Abstract
The snail Bithynia siamensis goniomphalos acts as the first intermediate host for the human liver fluke Opisthorchis viverrini, the major cause of cholangiocarcinoma (CCA) in Northeast Thailand. The undisputed link between CCA and O. viverrini infection has precipitated efforts to understand the molecular basis of host-parasite interactions with a view to ultimately developing new control strategies to combat this carcinogenic infection. To date most effort has focused on the interactions between the parasite and its human host, and little is known about the molecular relationships between the liver fluke and its snail intermediate host. In the present study we analyse the protein expression changes in different tissues of B. siamensis goniomphalos induced by infection with larval O. viverrini using iTRAQ labelling technology. We show that O. viverrini infection downregulates the expression of oxidoreductases and catalytic enzymes, while stress-related and motor proteins are upregulated. The present work could serve as a basis for future studies on the proteins implicated in the susceptibility/resistance of B. siamensis goniomphalos to O. viverrini, as well as studies on other pulmonate snail intermediate hosts of various parasitic flukes that infect humans.
Keywords: Bithynia siamensis goniomphalos, snail-borne trematodes, iTRAQ, liver fluke, Opisthorchis, cholangiocarcinoma, intermediate host, proteome, iQuantitator
1. Introduction
The liver fluke, Opisthorchis viverrini, represents a major public health problem in the Greater Mekong sub-region (Thailand, Lao PDR, Cambodia and southern Vietnam), with >10 million people estimated to be infected with this parasite. In addition to a spectrum of clinical signs associated with the infection, which include cholangitis, obstructive jaundice, hepatomegaly, periductal fibrosis, cholecystitis and cholelithiasis [1, 2], opisthorchiasis by O. viverrini is unequivocally associated with the development of cholangiocarcinoma (CCA) in infected individuals [3-5]. Incidence rates of CCA range from 93.8 to 317.6 per 100,000 people/year in some districts of Northeast Thailand alone and prognosis is poor [3, 6]. O. viverrini is characterised by a complex life cycle, involving developmental phases in the definitive human host as well as intermediate prosobranch snail and cyprinid fish hosts. Piscivorous mammals, including dogs, cats and humans, serve as definitive hosts and become infected by eating raw or fermented fish harbouring the metacercariae of the parasite [7]. Metacercariae then excyst in the duodenum and migrate as juvenile flukes to the intra-hepatic biliary tree, where they develop to adult hermaphrodite flukes within ∼4 weeks; mature flukes shed embryonated eggs into water through the faeces. Eggs are ingested by prosobranch snails of the genus Bithynia and hatch in the snail's digestive tract where the motile embryos (miracidia) develop into sporocysts. The sporocysts undergo asexual reproduction through the stages of rediae and cercariae, the latter of which exit the snail 6 to 8 weeks later and infect a cyprinid fish. In the fish host the parasite encysts in the muscle to form metacercariae, the stage that is infective to humans upon ingestion of raw or undercooked fish.
Despite the high prevalence of O. viverrini infection in humans and fish in endemic areas (i.e. up to 90% and 97%, respectively), prevalence in the snail intermediate host is surprisingly low (<1%) [8-12], and this observation has led to speculation that parasite infections may cause the activation of snail immune pathways aimed at eliminating and/or limiting the infection itself [13]. Indeed, both cellular and humoral factors have been reported to play important roles in ‘defending’ snails against trematode infections [14, 15]; fibrinogen-related proteins (FREPs) expressed by Biomphalaria glabrata, the intermediate snail host of Schistosoma mansoni, have been shown to precipitate parasite antigens, possibly playing a role in protective responses against parasite infections [16-19], and snail lectins and opsonins have also been shown to impact on trematode infections [20-23].
The biological interactions between trematodes and their intermediate hosts are crucial events that determine the success of a parasite's infective process; the study of such interactions is currently attracting significant attention, particularly in relation to the development of strategies aimed at interrupting parasite transmission [24-26]. Recently, we used RNA-Seq of cDNA libraries to characterize the entire transcriptome of B. siamensis goniomphalos [25], and investigated gene expression changes associated with O. viverrini infection [13]. Despite these advances, information on the proteome of B. siamensis goniomphalos, and consequently protein expression changes induced by fluke infection, is scarce. Since proteins represent the primary interface of molecular interactions between snails and trematode parasites, this information is crucial to assist future investigations of snail-focused approaches to parasite control. Herein we characterized the changes in protein expression of B. siamensis goniomphalos upon experimental infection with O. viverrini using a combination of quantitative and qualitative proteomic approaches. Knowledge of the molecular basis of immune processes that are regulated in B. siamensis goniomphalos after parasite infection could be of importance for the design of new control strategies against liver fluke infection and CCA.
2. Materials and Methods
2.1 Ethics statement
The protocols used for animal experimentation were approved by the Animal Ethics Committee of Khon Kaen University, based on the ethics of animal experimentation of the National Research Council of Thailand (Ethics clearance number AEKKU11/2555). All the snails and hamsters used in this study were maintained at the animal facilities at the Faculty of Medicine, Khon Kaen University, Thailand.
2.2 Snail preparation
Adult B. siamensis goniomphalos snails were collected from public freshwater ponds located in the Muang district, Khon Kaen Province, Thailand, kept in laboratory ceramic aquaria containing de-chlorinated tap water and fed with synthetic snail food [27]. Trematode-naïve snails, as confirmed by cercarial shedding once a week for 8 weeks, were used for experimental infections.
2.3 O. viverrini egg preparation
Syrian golden hamsters (Mesocricetus auratus) were experimentally infected with metacercariae of O. viverrini (50 metacercariae/animal) obtained from naturally infected cyprinid fish. After 4 months the infected animals were euthanized with ether and adult worms were recovered from the hamsters' livers and washed in 0.85% sodium chloride solution. The worms were subsequently dissected under a stereomicroscope to isolate eggs from the distal sections of the uteri as described previously [28]. Prior to experimental infection, the eggs were washed several times with distilled water and kept at room temperature for 2 weeks to undergo full maturation [29].
2.4 Experimental infection
Fully matured uterine-eggs of O. viverrini were fed to 40 (20 male and 20 female) B. siamensis goniomphalos maintained in the laboratory as previously described [29]. Briefly, snails were placed individually in transparent plastic containers with 6 ml of de-chlorinated tap water and exposed to 50 embryonated O. viverrini eggs for 24 h. After washing, the snails were placed in a new plastic container and kept at room temperature (RT) under dark and light in natural conditions and fed on synthetic snail food [27]. The plastic containers were checked daily and dead snails were removed. Each snail was subsequently examined for trematode infection by testing cercarial shedding and examination of hatched eggs in the snail faeces twice within a week as described previously [9, 30].
Four individuals (2 male and 2 female snails) were collected at 1, 7, 14, 28 and 56 days post-infection (p.i.), and 4 uninfected snails were used as controls. From all the collected snails, soft bodies were removed from their shells, separated into headfoot and body, snap frozen in liquid nitrogen and kept at -80 °C until use.
2.5 Sample preparation and protein extraction
Two biological replicates from each studied time point with two headfoot and body samples from two male and two female snails were pooled and placed in a 2 ml microcentrifuge tube with 600 μl of lysis buffer containing 5M urea, 2M thiourea, 0.1% SDS, 1% Triton X-100 and 40 mM Tris (pH 7.4). Each sample was ground with a TissueLyser II (QIAGEN) using a 5 mm stainless bead at 4°C for 10 min followed by incubation on ice for 30 min, and centrifugation at 12,000 g, at 4 °C for 20 min. The pellet was discarded and protein supernatant was subsequently precipitated with 10 volumes of cold methanol at -20°C overnight, centrifuged at 8,000 g for 10 min at 4°C, and air-dried for 5-10 min. Dried protein pellet was re-dissolved in buffer solution containing 0.5 M triethylammonium bicarbonate (TEAB) and 0.05% SDS, centrifuged at 12,000 g for 10 min at 4°C and protein content was determined by Bradford assay using BSA as a standard. One hundred (100) μg of protein was dried under vacuum before trypsin digestion. Protein extraction from the body portion was performed similarly. Headfoot and body samples from uninfected snails were used as controls and compared with experimentally infected tissues.
2.6 Protein digestion and iTRAQ labeling
Dried protein samples were re-suspended in 20 μl of dissolution buffer (0.5 TEAB) prior to reduction, alkylation, digestion and iTRAQ labeling according to the manufacturer's protocol (AB Sciex). Briefly, each protein sample was denatured with 2% SDS, reduced with 50 mM Tris-(2-carboxyethyl)-phosphine (TCEP) at 60°C for 1 h, and cysteine residues were alkylated with 10 mM methyl methanethiosulfate (MMTS) solution at RT for 10 min followed by tryptic digestion using 2 μg of trypsin (Sigma-Aldrich) at 37°C for 16 h. Digested peptide solutions were individually labeled with one vial of iTRAQ reagent at RT for 2 h. Each sample was labeled with different iTRAQ reagents having distinct isotopic compositions and all samples were subsequently combined into one tube for OFFGEL fractionation and LC-MS/MS analysis.
2.7 Peptide OFFGEL fractionation
A 3100 OFFGEL Fractionator (Agilent Technologies) with a 24 well setup was used for peptide separation based on pI. Prior to electrofocusing, desalting of samples was performed using a HiTrap SP HP column (GE Healthcare) and a Sep-Pak C18 cartridge (Waters) was used to remove excess of iTRAQ labeling according to the manufacturer's instructions. A total of 3.6 ml of OFFGEL peptide sample solution was used to dissolve the samples. The 24 cm long, 3-10 linear pH range IPG gel strips (GE Healthcare) were rehydrated with IPG Strip Rehydration Solution for 15 min, and 150 μl of dissolved sample was loaded in each well. The samples were focused with a maximum current of 50 μA until 50 kVh was reached. Every peptide fraction was harvested and each well rinsed with 150 μl of a solution of water/methanol/formic acid (49%/50%/1%). After 15 min, rinsing solutions were pooled with their corresponding peptide fraction and all fractions were evaporated using a vacuum concentrator. Prior to LC-MS/MS analysis, peptide fractions were desalted using ZipTip (Millipore) according to manufacturer's protocol followed by centrifugation under vacuum.
2.8 Reverse-Phase (RP) LC-MS/MS analysis
Each dried fraction was reconstituted in 12 μl of 5% formic acid and 3 μl of the resulting suspension was injected into a trap column (LC Packings, PepMap C18 pre-column; 5 mm 300 m i.d.; LC Packings) using an Ultimate 3000 HPLC (Dionex Corporation, Sunnyvalle, CA) via an isocratic flow of 0.1% formic acid in water at a rate of 20 μl/min for 3 minutes. Peptides were then eluted onto the PepMap C18 analytical column (15 cm 75 μm i.d.; LC Packings) at a flow rate of 300 nl/min and separated using a linear gradient of 4-80% solvent B over 120 min. The mobile phase consisted of solvent A (0.1% formic acid (aqueous)) and solvent B (0.1% formic acid (aqueous) in 90% acetonitrile). The column eluates were subsequently ionized using the NanoSpray II of a QSTAR Elite instrument (Applied Biosystems) operated in information-dependent acquisition mode, in which a 1-s TOF MS scan from 300-2000 m/z was performed, followed by 2-s product ion scans from 100-2000 m/z on the three most intense doubly or triply charged ions. Analyst 2.0 software was used for data acquisition and analysis.
2.9 Database searching and bioinformatics analysis
A predicted protein database containing transcriptome data for B. siamensis goniomphalos described previously [25] was used for amino acid sequence comparison. The database search was performed using Protein Pilot v4.0.8085 (Applied Biosystems) using the default parameters. Only proteins with a ProteinPilot unused scored above 1.3, which is equivalent to a protein confidence threshold greater than 95%, and for which there was at least one unique peptide match with a confidence >95% were selected. Under these conditions the calculated false discovery rate (FDR) using a reverse decoy database was <1%. The iQuantitator software was used to analyse the differentially expressed proteins in all replicates [31]. This software infers sample-dependent changes in protein expression using Markov Chain Monte Carlo and Bayesian statistical methods. Using iQuantitator, median and 95% confidence intervals were generated for each component peptide and integrating data across replicates. As described previously [31-33], for proteins whose iTRAQ ratios were downregulated in infected snails, the extent of downregulation was considered further if the null value of 1 was above the upper limit of credible interval. Conversely, for proteins whose iTRAQ ratios were upregulated in infected snails, the extent of upregulation was considered further if the lower limit of the credible interval had a value >1. The width of these credible intervals depends on the data available for a given protein. Since the number of peptides observed and the number of spectra used to quantify the change in expression for a given protein are taken into consideration, it is possible to detect small but significant changes in up- or downregulation when many peptides are available. For each protein and each peptide associated with a given protein, the mean, median, and 95% credible intervals were computed for each of the protein and peptide level treatment effects [32, 33]. In addition, only proteins with a fold change of at least 1.5 (log2=0.6) were considered for further analysis [34].
Proteins were classified according to GO categories using the program Blast2Go [35] and pie charts were generated using the second level of the GO hierarchy. Heatmaps representing the differentially expressed proteins in the headfoot and body of infected snails were generated in R using ggplot2 [36] and clustering was performed using Euclidean distances. Protein levels were compared in the heatmaps to gene expression levels obtained in previous studies [13]. The time points where proteins or genes presented no significant regulation are coloured in grey.
3. Results
Samples from the body and headfoot of infected and uninfected Bithynia snails were labeled with iTRAQ and subjected to LC-MS/MS analysis. Two different biological replicates from each sample were analysed and a total of 30,545 and 36,179 MS/MS spectra were acquired in body and headfoot samples, respectively, over all iTRAQ runs. From these, 16,359 and 21,673 spectra were used to assign unique peptides and unique proteins in body and headfoot samples, respectively. An analysis of the differential expression of the identified proteins in both replicates was performed using iQuantitator, which uses two different statistical methods to infer sample-dependent changes in protein expression. The total number of assigned unique peptides and their corresponding unique proteins together with the disallowed modifications and the R2 value of iQuantitator statistical model are reported in Table 1.
Table 1. Summary results from iQuantitator analysis.
The number of supplied, identified and unidentified spectra, together with the number of unique proteins and peptides in all time points from each sample is provided in this table. The model R2 is inferred from a Markov Chain Monte Carlo and a Bayesian statistical method.
| Day 1 | Day 7 | Day 14 | Day 28 | Day 56 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||
| Body | Head | Body | Head | Body | Head | Body | Head | Body | Head | |
|
| ||||||||||
| Supplied spectra | 30,545 | 36,179 | 30,545 | 36,179 | 30,545 | 36,179 | 30,545 | 36,179 | 30,545 | 36,179 |
| Identified spectra | 16,359 | 21,673 | 16,359 | 21,673 | 16,359 | 21,673 | 16,359 | 21,673 | 16,359 | 21,673 |
| Unidentified spectra | 14,186 | 14,506 | 14,186 | 14,506 | 14,186 | 14,506 | 14,186 | 14,506 | 14,186 | 14,506 |
| Disallowed modifications | 247 | 542 | 237 | 546 | 251 | 549 | 237 | 547 | 256 | 545 |
| Unique proteins | 814 | 655 | 800 | 657 | 820 | 656 | 809 | 657 | 824 | 653 |
| Unique peptides | 2,859 | 2,359 | 2,737 | 2,364 | 2,898 | 2,366 | 2,813 | 2,370 | 2,954 | 2,351 |
| Model R2 | 0.807 | 0.939 | 0.787 | 0.939 | 0.764 | 0.938 | 0.67 | 0.943 | 0.924 | 0.935 |
A total number of 945 and 746 different proteins from body and headfoot samples respectively were identified over all time points studied (confidence threshold >95%); of these, 452 proteins were common to both samples (Figure 1a). Of all the proteins identified, only those whose credible interval (from iQuantitator analysis) was above or below 1 and whose log2 fold-change was >0.6 or < -0.6 (for upregulated and downregulated proteins respectively), were considered for further investigation. A total of 108 significantly differentially expressed proteins were found in the body samples, whereas only 43 proteins were differentially expressed in the headfoot of the infected snails (Figure 1b). A comprehensive report was also generated with the iQuantitator software (Supplementary Files 1-4 in [37]).
Figure 1.
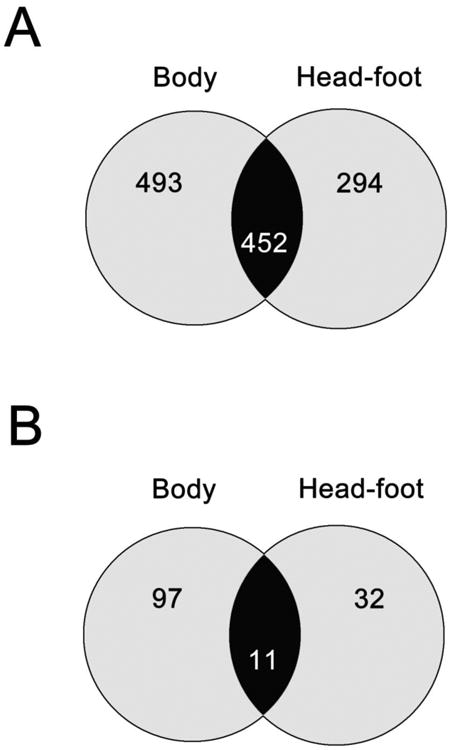
Venn diagram of all the identified (A) and significantly differentially expressed (B) proteins in the body and headfoot of Bithynia siamensis goniomphalos following infection with Opisthorchis viverrini.
A GO-enrichment analysis of significantly differentially expressed proteins from the body and the headfoot of infected snails was performed using Blast2GO [35]. The analysis revealed significant enrichment of the GO terms “binding” (13.2% and 14.4% in body and headfoot, respectively), “catalytic activity” (11.4% and 12.3%) and “protein binding” (8.3% and 9.8%) within “molecular function” (Figure 2a) and “single-organism cellular process” (9.3% and 8%), “regulation of biological process” (8.7% and 8%), “primary metabolic process” (8.1% and 9.3%) and “organic substance metabolic process” (8.1% and 9.3%) within “biological process” (Figure 2b). No significant differences were observed between enriched GO terms from body and headfoot of infected snails.
Figure 2.
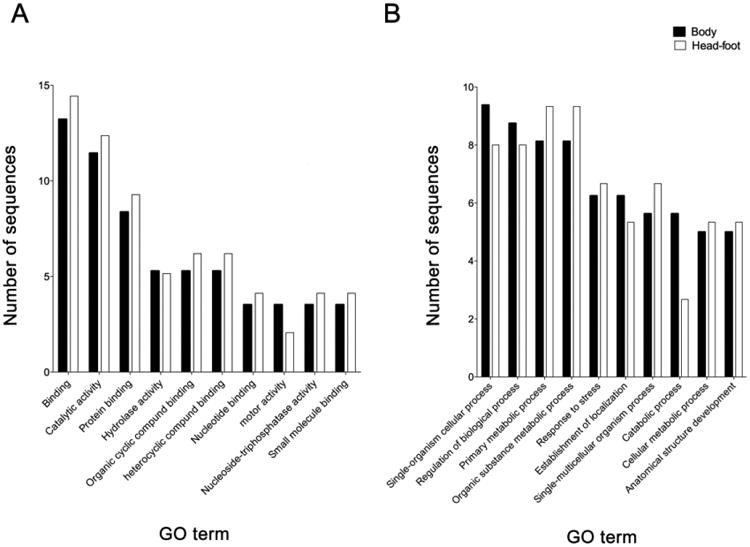
Enriched Gene Ontology (GO) terms assigned to significantly differentially expressed proteins in the body (closed bars) and the headfoot (open bars) of Opisthorchis viverrini-infected Bithynia siamensis goniomphalos snails, according to the categories “molecular function” (A) and “biological process” (B).
Significantly differentially expressed proteins from the headfoot of infected snails were grouped into 8 GO annotation categories and plotted in a clustered heatmap (Figure 3). Clustering was performed using Euclidean distances and dendrograms were reordered based on mean values. Proteins assigned to peptidase activity, and oxidoreductases (with the exception of 15-hydroxyprostaglandin dehydrogenase) together with proteins with a catalytic domain were significantly downregulated after infection with O. viverrini. Conversely, proteins involved in motor activity and structural proteins were upregulated in the headfoot of infected snails among the experiment (Figure 3).
Figure 3.
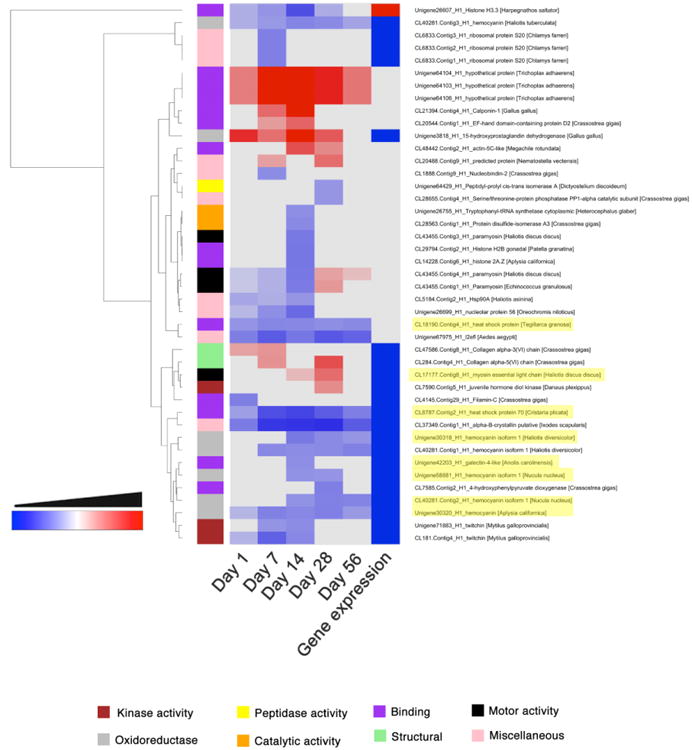
Clustered heatmap of the significantly regulated proteins and genes in the headfoot of Opisthorchis viverrini-infected Bithynia siamensis goniomphalos snails. Proteins were grouped into 8 different categories based on GO annotation and clustering was performed using Euclidean distances. The time points where proteins and genes presented no significant regulation are coloured in grey.
Significantly dysregulated proteins from the bodies of infected snails were grouped into 10 GO annotation categories and plotted in a clustered heatmap (Figure 4). Clustering was also performed using Euclidean distances and dendrograms were reordered based on mean values. The majority of differentially expressed proteins were identified at 28-56 dpi, and similar numbers of up- and downregulated proteins were detected. Proteins with kinase, motor and transporter activities were mostly upregulated (specially at 56 dpi) in the bodies of infected snails, whereas proteins with peptidase hydrolase and oxidoreductase activities were significantly downregulated in the bodies of infected snails (Figure 4).
Figure 4.
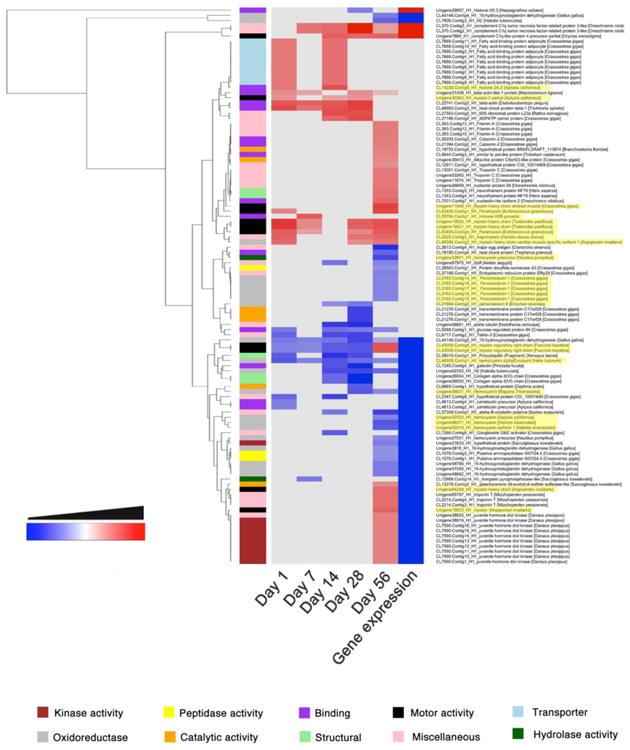
Clustered heatmap of the significantly regulated proteins and genes in the body of Opisthorchis viverrini-infected Bithynia siamensis goniomphalos snails. Proteins were grouped into 10 different categories based on GO annotation and clustering was performed using Euclidean distances. The time points where proteins and genes presented no significant regulation are coloured in grey.
4. Discussion
Despite the public health impact of infections with the carcinogenic liver fluke in Southeast Asia, and the significant advances in knowledge of the molecular and pathobiology of this infection in mammalian hosts [2, 4, 7, 38-40], little is known of the molecular interactions in the Bithynia-Opisthorchis system. We recently reported on the transcriptomic changes induced in snails following O. viverrini infection using next-generation RNA sequencing [13, 25]. In the present study we have monitored, for the first time, the effect that O. viverrini infection has on expression of proteins in the body and the headfoot of B. siamensis goniomphalos, throughout the period in which the parasite is developing within its snail intermediate host. Because of the difficulty in breeding snails in the laboratory, wild snails were collected from the field and checked for parasitic infections. Uninfected snails were infected with O. viverrini eggs and the infection was monitored by cercarial emission and examination of hatched eggs in the snail faeces twice each week over an 8 week period as described previously [9, 30].
Despite the similar number of proteins identified in the body and headfoot of B. siamensis goniomphalos (945 and 746 proteins respectively), the number of significantly differentially expressed proteins following infection by O. viverrini was significantly higher in the body than in the headfoot (108 and 43 proteins respectively). This difference could be associated with the developmental biology of O. viverrini in its intermediate host; indeed, unlike other trematodes such as S. mansoni whose eggs hatch in the water and miracidia actively penetrate the snail, O. viverrini eggs are eaten by B. siamensis goniomphalos and hatch in the snail's digestive system which is located within the gastropod body [41]; thus, it is likely that changes in protein expression in the body may be directly associated with parasite hatching and asexual reproduction, and localized to the immediate vicinity of the parasite [42]. Interestingly, the majority of differential (body) protein expression was observed at 28 dpi and particularly at 56 dpi, whereas no significant pattern of up- or downregulation was observed in the headfoot samples at the same time points. Given that the cercariae exit the snail within 6-8 weeks post infection [41], this observation could be linked to the parasite migration through the digestive glands within the body of the snail.
A GO analysis of differentially expressed proteins in the body and headfoot of B. siamensis goniomphalos following O. viverrini infection displayed an enrichment of proteins involved in “binding” and “catalytic” activities, which is consistent with previous transcriptomic studies [13]. For instance, heat shock proteins (HSPs) and histones, commonly linked to “stress-related responses”, were significantly differentially expressed in infected snails. In particular, expression levels of two different HSPs (HSP-70 and HSP) detected in the headfoot of infected snails were downregulated throughout the experiment. In previous studies, these stress-related proteins were upregulated following parasite infection and hypothesized to have an immunomodulatory role [43, 44]. Furthermore, increased levels of HSP-70 expression were observed in schistosome-susceptible Biomphalaria following experimental infection with S. mansoni [45]. Conversely, other studies have shown that HSPs are downregulated in hemocytes from susceptible and resistant snails infected with S. mansoni [46]. Despite the contradiction surrounding the role and differential expression of HSPs in the literature, our proteomic results from the body of the snail are in accordance with previous transcriptomic studies performed in naturally infected Bithynia, where mRNAs encoding HSPs were among the most highly upregulated in infected Bithynia [13]. Other proteins related to oxidative stress, like histones, were also upregulated in the body of infected Bithynia, supporting our earlier findings of other isoforms of histones at the RNA level [13] and those of others with S. mansoni infected B. glabrata [47, 48]. Despite the unclear role of histones in the response against parasitic infections, it has been speculated that an increase in transcription could trigger chromatin modifications in susceptible snails, contributing to the success of the infection [47].
Proteins functionally linked to motor activities were upregulated in both body and headfoot of infected Bithynia. Myosins were consistently upregulated throughout the study, with the exception of two myosin light chains in the body of infected Bithynia. These light chains are not usually considered “myosins” but regulatory components of the macromolecular complexes, and could not be related to motor activity [49, 50]. Consistent with these findings is the upregulation of actin, tropomyosin and paramyosin observed in the body of infected snails [13]. We recently hypothesized that actin-related gene expression in fluke-infected Bythinia is associated with the migration of circulating hemocytes and promoting phagocytosis and cell trafficking, which could assist in the defense of the snail against pathogens [13]. Moreover, a putative role for tropomyosin in host-parasite molecular mimicry has been suggested based on the unusually high degree of sequence similarity between S. mansoni and B. glabrata tropomyosins [51-53].
Oxidoreductases were also differentially expressed in the body and the headfoot of infected snails. This group of proteins includes all enzymes that catalyze the transfer of electrons from one molecule to another, thus playing a major role in aerobic and anaerobic metabolism. Peroxiredoxins are a family of enzymes playing protective roles against oxidative stress through the neutralization of reactive oxygen and nitrogen species that can damage cellular function. It has been shown previously that the expression levels of a peroxiredoxin from S. mansoni-resistant B. glabrata are increased following infection with S. mansoni; in contrast, expression levels of this enzyme were decreased in susceptible snails [54]. We detected significant downregulation of different Bithynia peroxiredoxins at 56 dpi, which may be related to a defense mechanism from the cercariae transiting through the body tissues to leave the snail. In this sense, the excretory/secretory (ES) products from the parasite could downregulate the expression levels of peroxiredoxins as a self-defense mechanism.
Only a few proteins playing putative roles in immunomodulation were identified as significantly differentially expressed in our experiment. Among these proteins, galectins were downregulated in the body of infected Bithynia. Galectins and C-type lectins are a family of glycan-binding proteins that are usually upregulated in infected snails [48, 55]. The B. glabrata galectin BgGal binds to hemocytes and the tegument of S. mansoni suggesting its role in parasite recognition [56]. Other immunomodulatory proteins include the hemocyanins, which rely on copper for the transport of oxygen throughout the body of gastropods and have been shown to be involved in defense mechanisms in invertebrates [48, 57, 58]. It has been hypothesized that the presence of hemocyanin in iron-containing hemoglobin of gastropods such as B. glabrata could be more related to defense mechanisms than to respiratory function [48]. The lack of key immunomodulatory proteins identified in this study could be related to the limited sensitivity of mass spectrometry instruments; however, many of the proteins of unknown function identified could also be playing an immunomodulatory role. In our previous transcriptomic study, there was a notable paucity of differentially expressed genes encoding immunomodulatory proteins in infected Bithynia [13]. The concordance between proteomic and transcriptomic data lends further credit to the hypothesis that O. viverrini may manipulate the snail by suppressing its immune responses, thus resulting in the inability of the hemocytes to recognize the parasite and/or the suppression of the snail humoral response against parasite invasion. In this sense, the model B. glabrata-Echinostoma spp with susceptible and resistant snails has been shown to be a good model to analyse the influence of parasites on the snail immune response [59-61]. The manipulation of the B. glabrata defense responses by Echinostoma paraensei has been well characterized in previous studies [48, 62]. DeGaffe and Loker [63] showed that susceptibility of B. glabrata to infection with E. paraensei is correlated with the ability of the ES products to interfere with the spreading behaviour of host hemocytes. Furthermore, the ES products of Echinostoma caproni have been shown to inhibit phagocytosis and adhesion mechanisms of susceptible B. glabrata snails [64].
Control and elimination of snail-borne diseases should not rely solely upon anti-parasite chemotherapy [65, 66], and integrated programs should be designed. Recently, a number of authors highlighted the importance of controlling snail-borne parasitic diseases by using integrated approaches aimed at eradicating the parasite from the definitive host (i.e. mass drug administration) as well as disrupting the life cycle in the intermediate host (i.e. use of molluscicides and health education) [24-26, 67]. The present study establishes a baseline for future investigations on host-parasite interactions in the Bithynia-Opisthorchis system aimed at dissecting the molecular mechanisms involved in the transmission of this carcinogenic infection by snails.
5. Conclusions
Our study compares for the first time the differentially expressed proteins in the body and the headfoot of the snail B. siamensis goniomphalos after infection with the liver fluke O. viverrini. In general, more proteins were differentially expressed after infection in the body of the snail, which could be related to the biology of the infection. Most notably, expression of oxidoreductases and catalytic enzymes was downregulated in infected snails, while motor and stress-related proteins were upregulated. This work provides new insights into the study of host-parasite interactions and could serve as a basis for the development of new strategies aimed at controlling parasite transmission.
Supplementary Material
Significance.
Despite the importance and high prevalence of opisthorchiasis in some regions of Southeast Asia and the direct relationship between infection by Opisthorchis viverrini and the incidence of cholangiocarcinoma, little is known of the modifications induced by this parasite in its snail intermediate hosts. This time-course study provides the first in-depth quantitative proteomic analysis of experimentally infected Bithynia siamensis goniomphalos. We show how motor and stress-related proteins are upregulated in infected snails, while O. viverrini infection downregulates the expression of oxidoreductases and catalytic enzymes. This work serves as a basis for the development of new strategies, focused on the invertebrate intermediate hosts, to control parasite transmission.
Highlights.
Comparison of body and headfoot proteins from infected B. siamensis goniomphalos.
More differentially expressed proteins were found in the body of infected snails.
Upregulation of motor and stress-related proteins in infected snails
Downregulation of oxidoreductases and catalytic enzymes in infected snails
The study of snails is important for controlling snail-borne parasitosis
Acknowledgments
This work was supported by project (613669) and program (1037304) grants from the National Health and Medical Research Council of Australia (NHMRC) and a Tropical Medicine Research Collaboration (TMRC) grant from the National Institute of Allergy and Infectious Disease, National Institutes of Health, USA (P50AI098639). AL is supported by a NHMRC principal research fellowship. Funding from Thailand Research Fund through the Royal Golden Jubilee Ph.D. Program (Grant No PHD/0027/2551) to SP and ST is also gratefully acknowledged. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vatanasapt V, Uttaravichien T, Mairiang EO, Pairojkul C, Chartbanchachai W, Haswell-Elkins M. Cholangiocarcinoma in north-east Thailand. Lancet. 1990;335:116–117. doi: 10.1016/0140-6736(90)90591-r. [DOI] [PubMed] [Google Scholar]
- 2.Sripa B, Kaewkes S, Sithithaworn P, Mairiang E, Laha T, Smout M, et al. Liver fluke induces cholangiocarcinoma. PLoS Med. 2007;4:e201. doi: 10.1371/journal.pmed.0040201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sithithaworn P, Andrews RH, Petney TN, Saijuntha W, Laoprom N. The systematics and population genetics of Opisthorchis viverrini sensu lato: implications in parasite epidemiology and bile duct cancer. Parasitol Int. 2012;61:32–37. doi: 10.1016/j.parint.2011.07.020. [DOI] [PubMed] [Google Scholar]
- 4.Smout MJ, Sripa B, Laha T, Mulvenna J, Gasser RB, Young ND, et al. Infection with the carcinogenic human liver fluke, Opisthorchis viverrini. Mol Biosyst. 2011;7:1367–1375. doi: 10.1039/c0mb00295j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sripa B, Bethony JM, Sithithaworn P, Kaewkes S, Mairiang E, Loukas A, et al. Opisthorchiasis and Opisthorchis-associated cholangiocarcinoma in Thailand and Laos. Acta Trop. 2011;120(1):158–168. doi: 10.1016/j.actatropica.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sriamporn S, Pisani P, Pipitgool V, Suwanrungruang K, Kamsa-ard S, Parkin DM. Prevalence of Opisthorchis viverrini infection and incidence of cholangiocarcinoma in Khon Kaen, Northeast Thailand. Trop Med Int Health. 2004;9:588–594. doi: 10.1111/j.1365-3156.2004.01234.x. [DOI] [PubMed] [Google Scholar]
- 7.Sripa B, Brindley PJ, Mulvenna J, Laha T, Smout MJ, Mairiang E, et al. The tumorigenic liver fluke Opisthorchis viverrini-multiple pathways to cancer. Trends Parasitol. 2012;28:395–407. doi: 10.1016/j.pt.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brockelman WY, Upatham ES, Viyanant V, Ardsungnoen S, Chantanawat R. Field studies on the transmission of the human liver fluke, Opisthorchis viverrini, in northeast Thailand: population changes of the snail intermediate host. Int J Parasitol. 1986;16:545–552. doi: 10.1016/0020-7519(86)90091-3. [DOI] [PubMed] [Google Scholar]
- 9.Prasopdee S, Kulsantiwong J, Piratae S, Khampoosa P, Thammasiri C, Suwannatrai A, et al. Temperature dependence of Opisthorchis viverrini infection in first intermediate host snail, Bithynia siamensis goniomphalos. Acta Trop. 2013;13 doi: 10.1016/j.actatropica.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 10.Upatham ES, Viyanant V, Kurathong S, Rojborwonwitaya J, Brockelman WY, Ardsungnoen S, et al. Relationship between prevalence and intensity of Opisthorchis viverrini infection, and clinical symptoms and signs in a rural community in north-east Thailand. Bulletin of the World Health Organization. 1984;62:451–61. [PMC free article] [PubMed] [Google Scholar]
- 11.Vichasri S, Viyanant V, Upatham ES. Opisthorchis viverrini: intensity and rates of infection in cyprinoid fish from an endemic focus in Northeast Thailand. Southeast Asian J Trop Med Public Health. 1982;13:138–141. [PubMed] [Google Scholar]
- 12.Wang YC. Examining landscape determinants of Opisthorchis viverrini transmission. EcoHealth. 2012;9:328–341. doi: 10.1007/s10393-012-0789-z. [DOI] [PubMed] [Google Scholar]
- 13.Prasopdee S, Sotillo J, Tesana S, Laha T, Kulsantiwong J, Nolan MJ, et al. RNA-Seq reveals infection-induced gene expression changes in the snail intermediate host of the carcinogenic liver fluke, Opisthorchis viverrini. PLoS Negl Trop Dis. 2014;8:e2765. doi: 10.1371/journal.pntd.0002765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Jong-Brink M. How schistosomes profit from the stress responses they elicit in their hosts. Adv Parasitol. 1995;35:177–256. doi: 10.1016/s0065-308x(08)60072-x. [DOI] [PubMed] [Google Scholar]
- 15.Vasquez RE, Sullivan JT. Hematopoietic tissue allografts in Biomphalaria glabrata (Mollusca: Pulmonata) induce humoral immunity to Schistosoma mansoni. Dev Comp Immunol. 2001;25:561–564. doi: 10.1016/s0145-305x(01)00022-2. [DOI] [PubMed] [Google Scholar]
- 16.Adema CM, Hertel LA, Miller RD, Loker ES. A family of fibrinogen-related proteins that precipitates parasite-derived molecules is produced by an invertebrate after infection. Proc Natl Acad Sci U S A. 1997;94:8691–8696. doi: 10.1073/pnas.94.16.8691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leonard PM, Adema CM, Zhang SM, Loker ES. Structure of two FREP genes that combine IgSF and fibrinogen domains, with comments on diversity of the FREP gene family in the snail Biomphalaria glabrata. Gene. 2001;269:155–165. doi: 10.1016/s0378-1119(01)00444-9. [DOI] [PubMed] [Google Scholar]
- 18.Zhang SM, Leonard PM, Adema CM, Loker ES. Parasite-responsive IgSF members in the snail Biomphalaria glabrata: characterization of novel genes with tandemly arranged IgSF domains and a fibrinogen domain. Immunogenetics. 2001;53:684–694. doi: 10.1007/s00251-001-0386-8. [DOI] [PubMed] [Google Scholar]
- 19.Zhang SM, Loker ES. Representation of an immune responsive gene family encoding fibrinogen-related proteins in the freshwater mollusc Biomphalaria glabrata, an intermediate host for Schistosoma mansoni. Gene. 2004;341:255–266. doi: 10.1016/j.gene.2004.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schoenberg DA, Cheng TC. Lectin-binding specificities of hemocytes from two strains of Biomphalaria glabrata as determined by microhemadsorption assays. Dev Comp Immunol. 1980;4:617–628. doi: 10.1016/s0145-305x(80)80064-4. [DOI] [PubMed] [Google Scholar]
- 21.Yoshino TP. Surface antigens of Biomphalaria glabrata (Gastropoda) hemocytes: occurrence of membrane-associated hemolymph-like factors antigenically related to snail hemoglobin. J Invertebr Pathol. 1983;41:310–320. doi: 10.1016/0022-2011(83)90248-3. [DOI] [PubMed] [Google Scholar]
- 22.Zelck U, Becker W. Lectin binding to cells of Schistosoma mansoni sporocysts and surrounding Biomphalaria glabrata tissue. J Invertebr Pathol. 1990;55:93–99. doi: 10.1016/0022-2011(90)90037-7. [DOI] [PubMed] [Google Scholar]
- 23.Zelck UE, Becker W, Bayne CJ. The plasma proteins of Biomphalaria glabrata in the presence and absence of Schistosoma mansoni. Dev Comp Immunol. 1995;19:181–194. doi: 10.1016/0145-305x(95)00012-i. [DOI] [PubMed] [Google Scholar]
- 24.Adema CM, Bayne CJ, Bridger JM, Knight M, Loker ES, Yoshino TP, et al. Will all scientists working on snails and the diseases they transmit please stand up? PLoS Negl Trop Dis. 2012;6:e1835. doi: 10.1371/journal.pntd.0001835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cantacessi C, Prasopdee S, Sotillo J, Mulvenna J, Tesana S, Loukas A. Coming out of the shell: building the molecular infrastructure for research on parasite-harbouring snails. PLoS Negl Trop Dis. 2013;7:e2284. doi: 10.1371/journal.pntd.0002284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gray DJ, McManus DP, Li Y, Williams GM, Bergquist R, Ross AG. Schistosomiasis elimination: lessons from the past guide the future. Lancet Infect Dis. 2010;10:733–736. doi: 10.1016/S1473-3099(10)70099-2. [DOI] [PubMed] [Google Scholar]
- 27.Sumethanurungkul P. MSc Thesis. Mahidol University; Bangkok, Thailand: 1970. Studies on physical effects on snail intermediate hosts of a liver fluke (Opisthorchis viverrini) [Google Scholar]
- 28.Khampoosa P, Jones MK, Lovas EM, Srisawangwong T, Laha T, Piratae S, et al. Light and electron microscopy observations of embryogenesis and egg development in the human liver fluke, Opisthorchis viverrini (Platyhelminthes, Digenea) Parasitol Res. 2012;110:799–808. doi: 10.1007/s00436-011-2557-3. [DOI] [PubMed] [Google Scholar]
- 29.Chanawong A, Waikagul J. Laboratory studies on host-parasite relationship of Bithynia snails and the liver fluke, Opisthorchis viverrini. Southeast Asian J Trop Med Public Health. 1991;22:235–239. [PubMed] [Google Scholar]
- 30.Kulsantiwong J, Prasopdee S, Ruangsittichai J, Ruangjirachuporn W, Boonmars T, Viyanant V, et al. DNA barcode identification of freshwater snails in the family Bithyniidae from Thailand. PloS One. 2013;8:e79144. doi: 10.1371/journal.pone.0079144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwacke JH, Hill EG, Krug EL, Comte-Walters S, Schey KL. iQuantitator: a tool for protein expression inference using iTRAQ. BMC Bioinformatics. 2009;10:342. doi: 10.1186/1471-2105-10-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Besson D, Pavageau AH, Valo I, Bourreau A, Belanger A, Eymerit-Morin C, et al. A quantitative proteomic approach of the different stages of colorectal cancer establishes OLFM4 as a new nonmetastatic tumor marker. Mol Cell Proteomics. 2011;10:M111 009712. doi: 10.1074/mcp.M111.009712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grant JE, Bradshaw AD, Schwacke JH, Baicu CF, Zile MR, Schey KL. Quantification of protein expression changes in the aging left ventricle of Rattus norvegicus. J Proteome Res. 2009;8:4252–4263. doi: 10.1021/pr900297f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kambiranda D, Katam R, Basha SM, Siebert S. iTRAQ-based quantitative proteomics of developing and ripening muscadine grape berry. J Proteome Res. 2014;13:555–569. doi: 10.1021/pr400731p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–6. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 36.Ginestet C. ggplot2: Elegant Graphics for Data Analysis. J R Stat Soc a Stat. 2011;174:245. [Google Scholar]
- 37.Prasopdee S, Tesana S, Cantacessi C, Laha T, Mulvenna J, Grams R, et al. Data set from the proteomic analysis of Bithynia siamensis goniomphalos snails upon infection with the carcinogenic liver-fluke Opisthorchis viverrini. Data in Brief. 2014 doi: 10.1016/j.dib.2014.09.005. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jex AR, Young ND, Sripa J, Hall RS, Scheerlinck JP, Laha T, et al. Molecular changes in Opisthorchis viverrini (Southeast Asian liver fluke) during the transition from the juvenile to the adult stage. PLoS Negl Trop Dis. 2012;6:e1916. doi: 10.1371/journal.pntd.0001916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Young ND, Campbell BE, Hall RS, Jex AR, Cantacessi C, Laha T, et al. Unlocking the transcriptomes of two carcinogenic parasites, Clonorchis sinensis and Opisthorchis viverrini. PLoS Negl Trop Dis. 2010;4:e719. doi: 10.1371/journal.pntd.0000719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Young ND, Nagarajan N, Lin SJ, Korhonen PK, Jex AR, Hall RS, et al. The Opisthorchis viverrini genome provides insights into life in the bile duct. Nature Commun. 2014;5:4378. doi: 10.1038/ncomms5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaewkes S. Taxonomy and biology of liver flukes. Acta Trop. 2003;88:177–86. doi: 10.1016/j.actatropica.2003.05.001. [DOI] [PubMed] [Google Scholar]
- 42.Lie KJ, Jeong KH, Heyneman D. Further characterization of acquired resistance in Biomphalaria glabrata. J Parasitol. 1982;68:529–531. [PubMed] [Google Scholar]
- 43.Oladiran A, Belosevic M. Trypanosoma carassii hsp70 increases expression of inflammatory cytokines and chemokines in macrophages of the goldfish (Carassius auratus L.) Dev Comp Immunol. 2009;33:1128–1136. doi: 10.1016/j.dci.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 44.Wang Z, Wu Z, Jian J, Lu Y. Cloning and expression of heat shock protein 70 gene in the haemocytes of pearl oyster (Pinctada fucata, Gould 1850) responding to bacterial challenge. Fish Shellfish Immunol. 2009;26:639–645. doi: 10.1016/j.fsi.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 45.Ittiprasert W, Miller A, Myers J, Nene V, El-Sayed NM, Knight M. Identification of immediate response genes dominantly expressed in juvenile resistant and susceptible Biomphalaria glabrata snails upon exposure to Schistosoma mansoni. Mol Biochem Parasitol. 2010;169:27–39. doi: 10.1016/j.molbiopara.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zahoor Z, Davies AJ, Kirk RS, Rollinson D, Walker AJ. Larval excretory-secretory products from the parasite Schistosoma mansoni modulate HSP70 protein expression in defence cells of its snail host, Biomphalaria glabrata. Cell Stress Chaperones. 2010;15:639–650. doi: 10.1007/s12192-010-0176-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bouchut A, Sautiere PE, Coustau C, Mitta G. Compatibility in the Biomphalaria glabrata/Echinostoma caproni model: Potential involvement of proteins from hemocytes revealed by a proteomic approach. Acta Trop. 2006;98:234–246. doi: 10.1016/j.actatropica.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 48.Hanington PC, Lun CM, Adema CM, Loker ES. Time series analysis of the transcriptional responses of Biomphalaria glabrata throughout the course of intramolluscan development of Schistosoma mansoni and Echinostoma paraensei. Int J Parasitol. 2010;40:819–831. doi: 10.1016/j.ijpara.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hooper SL, Thuma JB. Invertebrate muscles: muscle specific genes and proteins. Physiol Rev. 2005;85:1001–1060. doi: 10.1152/physrev.00019.2004. [DOI] [PubMed] [Google Scholar]
- 50.Trybus KM. Role of myosin light chains. J Muscle Res Cell Motil. 1994;15:587–594. doi: 10.1007/BF00121066. [DOI] [PubMed] [Google Scholar]
- 51.Damian RT. Molecular mimicry: parasite evasion and host defense. Curr Top Microbiol Immunol. 1989;415:101–115. doi: 10.1007/978-3-642-74594-2_9. [DOI] [PubMed] [Google Scholar]
- 52.Weston D, Allen B, Thakur A, LoVerde PT, Kemp WM. Invertebrate host-parasite relationships: convergent evolution of a tropomyosin epitope between Schistosoma sp., Fasciola hepatica, and certain pulmonate snails. Exp parasitol. 1994;78:269–278. doi: 10.1006/expr.1994.1028. [DOI] [PubMed] [Google Scholar]
- 53.Weston DS, Kemp WM. Schistosoma mansoni: comparison of cloned tropomyosin antigens shared between adult parasites and Biomphalaria glabrata. Exp Parasitol. 1993;76:358–370. doi: 10.1006/expr.1993.1044. [DOI] [PubMed] [Google Scholar]
- 54.Knight M, Raghavan N, Goodall C, Cousin C, Ittiprasert W, Sayed A, et al. Biomphalaria glabrata peroxiredoxin: effect of Schistosoma mansoni infection on differential gene regulation. Mol Biochem Parasitol. 2009;167:20–31. doi: 10.1016/j.molbiopara.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guillou F, Mitta G, Galinier R, Coustau C. Identification and expression of gene transcripts generated during an anti-parasitic response in Biomphalaria glabrata. Dev Comp Immunol. 2007;31:657–671. doi: 10.1016/j.dci.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 56.Yoshino TP, Dinguirard N, Kunert J, Hokke CH. Molecular and functional characterization of a tandem-repeat galectin from the freshwater snail Biomphalaria glabrata, intermediate host of the human blood fluke Schistosoma mansoni. Gene. 2008;411:46–58. doi: 10.1016/j.gene.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo D, Zhang Y, Zeng D, Wang H, Li X, Li Y, et al. Functional properties of hemocyanin from Oncomelania hupensis, the intermediate host of Schistosoma japonicum. Exp Parasitol. 2009;123:277–281. doi: 10.1016/j.exppara.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Y, Yan F, Hu Z, Zhao X, Min S, Du Z, et al. Hemocyanin from shrimp Litopenaeus vannamei shows hemolytic activity. Fish Shellfish Immunol. 2009;27:330–335. doi: 10.1016/j.fsi.2009.05.017. [DOI] [PubMed] [Google Scholar]
- 59.Ataev GL, Coustau C. Cellular response to Echinostoma caproni infection in Biomphalaria glabrata strains selected for susceptibility/resistance. Dev Comp Immunol. 1999;23:187–98. doi: 10.1016/s0145-305x(99)00023-3. [DOI] [PubMed] [Google Scholar]
- 60.Coustau C, Gourbal B, Mitta G, Adema CM. Echinostomes and snails: Exploring complex interactions. In: Fried B, Toledo R, editors. The Biology of Echinostomes. New York: Springer; 2009. pp. 35–59. [Google Scholar]
- 61.Toledo R, Munoz-Antoli C, Fried B. The use of echinostomes to study host-parasite relationships between larval trematodes and invertebrate and cold-blooded vertebrate hosts. Parasitol Res. 2007;100:1177–1785. doi: 10.1007/s00436-007-0470-6. [DOI] [PubMed] [Google Scholar]
- 62.Loker ES, Bayne CJ, Yui MA. Echinostoma paraensei: hemocytes of Biomphalaria glabrata as targets of echinostome mediated interference with host snail resistance to Schistosoma mansoni. Exp parasitol. 1986;62:149–154. doi: 10.1016/0014-4894(86)90018-4. [DOI] [PubMed] [Google Scholar]
- 63.DeGaffe G, Loker ES. Susceptibility of Biomphalaria glabrata to infection with Echinostoma paraensei: correlation with the effect of parasite secretory-excretory products on host hemocyte spreading. J Invertebr Pathol. 1998;71:64–72. doi: 10.1006/jipa.1997.4710. [DOI] [PubMed] [Google Scholar]
- 64.Humbert E, Coustau C. Refractoriness of host haemocytes to parasite immunosuppressive factors as a putative resistance mechanism in the Biomphalaria glabrata-Echinostoma caproni system. Parasitology. 2001;122:651–660. doi: 10.1017/s003118200100782x. [DOI] [PubMed] [Google Scholar]
- 65.Doenhoff MJ, Hagan P, Cioli D, Southgate V, Pica-Mattoccia L, Botros S, et al. Praziquantel: its use in control of schistosomiasis in sub-Saharan Africa and current research needs. Parasitology. 2009;136:825–835. doi: 10.1017/S0031182009000493. [DOI] [PubMed] [Google Scholar]
- 66.Geerts S, Coles GC, Gryseels B. Anthelmintic resistance in human helminths: Learning from the problems with worm control in livestock. Parasitol Today. 1997;13(56):149–151. doi: 10.1016/s0169-4758(97)01024-7. [DOI] [PubMed] [Google Scholar]
- 67.Tesana S, Thapsripair P, Thammasiri C, Prasopdee S, Suwannatrai A, Harauy S, et al. Effects of Bayluscide on Bithynia siamensis goniomphalos, the first intermediate host of the human liver fluke, Opisthorchis viverrini, in laboratory and field trials. Parasitol Int. 2012;61:52–55. doi: 10.1016/j.parint.2011.08.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


