods
We used unconditional logistic regression to evaluate the association of 96 single-nucleotide polymorphisms (SNPs) across twelve circadian-related genes with fatal prostate cancer in the AGES-Reykjavik cohort (n=24 cases), the Health Professionals Follow-Up Study (HPFS) (n=40 cases), and the Physicians’ Health Study (PHS) (n=105 cases). We used linear regression to evaluate the association between SNPs and morning void urinary 6-sulfatoxymelatonin levels in AGES-Reykjavik. We used a kernel machine test to evaluate whether multimarker SNP-sets in the pathway (gene based) were associated with our outcomes.
Results
None of the individual SNPs were consistently associated with fatal prostate cancer across the three cohorts. In each cohort, gene-based analyses showed that variation in the CRY1 gene was nominally associated with fatal prostate cancer (p-values = 0.01, 0.01, 0.05 for AGES-Reykjavik, HPFS, and PHS, respectively). In AGES-Reykjavik, SNPS in TIMELESS (4 SNPs), NPAS2 (6 SNPs), PER3 (2 SNPs) and CSNK1E (1 SNP) were nominally associated with 6-sulfatoxymelatonin levels.
Conclusion
We did not find a strong and consistent association between variation in core circadian clock genes and fatal prostate cancer risk, but observed nominally significant gene-based associations with fatal prostate cancer and 6-sulfatoxymelatonin levels.
Keywords: Circadian genes, prostate cancer, genetic polymorphisms
Introduction
The circadian system regulates the daily oscillations of a wide range of physiologic, metabolic, and behavioral processes [1, 2]. Disturbances of circadian rhythms impair physiological and biochemical processes and may result in tumorigenesis [3-5]. Several lines of evidence from epidemiological studies suggest conditions associated with alteration of circadian rhythms, such as jet lag, shift work, suppression of melatonin by exposure to light at night, and sleep disruption, are associated with increased prostate cancer risk[6-11].
In mammals, circadian rhythms are driven by a pacemaker located in the suprachiasmatic nucleus (SCN) of the hypothalamus that spontaneously generates a near 24-hour oscillation, which is then entrained primarily by the 24 hour light-dark cycle[1]. The hormone melatonin is secreted under the control of the SCN, and is the biochemical correlate of darkness. Thus, in a normally entrained individual, melatonin production reaches a peak at night and is virtually undetectable during the day. The molecular mechanism of the circadian system is based on a series of transcription-translation positive and negative feedback loops[1, 12, 13], regulated by a series of clock genes. The primary circadian clock genes in mammals have been proposed to be: CLOCK, neuronal PAS domain protein 2 (NPAS2), aryl hydrocarbon receptor nuclear translocator-like (ARNTL), cryptochrome 1 (CRY1), cryptochrome 2(CRY2), period 1 (PER1), period 2 (PER2), period 3 (PER3) and, casein kinase 1-epsilon (CSNK1E)[12]. The gene TIMELESS acts together with these genes and products of these processes and is involved in DNA damage checkpoint responses[14]. The MTNR1a and MTNR1b genes encode receptors for melatonin that are responsible for mediating downstream effects of melatonin, including melatonin receptors at the SCN[15].
Studies show that these circadian clock genes and their products interact with cancer-related biological pathways to help regulate and control expression of apoptosis, cell cycle genes, tumor suppressor genes, and genes encoding transcription factors[1, 2, 13, 16-19]. Mutations in the core clock genes have been shown to alter circadian rhythmicity in rodents, and have resulted in neoplastic growth, deficient DNA-damage response, and accelerated growth of malignant tumors in experimental models[12, 13, 20].
Epidemiological studies have reported associations between variation in ARNTL, CSNK1E, NPAS2, PER1, CRY2, and overall prostate cancer risk[2, 21]; variation in PER1, CLOCK and aggressive prostate cancer[2]; and, variation in CRY1 and prostate cancer-specific mortality[22]. The CGEMS project (a genome wide association study (GWAS) of 1,172 prostate cancer cases of European origin) found nominally significant (p-value <0.05) associations between SNPs in NPAS2, CSNK1E, CRY1, and CRY2; however, SNPs in the circadian genes have not been reported at genome-wide significance GWA studies. Additionally, lower levels of melatonin, as measured by the primary melatonin metabolite in urine 6-sulfatoxymelatonin, have been associated with increased risk of cancer, including prostate cancer[9]. To our knowledge, no study has looked at the association of circadian related genes with levels of 6-sulfatoxymelatonin, one of the potential mediators through which circadian disruption may act on risk of prostate cancer.
The primary goal of this study was to comprehensively assess the role of genetic variation in core circadian rhythm genes with the risk of fatal prostate cancer and morning void urinary 6-sulfatoxymelatonin levels, the primary metabolite of melatonin in the urine, within a prospective study of Icelandic men, and fatal prostate cancer in two independent U.S. cohorts.
Materials and Methods
Study Population
We utilized data from the AGES-Reykjavik GWAS, a subset of the AGES-Reykjavik study, a longitudinal population-based study in Iceland, described in detail elsewhere[22]. Protocols were approved by the National Bioethics Committee in Iceland (approval number VSN-00-063) and by the National Institute on Aging Intramural Institutional Review Board. A multistage consent is obtained to cover participation, use of specimens and DNA, and access to administrative records[23].
The data from the Health Professionals Follow-Up Study (HPFS) and Physicians’ Health Study (PHS) were obtained from previously conducted case-control studies of aggressive prostate cancer, as described elsewhere[24, 25]. The HPFS cases were matched to controls on year of birth, PSA test before blood draw, and time of day, season and year of blood draw; PHS cases were matched to controls on age at baseline, smoking status and follow-up time. The HPFS and PHS are approved by the institutional review board at the Harvard School of Public Health (HSPH) and Partners Healthcare, Boston, Massachusetts.
Outcome Ascertainment
Prostate Cancer AGES-Reykjavik
Prostate cancer diagnoses were identified from hospital records using ICD code (ICD9 code 185 and ICD10 code C61). Information on cause of death, including prostate cancer specific death and all-cause mortality, was obtained from linkage to the nationwide death registry by unique identification number[26]. Fatal prostate cancer was defined as death from prostate cancer. We identified 138 prostate cancer cases, 24 of which were fatal.
Prostate Cancer HPFS and PHS
Prostate cancer outcomes were initially obtained from self-report by participants or their next of kin on biennial questionnaires. Prostate cancer diagnoses are confirmed by medical record and pathology report review. Deaths are ascertained from family members and the National Death Index. Cause of death was assigned by an endpoints committee of study physicians through review of medical history, medical reports, registry information and death certificates. To be compatible with the AGES-Reykjavik analysis, we restricted to those cases who died of prostate cancer and identified 40 fatal cases in the HPFS and 105 in PHS.
6-sulfatoxymelatonin
Urinary 6-sulfatoxymelatonin levels in prediagnostic samples were available only for the AGES-Reykjavik study as described elsewhere[9]. Briefly, subjects were instructed to collect a first morning void urine sample and return it to the Icelandic Heart Association on the same day as collection at their baseline visit. The date and time of sample return were recorded, and all urinary samples were stored in 1.5-mL aliquots at −80°C. Urine samples were assayed for 6-sulfatoxymelatonin by laboratory personnel at the Icelandic Heart Association using the Melatonin-Sulfate ELISA (IBL International, Germany) according to manufacturer protocol. The minimal detectable concentration of this assay is 1.0 ng/mL. The coefficient of variation (CV) ranged from 5.4% to 8.5% across batches. 6% of the men with genetic and 6-sulfatoxymelatonin levels developed prostate cancer over follow-up with a mean time to diagnosis of 2.7 years (n=38/585); results were similar when we restricted to men who never developed prostate cancer (data not shown).
Gentoyping
We identified 96 single nucleotide polymorphisms (SNPs) across the twelve circadian related genes thought to regulate the molecular mechanism of the circadian system (CLOCK, NPAS2, ARNTL, CRY1, CRY2, PER1, PER2, PER3, CSNK1E, TIMELESS, MTNR1A, and MTNR1B)[1, 12, 13]. We selected SNPs to capture common genetic variation with R2>0.8 across each of the twelve circadian related genes. We also included those SNPs in circadian genes shown to be associated with prostate cancer risk in previous studies. Selection was restricted to those with a minor allele frequency of greater than 5% in the reference panel.
AGES-Reykjavik
Genotype data was obtained from 1,352 men in the AGES-Reykjavik cohort[22]. Briefly, DNA was genotyped using the Illumina 370 CNV BeadChip array. SNPs were excluded based on call rate (<97%), Hardy-Weinberg equilibrium (p-value <1E-6), and mismatched positions between Illumina, dbSNP and/or HapMap[27-29]. Imputation was done using MACH and the Phase II CEU HapMap data[30]. For this analysis 61 of the 96 SNPs were imputed and the quality of imputation ranged from 0.79-0.99.
HPFS and PHS
Genotyping for the original case-control studies was conducted at the Broad Institute (Boston, Massachusetts) using the Illumina Human 610-Quad platform[25] as a part of the NCI Breast and Prostate Cancer Cohort Consortium (BPC3) GWAS on aggressive prostate cancer. Imputation was done using MACH and the Phase II CEU HapMap data[25, 30]. Samples were excluded if genotyping call rate was <95% or autosomal heterozygosity was <0.25 or >0.35. Additional exclusions were made for Non-European ancestry and unexpected duplicates. We extracted the same 96 SNPs as were analyzed in the AGES-Reykjavik analysis. Of the 96 SNPs, 53 in HPFS and 54 in PHS were imputed.
Statistical Analysis
SNPs and Prostate Cancer
Among the 1,352 men in AGES-Reykjavik with genetic data available, we identified 138 prostate cancer cases, 24 of which were fatal, and 1,214 non-cases. We used unconditional logistic regression to analyze the associations for each of the individual SNPs and fatal prostate cancer risk. Using the additive model we calculated per-allele odds ratios (OR) and 95% confidence intervals (CI). Age at sample collection was included in all models.
We conducted similar analyses for the associations between each SNP and fatal prostate cancer in HPFS (n=40 cases; 204 controls) and in PHS (n=105 cases; 255 controls). Using unconditional logistic regression adjusted for the matching factors, we calculated per-allele ORs and 95% CIs for the same 96 SNPs evaluated in the AGES-Reykjavik study.
Using fixed-effects models, we calculated ORs and 95% CIs for the association between each SNP and fatal prostate cancer, including the results from AGES-Reykjavik, HPFS, and PHS. Heterogeneity across the cohorts was assessed using Cochran’s Q statistic[31, 32].
We used a kernel machine test to examine whether genetic variation across each of the genes or in sets across the pathway was associated with prostate cancer. This approach groups SNPs together into ‘SNP-sets’ based on biological significance (i.e. genes, pathways) and allows one to perform a multi-marker test for the entire SNP-set rather than evaluating SNPs individually[33]. We assessed global associations for each of the twelve genes individually and the nine core circadian rhythm genes together. The advantages of this approach are that it captures the joint effects of multiple SNPs and exploits multilocus linkage disequilibrium (LD) among the SNPs within sets to increase test power[33, 34]. The logistic kernel machine test was used to test the association of each SNP set with overall and fatal prostate cancer.
SNPs and 6-sulfatoxymelatonin levels
There were 547 men with both genetic and urinary 6-sulfatoxymelatonin measures available to assess the association between genetic variation and 6-sulfatoxymelatonin. We used linear regression to calculate betas and standard errors with 6-sulfatoxymelatonin as the dependent variable of interest. Age at sample collection was included in all models. We conducted a linear kernel machine test to assess the association between genetic variation across SNP sets with 6-sulfatoxymelatonin levels, utilizing the same SNP sets as above.
For all analyses, we report the nominal 2-sided P-values without adjustment for multiple comparisons. Analyses were conducted using SAS software version 9.2 (SAS Institute Inc., Cary, NC, USA) and R (http://www.r-project.org/) statistical packages.
Results
Characteristics of the study populations are shown in Table 1. The AGES-Reykjavik cases are older at sample collection and have shorter time from blood draw to diagnosis than HPFS and PHS. The HPFS cases have slightly shorter time from prostate cancer diagnosis to fatal disease than AGES-Reykjavik and PHS (Table 1).
Table 1. Characteristics of the study population - AGES-Reykjavik Study, Health Professionals Follow-up Study (HPFS) and Physicians’ Health Study (PHS).
| AGES-Reykjavik | HPFS | PHS | |||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Overall (n = 138) |
Fatal (n = 24) |
Controls (n = 1,214) |
Fatal (n = 40) |
Controls (n = 204) |
Fatal (n = 105) |
Controls (n=255) |
|
|
| |||||||
| Age at blood draw, y; mean (SD) | 76.7 (5.6) | 64.5 (7.7) | 59.0 (8.3) | ||||
| Prevalent cases | 79.0 (5.5) | 80.7 (6.4) | ---- | ---- | |||
| Incident cases | 75.5 (5.1) | 78.9 (5.1) | 67.9 (7.5) | 61.6 (7.9) | |||
|
| |||||||
| Cases Only | |||||||
|
| |||||||
| AGES-Reykjavik | HPFS | PHS | |||||
|
| |||||||
| Overall (n = 138) |
Fatal (n = 24) |
Fatal (n = 40) |
Fatal (n = 105) |
||||
|
| |||||||
| Time from blood draw to diagnosis, y; mean (SD)a |
2.6 (1.8) | 1.7 (1.8) | 3.9 (2.4) | 8.7 (5.1) | |||
|
| |||||||
| Time from diagnosis to blood draw, y; mean (SD)b |
6.1 (4.6) | 5.4 (4.1) | — | — | |||
|
| |||||||
| Time from diagnosis to fatal prostate cancer, y; mean (SD) |
---- | 6.8 (4.7) | 5.2 (2.8) | 6.2 (4.7) | |||
|
| |||||||
| Stage at diagnosisc | ---- | ---- | |||||
| T1/T2/T3a | 17 (43%) | 39 (38%) | |||||
| T3b | 6 (15%) | 17 (16%) | |||||
| T4 or N1 or M1 | 16 (40%) | 48 (46%) | |||||
|
| |||||||
| Gleason score at diagnosisc | ---- | ---- | |||||
| 2-6 | 0 | 14 (13%) | |||||
| 7 | 12 (30%) | 20 (19%) | |||||
| 8-10 | 24 (60%) | 58 (55%) | |||||
| Missing | 4 (10%) | 13 (13%) | |||||
Overall = total prostate cancer; fatal = death from prostate cancer
Incident cases (n=86)
Prevalent cases (n=52)
Stage and Gleason information was not available for the cases in AGES-Reykjavik
SNPs and Prostate Cancer
Full results for the associations between the SNPs evaluated and risk of fatal prostate cancer in each cohort and combined in Supplementary Table 1. None of the SNPs were statistically significantly associated with fatal prostate cancer across the three cohorts. In addition, meta-analysis failed to confirm associations and there was evidence of significant heterogeneity between the cohorts for some of the SNPs evaluated (Cochran Q p-value <0.05) (Supplementary Table 3).
Within the individual cohorts, two SNPs in CRY1, rs7297614 and rs1921126, were nominally associated with risk of fatal disease in both AGES-Reykjavik and HPFS; however, these findings were not replicated in the PHS cohort (Table 2). These SNPs are in LD with each other (r2>0.8). We also found rs12315175 in CRY1 was nominally associated with fatal prostate cancer in HPFS (OR: 0.43, 95% CI: 0.20-0.93, p-value 0.03) and PHS (OR: 1.73, 95% CI: 1.16-2.59, p-value 0.01); however, the direction of association differs between the cohorts.
Table 2. Nominally significant associations between SNPs and fatal prostate cancer in the AGES-Reykjavik cohort, the Health Professionals Follow-up Study (HPFS) or the Physicians’ Health Study (PHS).
| AGES Fatal | HPFS Fatal | PHS Fatal | |||||
|---|---|---|---|---|---|---|---|
| SNP | Gene | OR (95% CI) |
p-value | OR (95% CI) |
p-value | OR 95% CI |
p-value |
| rs7297614 | CRY1 |
2.31
(1.30, 4.13) |
0.004 |
1.75
(1.07, 2.85) |
0.02 | 0.89 (0.64, 1.24) |
0.49 |
| rs1921126 | CRY1 |
2.09
(1.15, 3.80) |
0.02 |
1.91
(1.16, 3.14) |
0.01 | 0.78 (0.56, 1.09) |
0.15 |
| rs12315175 | CRY1 | 1.20 (0.57, 2.53) |
0.63 |
0.43
(0.20, 0.93) |
0.03 |
1.73
(1.16, 2.59) |
0.01 |
| rs2289591 | PER1 |
2.54
(1.06, 6.07) |
0.04 | 1.20 (0.68, 2.12) |
0.53 | 1.01 (0.68, 1.51) |
0.95 |
| rs10462023 | PER2 |
0.55
(0.30, 0.99) |
0.04 | 1.18 (0.70, 2.00) |
0.54 | 0.80 (0.56, 1.15) |
0.22 |
| rs3754674 | NPAS2 |
0.40
(0.21, 0.73) |
0.003 | 1.04 (0.64, 1.69) |
0.89 | 1.12 (0.79, 1.59) |
0.53 |
| rs10206435 | NPAS2 | 1.06 (0.59, 1.92) |
0.84 |
0.62
(0.38, 1.01) |
0.05 |
1.42
(1.03, 1.96) |
0.03 |
| rs969485 | ARNTL |
0.52
(0.30, 0.91) |
0.02 | 1.10 (0.67, 1.79) |
0.71 | 1.06 (0.73, 1.53) |
0.77 |
Fatal = death from prostate cancer; Per-allele odds ratio and 95% confidence interval
CRY1 = cryptochrome 1; PER1 = period 1; PER2 = period 2; NPAS2 = neuronal PAS domain protein 2; ARNTL = aryl hydrocarbon receptor nuclear translocator-like
Table 3 displays pathway analysis results for the SNP-sets defined above and each of the study outcomes. Variation across CRY1 was significantly associated with fatal disease in AGES-Reykjavik (P=0.01) and HPFS (P=0.01), and was borderline significantly associated in PHS (P=0.05). These findings were largely driven by associations between fatal disease and rs1921126 and rs7297614 in AGES-Reykjavik and HPFS, and with rs12315175 in PHS. In AGES-Reykjavik, none of the gene sets evaluated were associated with overall risk of prostate cancer.
Table 3. Pathway analysis results by gene sets involved in the circadian rhythm in the AGES-Reykjavik cohort, the Health Professionals Follow-up Study (HPFS) and the Physicians’ Health Study (PHS).
| AGES-Reykjavik Cohort | HPFS Cohort | PHS Cohort | ||||||
|---|---|---|---|---|---|---|---|---|
| Gene | Chromosome | Size, kbp | Number of SNPs included |
Melatonin levels p-value a |
Overall prostate cancera p-value |
Fatal prostate cancer p-valuea |
Fatal prostate cancer p-valuea |
Fatal prostate cancer p-valuea |
| ARNTL | 11 | 109.50 | 13b | 0.78 | 0.22 | 0.09 | 0.71 | 0.83 |
| CLOCK | 4 | 114.30 | 4b | 0.44 | 0.51 | 0.98 | 0.33 | 0.24 |
| CRY1 | 12 | 102.20 | 3b | 0.78 | 0.29 | 0.01 | 0.01 | 0.05 |
| CRY2 | 11 | 35.77 | 6b | 0.23 | 0.67 | 0.46 | 0.48 | 0.81 |
| CSNK1E | 22 | 27.39 | 5b | 0.25 | 0.79 | 0.45 | 0.49 | 0.17 |
| MTNR1A | 4 | 21.73 | 4 | 0.72 | 0.48 | 0.46 | 0.52 | 0.82 |
| MTNR1B | 11 | 13.16 | 2 | 0.81 | 0.69 | 0.57 | 0.26 | 0.73 |
| NPAS2 | 2 | 176.7 | 40b | 0.05 | 0.60 | 0.42 | 0.80 | 0.53 |
| PER1 | 17 | 11.91 | 2b | 0.37 | 0.22 | 0.12 | 0.74 | 0.26 |
| PER2 | 2 | 44.53 | 3b | 0.23 | 0.29 | 0.05 | 0.34 | 0.36 |
| PER3 | 1 | 60.47 | 10b | 0.44 | 0.23 | 0.43 | 0.60 | 0.67 |
| TIMELESS | 12 | 32.26 | 4 | 0.02 | 0.64 | 0.92 | 0.76 | 0.66 |
| Core Genes | 86 | 0.11 | 0.53 | 0.13 | 0.68 | 0.53 | ||
| All 12 genes | 96 | 0.05 | 0.63 | 0.19 | 0.77 | 0.71 | ||
Melatonin levels = levels of 6-sulfatoxymelatonin in ng/mL; overall = total prostate cancer; fatal= death from prostate cancer
Global p-values
Included in the core circadian gene set
SNPs and 6-sulfatoxymelatonin
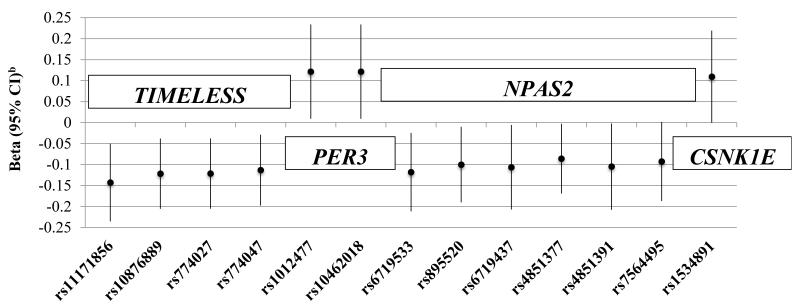
Full results for the associations between the SNPs and 6-sulfatoxymelatonin levels are shown in Supplementary Table 3. Individual polymorphisms in TIMELESS and NPAS2 were nominally associated with lower 6-sulfatoxymelatonin levels; and, polymorphisms in PER3 and CSNK1E were nominally associated with higher 6-sulfatoxymelatonin levels (Figure 1). All of the SNPs in TIMELESS are in LD with each other at r2>0.8, except rs11171856; similarly, the two SNPs in PER3 are in LD (r2>0.8). None of the SNPs nominally associated with fatal disease in AGES-Reykjavik were associated with 6-sulfatoxymelatonin levels. Variation across NPAS2 and TIMELESS was associated with 6-sulfatoxymelatonin levels (Table 3).
Figure 1.
Nominally significant associations between SNPs and 6-sulfatoxymelatonin levels (ng/mL) in the AGES-Reykjavik cohort
a6-sulfatoxymelatonin levels range: 0.40 ng/mL-97.41 ng/mL, mean: 21.09 mg/mL, and median: 17.09 ng/mL
bAge-adjusted betas and 95% confidence intervals
Discussion
We observed no strong association between genetic variation in circadian related genes and risk of prostate cancer. Within the individual cohorts, we found a nominally significant association between two SNPs in CRY1 and risk of fatal disease in AGES-Reykjavik and HPFS; however, we did not replicate the findings in our third cohort, the PHS. In AGES-Reykjavik, we also found thirteen different individual SNPs in four genes (TIMELESS, PER3, NPAS2, CSNK1E) were nominally associated with 6-sulfatoxymelatonin.
Prior studies of circadian related genes and risk of prostate cancer have been inconsistent. Associations between variation in PER1, CRY1, CRY2, CSNK1E, ARNTL, CLOCK, and NPAS2 and risk of overall or more aggressive prostate cancer have been reported[2, 21]. In our study, individual CRY1 SNPs rs7297614 and rs1921126 were associated with a nominally significant increased risk of fatal disease in AGES-Reykjavik and HPFS, but not in PHS. These SNPs are in LD (r2 > 0.80) with a SNP (rs8192440) predicted to affect splicing. Zhu et al in a study of Caucasian men (1,266 prostate cancer cases and 1,308 controls) did not find an association between any of the SNPs in CRY1 and risk of more aggressive prostate cancer; however, variation in rs12315175 was associated with an increased risk of less aggressive disease[2]. This SNP (rs12315175) was also recently associated with a reduced risk of diabetes[35]. In a survival analysis among prostate cancer cases, Lin et al found a significant association between rs10778534 in CRY1 and prostate-cancer specific mortality in a cohort of Caucasian men in Seattle (per-allele OR: 2.21, 95% CI: 1.19-4.12)[22]. This SNP was not in strong LD (r2=0.50) with the CRY1 SNPs we evaluated in our study. While the CGEMS project found a nominally significant (p-value 0.02) inverse association between overall prostate cancer and rs7297614 in CRY1, no GWAS of prostate cancer, overall or aggressive disease, have reported genome-wide significant findings for the circadian-related genes.
There is evidence for the role of the circadian clock as a tumor suppressor from experimental studies showing regulation of cell proliferation and apoptosis via expression of circadian-controlled genes[1]. PER1 and PER2 are required for maintenance of normal circadian function and interaction between CRYs and PERs are necessary for posttranslational regulation[1]. Overexpression of PER1 has been associated with significant growth inhibition and apoptosis in prostate cancer cells, and PER1 levels were down-regulated in prostate cancer tissue compared to normal prostate tissue[36]. Further, NPAS2 has been shown to affect pathways involved in DNA damage response[37]. Mutations in PER1, PER2, CSNK1E, CLOCK, CRY1, and CRY2 have been shown to alter circadian rhythmicity in rodents, and in PER2 have resulted in neoplastic growth and deficient DNA-damage response[1, 13].
Circadian genes may affect prostate cancer risk through their influence on sex hormone levels, metabolic processes, suppression of the circadian hormone melatonin, or sleep disruption[9, 38]. In the Icelandic cohort, we found men with 6-sulfatoxymelatonin levels lower than the median had an increased risk of advanced disease compared to men with high levels (HR=4.01, 95% CI: 1.25-12.50)[9]. We are unaware of any prior evaluation of the association between variation in circadian related genes and 6-sulfatoxymelatonin in men. We found polymorphisms in TIMELESS, NPAS2, PER3, and CSNK1E were nominally associated with 6-sulfatoxymelatonin levels. Variation in TIMELESS was previously associated with depression and early morning awakening in males[14]. Polymorphisms in PER2 have been associated with advanced sleep phase syndrome[39], whereas common variation in ARNTL and NPAS2 was significantly associated with later sleep and wake onset time[40]. These associations may reflect inter-individual differences in the timing of the circadian system, as there is wide variation in the phase of circadian rhythms, including melatonin, in relation to clock time and also in relation to each other[41]. In the current study, the morning void sampling cannot distinguish between differences in the level of circulating melatonin or differences in circadian timing. Consistent with this hypothesis, we found circadian clock genes associated with fatal prostate cancer and with urinary 6-sulfatoxymelatonin in AGES-Reykjavik, but these were not the same SNPs. Perhaps the associations are acting through different pathways or we lacked statistical power to observe an association.
The study was limited by small sample size, as each cohort included in our analysis involved few fatal cases. We were not able to replicate our findings across the three studies, and meta-analysis failed to confirm associations between any of the SNPs and fatal prostate cancer. The inconsistency in our results could not be explained by differences in minor allele frequencies, as they were similar for each SNP across the three cohorts.
The study is restricted to a homogeneous group of Caucasian men; thus, the relevance of these particular risk loci to other populations with higher (African-American men) or lower (Asian men) risk of fatal prostate cancer and different distributions of genetic variation in the selected genes is uncertain. Few studies have been conducted on this topic in non-Caucasian populations; however, a study in Chinese men showed some evidence of an association between circadian genes CRY2 and NPAS2 and risk of prostate cancer[21]. Finally, although we selected genes proposed to play a role in circadian rhythm regulation, this pathway is complex and we may not have captured all of the important genes.
A unique strength of the AGES-Reykjavik cohort is the diversity of molecular data available through biological specimens, including DNA and urine, and complete death information from nationwide registries. This allows us to not only evaluate the association between genetic variation, a urinary biomarker and fatal prostate cancer, but also to evaluate the contribution of different proposed pathways involving disruption of circadian rhythmicity on prostate cancer initiation and progression.
In summary, we found that variation in circadian clock genes was not associated with risk of fatal prostate cancer. We found a suggestion of an association between SNPs in the CRY1 gene and risk of fatal prostate cancer; however, we failed to confirm a strong and consistent association. Further studies are needed to elucidate the potential role of variation in core circadian clock genes and fatal prostate cancer risk.
Supplementary Material
Acknowledgements
We thank the study participants and the Icelandic Heart Association clinic staff for their invaluable contribution. We are grateful to the ongoing participation of men in the Health Professionals Follow-up Study (UM1 CA167552) and would like to acknowledge our colleagues (Lauren McLaughlin and Siobhan Saint-Surin) working on these studies for their valuable help. In addition we would like to thank the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY.
Financial Support: The AGES-Reykjavik is supported by Contract N01-AG- 12100 from the National Institutes on Aging Intramural Research Program; Hjartavernd (the Icelandic Heart Association); and the Althingi (the Icelandic Parliament). In addition, this study was supported by funding from the Harvard Catalyst Award, the Icelandic Cancer Society, the National Cancer Institute at the National Institutes of Health (P01 CA055075, CA133891, CA42182, CA34944, CA40360, CA141298, CA98233 and CA097193); the National Heart, Lung, and Blood Institute at the National Institutes of Health (HL26490, HL34595). National Cancer Institute at the National Institutes of Health Training Grant (R25 CA098566 and T32 CA09001-35 to SCM); and, the Prostate Cancer Foundation (to LAM and JRR); US Army Department of Defense Prostate Cancer Research Program Fellowship (to IMS). CAC was supported in part by NIH NIA grant P01-AG-009975.
Role of the sponsor: The funding agencies had no role in the design of the study, the collection of the data, or the data analysis.
Footnotes
Financial disclosure: Dr. Czeisler has received consulting fees from or served as a paid member of scientific advisory boards for: Bombardier, Inc., Boston Red Sox, Boston Celtics, Cephalon, Inc. (acquired by Teva Pharmaceutical Industries Ltd. October 2011), Michael Jackson’s mother and children, Koninklijke Philips Electronics, N.V., Novartis, United Parcel Service (UPS), Vanda Pharmaceuticals, Inc., and Zeo, Inc; owns an equity interest in Lifetrac, Inc., Somnus Therapeutics, Inc. and Vanda Pharmaceuticals, Inc.; received royalties from McGraw Hill, Pengu in Press/ Houghton Mifflin Harcourt, and Philips Respironics, Inc.; and has received research support from Cephalon, National Football League Charities, ResMed and Philips Respironics. Dr. Czeisler is the incumbent of an endowed professorship provided to Harvard University by Cephalon, Inc., holds a number of process patents in the field of sleep/circadian rhythms (e.g., photic resetting of the human circadian pacemaker), and since 1985, has also served as an expert witness on various legal matters related to sleep and/or circadian rhythms.
References
- 1.Fu L, Lee CC. The circadian clock: pacemaker and tumour suppressor. Nat Rev Cancer. 2003;3(5):350–61. doi: 10.1038/nrc1072. [DOI] [PubMed] [Google Scholar]
- 2.Zhu Y, et al. Testing the circadian gene hypothesis in prostate cancer: a population-based case-control study. Cancer Res. 2009;69(24):9315–22. doi: 10.1158/0008-5472.CAN-09-0648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis S, Mirick DK. Circadian disruption, shift work and the risk of cancer: a summary of the evidence and studies in Seattle. Cancer causes & control. 2006;17:539–545. doi: 10.1007/s10552-005-9010-9. [DOI] [PubMed] [Google Scholar]
- 4.Blask DE. Melatonin, sleep disturbance and cancer risk. Sleep Med Rev. 2009;13(4):257–64. doi: 10.1016/j.smrv.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 5.Costa G, Haus E, Stevens R. Shift work and cancer - considerations on rationale, mechanisms, and epidemiology. Scand J Work Environ Health. 2010;36(2):163–79. doi: 10.5271/sjweh.2899. [DOI] [PubMed] [Google Scholar]
- 6.Buja A, et al. Cancer incidence among male military and civil pilots and flight attendants: an analysis on published data. Toxicol Ind Health. 2005;21(10):273–82. doi: 10.1191/0748233705th238oa. [DOI] [PubMed] [Google Scholar]
- 7.Straif K, et al. Carcinogenicity of shift-work, painting, and fire-fighting. Lancet Oncol. 2007;8(12):1065–6. doi: 10.1016/S1470-2045(07)70373-X. [DOI] [PubMed] [Google Scholar]
- 8.Parent ME, et al. Night work and the risk of cancer among men. Am J Epidemiol. 2012;176(9):751–9. doi: 10.1093/aje/kws318. [DOI] [PubMed] [Google Scholar]
- 9.Sigurdardottir LG, et al. Urinary Melatonin Levels, Sleep Disruption, and Risk of Prostate Cancer in Elderly Men. Eur Urol. 2014 doi: 10.1016/j.eururo.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sigurdardottir LG, et al. Circadian disruption, sleep loss, and prostate cancer risk: a systematic review of epidemiologic studies. Cancer epidemiology, biomarkers & prevention. 2012;21:1002–1011. doi: 10.1158/1055-9965.EPI-12-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sigurdardottir LG, et al. Sleep disruption among older men and risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 2013;22(5):872–9. doi: 10.1158/1055-9965.EPI-12-1227-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kondratov RV, Gorbacheva VY, Antoch MP. The role of mammalian circadian proteins in normal physiology and genotoxic stress responses. Curr Top Dev Biol. 2007;78:173–216. doi: 10.1016/S0070-2153(06)78005-X. [DOI] [PubMed] [Google Scholar]
- 13.Young MW, Kay SA. Time zones: a comparative genetics of circadian clocks. Nat Rev Genet. 2001;2(9):702–15. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- 14.Utge SJ, et al. Systematic analysis of circadian genes in a population-based sample reveals association of TIMELESS with depression and sleep disturbance. PLoS One. 2010;5(2):e9259. doi: 10.1371/journal.pone.0009259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slominski RM, et al. Melatonin membrane receptors in peripheral tissues: distribution and functions. Mol Cell Endocrinol. 2012;351(2):152–66. doi: 10.1016/j.mce.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Antoch MP, Kondratov RV, Takahashi JS. Circadian clock genes as modulators of sensitivity to genotoxic stress. Cell Cycle. 2005;4(7):901–7. doi: 10.4161/cc.4.7.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gauger MA, Sancar A. Cryptochrome, circadian cycle, cell cycle checkpoints, and cancer. Cancer Res. 2005;65(15):6828–34. doi: 10.1158/0008-5472.CAN-05-1119. [DOI] [PubMed] [Google Scholar]
- 18.Borgs L, et al. Cell “circadian” cycle: new role for mammalian core clock genes. Cell Cycle. 2009;8(6):832–7. doi: 10.4161/cc.8.6.7869. [DOI] [PubMed] [Google Scholar]
- 19.Hua H, et al. Circadian gene mPer2 overexpression induces cancer cell apoptosis. Cancer Sci. 2006;97(7):589–96. doi: 10.1111/j.1349-7006.2006.00225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeBruyne JP, Weaver DR, Reppert SM. CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nat Neurosci. 2007;10(5):543–5. doi: 10.1038/nn1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chu LW, et al. Variants in circadian genes and prostate cancer risk: a population-based study in China. Prostate Cancer Prostatic Dis. 2008;11(4):342–8. doi: 10.1038/sj.pcan.4501024. [DOI] [PubMed] [Google Scholar]
- 22.Lin DW, et al. Genetic variants in the LEPR, CRY1, RNASEL, IL4, and ARVCF genes are prognostic markers of prostate cancer-specific mortality. Cancer Epidemiol Biomarkers Prev. 2011;20(9):1928–36. doi: 10.1158/1055-9965.EPI-11-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harris TB, et al. Age, Gene/Environment Susceptibility-Reykjavik Study: multidisciplinary applied phenomics. Am J Epidemiol. 2007;165(9):1076–87. doi: 10.1093/aje/kwk115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giovannucci E, et al. Nutritional predictors of insulin-like growth factor I and their relationships to cancer in men. Cancer Epidemiol Biomarkers Prev. 2003;12(2):84–9. [PubMed] [Google Scholar]
- 25.Schumacher FR, et al. Genome-wide association study identifies new prostate cancer susceptibility loci. Hum Mol Genet. 2011;20(19):3867–75. doi: 10.1093/hmg/ddr295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iceland S. 2011 cited 2014; Available from: http://www.statice.is/
- 27.Psaty BM, et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium Design of Prospective Meta-Analyses of Genome-Wide Association Studies From 5 Cohorts. Circulation: Cardiovascular Genetics. 2009;2(1):73–80. doi: 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murphy RA, et al. Candidate Gene Association Study of BMI-Related Loci, Weight, and Adiposity in Old Age. The journals of gerontology. Series A, Biological sciences and medical sciences. 2012 doi: 10.1093/gerona/gls227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nalls MA, et al. Multiple Loci Are Associated with White Blood Cell Phenotypes. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, et al. Genotype imputation. Annual review of genomics and human genetics. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cochran WG. The Combination of Estimates from Different Experiments. Biometrics. 1954;10 [Google Scholar]
- 32.Evangelou E, Ioannidis JPA. Meta-analysis methods for genome-wide association studies and beyond. Nature Reviews Genetics. 2013;14:379–389. doi: 10.1038/nrg3472. [DOI] [PubMed] [Google Scholar]
- 33.Wu MC, et al. Powerful SNP-set analysis for case-control genome-wide association studies. American journal of human genetics. 2010;86(6):929–942. doi: 10.1016/j.ajhg.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin X, et al. Kernel machine SNP-set analysis for censored survival outcomes in genome-wide association studies. Genetic epidemiology. 2011;35(7):620–631. doi: 10.1002/gepi.20610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelly MA, et al. Circadian Gene Variants and Susceptibility to Type 2 Diabetes: A Pilot Study. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0032670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao Q, et al. A Role for the Clock Gene Per1 in Prostate Cancer. Cancer Research. 2009;69(19):7619–7625. doi: 10.1158/0008-5472.CAN-08-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoffman AE, et al. The Circadian Gene NPAS2, a Putative Tumor Suppressor, Is Involved in DNA Damage Response. Molecular Cancer Research. 2008;6:1461–1468. doi: 10.1158/1541-7786.MCR-07-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu Y, et al. Does “clock” matter in prostate cancer? Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2006;15(1):3–5. doi: 10.1158/1055-9965.EPI-05-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carpen JD, et al. A single-nucleotide polymorphism in the 5′-untranslated region of the hPER2 gene is associated with diurnal preference. J Sleep Res. 2005;14(3):293–7. doi: 10.1111/j.1365-2869.2005.00471.x. [DOI] [PubMed] [Google Scholar]
- 40.Evans DS, et al. Common Genetic Variants in ARNTL and NPAS2 and at Chromosome 12p13 are Associated with Objectively Measured Sleep Traits in the Elderly. SLEEP. 2013 doi: 10.5665/sleep.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duffy JF, Wright KP. Entrainment of the Human Circadian System by Light. Journal of Biological Rhythms. 2005;20:326–338. doi: 10.1177/0748730405277983. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.