Abstract
C9orf72 promoter hypermethylation inhibits the accumulation of pathologies which have been postulated to be neurotoxic. We tested here whether C9orf72 hypermethylation is associated with prolonged disease in C9orf72 mutation carriers. C9orf72 methylation was quantified from brain or blood using methylation-sensitive restriction enzyme digest-qPCR in a cross-sectional cohort of 118 C9orf72 repeat expansion carriers and 19 non-carrier family members. Multivariate regression models were used to determine whether C9orf72 hypermethylation was associated with age at onset, disease duration, age at death, or hexanucleotide repeat expansion size. Permutation analysis was performed to determine whether C9orf72 methylation is heritable. We observed a high correlation between C9orf72 methylation across tissues including cerebellum, frontal cortex, spinal cord and peripheral blood. While C9orf72 methylation was not significantly different between ALS and FTD and did not predict age at onset, brain and blood C9orf72 hypermethylation was associated with later age at death in FTD (brain: β = 0.18, p = 0.006; blood: β = 0.15, p < 0.001), and blood C9orf72 hypermethylation was associated with longer disease duration in FTD (β = 0.03, p = 0.007). Furthermore, C9orf72 hypermethylation was associated with smaller hexanucleotide repeat length (β = −16.69, p = 0.033). Finally, analysis of pedigrees with multiple mutation carriers demonstrated a significant association between C9orf72 methylation and family relatedness (p < 0.0001). C9orf72 hypermethylation is associated with prolonged disease in C9orf72 repeat expansion carriers with FTD. The attenuated clinical phenotype associated with C9orf72 hypermethylation suggests that slower clinical progression in FTD is associated with reduced expression of mutant C9orf72. These results support the hypothesis that expression of the hexanucleotide repeat expansion is associated with a toxic gain of function.
Keywords: Neurodegeneration, Frontotemporal lobar degeneration, Frontotemporal dementia, Amyotrophic lateral sclerosis, Epigenetics
Introduction
Hexanucleotide repeat expansions in C9orf72 are the most frequent genetic cause of autosomal dominant amyotrophic lateral sclerosis (ALS) and frontotemporal degeneration (FTD) [15, 59]. The C9orf72 mutation is associated with highly variable clinical phenotypes including ALS, FTD, Alzheimer’s disease and others, and with a highly variable clinical course ranging from rapidly fatal motor neuron disease to a “slowly progressive” form of FTD [2, 6–8, 10, 14, 28, 29, 34–36, 38, 40, 45, 49, 56, 57, 61, 62]. Several studies have implicated C9orf72 repeat expansion size [2, 19, 20, 37, 65] and single-nucleotide polymorphisms in TMEM106B as disease modifiers in C9orf72 mutation carriers [26, 66]. However, the basis for much of the clinical heterogeneity amongst C9orf72 mutation carriers remains unknown.
Studies of other repeat expansion diseases indicate that DNA hypermethylation adjacent to trinucleotide repeat mutations is an epigenetic disease modifier, most notably in cases of Fragile X syndrome and Friedreich’s ataxia [9, 22, 33, 51, 52]. We and others have recently shown that the C9orf72 repeat expansion is associated with C9orf72 promoter hypermethylation and histone trimethylation which contribute to transcriptional silencing of mutant C9orf72 [4, 48, 68]. Moreover, we found that epigenetic silencing of mutant C9orf72 is associated with a decreased accumulation of neuropathologic inclusions associated with the C9orf72 mutation, namely RNA foci and dipeptide repeat (DPR) protein aggregates, raising the possibility that C9orf72 promoter hypermethylation mitigates disease pathogenesis [48].
We hypothesized that epigenetic silencing of mutant C9orf72 is associated with prolonged survival amongst C9orf72 hexanucleotide repeat expansion carriers. To test this hypothesis, we studied the relationship between C9orf72 promoter hypermethylation and clinical phenotype, disease onset, disease duration, age of death, and C9orf72 hexanucleotide repeat length in a cross-sectional cohort of C9orf72 repeat expansion carriers.
Materials and methods
Study cohort
Subjects were evaluated at the Penn Frontotemporal Degeneration Center, the ALS Center at Pennsylvania Hospital or the Penn Alzheimer’s Disease Center, or underwent autopsy at the Center for Neurodegenerative Disease Research [64]. The clinical diagnosis of ALS was made using the El Escorial criteria and FTD was diagnosed using established clinical criteria [30, 58]. In this study, patients presenting with ALS were defined as our ALS cohort and included subjects with mild cognitive impairment [63] and subjects who subsequently met criteria for FTD. Patients presenting with FTD were defined as our FTD cohort and included subjects who subsequently met criteria for ALS. Additional analyses in which the cohort was subdivided into ALS, ALS with mild cognitive impairment (ALS-MCI), ALS with FTD (ALS-FTD) and FTD are included in the Fig. 1e and supplementary materials. Detailed clinical characteristics were obtained from an integrated clinical and autopsy database [70] and by retrospective chart review of clinical visits. Age of onset data was unavailable for four subjects (1 ALS, 3 FTD). All clinical protocols were approved by the University of Pennsylvania Institutional Review Board.
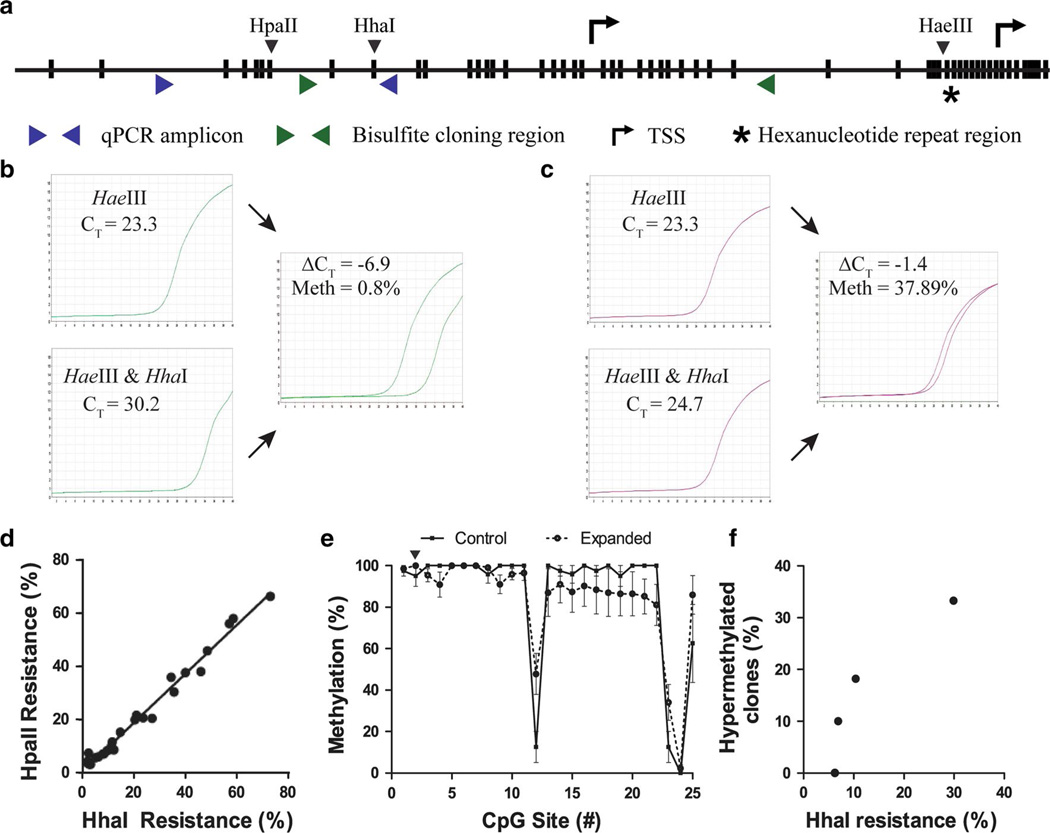
Fig. 1.
C9orf72 promoter methylation assay. a Schematic representation of the 5′ end of the C9orf72 gene in which individual CpG dinucleotides are represented by vertical bars, the transcriptional start sites (TSS) are designated by arrows, the hexanucleotide repeat region is designated by a star, the binding sites of the qPCR primers used in the C9orf72 promoter methylation assay are represented as blue triangles, and the binding sites of the primers used in bisulfite cloning are represented as green triangles. The HhaI and the HpaII restriction enzyme recognition sites within the qPCR amplicon as well as the first HaeIII restriction enzyme recognition site are shown. Representative qPCR amplification curves for HaeIII only versus HaeIII–HhaI double digested DNA for b a hypomethylated (0.8 %) and c a hypermethylated (37.89 %) case. The resulting ΔCT values are used to calculate the percent DNA resistance to HhaI digestion (2ΔCT × 100) as a measure of DNA methylation. d Scatterplot and linear regression of HpaII resistance versus HhaI resistance as measured by restriction enzyme digest-qPCR (n = 28). e Bisulfite cloning of HhaI resistant DNA. The percent methylation and standard error for each CpG site are plotted for repeat expanded cases (dashed line) and non-expanded controls (solid line). The HhaI site which is assessed by the digest-qPCR assay is marked with an inverted triangle. f Direct comparison of HhaI-qPCR and bisulfite cloning. The percentage of bisulfite-treated DNA clones exhibiting hypermethylation (methylation of at least 10 of 25 CpG sites) is plotted as a function of HhaI resistance (n = 4)
C9orf72 genotyping and Southern blotting
Genomic DNA from blood was extracted with the Quick-Gene-610L kit (AutoGen, Holliston, MA, USA), while genomic DNA from brain and other tissues was extracted with the QIAamp DNA mini kit or the DNeasy Blood & Tissue kit (Qiagen, Valencia, CA, USA). C9orf72 genotyping with repeat-primed PCR was performed as described previously [7]. Southern blot hybridization of AluI- and DdeI-digested genomic DNA was performed to determine the hexanucleotide repeat length of C9orf72 using a digoxigenin-labeled (GGGGCC)5 oligonucleotide probe as previously described [2]. Southern blot images were analyzed in ImageJ to determine the modal hexanucleotide repeat size defined as the point of highest density.
Bisulfite conversion-based mapping of CpG methylation
Genomic DNA was extracted from two C9orf72 patient-derived lymphoblastoid cell lines (ND10966 and ND14442, Coriell NINDS Repository, Camden, NJ, USA), five cerebella from C9orf72 expansion carriers and four cerebella from control cases. DNA was digested for 6 h with 10 U of HhaI (New England Biolabs, Ipswich, MA, USA) and purified by phenol:chloroform:isoamyl alcohol extraction. Purified DNA was quantified by NanoDrop (Thermo Fisher Scientific, Wilmington, DE, USA) and 100 ng of DNA was bisulfite converted using the EpiTect Bisulfite Kit (Qiagen). Bisulfite-converted DNA was subjected to PCR to amplify the marked C9orf72 promoter region (Fig. 1a), and the PCR product was cloned into pGEM as described previously [48]. Six to sixteen individual clones from each case were Sanger sequenced using a T7 promoter sequencing primer.
Bisulfite cloning
Bisulfite cloning was performed on cerebellar genomic DNA from four C9orf72 repeat expansion cases as described previously [48]. Five clones per case were analyzed, in addition to the five or six individual clones per case previously published in Liu et al. [48] to yield a total of 9–11 clones per case.
C9orf72 promoter methylation assay
For quantitative assessment of methylation levels, we used methylation-sensitive restriction enzyme DNA digestion coupled with quantitative PCR (Fig. 1) as described previously [48]. Briefly, 100 ng of genomic DNA was digested for 6 h with 2 units of HhaI (New England Biolabs, Ipswich, MA, USA) and 2 units of HaeIII (NEB) or only with 2 units of HaeIII followed by heat inactivation. qPCR was done using 12.5 ng of digested DNA per reaction with 2× FastStart SYBR Green Master mix (Roche Applied Science, Indianapolis, IN, USA) using primers amplifying the differentially methylated C9orf72 promoter region (5′-CAGTGTGAAAATCATGCTTGAGAGA-3′ and 5′-TTTGTGCTTGGTAGGCAGTG-3′). The difference in the number of cycles to threshold amplification between double versus single digested DNA was used as measure of CpG methylation. HaeIII was added to the DNA digestion reactions to digest the hexanucleotide repeat as we found empirically that the presence of the hexanucleotide repeat expansion mutation resulted in a slight reduction in PCR efficiency.
Statistical analysis
We applied Wilcoxon rank-sum tests and Kruskal–Wallis non-parametric ANOVA to compare C9orf72 hypermethylation according to C9orf72 mutation status, clinical diagnosis, site of motor neuron disease onset, and FTD clinical phenotypes. Multivariate linear regression analysis was performed to determine the relationship between C9orf72 promoter hypermethylation and age at onset, age at death, disease duration, or hexanucleotide repeat length (mode). Because disease duration values were not normally distributed, disease duration was natural log (ln) transformed when used as dependent variable in linear regression analyses. Spearman’s test of correlation or univariate linear regressions were used to assess the association between C9orf72 methylation and age at collection as well as to compare C9orf72 methylation in cerebellum and peripheral blood, frontal cortex or spinal cord. One-way ANOVA was used to compare linear regression models. Statistical analysis of the association of repeat size and promoter methylation within families was done with a linear mixed-effects model where the fixed effects were age at sample collection and C9orf72 promoter methylation, and the random effect was family group [43]. For both multiple linear regression models and linear mixed-effects models, we report the β value for each predictor, which represents the change in outcome (in years for age at onset and age at death, in natural log years for disease duration and number of repeats for repeat length) associated with a unit change in the predictor (e.g., methylation increases by 1 %). A permutation test was used to analyze C9orf72 promoter hypermethylation within families as described below. Unadjusted p values are reported as given hypotheses were made a priori. Notably, for analyses where multiple comparisons are made (pairwise comparisons of methylation in cerebellum versus blood, frontal cortex and spinal cord; linear regression models involving age at onset, age at death and disease duration), p values less than 0.0167 would be considered significant after using a Bonferroni correction, and so the reported unadjusted p values would remain significant. The R Statistical package (v3.0.1) was used for statistical analyses and GraphPad Prism Software (v5.02, GraphPad, San Diego, CA, USA) was used for data visualization.
Results
Study cohort characteristics
C9orf72 promoter methylation was analyzed in C9orf72 hexanucleotide repeat expansion carriers and non-expanded family members. Genomic DNA was extracted from cerebellum (n = 22), peripheral blood (n = 99) or both (n = 16). The cohort included 118 individuals with the C9orf72 repeat expansion as identified by repeat-primed PCR and Southern blotting. 60 mutation carriers presented with motor neuron disease, while 45 individuals presented with FTD. 13 repeat expansion carriers had no clinical phenotype (i.e., prodromal cases). The median age at onset, age at death and disease duration was higher in FTD relative to ALS (Table 1).
Table 1.
Clinical characteristics of C9orf72 repeat expansion carriers
| Overall | ALS | FTD | |
|---|---|---|---|
| Cerebellum, n | 38 | 22 | 16 |
| Gender, no. male (%) | 23 (60.53) | 15 (68.18) | 8 (50) |
| Age at onset, year (IQR) | 55.89 (52–64.63) | 54.85 (52–63.25) | 61 (51–66) |
| Age at death, year (IQR) | 59.98 (55–70.66) | 57.81 (54.92–65.67) | 68.22 (56.82–74.7) |
| Disease duration, year (IQR) | 2.87 (2.09–7.11) | 2.36 (1.61–2.92) | 7.23a (5.53–10.11) |
| Methylation, pct (IQR) | 6.19 (4.64–10.37) | 6.09 (4.76–13.18) | 6.41 (4.33–9.94) |
| Blood, n | 96 | 50 | 33 |
| Deaths | 48 | 37 | 11 |
| Gender, no. male (%) | 52 (54.17) | 30 (60.0) | 21 (63.64) |
| Age at onset, year (IQR) | 55.95 (51.15–62.15) | 55.12 (51.30–63.5) | 58.0 (50.73–62.0) |
| Age at death, year (IQR) | 61.69 (55.16–67.42) | 59.89 (55.05–67.71) | 62.75 (55.56–67.48) |
| Disease duration, year (IQR) | 2.68 (2.03–4.08) | 2.53 (1.95–3.31) | 5.29b (2.87–9.98) |
| Methylation, pct (IQR) | 6.04 (2.17–20.13) | 5.74 (1.71–10.5) | 6.22 (1.82–29.35) |
Data are shown as median and interquartile range (IQR) unless otherwise stated
p < 0.001,
p = 0.0012 compared to ALS, Wilcoxon rank-sum test
Cerebellar DNA was available for 38 mutation carriers including 22 presenting with ALS and 16 presenting with FTD. Blood DNA was available for 96 mutation carriers including 50 with ALS and 33 with FTD. Among the subjects for whom we had blood DNA, 70 were unrelated while 26 were members of one of 9 families. In addition to the repeat expansion carriers, we also measured C9orf72 methylation in 19 non-expanded, unaffected family members as controls.
Validation of HhaI-qPCR methylation assay for C9orf72 promoter region
C9orf72 promoter methylation was quantified using methylation-sensitive restriction enzyme digestion with qPCR [48]. HhaI is a restriction enzyme which is able to cut the C9orf72 promoter only in the absence of CpG methylation, and so qPCR was used to quantify the percent DNA resistant to HhaI digestion as a measure of C9orf72 methylation (Fig. 1a–c). To validate this restriction enzyme-based assay, we sought to determine whether CpG methylation at the HhaI cleavage site was associated with methylation of adjacent CpG nucleotides.
Adjacent to the HhaI cut site within the qPCR amplicon, there is also an HpaII cut site (Fig. 1a). HpaII also specifically cuts unmethylated CpGs. In a subset of 28 peripheral blood DNA samples, we quantified the resistance against HhaI or HpaII digest compared to mock digested DNA using qPCR. Both enzymes gave highly comparable results (Fig. 1d; R2 = 0.984, p < 0.0001), suggesting that CpG methylation at these two sites is highly concordant.
To further determine if other CpG sites within the C9orf72 promoter region are also methylated when the HhaI digest site is methylated, genomic DNA from C9orf72 repeat expansion carriers was HhaI digested (n = two C9orf72 patient-derived lymphoblastoid cell lines and five cerebella of C9orf72 expansion carriers) followed by bisulfite cloning to assess CpG methylation downstream of the HhaI cut site. The cloned DNA included 25 CpG sites with the second CpG corresponding to the HhaI cut site assessed by our digest-qPCR assay (Fig. 1a). This procedure demonstrated that HhaI resistant DNA is densely methylated across the entire C9orf72 promoter (Fig. 1e and S1), indicating that HhaI resistance is a proxy for dense promoter hypermethylation. Notably, CpG sites 12, 23 and 24 remained hypomethylated (Fig. 1e). Interestingly, there are exactly 146 bp of DNA between CpG sites 12 and 24 which corresponds to the length of DNA within a single nucleosome. Therefore, these sites are likely protected from CpG methylation due to nucleosomal positioning which is known to be present around transcription start sites and is known to hinder accessibility of DNA methyltransferases [25].
A similar analysis was performed on genomic DNA from non-expanded controls (n = four cerebella). Although amplification was low due to the low level of C9orf72 promoter methylation in controls, HhaI resistant DNA from non-expanded controls also demonstrated dense promoter hypermethylation (Fig. 1e and S1). This indicates that the low level of HhaI resistance in non-expanded genomic DNA is also associated with dense promoter hypermethylation.
Finally, HhaI-based methylation measurements were directly compared to the gold standard bisulfite cloning. Bisulfite cloning (without predigestion with HhaI) was performed on cerebellar DNA from four C9orf72 repeat expansion carriers to determine the percentage of hypermethylated clones. Both bisulfite cloning and digest-qPCR assays produced highly comparable results, further validating our restriction enzyme-based qPCR quantification assay (Fig. 1f and S2).
C9orf72 promoter hypermethylation in brain is associated with later age at death in FTD patients
We have recently found that C9orf72 promoter hypermethylation is associated with a reduction in pathologic RNA foci and DPR aggregates, suggesting that epigenetic silencing of mutant C9orf72 may protect against disease pathogenesis [48]. Based on these findings, we hypothesized that C9orf72 hypermethylation is associated with longer survival. To test this hypothesis, we first analyzed brain DNA from individuals harboring the C9orf72 mutation as a discovery cohort to determine whether C9orf72 methylation affects clinical disease phenotype, age at death, disease duration, or age at onset.
There was no difference in C9orf72 methylation in individuals who presented with ALS (n = 22; Mdn = 6.09) versus FTD (n = 16; Mdn = 6.41; r = −0.08, p = 0.6049, Wilcoxon rank-sum test). We also detected no difference in C9orf72 promoter methylation in the cerebellum when comparing ALS patients with different sites of onset (bulbar, cervical or lumbosacral; χ2(2) = 0.25, p = 0.8842, Kruskal–Wallis ANOVA).
A multivariate linear regression model correcting for gender and age at onset (n = 37 unrelated C9orf72 mutation carriers after removing one autopsy case for which complete clinical data were not available) showed a highly significant interaction between C9orf72 methylation and disease diagnosis such that increased C9orf72 promoter hypermethylation in individuals with FTD was associated with a later age at death (β = 0.18, p = 0.006; Table 2). This corresponds to a 1.8-year later age at death for every 10 % increase in C9orf72 promoter methylation. To demonstrate that C9orf72 promoter hypermethylation contributed significantly to this statistical model, an alternate multivariate linear regression model was generated in which C9orf72 methylation was omitted. This alternate model did significantly worse at predicting age at death (ANOVA F(2,31) = 6.97, p = 0.0032). Additional multivariate regression analyses failed to reveal a significant relationship between cerebellar C9orf72 hypermethylation and either disease duration or age at onset (Table 2).
Table 2.
Cerebellar C9orf72 promoter methylation and age at onset, age at death and disease duration
| Age at onset (n = 37) | Age at death (n = 37) | Disease duration (ln transformed, n = 37) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| β (SE) | 95 % CI | p value | β (SE) | 95 % CI | p value | β (SE) | 95 % CI | p value | |
| Intercept | 61.07 (3.21) | 54.43 to 67.61 | <0.001 | 0.13 (2.53) | −5.02 to 5.28 | 0.960 | 0.27 (0.66) | −1.08 to 1.61 | 0.690 |
| Diagnosis (FTD) | −0.99 (4.03) | −9.20 to 7.24 | 0.806 | 3.39 (0.90) | 1.55 to 5.24 | <0.001 | 0.87 (0.24) | 0.39 to 1.35 | <0.001 |
| Gender (male) | −5.61 (2.90) | −11.51 to 0.29 | 0.062 | −0.02 (0.69) | −1.42 to 1.38 | 0.980 | −0.03 (0.18) | −0.40 to 0.33 | 0.865 |
| Age at onset | n.a. | n.a. | n.a. | 1.04 (0.04) | 0.96 to 1.12 | <0.001 | 0.01 (0.01) | −0.01 to 0.03 | 0.341 |
| Methylation | −0.03 (0.19) | −0.41 to 0.35 | 0.884 | −0.01 (0.04) | −0.10 to 0.07 | 0.759 | −0.01 (0.01) | −0.03 to 0.02 | 0.604 |
| Methylation × diagnosis (FTD) | 0.11 (0.27) | −0.44 to 0.67 | 0.684 | 0.18 (0.06) | 0.06 to 0.31 | 0.006 | 0.03 (0.02) | −0.01 to 0.06 | 0.097 |
| R2 | 0.116 | 0.967 | 0.644 | ||||||
SE standard error, CI confidence interval, ln natural log, n.a. not applicable
C9orf72 promoter hypermethylation in peripheral blood
C9orf72 methylation was then measured in peripheral blood DNA of 96 expansion carriers (Mdn = 6.04), and 19 related, non-expanded unaffected controls (Mdn = 0.97, IQR = 0.69–1.63). C9orf72 promoter methylation levels were significantly higher in repeat expansion carriers compared to non-expanded controls (p < 0.001, Wilcoxon rank-sum test; Fig. 2a). Overall, 33.3 % (n = 32) of the C9orf72 repeat expansion carriers exhibited C9orf72 promoter methylation values greater than 10 %, in contrast with the non-expanded control group in which C9orf72 promoter methylation values were all less than 10 %. Similar to what was observed using cerebellar DNA, peripheral blood C9orf72 promoter methylation did not differ between patients who presented with ALS (n = 50; Mdn = 6.062) versus FTD (n = 33; Mdn = 6.41, r = −0.106, p = 0.3155, Wilcoxon rank-sum test). Similar results were observed when further subdividing the cohort into four disease subcategories (ALS, ALS-MCI, ALS-FTD and FTD, Table S1, χ2(3) = 2.65, p = 0.4493, Kruskal–Wallis non-parametric one-way ANOVA). Furthermore, no difference in blood C9orf72 promoter methylation was observed when subdividing individuals with motor neuron disease based on site of onset (bulbar, cervical and lumbosacral; χ2(2) = 1.139, p = 0.5659, Kruskal–Wallis ANOVA) or when subdividing individuals with cognitive impairment into individuals with behavioral variant FTD versus primary progressive aphasia (p = 0.9422, Wilcoxon rank-sum test). Overall these results suggest that C9orf72 methylation was similar between different clinical phenotypes.
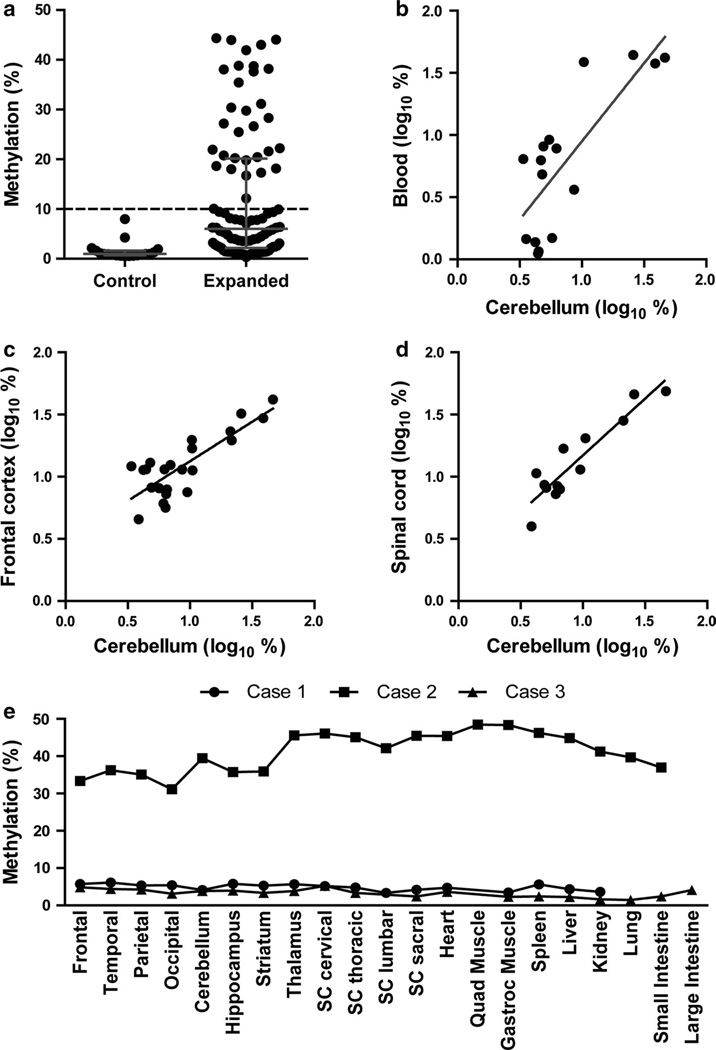
Fig. 2.
C9orf72 promoter methylation in peripheral tissue and cerebellum. a C9orf72 methylation as measured by restriction enzyme digestqPCR of peripheral blood from controls (n = 19) or repeat expansion cases (n = 96) are shown. Horizontal gray line represents median and error bars show interquartile range. The dashed line represents 10 % methylation. Wilcoxon rank-sum test: p < 0.001. b Relationship between log10-transformed C9orf72 methylation from peripheral blood versus cerebellum (n = 16 repeat expansion carriers, R2 = 0.621, p = 0.0003). c Relationship between log10-transformed C9orf72 methylation from frontal cortex versus cerebellum (n = 23 repeat expansion carriers, R2 = 0.659, p < 0.0001). d Relationship between log10-transformed C9orf72 methylation from spinal cord versus cerebellum (n = 13 repeat expansion carriers, R2 = 0.853, p < 0.0001). e C9orf72 promoter methylation across central and peripheral tissues. Percent methylation for each available brain, spinal cord (SC) or peripheral tissue is shown for three C9orf72 repeat expansion cases
Promoter methylation is stable across time and tissue types
C9orf72 methylation in repeat expansion carriers was not associated with age at peripheral blood collection (rs = −0.166, p = 0.1206, Spearman rank correlation) suggesting that C9orf72 methylation is stable over time. This is in contrast with C9orf72 repeat length which has been reported as being dependent on age of collection [65]. Additionally, we identified 16 repeat expansion carriers for which we had DNA available from both cerebellum and peripheral blood, allowing for a direct comparison of C9orf72 promoter methylation in peripheral blood and brain. C9orf72 promoter methylation between peripheral blood and cerebellum was highly consistent (R2 = 0.621, p = 0.0003; Fig. 2b). The time between blood and brain collection ranged between 0.4 and 9.5 years. There was no correlation between differences in C9orf72 methylation between blood and cerebellum and the collection time interval (rs = 0.14, p = 0.6174, Spearman rank correlation). This further supports that C9orf72 promoter methylation is stable across time.
Genomic DNA was available from both cerebellum and frontal cortex from 23 C9orf72 repeat expansion carriers, and from both cerebellum and spinal cord for 13 patients. C9orf72 promoter methylation in frontal cortex and cerebellum (Fig. 2c) as well as in spinal cord and cerebellum (Fig. 2d) was highly concordant (frontal cortex: R2 = 0.659, p < 0.0001; spinal cord: R2 = 0.853, p < 0.0001). Tissues from three additional autopsies were available from which genomic DNA was extracted from multiple central and peripheral tissues. C9orf72 promoter methylation was measured in 8 different brain regions, in 3–4 spinal cord sections and in up to 8 different peripheral tissues (Fig. 2e). One of the three cases was hypermethylated (Case 2: Mdn = 41.69), while the other two cases were hypomethylated (Case 1: Mdn = 5.20; Case 3: Mdn = 3.39). While there was some variability between tissues, C9orf72 promoter methylation was overall highly consistent across all analyzed tissues types in these three cases (Case 1: IQR = 4.18–5.67; Case 2: IQR = 35.98–45.56; Case 3: IQR = 2.40–4.14). These results indicate that C9orf72 promoter methylation is stable across a wide variety of tissues.
C9orf72 promoter hypermethylation in FTD patients is associated with longer survival
Given the strong correlation between peripheral blood and brain C9orf72 methylation, we sought to replicate our autopsy cohort results by determining whether C9orf72 hypermethylation in peripheral blood of repeat expansion carriers is associated with age at death. Complete clinical information including age at death and disease duration was available for 47 unrelated C9orf72 mutation carriers. A multivariate linear regression model correcting for gender, age at onset and clinical diagnosis revealed a significant interaction such that increased peripheral blood C9orf72 methylation in individuals with FTD was associated with a later age at death (β = 0.15, p < 0.001, Table 3). This corresponds to a 1.5-year later age at death for every 10 % increase in methylation. These findings based on peripheral blood DNA were highly similar to what was observed using cerebellar DNA. To demonstrate that C9orf72 promoter methylation contributed significantly to this statistical model, an alternate linear regression model was generated in which C9orf72 methylation was omitted. This alternate model did significantly worse at predicting age at death (ANOVA F(2,41) = 10.23, p = 0.0003).
Table 3.
Peripheral blood C9orf72 promoter methylation and age at onset, age at death and disease duration (two disease subgroups)
| Age at onset (n = 77) | Age at death (n = 47) | Disease duration (ln transformed, n = 47) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| β (SE) | 95 % CI | p value | β (SE) | 95 % CI | p value | β (SE) | 95 % CI | p value | |
| Intercept | 56.57 (1.89) | 52.81 to 60.34 | <0.001 | 2.86 (1.85) | −0.86 to 6.59 | 0.129 | 0.93 (0.50) | 0.03 to 2.04 | 0.071 |
| Diagnosis (FTD) | −0.70 (2.61) | −5.90 to 4.51 | 0.791 | 0.53 (0.88) | −1.25 to 2.31 | 0.553 | 0.19 (0.24) | −0.32 to 0.65 | 0.440 |
| Gender (male) | 2.79 (1.89) | −0.97 to 6.55 | 0.144 | −0.07 (0.51) | −1.10 to 0.97 | 0.899 | −0.10 (0.14) | −0.41 to 0.15 | 0.460 |
| Age at onset | n.a. | n.a. | n.a. | 1.00 (0.03) | 0.94 to 1.06 | <0.001 | 0.00 (0.01) | −0.02 to 0.02 | 0.889 |
| Methylation | −0.16 (0.11) | −0.38 to 0.06 | 0.151 | −0.01 (0.03) | −0.06 to 0.04 | 0.661 | −0.01 (0.01) | −0.02 to 0.01 | 0.411 |
| Methylation × diagnosis (FTD) | 0.13 (0.15) | −0.16 to 0.42 | 0.379 | 0.15 (0.04) | 0.07 to 0.22 | <0.001 | 0.03 (0.01) | 0.01 to 0.05 | 0.007 |
| R2 | 0.054 | 0.968 | 0.453 | ||||||
SE standard error, CI confidence interval, ln natural log, n.a. not applicable
In addition to replicating the association between C9orf72 hypermethylation and age at death in FTD, multivariate linear regression analysis correcting for gender, age at onset and diagnosis also revealed that increased blood C9orf72 methylation in FTD was associated with longer disease duration (β = 0.03, p = 0.007, Table 3). Because disease duration was ln transformed, an increase of 1 % in C9orf72 promoter methylation would result in a 3 % increase of disease duration. Once again, a regression model in which C9orf72 methylation was omitted resulted in a model which performed significantly worse relative to the linear regression model containing C9orf72 methylation (ANOVA F(2,41) = 5.03, p = 0.0111). Age of onset data was available for 77 unrelated C9orf72 mutation carriers from which no association was observed between peripheral blood C9orf72 hypermethylation and age at onset (Table 3).
Similar results were obtained upon subdividing the cohort into four disease subtypes (ALS, ALS-MCI, ALS-FTD, FTD). Again no association was observed between peripheral blood C9orf72 hypermethylation and age at onset. However, there was a significant association between C9orf72 hypermethylation and both age at death and disease duration in FTD and/or ALS-FTD (Table S2).
Promoter methylation levels are heritable
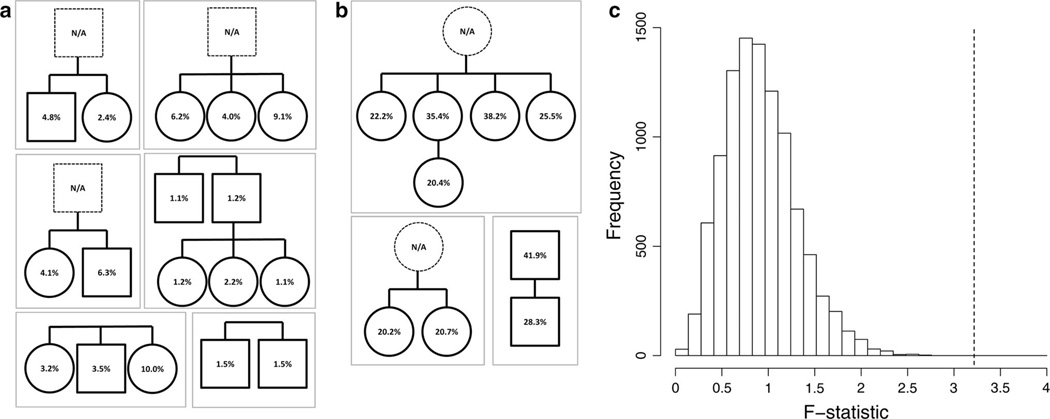
The mechanisms regulating differential C9orf72 promoter methylation in repeat expansion carriers are not known. However, prior studies have suggested that C9orf72 methylation levels are similar amongst closely related family members [68]. We analyzed the nine families in our study for which peripheral blood C9orf72 promoter hypermethylation measurements were available from at least two repeat expansion carrier family members. Figure 3a highlights six families in which all individuals within these families exhibited low C9orf72 hypermethylation ranging from 1.1 to 10.0 %. In contrast, there were three families, including one family with five first-degree relatives harboring the hexanucleotide repeat expansion, where individuals exhibited high levels of C9orf72 methylation ranging from 20.2 to 41.9 %, suggesting that C9orf72 hypermethylation is heritable (Fig. 3b). Overall, it appeared that C9orf72 methylation was stable both within generations and across different generations. Non-expanded, unaffected family members are not shown because C9orf72 hypermethylation is only observed in repeat expansion carriers, and to better preserve anonymity of the research participants. Many repeat expansion carriers in the nine pedigrees were clinically unaffected at sample collection. Four pedigrees had multiple affected individuals. Of those four pedigrees, individuals within two of the families exhibited concordant clinical phenotypes (all motor neuron disease, or all FTD), while individuals within the other two families exhibited discordant clinical phenotypes (mixtures of both motor neuron disease and FTD). We observed no relationship between C9orf72 methylation and concordance of clinical diagnosis in these families.
Fig. 3.
C9orf72 promoter methylation within family pedigrees. C9orf72 promoter methylation values are shown for individuals from families with (a) low or (b) high peripheral blood C9orf72 methylation. Male family members are represented as squares and females as circles. Dotted squares or circles represent inferred repeat expansion carriers for which no DNA was available. Non-expanded family members are not shown. c Frequency histogram of F statistics from 10,000 randomly permutated linear models. Dotted line represents the F statistic obtained with the true linear model (permutation test p < 0.0001)
To more rigorously test whether there is a significant relationship between C9orf72 methylation and family relatedness, we used a permutation test to determine if affiliation within a certain family is associated with C9orf72 methylation. First, a univariate linear regression model was generated relating different families to C9orf72 methylation. Then, each of the 26 studied family members was randomly assigned to one of the nine families, while the number of family members within each family was kept constant; thereby an alternate linear regression model was generated in which the relationship between the dependent and independent variables was broken. This process was repeated 10,000 times to generate a random set of linear regression models. The true linear regression model revealed an F statistic of 3.41, while the 10,000 random linear regression models were associated with F statistics ranging from 0.049 to 2.711 (Fig. 3c). Thus, maintaining the true relationship between family membership and C9orf72 methylation predicted C9orf72 methylation significantly better than when the relationship between family membership and C9orf72 methylation was randomly broken (p < 0.0001). These results suggest that C9orf72 methylation is heritable and raises the possibility that there may be a latent polymorphism which promotes C9orf72 methylation in a subset of C9orf72 repeat expansion carriers.
C9orf72 promoter hypermethylation is associated with a shorter repeat size
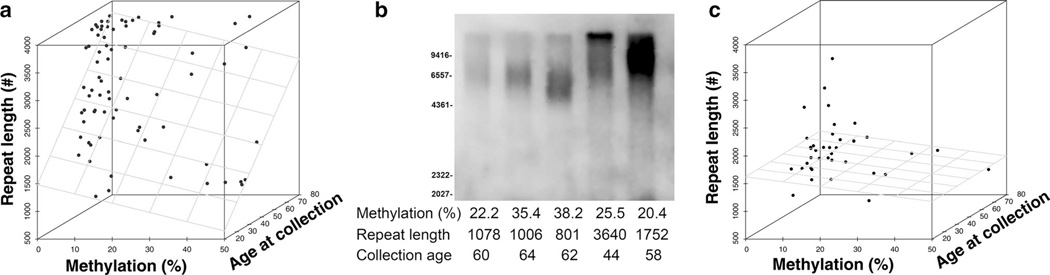
Finally, DNA methylation has been shown to affect repeat expansion size in several repeat expansion-associated diseases [12, 17, 18, 47]. To determine whether C9orf72 promoter methylation affects hexanucleotide repeat expansion size, hexanucleotide repeat expansion size was measured by Southern blot of peripheral blood of 93 repeat expansion carriers. A multivariate regression analysis demonstrated that both age at collection and C9orf72 promoter methylation influenced repeat expansion size (n = 76 unrelated repeat expansion carriers, Fig. 4a and Table 4). As has been previously noted by others [65], later age of sample collection was associated with longer repeat sizes (β = 38.48, p = 0.001). This corresponds to an increase of 38.5 repeats per 1 year increase in age at collection. In contrast, increasing C9orf72 promoter methylation levels are associated with smaller repeat size (β = −16.69, p = 0.033), i.e., an increase of 10 % in C9orf72 promoter methylation results in a decrease of repeat size by 167 repeats. To further explore the relationship between repeat size and promoter methylation, 24 individuals were identified within eight families for which both peripheral blood hexanucleotide repeat expansion size and C9orf72 promoter methylation values were available from at least two family members. A representative Southern blot of DNA from five individuals from a single family is shown in Fig. 4b. We observed that within families, increased C9orf72 methylation was associated with reduced hexanucleotide repeat expansion size. To verify this observation, a linear mixed-effects model was generated to control for family relatedness which again revealed that C9orf72 promoter methylation was associated with reduced hexanucleotide repeat expansion size (β = −51.44, p = 0.010, Table 4). This corresponds to a decrease in repeat size by 51.4 repeats with every 1 % increase of C9orf72 promoter methylation within family members. We also analyzed the relationship between repeat size, methylation and age at collection in cerebellum (n = 38, Fig. 4c). There was no significant effect of methylation or age of collection on repeat size in cerebellum (Table 4), perhaps due to the smaller sample size compared to the peripheral blood dataset, or due to the fact that the C9orf72 repeat expansion may be more stable in cerebellum relative to other tissues [20, 65].
Fig. 4.
C9orf72 promoter hypermethylation is associated with shorter repeat length. a Scatterplot of the relationship between repeat length, promoter methylation and age at sample collection of peripheral blood DNA (n = 77). The results of the multivariate regression model (Table 4) are overlaid as a gray-colored plane. b Representative Southern blot for the C9orf72 hexanucleotide repeat expansion from peripheral blood DNA of five repeat expansion carriers from a single family. Values for C9orf72 promoter methylation, hexanucleotide repeat length, and age at DNA collection are shown. c Scatterplot of the relationship between repeat length, promoter methylation and age at sample collection of cerebellar DNA (n = 38). The results of the multivariate regression model (Table 4) are overlaid as a gray-colored plane
Table 4.
C9orf72 promoter methylation and hexanucleotide repeat expansion length
| Repeat length, blooda (n = 76 unrelated individuals) |
Repeat length, bloodb (n = 24 individuals in eight families) |
Repeat length, brainc (n = 38 unrelated individuals) |
||||
|---|---|---|---|---|---|---|
| β (SE) | p value | β (SE) | p value | β (SE) | p value | |
| Intercept | 713.24 (701.41) | 0.313 | 2,408.56 (1,080.22) | 0.043 | 1,615.56 (514.98) | 0.004 |
| Age at collection | 38.48 (11.62) | 0.001 | 15.15 (20.26) | 0.467 | 0.38 (8.11) | 0.963 |
| C9orf72 methylation | −16.69 (7.68) | 0.033 | −51.44 (17.32) | 0.010 | −8.89 (7.83) | 0.264 |
SE standard error
Linear multivariate regression model (R2 = 0.189)
Linear mixed-effects model using age at sample collection and methylation as fixed effects and family group as random effect
Linear multivariate regression model (R2 = 0.036)
Discussion
The hexanucleotide repeat expansion in C9orf72 is associated with a variable clinical phenotype including ALS, ALS with FTD, FTD, Alzheimer’s disease and others [2, 6–8, 10, 14, 28, 29, 34–36, 38, 40, 45, 49, 56, 57, 61, 62]. Additionally, both ALS and FTD are characterized by clinical heterogeneity in terms of age at onset and disease duration [2, 6–8, 10, 14, 28, 29, 34–36, 38, 40, 45, 49, 56, 57, 61, 62]. The clinical heterogeneity associated with the C9orf72 mutation indicates that there are likely several environmental or genetic disease modifiers which remain to be discovered.
We hypothesized that epigenetic silencing of mutant C9orf72 associated with DNA methylation prolongs survival. We found that C9orf72 methylation appears to be stable across a diverse set of central and peripheral tissues as well as over time. Additionally, C9orf72 hypermethylation in both cerebellum and peripheral blood is associated with later age at death in C9orf72 mutation carriers with FTD. Moreover, C9orf72 hypermethylation in blood is associated with longer disease duration and shorter C9orf72 hexanucleotide repeat length. These results provide evidence that C9orf72 hypermethylation is associated with prolonged survival in C9orf72 repeat expansion carriers with FTD, supporting the hypothesis that epigenetic silencing of mutant C9orf72 may be protective.
We did not see an effect of C9orf72 promoter hypermethylation on disease duration or age at death in ALS patients. The pyramidal motor system may have less functional reserve compared to the CNS networks which regulate executive or language function, resulting in more rapid progression to death in ALS such that C9orf72 hypermethylation may not lead to a significant modifying effect (see Table 1). Indeed, ALS is known to exhibit a rapid clinical course compared to FTD as seen in the cohort reported here (Table 1), compounded by the fact that the C9orf72 mutation is associated with even more rapid clinical decline relative to cases without the C9orf72 mutation [38].
A previous study examined C9orf72 promoter methylation in repeat expanded ALS patients, non-expanded ALS patients and normal controls [68]. Similar to us, they found that C9orf72 promoter methylation in blood is significantly higher in C9orf72 repeat expansion carriers than in non-expanded controls, and that there is no correlation between C9orf72 methylation and age at onset or age at sample collection [68]. These results were recently confirmed in a cohort of FTD patients [69]. We and others have observed faster disease progression in cases with the C9orf72 mutation relative to non-expanded cases [6, 7, 10, 14, 36, 38, 49, 61]. Therefore, there is an overall inverse relationship between C9orf72 methylation and disease duration due to the fact that C9orf72 hypermethylation is seen only in individuals with the hexanucleotide repeat expansion [48, 68, 69]. Rather than studying C9orf72 hypermethylation in a cohort of both repeat expanded and non-expanded cases, our goal here was to determine whether epigenetic silencing of mutant C9orf72 is a disease modifier within C9orf72 carriers. In this context, we found that increased C9orf72 methylation levels in FTD patients are associated with prolonged survival.
We also observed that C9orf72 methylation is stable within and across generations. We demonstrated statistically a significant relationship between C9orf72 methylation and family relatedness. These results are consistent with a previous report that suggested that C9orf72 methylation is heritable [68]. Genetic polymorphisms are known to influence DNA methylation [3, 27, 39]. Our results raise the possibility that there may be a latent polymorphism which promotes C9orf72 hypermethylation in C9orf72 repeat expansion carriers. TMEM106B polymorphisms have been reported to be genetic modifiers in FTD individuals with the C9orf72 repeat expansion [26, 66]. Importantly, we found no relationship between TMEM106B genotype and C9orf72 promoter methylation (data not shown).
The factors which regulate C9orf72 methylation are not known. Interestingly, several repeat expansion disease loci are associated with local heterochromatin formation including FMR1 and FXN [23]. FMR1 silencing occurs very early during embryogenesis and has been linked to the formation of RNA–DNA duplexes within the FMR1 promoter [13, 16]. Whether a similar mechanism causes silencing of mutant C9orf72 remains to be seen. Although the molecular association between repeat expansions and local epigenetic silencing is remarkably consistent across several diseases, the clinical consequences of epigenetic silencing are divergent. Thus, in contrast to what we observe for C9orf72, DNA hypermethylation in Friedreich’s ataxia and Fragile X syndrome is associated with faster disease progression or more severe disease phenotype [9, 22, 33, 51, 52]. Both Friedreich’s ataxia and Fragile X syndrome are caused by a loss of gene function. Our results imply that the C9orf72 mutation does not cause disease through a loss of function.
C9orf72 repeat expansion length has also been studied as a potential disease modifier. One study found that a threshold C9orf72 repeat length from cerebellar DNA is associated with longer survival in individuals with FTD [65]. Three studies reported a positive correlation between repeat size and age at onset [2, 37, 65]. Two other studies failed to observe an association between C9orf72 repeat length and age at onset [19, 20]. However, these associations are confounded by the fact that the C9orf72 hexanucleotide repeat exhibits somatic instability such that repeat length in peripheral blood does not correlate with repeat length in brain [65]. Moreover, C9orf72 repeat length changes according to age at collection, confounding potential associations between repeat length and clinical disease [65]. In contrast, C9orf72 methylation did not vary according to age at sample collection and thereby may represent a more stable disease biomarker.
C9orf72 hexanucleotide repeat expansions characteristically exhibit a high degree of somatic instability [2, 37, 65]. The molecular mechanisms that lead to GGGGCC repeat instability are not known. However, DNA methylation, by virtue of altering DNA transcription or replication dynamics, has been shown in most cases to stabilize microsatellite repeats [12, 17, 18, 31, 47]. The significant inverse correlation between C9orf72 promoter methylation and hexanucleotide repeat expansion length is thus another example of epigenetic regulation of microsatellite instability.
The C9orf72 hexanucleotide repeat expansion has been postulated to cause disease due to a gain of toxic function linked to the accumulation of RNA foci [15]. RNA foci are predominantly intranuclear accumulations of hexanucleotide repeat containing RNA which have been reported to bind to several RNA-binding proteins including Pur α, hnRNPA3, hnRNP-H, ADARB2 and nucleolin [21, 32, 44, 54, 60, 71]. The sequestration of RNA-binding proteins within RNA foci has been postulated to lead to transcriptomic abnormalities which lead to neurodegeneration [21, 32, 42, 44, 54, 60, 71]. Additionally, repeat expanded RNA can be translated into DPR proteins which aggregate within neurons and may be toxic [1, 41, 50, 53, 55].
Alternatively, the C9orf72 mutation has been suggested to result in haploinsufficiency linked to epigenetic silencing of mutant C9orf72. The repeat expansion mutation is associated with reduced C9orf72 mRNA and protein [4, 48, 67, 68]. C9orf72 protein regulates endosomal trafficking, consistent with bioinformatics-based analysis which indicated that C9orf72 is structurally related to the DENN family of proteins which regulate organelle trafficking [24, 46, 72].
Epigenetic silencing of mutant C9orf72 leads to reduced expression of C9orf72 mRNA [4, 5, 48, 68, 69]. Moreover, knockdown of the C9orf72 homolog in zebrafish results in motor neuron axonal degeneration [11]. However, we have found that C9orf72 hypermethylation is associated with a protective phenotype in C9orf72 repeat expanded lymphoblast cells, and that at time of autopsy, brain tissue from individuals within the C9orf72 mutation carrier cohort reported here demonstrates that C9orf72 hypermethylation is associated with reduced RNA foci and DPR aggregate burden [48]. Thus, it was unclear whether epigenetic silencing of mutant C9orf72 is deleterious or protective against disease. The association between C9orf72 hypermethylation and prolonged survival and smaller repeat length that we report here supports the toxic gain of function hypothesis, and indicates that transcriptional silencing of mutant C9orf72 may be protective, at least in terms of disease progression after onset in FTD cases. Importantly, we did not observe a significant relationship between C9orf72 hypermethylation and age at onset or clinical phenotype (ALS versus FTD, site of onset, FTD phenotype). The factors that influence disease onset and clinical phenotypes remain unknown.
Our results have implications in terms of developing therapies for C9orf72 mutation carriers. While haploinsufficiency is not entirely excluded as a disease mechanism, therapies to increase C9orf72 gene expression may be detrimental, particularly if these therapies increase expression of the mutant allele. Conversely, enhancing epigenetic silencing of mutant C9orf72 or other methods to suppress mutant C9orf72 may be beneficial, including recent efforts to develop antisense oligonucleotides to target mutant C9orf72 RNA for degradation [21, 42, 60].
The strengths of this study include a well-characterized cohort of C9orf72 mutation carriers, the use of DNA from multiple different sources, and the use of a highly quantitative methylation assay. Replication of our results with independent C9orf72 mutation carrier cohorts would be desirable. Moreover, because of our retrospective study design, prospective longitudinal studies are necessary to validate the predictive power of C9orf72 promoter methylation in terms of clinical disease progression. Furthermore, a more thorough analysis of C9orf72 methylation over the entire C9orf72 promoter from multiple DNA sources including multiple brain regions is needed to further elucidate potential tissue-specific differences in C9orf72 methylation.
Supplementary Material
Acknowledgments
We thank the patients and patients’ families who made this research possible. We acknowledge Drs. J. Q. Trojanowski and V. M.-Y. Lee and the Center for Neurodegenerative Disease Research for their support. This study was supported in part by a grant from the Judith & Jean Pape Adams Foundation and by the National Institutes of Health (K08AG039510, P30AG10124, P01AG017586, P01AG032953, P50AG005681). EBL is supported by a Doris Duke Charitable Foundation Clinical Scientist Development Award.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00401-014-1365-0) contains supplementary material, which is available to authorized users.
Contributor Information
Jenny Russ, Translational Neuropathology Research Laboratory, Department of Pathology and Laboratory Medicine, Perelman School of Medicine at the University of Pennsylvania, 605B Stellar Chance Laboratories, 422 Curie Blvd, Philadelphia, PA 19104, USA.
Elaine Y. Liu, Translational Neuropathology Research Laboratory, Department of Pathology and Laboratory Medicine, Perelman School of Medicine at the University of Pennsylvania, 605B Stellar Chance Laboratories, 422 Curie Blvd, Philadelphia, PA 19104, USA
Kathryn Wu, Translational Neuropathology Research Laboratory, Department of Pathology and Laboratory Medicine, Perelman School of Medicine at the University of Pennsylvania, 605B Stellar Chance Laboratories, 422 Curie Blvd, Philadelphia, PA 19104, USA.
Donald Neal, Center for Neurodegenerative Disease Research, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, USA.
EunRan Suh, Center for Neurodegenerative Disease Research, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, USA.
David J. Irwin, Center for Neurodegenerative Disease Research, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, USA Department of Neurology, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, USA.
Corey T. McMillan, Department of Neurology, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, USA
Matthew B. Harms, Department of Neurology, Washington University School of Medicine, St. Louis, MO, USA
Nigel J. Cairns, Department of Neurology, Washington University School of Medicine, St. Louis, MO, USA
Elisabeth M. Wood, Center for Neurodegenerative Disease Research, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, USA
Sharon X. Xie, Department of Biostatistics and Epidemiology, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, USA
Lauren Elman, Department of Neurology, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, USA.
Leo McCluskey, Department of Neurology, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, USA.
Murray Grossman, Department of Neurology, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, USA.
Vivianna M. Van Deerlin, Center for Neurodegenerative Disease Research, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, USA
Edward B. Lee, Email: edward.lee@uphs.upenn.edu, Translational Neuropathology Research Laboratory, Department of Pathology and Laboratory Medicine, Perelman School of Medicine at the University of Pennsylvania, 605B Stellar Chance Laboratories, 422 Curie Blvd, Philadelphia, PA 19104, USA.
References
- 1.Ash PE, Bieniek KF, Gendron TF, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beck J, Poulter M, Hensman D, et al. Large C9orf72 hexanucleotide repeat expansions are seen in multiple neurodegenerative syndromes and are more frequent than expected in the UK population. Am J Hum Genet. 2013;92:345–353. doi: 10.1016/j.ajhg.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell JT, Pai AA, Pickrell JK, et al. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 2011;12:R10. doi: 10.1186/gb-2011-12-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belzil VV, Bauer PO, Prudencio M, et al. Reduced C9orf72 gene expression in c9FTD/ALS is caused by histone trimethylation, an epigenetic event detectable in blood. Acta Neuropathol. 2013;126:895–905. doi: 10.1007/s00401-013-1199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belzil VV, Bauer PO, Gendron TF, Murray ME, Dickson D, Petrucelli L. Characterization of DNA hypermethylation in the cerebellum of c9FTD/ALS patients. Brain Res. 2014 doi: 10.1016/j.brainres.2014.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boeve BF, Boylan KB, Graff-Radford NR, et al. Characterization of frontotemporal dementia and/or amyotrophic lateral sclerosis associated with the GGGGCC repeat expansion in C9ORF72. Brain. 2012;135:765–783. doi: 10.1093/brain/aws004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brettschneider J, Van Deerlin VM, Robinson JL, et al. Pattern of ubiquilin pathology in ALS and FTLD indicates presence of C9ORF72 hexanucleotide expansion. Acta Neuropathol. 2012;123:825–839. doi: 10.1007/s00401-012-0970-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Byrne S, Elamin M, Bede P, et al. Cognitive and clinical characteristics of patients with amyotrophic lateral sclerosis carrying a C9orf72 repeat expansion: a population-based cohort study. Lancet Neurol. 2012;11:232–240. doi: 10.1016/S1474-4422(12)70014-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castaldo I, Pinelli M, Monticelli A, et al. DNA methylation in intron 1 of the frataxin gene is related to GAA repeat length and age of onset in Friedreich ataxia patients. J Med Genet. 2008;45:808–812. doi: 10.1136/jmg.2008.058594. [DOI] [PubMed] [Google Scholar]
- 10.Chio A, Borghero G, Restagno G, et al. Clinical characteristics of patients with familial amyotrophic lateral sclerosis carrying the pathogenic GGGGCC hexanucleotide repeat expansion of C9ORF72. Brain. 2012;135:784–793. doi: 10.1093/brain/awr366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ciura S, Lattante S, Le Ber I, et al. Loss of function of C9orf72 causes motor deficits in a zebrafish model of Amyotrophic Lateral Sclerosis. Ann Neurol. 2013 doi: 10.1002/ana.23946. [DOI] [PubMed] [Google Scholar]
- 12.Cleary JD, Tome S, Lopez Castel A, et al. Tissue- and age-specific DNA replication patterns at the CTG/CAG-expanded human myotonic dystrophy type 1 locus. Nat Struct Mol Biol. 2010;17:1079–1087. doi: 10.1038/nsmb.1876. [DOI] [PubMed] [Google Scholar]
- 13.Colak D, Zaninovic N, Cohen MS, et al. Promoter-bound trinucleotide repeat mRNA drives epigenetic silencing in fragile X syndrome. Science. 2014;343:1002–1005. doi: 10.1126/science.1245831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooper-Knock J, Hewitt C, Highley JR, et al. Clinico-pathological features in amyotrophic lateral sclerosis with expansions in C9ORF72. Brain. 2012;135:751–764. doi: 10.1093/brain/awr365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devys D, Biancalana V, Rousseau F, Boue J, Mandel JL, Oberle I. Analysis of full fragile X mutations in fetal tissues and monozygotic twins indicate that abnormal methylation and somatic heterogeneity are established early in development. Am J Med Genet. 1992;43:208–216. doi: 10.1002/ajmg.1320430134. [DOI] [PubMed] [Google Scholar]
- 17.Dion V, Lin Y, Hubert L, Jr, Waterland RA, Wilson JH. Dnmt1 deficiency promotes CAG repeat expansion in the mouse germline. Hum Mol Genet. 2008;17:1306–1317. doi: 10.1093/hmg/ddn019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dion V, Wilson JH. Instability and chromatin structure of expanded trinucleotide repeats. Trends Genet. 2009;25:288–297. doi: 10.1016/j.tig.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dobson-Stone C, Hallupp M, Loy CT, et al. C9ORF72 repeat expansion in Australian and Spanish frontotemporal dementia patients. PLoS One. 2013;8:e56899. doi: 10.1371/journal.pone.0056899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dols-Icardo O, Garcia-Redondo A, Rojas-Garcia R, et al. Characterization of the repeat expansion size in C9orf72 in amyotrophic lateral sclerosis and frontotemporal dementia. Hum Mol Genet. 2014;23:749–754. doi: 10.1093/hmg/ddt460. [DOI] [PubMed] [Google Scholar]
- 21.Donnelly CJ, Zhang PW, Pham JT, et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80:415–428. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evans-Galea MV, Carrodus N, Rowley SM, et al. FXN methylation predicts expression and clinical outcome in Friedreich ataxia. Ann Neurol. 2012;71:487–497. doi: 10.1002/ana.22671. [DOI] [PubMed] [Google Scholar]
- 23.Evans-Galea MV, Hannan AJ, Carrodus N, Delatycki MB, Saffery R. Epigenetic modifications in trinucleotide repeat diseases. Trends Mol Med. 2013;19:655–663. doi: 10.1016/j.molmed.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 24.Farg MA, Sundaramoorthy V, Sultana JM, et al. C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum Mol Genet. 2014 doi: 10.1093/hmg/ddu068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Felle M, Hoffmeister H, Rothammer J, Fuchs A, Exler JH, Langst G. Nucleosomes protect DNA from DNA methylation in vivo and in vitro. Nucleic Acids Res. 2011;39:6956–6969. doi: 10.1093/nar/gkr263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gallagher MD, Suh E, Grossman M, et al. TMEM106B is a genetic modifier of frontotemporal lobar degeneration with C9orf72 hexanucleotide repeat expansions. Acta Neuropathol. 2014;127:407–418. doi: 10.1007/s00401-013-1239-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gibbs JR, van der Brug MP, Hernandez DG, et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6:e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gijselinck I, Van Langenhove T, van der Zee J, et al. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: a gene identification study. Lancet Neurol. 2012;11:54–65. doi: 10.1016/S1474-4422(11)70261-7. [DOI] [PubMed] [Google Scholar]
- 29.Goldman JS, Quinzii C, Dunning-Broadbent J, et al. Multiple system atrophy and amyotrophic lateral sclerosis in a family with hexanucleotide repeat expansions in C9orf72. JAMA Neurol. 2014 doi: 10.1001/jamaneurol.2013.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorno-Tempini ML, Hillis AE, Weintraub S, et al. Classification of primary progressive aphasia and its variants. Neurology. 2011;76:1006–1014. doi: 10.1212/WNL.0b013e31821103e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goula AV, Stys A, Chan JP, Trottier Y, Festenstein R, Merienne K. Transcription elongation and tissue-specific somatic CAG instability. PLoS Genet. 2012;8:e1003051. doi: 10.1371/journal.pgen.1003051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haeusler AR, Donnelly CJ, Periz G, et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507:195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hagerman RJ, Hull CE, Safanda JF, et al. High functioning fragile X males: demonstration of an unmethylated fully expanded FMR-1 mutation associated with protein expression. Am J Med Genet. 1994;51:298–308. doi: 10.1002/ajmg.1320510404. [DOI] [PubMed] [Google Scholar]
- 34.Harms M, Benitez BA, Cairns N, et al. C9orf72 hexanucleotide repeat expansions in clinical Alzheimer disease. JAMA Neurol. 2013;70:736–741. doi: 10.1001/2013.jamaneurol.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hensman Moss DJ, Poulter M, Beck J, et al. C9orf72 expansions are the most common genetic cause of Huntington disease phenocopies. Neurology. 2014;82:292–299. doi: 10.1212/WNL.0000000000000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsiung GY, DeJesus-Hernandez M, Feldman HH, et al. Clinical and pathological features of familial frontotemporal dementia caused by C9ORF72 mutation on chromosome 9p. Brain. 2012;135:709–722. doi: 10.1093/brain/awr354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hubers A, Marroquin N, Schmoll B, et al. Polymerase chain reaction and Southern blot-based analysis of the C9orf72 hexanucleotide repeat in different motor neuron diseases. Neurobiol Aging. 2014;35(1214):e1211–e1216. doi: 10.1016/j.neurobiolaging.2013.11.034. [DOI] [PubMed] [Google Scholar]
- 38.Irwin DJ, McMillan CT, Brettschneider J, et al. Cognitive decline and reduced survival in C9orf72 expansion frontotemporal degeneration and amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry. 2013;84:163–169. doi: 10.1136/jnnp-2012-303507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kerkel K, Spadola A, Yuan E, et al. Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat Genet. 2008;40:904–908. doi: 10.1038/ng.174. [DOI] [PubMed] [Google Scholar]
- 40.Khan BK, Yokoyama JS, Takada LT, et al. Atypical, slowly progressive behavioural variant frontotemporal dementia associated with C9ORF72 hexanucleotide expansion. J Neurol Neurosurg Psychiatry. 2012;83:358–364. doi: 10.1136/jnnp-2011-301883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwon I, Xiang S, Kato M, et al. Poly-dipeptides encoded by the C9ORF72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science. 2014 doi: 10.1126/science.1254917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lagier-Tourenne C, Baughn M, Rigo F, et al. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc Natl Acad Sci. 2013;110:E4530–E4539. doi: 10.1073/pnas.1318835110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laird NM, Ware JH. Random-effects models for longitudinal data. Biometrics. 1982;38:963–974. [PubMed] [Google Scholar]
- 44.Lee YB, Chen HJ, Peres JN, et al. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep. 2013;5:1178–1186. doi: 10.1016/j.celrep.2013.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lesage S, Le Ber I, Condroyer C, et al. C9orf72 repeat expansions are a rare genetic cause of parkinsonism. Brain. 2013;136:385–391. doi: 10.1093/brain/aws357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levine TP, Daniels RD, Gatta AT, Wong LH, Hayes MJ. The product of C9orf72, a gene strongly implicated in neurodegeneration, is structurally related to DENN Rab-GEFs. Bioinformatics. 2013;29:499–503. doi: 10.1093/bioinformatics/bts725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Libby RT, Hagerman KA, Pineda VV, et al. CTCF cis-regulates trinucleotide repeat instability in an epigenetic manner: a novel basis for mutational hot spot determination. PLoS Genet. 2008;4:e1000257. doi: 10.1371/journal.pgen.1000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu EY, Russ J, Wu K, et al. C9orf72 hypermethylation protects against repeat expansion-associated pathology in ALS/FTD. Acta Neuropathol. 2014 doi: 10.1007/s00401-014-1286-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mahoney CJ, Beck J, Rohrer JD, et al. Frontotemporal dementia with the C9ORF72 hexanucleotide repeat expansion: clinical, neuroanatomical and neuropathological features. Brain. 2012;135:736–750. doi: 10.1093/brain/awr361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.May S, Hornburg D, Schludi MH, et al. C9orf72 FTLD/ALS-associated Gly-Ala dipeptide repeat proteins cause neuronal toxicity and Unc119 sequestration. Acta Neuropathol. 2014 doi: 10.1007/s00401-014-1329-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McConkie-Rosell A, Lachiewicz AM, Spiridigliozzi GA, et al. Evidence that methylation of the FMR-I locus is responsible for variable phenotypic expression of the fragile X syndrome. Am J Hum Genet. 1993;53:800–809. [PMC free article] [PubMed] [Google Scholar]
- 52.Merenstein SA, Sobesky WE, Taylor AK, Riddle JE, Tran HX, Hagerman RJ. Molecular-clinical correlations in males with an expanded FMR1 mutation. Am J Med Genet. 1996;64:388–394. doi: 10.1002/(SICI)1096-8628(19960809)64:2<388::AID-AJMG31>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 53.Mizielinska S, Gronke S, Niccoli T, et al. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science. 2014 doi: 10.1126/science.1256800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mori K, Lammich S, Mackenzie IR, et al. hnRNP A3 binds to GGGGCC repeats and is a constituent of p62-positive/TDP43-negative inclusions in the hippocampus of patients with C9orf72 mutations. Acta Neuropathol. 2013;125:413–423. doi: 10.1007/s00401-013-1088-7. [DOI] [PubMed] [Google Scholar]
- 55.Mori K, Weng SM, Arzberger T, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339:1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- 56.Murray ME, DeJesus-Hernandez M, Rutherford NJ, et al. Clinical and neuropathologic heterogeneity of c9FTD/ALS associated with hexanucleotide repeat expansion in C9ORF72. Acta Neuropathol. 2011;122:673–690. doi: 10.1007/s00401-011-0907-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murray ME, Bieniek KF, Banks Greenberg M, et al. Progressive amnestic dementia, hippocampal sclerosis, and mutation in C9ORF72. Acta Neuropathol. 2013;126:545–554. doi: 10.1007/s00401-013-1161-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rascovsky K, Hodges JR, Knopman D, et al. Sensitivity of revised diagnostic criteria for the behavioural variant of frontotemporal dementia. Brain. 2011;134:2456–2477. doi: 10.1093/brain/awr179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Renton AE, Majounie E, Waite A, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sareen D, O’Rourke JG, Meera P, et al. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci Transl Med. 2013;5:208ra149. doi: 10.1126/scitranslmed.3007529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Simon-Sanchez J, Dopper EG, Cohn-Hokke PE, et al. The clinical and pathological phenotype of C9ORF72 hexanucleotide repeat expansions. Brain. 2012;135:723–735. doi: 10.1093/brain/awr353. [DOI] [PubMed] [Google Scholar]
- 62.Snowden JS, Rollinson S, Thompson JC, et al. Distinct clinical and pathological characteristics of frontotemporal dementia associated with C9ORF72 mutations. Brain. 2012;135:693–708. doi: 10.1093/brain/awr355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Strong MJ, Grace GM, Freedman M, et al. Consensus criteria for the diagnosis of frontotemporal cognitive and behavioural syndromes in amyotrophic lateral sclerosis. Amyotroph Lateral Scler. 2009;10:131–146. doi: 10.1080/17482960802654364. [DOI] [PubMed] [Google Scholar]
- 64.Toledo JB, Van Deerlin VM, Lee EB, et al. A platform for discovery: the University of Pennsylvania integrated neurodegenerative disease biobank. Alzheimers Dement. 2013 doi: 10.1016/j.jalz.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Blitterswijk M, DeJesus-Hernandez M, Niemantsverdriet E, et al. Association between repeat sizes and clinical and pathological characteristics in carriers of C9ORF72 repeat expansions (Xpansize-72): a cross-sectional cohort study. Lancet Neurol. 2013;12:978–988. doi: 10.1016/S1474-4422(13)70210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Blitterswijk M, Mullen B, Nicholson AM, et al. TMEM106B protects C9ORF72 expansion carriers against frontotemporal dementia. Acta Neuropathol. 2014;127:397–406. doi: 10.1007/s00401-013-1240-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Waite AJ, Baumer D, East S, et al. Reduced C9orf72 protein levels in frontal cortex of amyotrophic lateral sclerosis and frontotemporal degeneration brain with the C9ORF72 hexanucleotide repeat expansion. Neurobiol Aging. 2014;35:1779 e1775–1779 e1713. doi: 10.1016/j.neurobiolaging.2014.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xi Z, Zinman L, Moreno D, et al. Hypermethylation of the CpG island near the G4C2 repeat in ALS with a C9orf72 expansion. Am J Hum Genet. 2013;92:981–989. doi: 10.1016/j.ajhg.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xi Z, Rainero I, et al. Hypermethylation of the CpG-island near the C9orf72 G4C2-repeat expansion in FTLD patients. Hum Mol Genet. 2014 doi: 10.1093/hmg/ddu279. [DOI] [PubMed] [Google Scholar]
- 70.Xie SX, Baek Y, Grossman M, et al. Building an integrated neurodegenerative disease database at an academic health center. Alzheimers Dement. 2011;7(4):e84–e93. doi: 10.1016/j.jalz.2010.08.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu Z, Poidevin M, Li X, et al. Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proc Natl Acad Sci. 2013;110:7778–7783. doi: 10.1073/pnas.1219643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang D, Iyer LM, He F, Aravind L. Discovery of novel DENN proteins: implications for the evolution of eukaryotic intracellular membrane structures and human disease. Front Genet. 2012;3:283. doi: 10.3389/fgene.2012.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.