Abstract
It is well known that atmospheric concentrations of carbon dioxide (CO2) (and other greenhouse gases) have increased markedly as a result of human activity since the industrial revolution. It is perhaps less appreciated that natural and managed soils are an important source and sink for atmospheric CO2 and that, primarily as a result of the activities of soil microorganisms, there is a soil-derived respiratory flux of CO2 to the atmosphere that overshadows by tenfold the annual CO2 flux from fossil fuel emissions. Therefore small changes in the soil carbon cycle could have large impacts on atmospheric CO2 concentrations. Here we discuss the role of soil microbes in the global carbon cycle and review the main methods that have been used to identify the microorganisms responsible for the processing of plant photosynthetic carbon inputs to soil. We discuss whether application of these techniques can provide the information required to underpin the management of agro-ecosystems for carbon sequestration and increased agricultural sustainability. We conclude that, although crucial in enabling the identification of plant-derived carbon-utilising microbes, current technologies lack the high-throughput ability to quantitatively apportion carbon use by phylogentic groups and its use efficiency and destination within the microbial metabolome. It is this information that is required to inform rational manipulation of the plant–soil system to favour organisms or physiologies most important for promoting soil carbon storage in agricultural soil.
Keywords: carbon cycling, rhizosphere carbon flow, decomposition, soil microbial respiration, climate change, methods, carbon tracking, agro-ecosystem management
THE SOIL CARBON CYCLE AND MICROBIAL DECOMPOSERS: FUNDAMENTAL PRINCIPLES
All living organisms depend on the supply of necessary elements from the Earth. Since the Earth is a closed system with a finite supply of essential elements such as hydrogen (H), oxygen (O), carbon (C), nitrogen (N), sulfur (S) and phosphorus (P), recycling of these elements is fundamental to avoid exhaustion. Microbes are critical in the process of breaking down and transforming dead organic material into forms that can be reused by other organisms. This is why the microbial enzyme systems involved are viewed as key ‘engines’ that drive the Earth's biogeochemical cycles.1
The terrestrial carbon cycle is dominated by the balance between photosynthesis and respiration.2 Carbon is transferred from the atmosphere to soil via ‘carbon-fixing’ autotrophic organisms, mainly photosynthesising plants and also photo- and chemoautotrophic microbes,3,4 that synthesise atmospheric carbon dioxide (CO2) into organic material (Fig. 1). Fixed carbon is then returned to the atmosphere by a variety of different pathways that account for the respiration of both autotrophic and heterotrophic organisms4 (Fig. 1). The reverse route includes decomposition of organic material by ‘organic carbon-consuming’ heterotrophic microorganisms that utilise the carbon of either plant, animal or microbial origin as a substrate for metabolism, retaining some carbon in their biomass and releasing the rest as metabolites or as CO2 back to the atmosphere (Figs 1 and 2).5 Globally, most soils are unsaturated and oxic, so CO2 is the main respiration flux. In waterlogged anoxic soils such as rice paddies and peatlands, CO2 is reduced by hydrogenotrophic archaea in methanogenesis,3,6 with the net flux of the methane produced dependent on the relative activity of methanogens (including those fermenting acetate) versus the activity of aerobic methane-oxidising bacteria7–9 (methanotrophs) residing in the surface, oxic layers of soil of such wetland systems and also probably the microbial anaerobic oxidation of methane in anoxic layers.10
Figure 1.
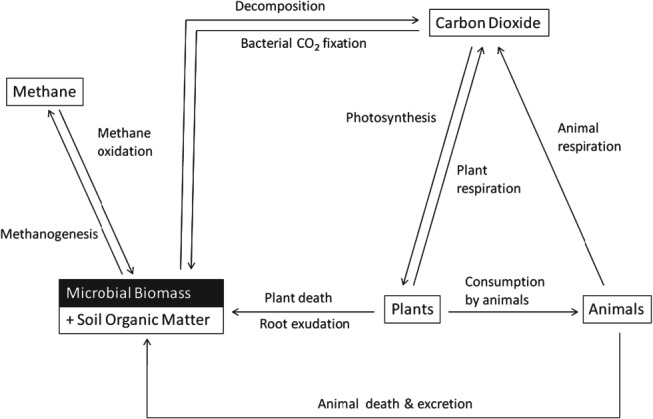
The terrestrial carbon cycle with the major processes mediated by soil microorganisms (adapted from Prosser125).
Figure 2.
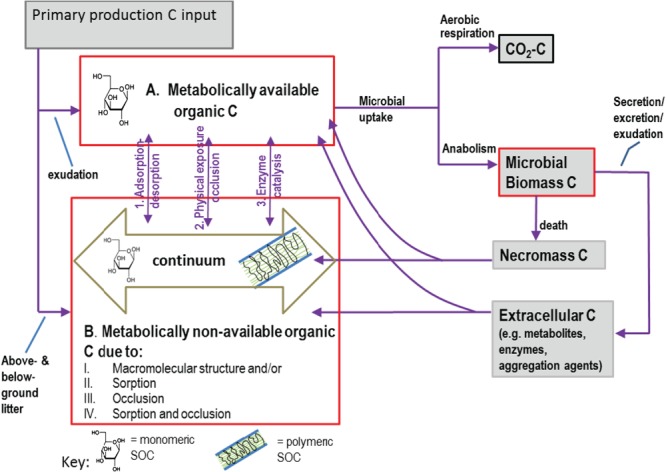
Fate of primary production inputs to soil. Plant-derived organic carbon (after appropriate extracellular depolymerisation) is processed by soil microorganisms to CO2, microbial biomass and extracellular substances. Microbial necromass and metabolites are the precursors for stable soil organic matter, while extracellular microbial carbon may also influence the stability of soil organic carbon (SOC). Enzymes may catalyse the depolymerisation of soil macromolecular constituents, while other extracellular substances may promote aggregation and the physical protection of SOC. SOC (red boxes) is depicted as a continuum of structures derived from the progressive decomposition of litter and exudates and includes the microbial biomass carbon. Dissolved and exposed organic carbon (A) is available for microbial cellular uptake and metabolism (catabolism + anabolism) to produce CO2 and new biomass respectively. Macromolecular or sorbed or occluded SOC is metabolically non-available (B) but may become available via enzymatic depolymerisation, desorption or exposure (I–III respectively), assuming adequate water, electron acceptors, heat, pH and nutrients for microbial activity.
Soil microbes essentially transfer carbon between environmental compartments to fulfil their fundamental goal: survival through reproduction. Thus, microbes utilise different organic and inorganic forms of carbon as carbon and energy sources. However, the C cycle does not operate independently; it is closely coupled with that of other essential elements for microbial metabolism. This linkage occurs either via the use of the other elements as electron donors and acceptors (e.g. N species ranging from the most reduced, 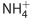 , to the most oxidised,
, to the most oxidised, 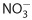 ) in energy transduction or via their immobilisation and mineralisation as part of multiple essential element-containing biomolecules (e.g. proteins, DNA). Hence the availability of other key elements essential for life, particularly N and P, and other environmental factors such as pH, soil texture and mineralogy, temperature and soil water content control the rate at which microbes consume and respire carbon.11 It is these interactions between environmental conditions and biological processes, including primary production, that are chief in controlling the unequal distribution of organic matter across the world's soils.12 The largest global concentration of carbon can be found in wet and cool areas in the northern hemisphere, dominated by deep accumulations of peat and permafrost soils,13 whereas soils with the lowest carbon content tend to be those of desert biomes,14 where low mean annual precipitation limits primary production and encourages the prevalence of aerobic soil conditions.
) in energy transduction or via their immobilisation and mineralisation as part of multiple essential element-containing biomolecules (e.g. proteins, DNA). Hence the availability of other key elements essential for life, particularly N and P, and other environmental factors such as pH, soil texture and mineralogy, temperature and soil water content control the rate at which microbes consume and respire carbon.11 It is these interactions between environmental conditions and biological processes, including primary production, that are chief in controlling the unequal distribution of organic matter across the world's soils.12 The largest global concentration of carbon can be found in wet and cool areas in the northern hemisphere, dominated by deep accumulations of peat and permafrost soils,13 whereas soils with the lowest carbon content tend to be those of desert biomes,14 where low mean annual precipitation limits primary production and encourages the prevalence of aerobic soil conditions.
The objectives of this review are (i) to outline the significance of soil microbial communities to global environmental issues of soil organic matter persistence and climate change through carbon cycle feedbacks, (ii) to briefly describe the main techniques that can be used to apportion below-ground utilisation of plant-derived carbon to specific microbial groups and (iii) to discuss whether application of these techniques can provide the information required to underpin the management of agro-ecosystems for carbon sequestration and increased agricultural sustainability.
THE SOIL CARBON CYCLE AND MICROBIAL DECOMPOSERS: SIGNIFICANCE AND ENVIRONMENTAL IMPLICATIONS
Relationship between soil and atmospheric carbon pools
Estimates suggest that global soil organic carbon stocks are equivalent to at least three times the amount of carbon stored in the atmosphere (Table1). About 8% of the total atmospheric carbon pool is exchanged annually between terrestrial ecosystems and the atmosphere via net primary production and terrestrial heterotrophic (predominantly microbial) respiration (Table1). In other words, if soil (microbial) respiration ceased, it would only take about 12 years of primary production at current rates to exhaust atmospheric CO2 stocks (if all other components of the carbon cycle are ignored, e.g. oceanic CO2 exchange).15
Table 1.
Estimates of the magnitude of soil carbon pools in relation to the atmospheric carbon pool and annual fluxes
| Carbon (Gt or Gt year−1) | |
|---|---|
| Pool | |
| Global soil organic carbon (0–300 cm depth) | 2344a |
| Northern circumpolar permafrost region soil organic carbon (0–300 cm) | 1024b |
| Cropland soil organic carbon (0–300 cm) | 248a |
| CO2-C in atmosphere | 762c |
| Annual flux | |
| Net primary production (photo- and chemosynthesis minus autotrophic respiration) | 60d |
| Terrestrial heterotrophic respiration | 55d |
| Anthropogenic CO2-C (fossil, cement, land-use change) | 8c |
Jobbágy and Jackson (2000).14
Tarnocai et al. (2009)13 – a new estimate suggesting significantly more organic carbon in this northern latitude region than reported in previous analysis, e.g. tundra 144 Gt and boreal 150 Gt, by Jobbágy and Jackson (2000).14
Solomon et al. (2007)126 – estimated for the 1990s.
Prentice et al. (2001)2 – estimated for the 1980s.
At present, terrestrial ecosystems fix, globally, more atmospheric CO2 by photosynthesis than they return to the atmosphere through respiration, which includes removing around 25% of global fossil fuel emissions annually.16 However, net carbon sequestration varies between locations and is significantly affected by land management. Estimates suggest that 42–78 Gt of carbon have been lost from the world's degraded and agricultural soils owing to human activity in both pre- and post-industrial times (Fig. 3), and land remediation to ‘restore’ some of this lost carbon could make a significant contribution to offsetting fossil fuel emissions.17
Figure 3.

Cereal crop production, Warwickshire, UK (image courtesy of JM Clark). Soils are often seen as simply ‘growing media’ for food crops; however, functioning of microbiological communities within soils not only influences the supply of essential plant nutrients but also has great implications for the global carbon cycle and climate system that determines whether environmental conditions are favourable for crop growth.
Microbial decomposition of plant-derived carbon and persistence of soil organic matter
There are two main routes of input for plant organic carbon to the soil system: (i) above-ground plant litter and its leachates, i.e. dissolved organic carbon washed into the soil from plant material by infiltrating rainfall, and (ii) below-ground root litter and exudation, collectively known as rhizodeposition. The relative magnitude of the various inputs from above and below ground will depend upon plant species and, in soils under agriculture, crop management. Rhizodeposition consists of a continuous flow of carbon-containing compounds from roots to soil. Simple molecules such as sugars, amino acids, sugar alcohols, organic acids and more structurally complex secondary metabolites are among the chemical groups that make up the plethora of root exudates18 that can be rapidly (hours to days) respired following their deposition to soil.19 By contrast, polymers such as lignin, cellulose and hemicellulose (the typical structural constituents of plant cells20) require depolymerisation by extracellular enzymes before they can be taken into the microbial cell and metabolised (Fig. 2).21 Of particular note in soil carbon cycling is the role of the mycorrhizal fungi. These range from obligate symbionts that can only obtain carbon from the host plant, i.e. the arbuscular mycorrhizal fungi (AMF), to facultative symbionts that can also mineralise organic carbon, e.g. the ectomycorrhizal fungi (ECM). The AMF symbiosis is found in about 85% of all plant families (typically herbaceous, including many crop species, but also woody),22 and experimental evidence suggests that up to 20%23–25 or even 30%26 of total carbon assimilated by plants may be transferred to the fungal partner, with the symbiosis having profound effects on rhizodeposition.27 A proportion of the plant carbon that is transferred to the mycelia is very quickly respired back to the atmosphere, and this represents a short-circuit of the soil carbon cycle.23,25,26,28,29
Soil organic matter (SOM) consists of the continuum from fresh to progressively decomposing plant, microbial and faunal-derived debris and exudates, including the microbial biomass that is responsible for the primary decomposition of the exudate and detrital inputs (Fig. 2).30 Traditionally, this continuum has been divided into a series of pools with varying decomposition kinetics, ranging from ‘active’ pools that turn over in months to ‘passive’ pools that turn over in thousands of years. In addition to containing fire-derived ‘black carbon’, the ‘passive’ pool has long been thought to be composed of constituents that get their resistance to decomposition from their humified nature, with the formation of the humified substances resulting from spontaneous condensation reactions between reactive microbial products and biochemically altered structural biomolecules.31 However, recent evidence suggests that environmental and biological factors may exert a far greater control on the long-term persistence of SOM than the molecular structure of plant litter inputs and subsequent formation of humus, forcing a re-evaluation of the concept of ‘recalcitrant’ soil humic substances that underpins predictive models of carbon turnover.12,30 Direct, in situ observations have not been able to verify the existence of humic macromolecules in soil, suggesting that the extraction of humic substances from soil may be an artefact of the method used to extract them.32 Instead, it is suggested that SOM consists of partially decomposed litter and a significant proportion of microbial necromass (i.e. dead biomass residues)33 and that SOM persists or is ‘passive’ owing to its physical disconnection from, or inaccessibility to, the extracellular enzymes, microorganisms and the optimal environmental conditions (e.g. electron acceptors, water, inorganic nutrients) needed for decomposition as a result of entrapment within soil aggregates and/or sorption to soil mineral phases. Therefore, in addition to their role in the breakdown and release of CO2 from organic matter, soil microbes contribute to the formation of persistent SOM via their necromass. Additionally, the activities of soil microorganisms may contribute to the stabilisation of soil organic carbon through promotion of the formation of microaggregates, within which SOM may be physically protected from decomposition.34,35 In particular, glomalin, a glycoprotein produced by the hyphae of AMF, has received much attention with respect to its suggested role in the stabilisation of microaggregates.36,37 Similarly, other hydrophobic proteins produced by mycorrhizal fungi and filamentous bacteria, such as hydrophobins and chaplins, have been associated with microaggregate formation and stabilisation.36,37
Soil carbon cycle, microbial decomposers and climate change
As mentioned, humans have heavily perturbed the carbon cycle during the industrial period through inputs of CO2 to the atmosphere, mainly via combustion of fossil fuel and conversion of natural ecosystems to agricultural land (Table1).2,38 The consequences of human actions for the global climate are still uncertain, partly owing to our limited understanding about soil respiration and its representation in Earth system models.38–40 Microbial contributions to climate change through carbon cycle feedbacks are far from straightforward, complicated by direct and indirect effects and interactions with other factors41 (also reviewed in Bardgett et al.42 and Singh et al.43).
An example of a simplified direct positive feedback to global warming is that microbial activity, and therefore organic carbon decomposition and CO2 released by respiration, may be accelerated in response to an increase in temperature.11,44–50 Analyses of field observations made across the globe point to a link between increased respiration flux from land and increased temperatures.51
An example of an indirect positive feedback to elevated CO2 is a consequence of the carbon fertilisation of primary (photosynthetic) production, whereby increased atmospheric CO2 stimulates photosynthesis52,53 and the release of root exudates, which in turn means more labile carbon available for microbial decomposition and respiration.53–57 Moreover, increased root deposition of easily available exudates may ‘prime’ the turnover of less available SOM constituents that otherwise would not be subject to decomposition.58–63
Understanding how the balance between terrestrial ecosystem sinks (i.e. photosynthesis) and sources (i.e. respiration, including microbial respiration) of atmospheric CO2 will be affected in an elevated CO2 world is still one of the main uncertainties in understanding the coupled carbon–climate system.4,39,41,47,57 The uncertainty increases when soil nitrogen and its availability to plants are taken into consideration, with contradicting indications.64 Some modelling studies have suggested that there may be a point during this century where terrestrial ecosystems shift from a net sink to a net source of atmospheric CO2,45,54,65 possibly reflecting the scenarios of increased microbial respiration;45 however, these models are still in an early stage of development.55 Questions still remain about the actual temperature sensitivity of soil (microbial) respiration and how this sensitivity is modified by other environmental factors such as changes in soil moisture during droughts and nutrient limitations and physical protection of organic matter in aggregates or by sorption.4,11,30,66,67 This problem is exacerbated by the diversity of soil ecosystems across the world, which vary in their function owing to differences in their forming factors: parent material, topography, climate, organisms and time. Particular concerns have been raised about peatlands and permafrost soils, where climatic conditions that cause the accumulation or preservation of organic material may not be favourable under a future climate, resulting in the release of significant quantities of carbon to the atmosphere.11,39 Further research in this area is an urgent priority if we are to be able to predict impacts and feedbacks between climate change and the global carbon cycle.39,67
METHODOLOGIES FOR TRACKING CARBON FLOW BELOW GROUND TO MICROBIAL GROUPS
Despite the important role of soil microbes in the carbon cycle and the environmental implications of carbon cycle–climate change feedbacks, most carbon cycle models treat the soil microbial biomass as a black box.68 The majority of models calculate soil respiration using first-order kinetics, where decomposition (and CO2 flux) is proportional to the size of the carbon pool defined by an empirically derived decomposition rate constant that captures the net effect of microbial activity under specific conditions.60,69 Pools typically represent ‘slow’ and ‘passive’ organic matter fractions; and models differ in terms of the number of conceptual pools used (one to nine) and addition of rate modifiers that account for changes in temperature, moisture, etc. (see e.g. Freidlingstein et al.55). Recent studies have called for modifications to these traditional ‘black box’ SOM models to include more explicit representations of microbial community and functions that control decomposition. However, much of this detail is absent because the basic processes are still poorly understood.11
In empirical microbial ecology we are now beginning to be able to unpack the microbial biomass ‘black box’ and to identify which microbial groups are responsible for the turnover of plant-derived inputs to soil. The methods available to do this generally involve the combination of either stable (SIP) or radioactive (RIP) isotope probing and molecular ecology techniques (for a thorough review on methodologies for linking microbial identity to environmental processes, see Gutierrez-Zamora and Manefield70). In SIP or RIP the stable or radioactive isotope (13C or 14C respectively) is used to track the microbial fate of a labelled carbon source (or sources). As a result of anabolic processes, the carbon label becomes incorporated into the biomolecules of those microbes actively decomposing the carbon source of interest. In nucleic acid-SIP the resulting ‘heavier’ 13C DNA or RNA is fractionated by isopycnic density gradient ultracentrifugation from unlabelled nucleic acids and used as a template for downstream analysis. When 14C is used as the label, RIP can be detected with microautoradiography (MAR) and combined with fluorescence in situ hybridisation (FISH), termed FISH-MAR, to enable allocation of carbon utilisation to specific microbial ribotypes with the use of fluorescent rRNA-targeted oligonucleotide probes. The use of an autoradiographic emulsion causes silver grains to form in areas immediately adjacent to radioactive cells (proof of 14C substrate incorporation into the microbial biomass), whereas cell fluorescence after exposure of slides to FISH probes under hybridisation conditions is used for the phylogenetic identification.
Nucleic acid-SIP was first applied over a decade ago, and the majority of studies have focused on non-planted systems. Methanol-utilising microorganisms in soil were studied first with DNA-SIP,71 and subsequent studies have built on this work and used DNA-SIP to link function and identity with respect to the cycling of methane6,7,72,73 and the biodegradation of organic pollutants.74–77
Phenol-utilising microorganisms in wastewater treatment plants were first studied with rRNA-SIP.78 Since then, RNA-SIP has successfully been used in numerous studies to track the carbon utilisation in specific microbial groups in diverse ecosystems (e.g. rivers, tidal flats, aquifers, groundwater) with respect to carbon biogeochemical processes such as methanotrophy,9,72,79 degradation of xenobiotics80–84 and other ecosystem functions.79,84,85
The application of nucleic acid-SIP to trace, in situ, the microbial fate of rhizodeposit carbon in soil involves the growth of plants in a 13C-CO2 atmosphere to promote 13C labelling of photosynthate and therefore rhizodeposition. For successful nucleic acid-SIP, a high proportion of 13C-labelled substrate incorporation is required in order to achieve sufficient separation of ‘heavy’ and ‘light’ nucleic acids. With the exception of the use of DNA-SIP to study endophytes that are, by definition, in intimate association with plant root systems,86 this level of labelling is sometimes difficult to achieve when tracking the microbial fate of plant-derived carbon in the rhizosphere because of dilution with plant 12C and the native 12C SOM.87 This sensitivity problem was demonstrated in a study88 where, although the labelling of bacterial nucleic acids with 13C following 13CO2 incubation of grassland turfs occurred, the amount of labelling was indeed too low, preventing the separation of 13C- from 12C-nucleic acids (for reviews on methodological considerations, see Manefield et al.87 and Neufeld et al.89). As rRNA is turned over independently of cell replication with a high copy number within the microbial cell, rRNA-SIP has a greater sensitivity than DNA-SIP (reviewed in Whiteley et al.90) and has therefore been used more successfully to directly track plant-derived carbon to microorganisms in the rhizosphere.3,26,29,91–93 The promise of mRNA-SIP has also been recently explored for understanding the links between root exudation and bacterial gene expression in the rhizosphere.94 The SIP approach has been complemented and extended to track plant-derived carbon into biochemical markers other than nucleic acids, such as proteins (protein-SIP;95 for reviews, see Seifert et al.96 and von Bergen et al.97) and phospolipid fatty acids (PLFA-SIP).98–100 Drigo et al.,26 Hannula et al.92 and Dias et al. (2013)101 have recently combined RNA-SIP, neutral lipid fatty acid (NLFA)-SIP with neutral (NLFA) and phospholipid (PLFA) lipid fatty acids biomarker analyses and/or PLFA-SIP with real-time polymerase chain reaction (PCR) and community fingerprinting techniques to examine how elevated CO2 or plant genetic modification alters the destination of photosynthetically fixed carbon with respect to its utilisation by AMF and mycorrhizosphere bacterial and fungal species.
FISH-MAR was first demonstrated in 1999 by two separate research groups that managed to visualise the incorporation of 14C-labelled substrates in probe-detected bacteria under the microscope.102,103 Since then, it has been used mainly to study in situ physiology of bacteria in biofilms104 and activated sewage sludge with enhanced biological phosphorus removal.105–107 Although FISH-MAR has greater sensitivity than DNA- or RNA-SIP, as detection of substrate incorporation is not restricted to analysis of a specific biomolecule (in contrast to nucleic acid-SIP), it is limited, firstly because the microbial groups to be targeted need to be known (and therefore selected with the use of appropriate molecular probes) in advance and secondly owing to the fact that its application is restricted to either single or small clusters of cells (reviewed in Wagner et al.108). Limitations on the number of different fluorophores that can be detected simultaneously also restrict the number of microbial groups that can be targeted at the same time in FISH-MAR.109 However, the development of radioactively labelled RNA-targeted isotope arrays to study multiple microbial populations for their ability to consume a radioactive substrate in activated sludge samples has given promising results.110 The isotope array concept has recently been expanded in isotope rRNA-targeted oligonucleotide microarrays (PhyloChips), containing a much larger number of probes, to reveal substrate consumption profiles of Rhodocyclales spp. in activated sludge.111 Isotope arrays have, to our knowledge, not yet been used in soil-based studies.
Returning to in situ hybridisation-based techniques, FISH-MAR has been used in combination with catalysed reporter deposition112 to improve signal detection in oligotrophic prokaryotes, and quantitative (Q)-FISH-MAR104 has been developed to quantify cell-specific carbon uptake in probe-targeted bacterial groups. In addition, the combination of FISH with other methodologies has given birth to further hyphenated techniques such as FISH-SIMS113,114 (the combination of FISH with secondary ion mass spectrometry) and FISH-RAMAN115 (the combination of FISH with Raman microspectroscopy). Both SIMS and Raman microspectroscopy can be applied to characterise cellular incorporation of 13C-labelled substrates, negating the requirement for experiments using 14C-CO2 pulse chase and attendant safety concerns with respect to use of radioactivity. In FISH-SIMS the FISH-probed identification of microbial cells is coupled to SIMS, which determines the isotopic composition/incorporation of the targeted cells after a caesium ion beam is directed on their surface. In FISH-RAMAN the FISH identification is coupled to highly resolved Raman confocal spectra, and cells that are 13C-labelled through anabolic incorporation of the isotope exhibit key ‘red-shifted’ spectral peaks highly correlated with their 13C content. The FISH-RAMAN method was initially used in naphthalene-degrading groundwater samples115 and can be quantitative with appropriate calibration. The same authors expanded their research by combining RNA-SIP and FISH-RAMAN to fully explore naphthalene degradation of the polluted groundwater.83 Although not yet applied to the root zone, to our knowledge, both FISH-SIMS and FISH-RAMAN have considerable promise for use in rhizosphere carbon flow tracking experiments.
POTENTIAL FOR MANIPULATING CARBON DYNAMICS IN AGRICULTURAL SYSTEMS
The intensive cultivation of soils under agriculture results in the loss of soil carbon due to (i) the acceleration of decomposition through improved aeration and the exposure of physically protected organic matter as a result of tillage and drainage and (ii) the reduction of primary production inputs to soil through the removal of plant biomass during harvest. As already mentioned, it is estimated that somewhere in the range of 42–78 Gt of carbon17 that was historically stored in the soil system has been lost as a result of the intensive cultivation of soils, and the capacity for agricultural soils to regain this lost carbon is currently being discussed as one potential contribution to atmospheric carbon remediation and mitigation of climate change.17,116
The size of the C store in soil depends on the interactions between (i) the quantity and quality of primary production inputs and (ii) the fate of these inputs once they have entered the soil in the short and long term. Strategies with respect to the management of soil for C sequestration therefore involve increasing the quantity of primary production, and indeed other organic inputs into the system; this possibility has been widely debated with respect to breeding crop plants with more extensive root systems117 or altered physiological traits, cover- and inter-cropping, increasing the return of crop residues to soil118 and addition of amendments such as compost or biochar.119–121 However, the addition of crop residues and other amendments, while increasing soil organic C, does not usually transfer C additional to that already fixed from the atmosphere to land (depending on the alternative fate of the amendment) and therefore the end point of such practice does not necessarily qualify as ‘soil C sequestration’ under the strictest definitions of this term.118 A second strategy involves manipulating the fate of the inputs once added to the soil. On entering soil, inputs may (after extracellular depolymerisation in the case of macromolecular constituents) be taken up by the soil microbial biomass and the C partitioned for use in the production of biomass (subsequently necromass), excretions and secretions (e.g. extracellular enzymes) and respiration (Fig. 2). The aim of this second strategy is to encourage the processing of plant-derived C to biomass and metabolite precursors of soil organic matter or to secretions that promote the physical protection of C substrates against decomposition rather than to CO2; in other words, to increase the C use efficiency of the microbial biomass potentially by manipulation of the quality of rhizodeposit inputs or edaphic environmental conditions that have a moderating effect on soil microbial physiology.
We know that climatic and abiotic soil factors (e.g. clay content) influence soil C cycling; however, the identity of soil microorganisms, as the primary decomposers of plant-derived C, is likely to significantly influence the fate of C inputs to soil. The extent to which climatic effects on soil C cycling are confounded with microbial adaptation to certain environmental niches is currently unknown. There is evidence (reviewed in Six et al.122) to suggest that the relative abundance of fungi and bacteria may be important, with more stable carbon being formed in soils with high fungal/bacterial biomass ratios. That fungi have a higher C use efficiency than bacteria and therefore form more biomass per unit of C utilised and also a biomass (subsequently necromass) of a more recalcitrant nature are the suggested mechanisms for the greater accumulation of fungal SOM, although both these mechanisms require further study.122 There have also been some studies (reviewed in Nielsen et al.123) that have reported relationships (positive and negative) between soil biodiversity and C cycling processes such as respiration, but these have generally focused on total species richness as the biodiversity measure and not the richness or identity of those species processing the carbon in situ.
At this stage it is not clear how the diversity and identity of those microorganisms using plant C influence the fate of that C. In addition, the edaphic abiotic factors controlling microbial C utilisation efficiency have not been thoroughly characterised.124 Ultimately it is not clear to what extent rhizosphere microbes within agricultural systems can be manipulated for C sequestration. If the community structure of plant C-utilising microbes is important for C fate, then the next step is to understand which are the most important groups that control soil storage with respect to expression of specific functions (e.g. metabolite production) and the proportion of the plant C inputs they are responsible for processing. This information is essential for both conceptualisation and parametrisation of the next generation of SOM models. To do this, we need to be able to quantitatively apportion plant C to specific microbial groups in situ and to partition its use (i.e. for biomass/metabolite/CO2 production) within that group.
We conclude that the methodologies outlined in this paper, although crucial in enabling the identification of plant-derived carbon-utilising microbes, lack the high-throughput ability to do this because of their reliance on extracted biomolecules (nucleic acid-SIP, protein-SIP, PLFA-SIP), precluding the ability to study the partitioning of carbon at the whole cell level, or because they are limited to the study of a small number of cells (FISH-SIMS, FISH-RAMAN). The challenge is the development of new methodologies that allow quantification of microbial use efficiency and destination of plant carbon (within phylogenetic groups and the metabolome) to enable a step-change level of understanding. The ultimate benefits from this investment will be the knowledge to inform manipulation of the plant–soil system to favour organisms or physiologies most important for promoting soil carbon storage across the diverse conditions present in the global agricultural land.
Acknowledgments
The authors would like to thank the Biotechnology and Biological Sciences Research Council (BBSRC) in the United Kingdom (contract BB/F000251/1) for supporting this work.
REFERENCES
- 1.Falkowski PG, Fenchel T, Delong EF. The microbial engines that drive Earth's biogeochemical cycles. Science. 2008;320:1034–1039. doi: 10.1126/science.1153213. [DOI] [PubMed] [Google Scholar]
- 2.Prentice IC, Farquhar GD, Fasham MJR, Goulden MI, Heinmann M, Jaramillo VJ. The carbon cycle and atmospheric carbon dioxide. In: Houghton JT, editor. Climate Change 2001. The Scientific Basis. Contribution of Working Group I to the Third Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge: Cambridge University Press; 2001. pp. 183–238. [Google Scholar]
- 3.Lu Y, Conrad R. In situ stable isotope probing of methanogenic archaea in the rice rhizosphere. Science. 2005;309:1088–1090. doi: 10.1126/science.1113435. [DOI] [PubMed] [Google Scholar]
- 4.Trumbore S. Carbon respired by terrestrial ecosystems – recent progress and challenges. Global Change Biol. 2006;12:141–153. [Google Scholar]
- 5.Liang C, Balser TC. Microbial production of recalcitrant organic matter in global soils: implications for productivity and climate policy. Nat Rev Microbiol. 2011;9:75. doi: 10.1038/nrmicro2386-c1. [DOI] [PubMed] [Google Scholar]
- 6.Lu Y, Lueders T, Friedrich MW, Conrad R. Detecting active methanogenic populations on rice roots using stable isotope probing. Environ Microbiol. 2005;7:326–336. doi: 10.1111/j.1462-2920.2005.00697.x. [DOI] [PubMed] [Google Scholar]
- 7.Chen Y, Dumont MG, Neufeld JD, Bodrossy L, Stralis-Pavese N, McNamara NP, et al. Revealing the uncultivated majority: combining DNA stable-isotope probing, multiple displacement amplification and metagenomic analyses of uncultivated Methylocystis in acidic peatlands. Environ Microbiol. 2008;10:2609–2622. doi: 10.1111/j.1462-2920.2008.01683.x. [DOI] [PubMed] [Google Scholar]
- 8.Chistoserdova L, Vorholt JA, Lidstrom ME. A genomic view of methane oxidation by aerobic bacteria and anaerobic archaea. Genome Biol. 2005;6:208. doi: 10.1186/gb-2005-6-2-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qiu QF, Noll M, Abraham W-R, Lu YH, Conrad R. Applying stable isotope probing of phospholipid fatty acids and rRNA in a Chinese rice field to study activity and composition of the methanotrophic bacterial communities in situ. ISME J. 2008;2:602–614. doi: 10.1038/ismej.2008.34. [DOI] [PubMed] [Google Scholar]
- 10.Gupta V, Smemo KA, Yavitt JB, Fowle D, Branfireun B, Basiliko N. Stable isotopes reveal widespread anaerobic methane oxidation across latitude and peatland type. Environ Sci Technol. 2013;47:8273–8279. doi: 10.1021/es400484t. [DOI] [PubMed] [Google Scholar]
- 11.Davidson EA, Janssens IA. Temperature sensitivity of soil carbon decomposition and feedbacks to climate change. Nature. 2006;440:165–173. doi: 10.1038/nature04514. [DOI] [PubMed] [Google Scholar]
- 12.Schmidt MWI, Torn MS, Abiven S, Dittmar T, Guggenberger G, Janssens IA, et al. Persistence of soil organic matter as an ecosystem property. Nature. 2011;478:49–56. doi: 10.1038/nature10386. [DOI] [PubMed] [Google Scholar]
- 13.Tarnocai C, Canadell JG, Schuur EAG, Kuhry P, Mazhitova G, Zimov S. Soil organic carbon pools in the northern circumpolar permafrost region. Global Biogeochem Cycles. 2009;23:GB2023. [Google Scholar]
- 14.Jobbágy EG, Jackson RB. The vertical distribution of soil organic carbon and its relation to climate and vegetation. Ecol Appl. 2000;10:423–436. [Google Scholar]
- 15.Sylvia DM, Fuhrmann JJ, Hartel PG, Zuberer DA, editors. Principles and Applications of Soil Microbiology. Upper Saddle River, NJ: Pearson Education; 2005. [Google Scholar]
- 16.Le Quere C, Raupach MR, Canadell JG, Marland G, Bopp L, Ciais P, et al. Trends in the sources and sinks of carbon dioxide. Nature Geosci. 2009;2:831–836. [Google Scholar]
- 17.Lal R. Soil carbon sequestration impacts on global climate change and food security. Science. 2004;304:1623–1627. doi: 10.1126/science.1097396. [DOI] [PubMed] [Google Scholar]
- 18.Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol. 2006;57:233–266. doi: 10.1146/annurev.arplant.57.032905.105159. [DOI] [PubMed] [Google Scholar]
- 19.Muller S, Vandermerwe A, Schildknecht H, Visser JH. An automated-system for large-scale recovery of germination stimulants and other root exudates. Weed Sci. 1993;41:138–143. [Google Scholar]
- 20.Koegel-Knabner I. The macromolecular organic composition of plant and microbial residues as inputs to soil organic matter. Soil Biol Biochem. 2002;34:139–162. [Google Scholar]
- 21.Wallenstein MD, Weintraub MN. Emerging tools for measuring and modeling the in situ activity of soil extracellular enzymes. Soil Biol Biochem. 2008;40:2098–2106. [Google Scholar]
- 22.Smith SE, Read DJ. The Mycorrhizal Symbiosis. San Diego, CA: Academic Press; 2008. [Google Scholar]
- 23.Johnson D, Leake JR, Read DJ. Transfer of recent photosynthate into mycorrhizal mycelium of an upland grassland: short-term respiratory losses and accumulation of 14C. Soil Biol Biochem. 2002;34:1521–1524. [Google Scholar]
- 24.Nakano-Hylander A, Olsson PA. Carbon allocation in mycelia of arbuscular mycorrhizal fungi during colonisation of plant seedlings. Soil Biol Biochem. 2007;39:1450–1458. [Google Scholar]
- 25.Gavito ME, Olsson PA. Allocation of plant carbon to foraging and storage in arbuscular mycorrhizal fungi. FEMS Microbiol Ecol. 2003;45:181–187. doi: 10.1016/S0168-6496(03)00150-8. [DOI] [PubMed] [Google Scholar]
- 26.Drigo B, Pijl AS, Duyts H, Kielak AM, Gamper HA, Houtekamer MJ, et al. Shifting carbon flow from roots into associated microbial communities in response to elevated atmospheric CO2. Proc Natl Acad Sci. 2010;107:10938–10942. doi: 10.1073/pnas.0912421107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones DL, Hodge A, Kuzyakov Y. Plant and mycorrhizal regulation of rhizodeposition. New Phytol. 2004;163:459–480. doi: 10.1111/j.1469-8137.2004.01130.x. [DOI] [PubMed] [Google Scholar]
- 28.Talbot JM, Allison SD, Treseder KK. Decomposers in disguise: mycorrhizal fungi as regulators of soil C dynamics in ecosystems under global change. Funct Ecol. 2008;22:953–963. [Google Scholar]
- 29.Vandenkoornhuyse P, Mahé S, Ineson P, Staddon P, Ostle N, Cliquet J-B, et al. Active root-inhabiting microbes identified by rapid incorporation of plant-derived carbon into RNA. Proc Natl Acad Sci. 2007;104:16970–16975. doi: 10.1073/pnas.0705902104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dungait JAJ, Hopkins DW, Gregory AS, Whitmore AP. Soil organic matter turnover is governed by accessibility not recalcitrance. Global Change Biol. 2012;18:1781–1796. [Google Scholar]
- 31.Wolf DC, Wagner GH. Carbon transformations and soil organic matter formation. In: Sylvia DM, Fuhrmann JJ, Hartel PG, Zuberer DA, editors. Principles and Applications of Soil Microbiology. Upper Saddle River, NJ: Pearson Education; 2005. pp. 285–332. [Google Scholar]
- 32.Kleber M, Johnson MG. Advances in understanding the molecular structure of soil organic matter: implications for interactions in the environment. Adv Agron. 2010;106:77–142. [Google Scholar]
- 33.Kindler R, Miltner A, Thullner M, Richnow H-H, Kästner M. Fate of bacterial biomass derived fatty acids in soil and their contribution to soil organic matter. Org Geochem. 2009;40:29–37. [Google Scholar]
- 34.Tisdall JM. Possible role of soil microorganisms in aggregation in soils. Plant Soil. 1994;159:115–121. [Google Scholar]
- 35.Six J, Bossuyt H, Degryze S, Denef K. A history of research on the link between (micro)aggregates, soil biota, and soil organic matter dynamics. Soil Tillage Res. 2004;79:7–31. [Google Scholar]
- 36.Miller RM, Jastrow JD. Mycorrhizal fungi influence soil structure. In: Kapulnik Y, Douds D Jr, editors. Arbuscular Mycorrhizas: Physiology and Function. The Netherlands: Springer, Kluwer; 2000. pp. 3–18. [Google Scholar]
- 37.Rillig MC, Mummey DL. Mycorrhizas and soil structure. New Phytol. 2006;171:41–53. doi: 10.1111/j.1469-8137.2006.01750.x. [DOI] [PubMed] [Google Scholar]
- 38.Canadell JG, Le Quere C, Raupach MR, Field CB, Buitenhuis ET, Ciais P, et al. Contributions to accelerating atmospheric CO2 growth from economic activity, carbon intensity, and efficiency of natural sinks. Proc Natl Acad Sci. 2007;104:18866–18870. doi: 10.1073/pnas.0702737104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith P, Fang C. Carbon cycle: a warm response by soils. Nature. 2010;464:499–500. doi: 10.1038/464499a. [DOI] [PubMed] [Google Scholar]
- 40.Canadell JG, Kirschbaum MUF, Kurz WA, Sanz MJ, Schlamadinger B, Yamagata Y. Factoring out natural and indirect human effects on terrestrial carbon sources and sinks. Environ Sci Pol. 2007;10:370–384. [Google Scholar]
- 41.Jin VL, Evans RD. Elevated CO2 increases microbial carbon substrate use and nitrogen cycling in Mojave Desert soils. Global Change Biol. 2007;13:452–465. [Google Scholar]
- 42.Bardgett RD, Freeman C, Ostle NJ. Microbial contributions to climate change through carbon cycle feedbacks. ISME J. 2008;2:805–814. doi: 10.1038/ismej.2008.58. [DOI] [PubMed] [Google Scholar]
- 43.Singh BK, Bardgett RD, Smith P, Reay DS. Microorganisms and climate change: terrestrial feedbacks and mitigation options. Nat Rev Microbiol. 2010;8:779–790. doi: 10.1038/nrmicro2439. [DOI] [PubMed] [Google Scholar]
- 44.Katterer T, Reichstein M, Andren O, Lomander A. Temperature dependence of organic matter decomposition: a critical review using literature data analyzed with different models. Biol Fertil Soils. 1998;27:258–262. [Google Scholar]
- 45.Cox PM, Betts RA, Jones CD, Spall SA, Totterdell IJ. Acceleration of global warming due to carbon-cycle feedbacks in a coupled climate model. Nature. 2000;408:184–187. doi: 10.1038/35041539. [DOI] [PubMed] [Google Scholar]
- 46.Knorr W, Prentice IC, House JI, Holland EA. Long-term sensitivity of soil carbon turnover to warming. Nature. 2005;433:298–301. doi: 10.1038/nature03226. [DOI] [PubMed] [Google Scholar]
- 47.Lopez-Gutierrez JC, Malcolm GM, Koide RT, Eissenstat DM. Ectomycorrhizal fungi from Alaska and Pennsylvania: adaptation of mycelial respiratory response to temperature? New Phytol. 2008;180:741–744. doi: 10.1111/j.1469-8137.2008.02655.x. [DOI] [PubMed] [Google Scholar]
- 48.Vargas R, Baldocchi DD, Querejeta JI, Curtis PS, Hasselquist NJ, Janssens IA, et al. Ecosystem CO2 fluxes of arbuscular and ectomycorrhizal dominated vegetation types are differentially influenced by precipitation and temperature. New Phytol. 2010;185:226–236. doi: 10.1111/j.1469-8137.2009.03040.x. [DOI] [PubMed] [Google Scholar]
- 49.Hawkes CV, Hartley IP, Ineson P, Fitter AH. Soil temperature affects carbon allocation within arbuscular mycorrhizal networks and carbon transport from plant to fungus. Global Change Biol. 2008;14:1181–1190. [Google Scholar]
- 50.Malcolm GM, López-Gutiérrez JC, Koide RT, Eissenstat DM. Acclimation to temperature and temperature sensitivity of metabolism by ectomycorrhizal fungi. Global Change Biol. 2008;14:1169–1180. [Google Scholar]
- 51.Bond-Lamberty B, Thomson A. Temperature-associated increases in the global soil respiration record. Nature. 2010;464:579–582. doi: 10.1038/nature08930. [DOI] [PubMed] [Google Scholar]
- 52.Dijkstra FA, Hobbie SE, Reich PB, Knops JMH. Divergent effects of elevated CO2, N fertilization, and plant diversity on soil C and N dynamics in a grassland field experiment. Plant Soil. 2005;272:41–52. [Google Scholar]
- 53.Hungate BA, Johnson DW, Dijkstra P, Hymus G, Stiling P, Megonigal JP, et al. Nitrogen cycling during seven years of atmospheric CO2 enrichment in a scrub oak woodland. Ecology. 2006;87:26–40. doi: 10.1890/04-1732. [DOI] [PubMed] [Google Scholar]
- 54.Rayner PJ, Scholze M, Knorr W, Kaminski T, Giering R, Widmann H. Two decades of terrestrial carbon fluxes from a carbon cycle data assimilation system (CCDAS) Global Biogeochem Cycles. 2005;19:GB2026. [Google Scholar]
- 55.Friedlingstein P, Cox P, Betts R, Bopp L, von Bloh W, Brovkin V, et al. Climate–carbon cycle feedback analysis: results from the C4MIP model intercomparison. J Clim. 2006;19:3337–3353. [Google Scholar]
- 56.Ainsworth EA, Long SP. What have we learned from 15 years of free-air CO2 enrichment (FACE)? A meta-analytic review of the responses of photosynthesis, canopy properties and plant production to rising CO2. New Phytol. 2005;165:351–372. doi: 10.1111/j.1469-8137.2004.01224.x. [DOI] [PubMed] [Google Scholar]
- 57.Heath J, Ayres E, Possell M, Bardgett RD, Black HI, Grant H, et al. Rising atmospheric CO2 reduces sequestration of root-derived soil carbon. Science. 2005;309:1711–1713. doi: 10.1126/science.1110700. [DOI] [PubMed] [Google Scholar]
- 58.Blagodatskaya E, Kuzyakov Y. Mechanisms of real and apparent priming effects and their dependence on soil microbial biomass and community structure: critical review. Biol Fertil Soils. 2008;45:115–131. [Google Scholar]
- 59.Fontaine S, Bardoux G, Abbadie L, Mariotti A. Carbon input to soil may decrease soil carbon content. Ecol Lett. 2004;7:314–320. [Google Scholar]
- 60.Fontaine S, Barot S. Size and functional diversity of microbe populations control plant persistence and long-term soil carbon accumulation. Ecol Lett. 2005;8:1075–1087. [Google Scholar]
- 61.Langley JA, McKee KL, Cahoon DR, Cherry JA, Megonigal JP. Elevated CO2 stimulates marsh elevation gain, counterbalancing sea-level rise. Proc Natl Acad Sci. 2009;106:6182–6186. doi: 10.1073/pnas.0807695106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schleppi P, Bucher-Wallin I, Hagedorn F, Körner C. Increased nitrate availability in the soil of a mixed mature temperate forest subjected to elevated CO2 concentration (canopy FACE) Global Change Biol. 2012;18:757–768. [Google Scholar]
- 63.Zhu B, Cheng W. Rhizosphere priming effect increases the temperature sensitivity of soil organic matter decomposition. Global Change Biol. 2011;17:2172–2183. [Google Scholar]
- 64.Langley JA, Megonigal JP. Ecosystem response to elevated CO2 levels limited by nitrogen-induced plant species shift. Nature. 2010;466:96–99. doi: 10.1038/nature09176. [DOI] [PubMed] [Google Scholar]
- 65.Denman KL, Brasseur G, Chidthaisong A, Ciais P, Cox PM. Coupling between changes in the climate system and biogeochemistry. In: Solomon S, Qin D, Manning M, Chen Z, Marquis M, Averyt KB, et al., editors. Climate Change 2007: The Physical Science Basis. Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge: Cambridge University Press; 2007. pp. 499–588. [Google Scholar]
- 66.Schimel J, Balser TC, Wallenstein M. Microbial stress-response physiology and its implications for ecosystem function. Ecology. 2007;88:1386–1394. doi: 10.1890/06-0219. [DOI] [PubMed] [Google Scholar]
- 67.King GM. Enhancing soil carbon storage for carbon remediation: potential contributions and constraints by microbes. Trends Microbiol. 2011;19:75–84. doi: 10.1016/j.tim.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 68.Ostle NJ, Smith P, Fisher R, Woodward FI, Fisher JB, Smith JU, et al. Integrating plant–soil interactions into global carbon cycle models. J Ecol. 2009;97:851–863. [Google Scholar]
- 69.Smith P, Fang CM, Dawson JJC, Moncrieff JB. Impact of global warming on soil organic carbon. Adv Agron. 2008;97:1–43. [Google Scholar]
- 70.Gutierrez-Zamora M-L, Manefield M. An appraisal of methods for linking environmental processes to specific microbial taxa. Rev Environ Sci Biotechnol. 2010;9:153–185. [Google Scholar]
- 71.Radajewski S, Ineson P, Parekh NR, Murrell JC. Stable-isotope probing as a tool in microbial ecology. Nature. 2000;403:646–649. doi: 10.1038/35001054. [DOI] [PubMed] [Google Scholar]
- 72.Hery M, Singer AC, Kumaresan D, Bodrossy L, Stralis-Pavese N, Prosser JI, et al. Effect of earthworms on the community structure of active methanotrophic bacteria in a landfill cover soil. ISME J. 2008;2:92–104. doi: 10.1038/ismej.2007.66. [DOI] [PubMed] [Google Scholar]
- 73.Qiu Q, Conrad R, Lu Y. Cross-feeding of methane carbon among bacteria on rice roots revealed by DNA-stable isotope probing. Environ Microbiol Rep. 2009;1:355–361. doi: 10.1111/j.1758-2229.2009.00045.x. [DOI] [PubMed] [Google Scholar]
- 74.Aitken MD, Jones MD, Singleton DR, Carstensen DP, Powell SN, Swanson JS, et al. Effect of incubation conditions on the enrichment of pyrene-degrading bacteria identified by stable-isotope probing in an aged, PAH-contaminated soil. Microb Ecol. 2008;56:341–349. doi: 10.1007/s00248-007-9352-9. [DOI] [PubMed] [Google Scholar]
- 75.Cupples AM, Luo CL, Xie SG, Sun WM, Li XD. Identification of a novel toluene-degrading bacterium from the candidate phylum TM7, as determined by DNA stable isotope probing. Appl Environ Microbiol. 2009;75:4644–4647. doi: 10.1128/AEM.00283-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jones MD, Singleton DR, Sun W, Aitken MD. Multiple DNA extractions coupled with stable-isotope probing of anthracene-degrading bacteria in contaminated soil. Appl Environ Microbiol. 2011;77:2984–2991. doi: 10.1128/AEM.01942-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Uhlik O, Jecna K, Mackova M, Vlcek C, Hroudova M, Demnerova K, et al. Biphenyl-Metabolizing Bacteria in the Rhizosphere of Horseradish and Bulk Soil Contaminated by Polychlorinated Biphenyls as Revealed by Stable Isotope Probing. Appl Environ Microbiol. 2009;75:6471–6477. doi: 10.1128/AEM.00466-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Manefield M, Whiteley AS, Griffiths RI, Bailey MJ. RNA stable isotope probing, a novel means of linking microbial community function to phylogeny. Appl Environ Microbiol. 2002;68:5367–5373. doi: 10.1128/AEM.68.11.5367-5373.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Noll M, Frenzel P, Conrad R. Selective stimulation of type I methanotrophs in a rice paddy soil by urea fertilization revealed by RNA-based stable isotope probing. FEMS Microbiol Ecol. 2008;65:125–132. doi: 10.1111/j.1574-6941.2008.00497.x. [DOI] [PubMed] [Google Scholar]
- 80.Kittelmann S, Friedrich MW. Identification of novel perchloroethene-respiring microorganisms in anoxic river sediment by RNA-based stable isotope probing. Environ Microbiol. 2008;10:31–46. doi: 10.1111/j.1462-2920.2007.01427.x. [DOI] [PubMed] [Google Scholar]
- 81.Kittelmann S, Friedrich MW. Novel uncultured Chloroflexi dechlorinate perchloroethene to trans-dichloroethene in tidal flat sediments. Environ Microbiol. 2008;10:1557–1570. doi: 10.1111/j.1462-2920.2008.01571.x. [DOI] [PubMed] [Google Scholar]
- 82.Aburto A, Ball AS. Bacterial population dynamics and separation of active degraders by stable isotope probing during benzene degradation in a BTEX-impacted aquifer. Rev Int Contaminacion Ambiental. 2009;25:147–156. [Google Scholar]
- 83.Huang WE, Ferguson A, Singer AC, Lawson K, Thompson IP, Kalin RM, et al. Resolving genetic functions within microbial populations: in situ analyses using rRNA and mRNA stable isotope probing coupled with single-cell Raman-fluorescence in situ hybridization. Appl Environ Microbiol. 2009;75:234–241. doi: 10.1128/AEM.01861-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Langenheder S, Prosser JI. Resource availability influences the diversity of a functional group of heterotrophic soil bacteria. Environ Microbiol. 2008;10:2245–2256. doi: 10.1111/j.1462-2920.2008.01647.x. [DOI] [PubMed] [Google Scholar]
- 85.Drake HL, Degelmann DM, Kolb S, Dumont M, Murrell JC. Enterobacteriaceae facilitate the anaerobic degradation of glucose by a forest soil. FEMS Microbiol Ecol. 2009;68:312–319. doi: 10.1111/j.1574-6941.2009.00681.x. [DOI] [PubMed] [Google Scholar]
- 86.Rasche F, Lueders T, Schloter M, Schaefer S, Buegger F, Gattinger A, et al. DNA-based stable isotope probing enables the identification of active bacterial endophytes in potatoes. New Phytol. 2009;181:802–807. doi: 10.1111/j.1469-8137.2008.02744.x. [DOI] [PubMed] [Google Scholar]
- 87.Manefield M, Whiteley AS, Ostle N, Ineson P, Bailey MJ. Technical considerations for RNA-based stable isotope probing: an approach to associating microbial diversity with microbial community function. Rapid Commun Mass Spectrom. 2002;16:2179–2183. doi: 10.1002/rcm.782. [DOI] [PubMed] [Google Scholar]
- 88.Griffiths RI, Manefield M, Ostle N, McNamara N, O'Donnell AG, Bailey MJ, et al. 13CO2 pulse labelling of plants in tandem with stable isotope probing: methodological considerations for examining microbial function in the rhizosphere. J Microbiol Meth. 2004;58:119–129. doi: 10.1016/j.mimet.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 89.Neufeld JD, Dumont MG, Vohra J, Murrell JC. Methodological considerations for the use of stable isotope probing in microbial ecology. Microb Ecol. 2007;53:435–442. doi: 10.1007/s00248-006-9125-x. [DOI] [PubMed] [Google Scholar]
- 90.Whiteley AS, Manefield M, Lueders T. Unlocking the ‘microbial black box’ using RNA-based stable isotope probing technologies. Curr Opin Biotechnol. 2006;17:67–71. doi: 10.1016/j.copbio.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 91.Rangel-Castro JI, Killham K, Ostle N, Nicol GW, Anderson IC, Scrimgeour CM, et al. Stable isotope probing analysis of the influence of liming on root exudate utilization by soil microorganisms. Environ Microbiol. 2005;7:828–838. doi: 10.1111/j.1462-2920.2005.00756.x. [DOI] [PubMed] [Google Scholar]
- 92.Hannula SE, Boschker HTS, de Boer W, van Veen JA. 13C pulse-labeling assessment of the community structure of active fungi in the rhizosphere of a genetically starch-modified potato (Solanum tuberosum) cultivar and its parental isoline. New Phytol. 2012;194:784–799. doi: 10.1111/j.1469-8137.2012.04089.x. [DOI] [PubMed] [Google Scholar]
- 93.Lu Y, Rosencrantz D, Liesack W, Conrad R. Structure and activity of bacterial community inhabiting rice roots and the rhizosphere. Environ Microbiol. 2006;8:1351–1360. doi: 10.1111/j.1462-2920.2006.01028.x. [DOI] [PubMed] [Google Scholar]
- 94.Haichar FZ, Roncato MA, Achouak W. Stable isotope probing of bacterial community structure and gene expression in the rhizosphere of Arabidopsis thaliana. FEMS Microbiol Ecol. 2012;81:291–302. doi: 10.1111/j.1574-6941.2012.01345.x. [DOI] [PubMed] [Google Scholar]
- 95.Jehmlich N, Schmidt F, Hartwich M, von Bergen M, Richnow HH, Vogt C. Incorporation of carbon and nitrogen atoms into proteins measured by protein-based stable isotope probing (Protein-SIP) Rapid Commun Mass Spectrom. 2008;22:2889–2897. doi: 10.1002/rcm.3684. [DOI] [PubMed] [Google Scholar]
- 96.Seifert J, Taubert M, Jehmlich N, Schmidt F, Völker U, Vogt C, et al. Protein-based stable isotope probing (protein-SIP) in functional metaproteomics. Mass Spectrom Rev. 2012;31:683–697. doi: 10.1002/mas.21346. [DOI] [PubMed] [Google Scholar]
- 97.Von Bergen M, Jehmlich N, Taubert M, Vogt C, Bastida F, Herbst F-A, et al. Insights from quantitative metaproteomics and protein-stable isotope probing into microbial ecology. ISME J. 2013;7:1877–1885. doi: 10.1038/ismej.2013.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Treonis AM, Ostle NJ, Stott AW, Primrose R, Grayston SJ, Ineson P. Identification of groups of metabolically active rhizosphere microorganisms by stable isotope probing of PLFAs. Soil Biol Biochem. 2004;36:533–537. [Google Scholar]
- 99.Paterson E, Gebbing T, Abel C, Sim A, Telfer G. Rhizodeposition shapes rhizosphere microbial community structure in organic soil. New Phytol. 2007;173:600–610. doi: 10.1111/j.1469-8137.2006.01931.x. [DOI] [PubMed] [Google Scholar]
- 100.Shrestha M, Abraham W-R, Shrestha PM, Noll M, Conrad R. Activity and composition of methanotrophic bacterial communities in planted rice soil studied by flux measurements, analyses of pmoA gene and stable isotope probing of phospholipid fatty acids. Environ Microbiol. 2008;10:400–412. doi: 10.1111/j.1462-2920.2007.01462.x. [DOI] [PubMed] [Google Scholar]
- 101.Dias ACF, Dini-Andreote F, Hannula SE, Andreote FD, Pereira e Silva MdC, Salles JF, de Boer W, van Veen J, van Elsas JD. Different Selective Effects on Rhizosphere Bacteria Exerted by Genetically Modified versus Conventional Potato Lines. PLoS ONE. 2013;8:e67948. doi: 10.1371/journal.pone.0067948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee N, Nielsen PH, Andreasen KH, Juretschko S, Nielsen JL, Schleifer KH, et al. Combination of fluorescent in situ hybridization and microautoradiography – a new tool for structure–function analyses in microbial ecology. Appl Environ Microbiol. 1999;65:1289–1297. doi: 10.1128/aem.65.3.1289-1297.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ouverney CC, Fuhrman JA. Combined microautoradiography–16S rRNA probe technique for determination of radioisotope uptake by specific microbial cell types in situ. Appl Environ Microbiol. 1999;65:1746–1752. doi: 10.1128/aem.65.4.1746-1752.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nielsen JL, Christensen D, Kloppenborg M, Nielsen PH. Quantification of cell-specific substrate uptake by probe-defined bacteria under in situ conditions by microautoradiography and fluorescence in situ hybridization. Environ Microbiol. 2003;5:202–211. doi: 10.1046/j.1462-2920.2003.00402.x. [DOI] [PubMed] [Google Scholar]
- 105.Thomsen TR, Kong Y, Nielsen PH. Ecophysiology of abundant denitrifying bacteria in activated sludge. FEMS Microbiol Ecol. 2007;60:370–382. doi: 10.1111/j.1574-6941.2007.00309.x. [DOI] [PubMed] [Google Scholar]
- 106.Kong Y, Nielsen JL, Nielsen PH. Microautoradiographic study of Rhodocyclus-related polyphosphate-accumulating bacteria in full-scale enhanced biological phosphorus removal plants. Appl Environ Microbiol. 2004;70:5383–5390. doi: 10.1128/AEM.70.9.5383-5390.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nguyen HT, Le VQ, Hansen AA, Nielsen JL, Nielsen PH. High diversity and abundance of putative polyphosphate-accumulating Tetrasphaera-related bacteria in activated sludge systems. FEMS Microbiol Ecol. 2011;76:256–267. doi: 10.1111/j.1574-6941.2011.01049.x. [DOI] [PubMed] [Google Scholar]
- 108.Wagner M, Nielsen PH, Loy A, Nielsen JL, Daims H. Linking microbial community structure with function: fluorescence in situ hybridization–microautoradiography and isotope arrays. Curr Opin Biotechnol. 2006;17:83–91. doi: 10.1016/j.copbio.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 109.Amann R, Snaidr J, Wagner M, Ludwig W, Schleifer KH. In situ visualization of high genetic diversity in a natural microbial community. J Bacteriol. 1996;178:3496–3500. doi: 10.1128/jb.178.12.3496-3500.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Adamczyk J, Hesselsoe M, Iversen N, Horn M, Lehner A, Nielsen PH, et al. The isotope array, a new tool that employs substrate-mediated labeling of rRNA for determination of microbial community structure and function. Appl Environ Microbiol. 2003;69:6875–6887. doi: 10.1128/AEM.69.11.6875-6887.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hesselsoe M, Fureder S, Schloter M, Bodrossy L, Iversen N, Roslev P, et al. Isotope array analysis of Rhodocyclales uncovers functional redundancy and versatility in an activated sludge. ISME J. 2009;3:1349–1364. doi: 10.1038/ismej.2009.78. [DOI] [PubMed] [Google Scholar]
- 112.Teira E, Reinthaler T, Pernthaler A, Pernthaler J, Herndl GJ. Combining catalyzed reporter deposition-fluorescence in situ hybridization and microautoradiography to detect substrate utilization by bacteria and archaea in the deep ocean. Appl Environ Microbiol. 2004;70:4411–4414. doi: 10.1128/AEM.70.7.4411-4414.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Orphan VJ, House CH, Hinrichs KU, McKeegan KD, DeLong EF. Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science. 2001;293:484–487. doi: 10.1126/science.1061338. [DOI] [PubMed] [Google Scholar]
- 114.Orphan VJ, House CH, Hinrichs KU, McKeegan KD, DeLong EF. Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc Natl Acad Sci. 2002;99:7663–7668. doi: 10.1073/pnas.072210299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Huang WE, Stoecker K, Griffiths R, Newbold L, Daims H, Whiteley AS, et al. Raman-FISH: combining stable-isotope Raman spectroscopy and fluorescence in situ hybridization for the single cell analysis of identity and function. Environ Microbiol. 2007;9:1878–1889. doi: 10.1111/j.1462-2920.2007.01352.x. [DOI] [PubMed] [Google Scholar]
- 116.Smith P, Olesen JE. Synergies between the mitigation of, and adaptation to, climate change in agriculture. J Agric Sci. 2010;148:543–552. [Google Scholar]
- 117.Kell DB. Breeding crop plants with deep roots: their role in sustainable carbon, nutrient and water sequestration. Ann Bot. 2011;108:407–418. doi: 10.1093/aob/mcr175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Powlson DS, Whitmore AP, Goulding KWT. Soil carbon sequestration to mitigate climate change: a critical re-examination to identify the true and the false. Eur J Soil Sci. 2011;62:42–55. [Google Scholar]
- 119.Sohi SP, Krull E, Lopez-Capel E, Bol R. A review of biochar and its use and function in soil. Adv Agron. 2010;105:47–82. [Google Scholar]
- 120.Lal R. Soil carbon sequestration to mitigate climate change. Geoderma. 2004;123:1–22. [Google Scholar]
- 121.Smith P. Carbon sequestration in croplands: the potential in Europe and the global context. Eur J Agron. 2004;20:229–236. [Google Scholar]
- 122.Six J, Frey SD, Thiet RK, Batten KM. Bacterial and fungal contributions to carbon sequestration in agroecosystems. Soil Sci Soc Am J. 2006;70:555–569. [Google Scholar]
- 123.Nielsen UN, Ayres E, Wall DH, Bardgett RD. Soil biodiversity and carbon cycling: a review and synthesis of studies examining diversity–function relationships. Eur J Soil Sci. 2011;62:105–116. [Google Scholar]
- 124.Cotrufo MF, Wallenstein MD, Boot CM, Denef K, Paul E. The Microbial Efficiency-Matrix Stabilization (MEMS) framework integrates plant litter decomposition with soil organic matter stabilization: do labile plant inputs form stable soil organic matter? Global Change Biol. 2013;19:988–995. doi: 10.1111/gcb.12113. [DOI] [PubMed] [Google Scholar]
- 125.Prosser JI. Microorganisms cycling soil nutrients and their diversity. In: Van Elsas JD, Jansson JK, Trevors JT, editors. Modern Soil Microbiology. New York, NY: CRC Press; 2007. pp. 237–261. [Google Scholar]
- 126.Solomon S, Qin D, Manning M, Chen Z, Marquis M, Averyt K, et al., editors. Climate Change 2007. The Physical Science Basis. Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge: Cambridge University Press; 2007. [Google Scholar]


