SUMMARY
To maintain genome function and stability, DNA sequence and its organization into chromatin must be duplicated during cell division. Understanding how entire chromosomes are copied remains a major challenge. Here, we use Nascent Chromatin Capture (NCC) to profile chromatin proteome dynamics during replication in human cells. NCC relies on biotin-dUTP labelling of replicating DNA, affinity-purification and quantitative proteomics. Comparing nascent chromatin with mature post-replicative chromatin, we provide association dynamics for 3995 proteins. The replication machinery and 485 chromatin factors like CAF-1, DNMT1, SUV39h1 are enriched in nascent chromatin, whereas 170 factors including histone H1, DNMT3, MBD1-3 and PRC1 show delayed association. This correlates with H4K5K12diAc removal and H3K9me1 accumulation, while H3K27me3 and H3K9me3 remain unchanged. Finally, we combine NCC enrichment with experimentally derived chromatin probabilities to predict a function in nascent chromatin for 93 uncharacterized proteins and identify FAM111A as a replication factor required for PCNA loading. Together, this provides an extensive resource to understand genome and epigenome maintenance.
INTRODUCTION
Mammalian genomes are replicated by the simultaneous progression of thousands of replication forks1. In this process, chromatin organization is disrupted ahead of the replication machinery and restored behind, on the two daughter strands2, 3. This genome-wide disruption of chromatin raises fundamental questions about how DNA replication is integrated with chromatin dynamics and how specific chromatin structures are transmitted through mitotic cell division. These questions underpin how cells maintain epigenetically defined gene-expression patterns and thus their identity, which is central to development and disease avoidance2-4.
Chromatin restoration on newly synthesized DNA is a multi-layered process, including nucleosome assembly and remodelling, restoration of DNA methylation and histone marks, deposition of histone variants and establishment of higher order chromosomal structures including sister-chromatid cohesion2, 3. Correspondingly, a large number of proteins are involved, for many of which timing and action are yet to be elucidated or even their identity to be revealed. Pioneering work using nucleases to probe chromatin assembly, argues for a time window of approximately 15-20 minutes for new DNA to be packaged into a structure with nucleosomal periodicity and nuclease resistance similar to bulk chromatin5, 6. This process is referred to as chromatin maturation6, 7. Restoration of nucleosomal density is rapid and relies on recycling of old histones combined with addition of new ones8. Old parental histones are thought to maintain their marks, which are thereby transmitted to the daughter strands. In contrast, new histones H3-H4 are acetylated, principally at lysine 5 and 12 of histone H4 (K5K12diAc)9. These histone H4 acetylations are removed shortly after replication, as nascent (immature) chromatin matures into a nuclease resistant state6, 10, 11.
The sliding clamp PCNA plays a central role in coupling replication with chromatin restoration through its ability to recruit the nucleosome assembly factor CAF-1, the maintenance DNA methyltransferase DNMT1, several chromatin remodellers and lysine deacetylases and methyl transferases2, 3. Still it remains largely unknown when and how the vast majority of chromatin constituents and modifiers are recruited. As a discovery approach, proteomics has been used to define the composition of purified telomeres12 and of mitotic chromosomes13. The latter study revealed a surprising complexity of whole chromosomes and a machine learning approach, multi-classifier combinatorial proteomics, MCCP, was used to predict functional significance of associated proteins13. Given the power of these tools in defining chromosomes, we applied them to study chromatin replication.
We profile DNA replication and chromatin maturation in human cells by nascent chromatin capture (NCC), a biochemical approach to isolate newly synthesized DNA for quantitative proteomic analysis. By determining the dynamic association of 3995 proteins with nascent newly synthesized chromatin and mature post-replicative chromatin, we reveal three classes of factors: i) enriched in nascent chromatin, ii) present in both nascent and mature chromatin and iii) enriched in mature chromatin. This rich data set offers unprecedented insight into the process of building chromatin and maintaining epigenetic information. Moreover, our proteomic analysis provides a discovery approach to identify proteins recruited to replication forks and/or nascent chromatin. Applying a MCCP-derived chromatin-probability list to our data, we predict a role for 93 uncharacterized proteins at replication forks or in newly replicated chromatin behind the fork, and identify FAM111A as a PCNA interaction partner required for DNA replication and S phase entry.
RESULTS
Isolation of replication forks and nascent chromatin by NCC
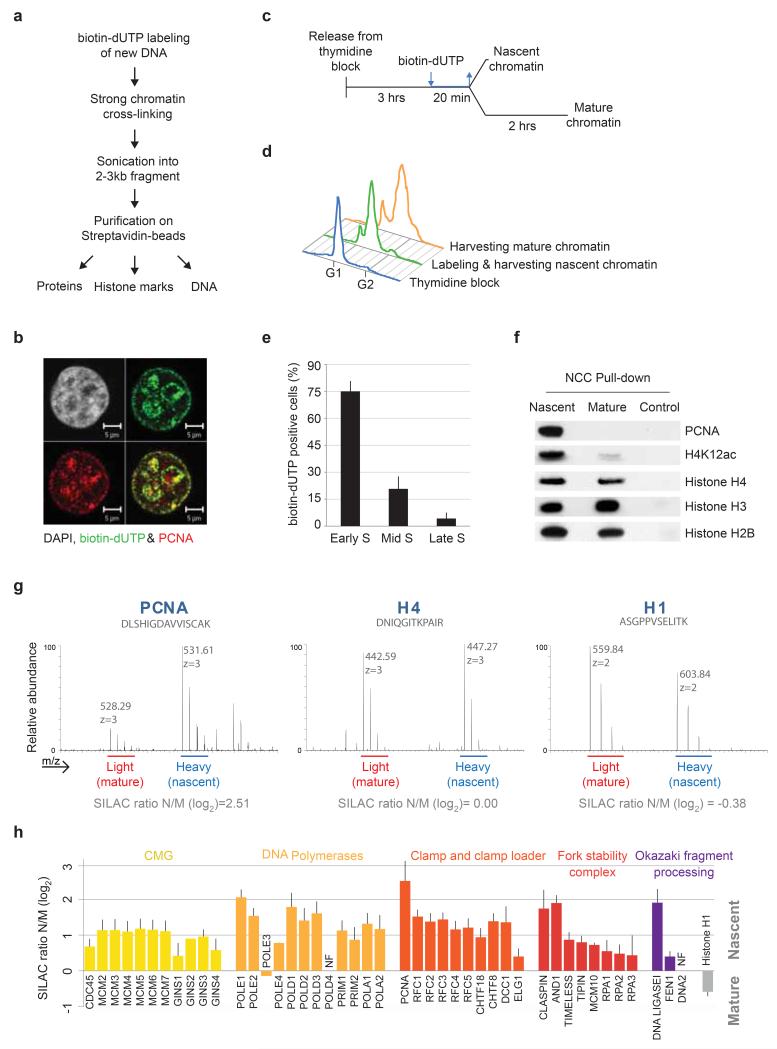
Mamalian replication and chromatin assembly factors are commonly recognized by co-localization with newly synthesized DNA labelled by nucleotide analogs. We have developed an analogous biochemical method, termed Nascent Chromatin Capture (NCC), for isolation of protein complexes present on newly replicated DNA (Fig. 1a). This method involves incorporation of a biotin-dUTP14, 15 (Fig. 1b), strong chromatin cross-linking and sonication followed by affinity purification of tagged chromatin (see online methods). Biotin-dUTP can be introduced by a short hypotonic shift of HeLa S3 spinner cultures without affecting S-phase progression or triggering DNA damage (Supplementary Fig. 1a and 1b).
Figure 1. Isolation of replication forks and nascent chromatin by NCC technology.
(a) Outline of the NCC protocol. See online methods for details. (b) Biotin-labelled newly synthesized DNA co-localizes with PCNA in replication foci. (c) Experimental design for comparison of nascent and mature chromatin. Cells released from a single thymidine block were labelled with biotin-dUTP in early-mid S-phase and harvested directly (nascent) or after 2 hours chase without biotin-dUTP (mature). (d) Cell cycle profiles representative of experimental design in c. (e) S-phase distribution of cells labelled according to the experimental design in c. Cells were scored according to their PCNA pattern49. Error bars represent standard deviation (s.d.), n=4 with 300 cells counted in total. (f) NCC pull-down analysed by western blot. Control, no addition of biotin-dUTP. (g) Mass spectra showing the SILAC intensities for peptides of PCNA, H4 and H1. (h) Nascent chromatin enrichment of core replication fork components. Enrichments are given as the median, from three independent experiments, of log2 of the SILAC ratios of nascent (heavy) over mature (light) (N/M). Error bars represent standard error of the mean (s.e.m.), and indicate the precision of ratio measurements for each protein. POLE1 and histone H1 are included as a reference in all enrichment plots.
To follow DNA synthesis and its assembly into chromatin, we compared nascent chromatin pulse-labelled for 20 minutes with post-replicative chromatin allowed to mature during a 2 hours chase (Fig. 1c, 1d, and Supplementary Fig. 1c). Given a fork speed of 1-2 kb/min1, 20 to 40 kb of DNA could be labelled behind each replication fork, in total corresponding to 5 % of the genome. We thus isolate replication forks and nascent (immature) chromatin behind, but shorter labelling times can be applied to focus on DNA replication (Supplementary Fig. 1d, e). Synchronized cells were labelled in early-mid S phase (Fig. 1d and 1e), including both euchromatin and heterochromatin regions in our analysis. Western blot of NCC pull-downs confirmed enrichment of PCNA and new histone H4 acetylated at lysine 12 in nascent chromatin, whereas canonical histones H3, H4 and H2B were present in both nascent and mature chromatin (Fig. 1f). Histones and other factors were not detected in a negative control without biotin-dUTP (Fig. 1f).
To quantitatively compare the composition of nascent and mature chromatin, we combined NCC and stable isotope labelling by amino acids in cell culture (SILAC)16 (Supplementary Fig. 2a, 2b, 2c). A low fold difference for most proteins between three independent biological replicates illustrated high reproducibility and robustness of the technology (Supplementary Fig. 2d and 2e). We therefore present the full combined data set, normalized to histone H4 to adjust for variations in yield between independent experiments (Supplementary Table 1). We quantified in total 3995 proteins providing a comprehensive view of i) factors enriched in nascent chromatin (e.g. PCNA), ii) factors present in both nascent and mature chromatin (e.g. histone H4), and iii) factors enriched in mature chromatin (e.g. histone H1) (Fig. 1g). Given that the SILAC ratio generally reflects relative protein abundance, these groups typically represents replication-associated factors (Fig. 1g, left), and early (Fig. 1g, middle) or late arriving chromatin components (Fig. 1g, right). However, as cross-linking efficiency can be context dependent, extreme SILAC ratios may also indicate changes in the environment or conformation of a protein. Importantly, we identified 41 of the 44 known core replication fork components enriched on newly replicated DNA (Fig. 1h), arguing that we successfully isolated active human replication forks.
Identification of accessory fork components
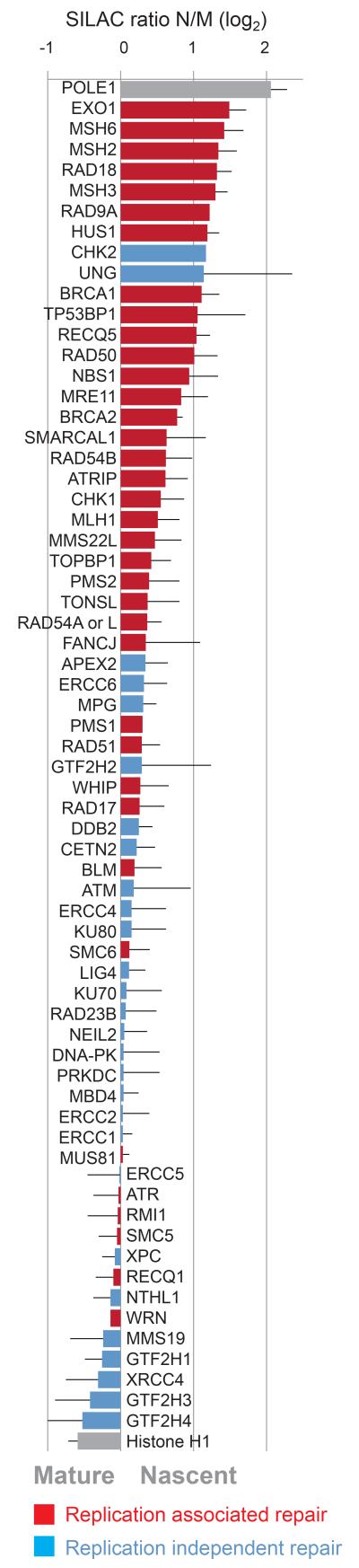
We also identified a number of accessory factors dealing with DNA-RNA duplexes, protein degradation and DNA repair at the fork (Fig. 2 and Supplementary Fig. 3a), whereas factors primarily involved in origin recognition and licensing were either not identified or did not show strong enrichment (Supplementary Fig. 3b). Ranking replication-associated and replication-independent repair and checkpoint factors according to NCC-SILAC ratio illustrates that the former group is highly overrepresented in nascent chromatin (Fig. 2). This corroborates other reports that several genome caretakers serve functions and localize to sites of ongoing replication (e.g. MRN17, SMARCAL118, TONSL-MMS22L19).
Figure 2. Genome maintenance factors at the fork.
Replication-associated and replication-independent repair pathways annotated according to literature are highlighted in red and blue, respectively. Enrichments are presented as in Figure 1h.
Several accessory factors are recruited via PCNA (e.g. RNAse H220, EXO121, and MSH2/3/622), and this prompted us to evaluate the coverage of known PCNA interactors in our analysis23. Whereas DNA replication and chromatin restoration factors are comprehensively represented and enriched in nascent chromatin, many factors involved in cell cycle control and survival are not identified, and those we find are not enriched (Supplementary Fig. 3c, Supplementary Table 2). Furthermore, factors carrying a PIP-degron, which undergo PCNA-dependent degradation in S phase, were not found. The NCC data thus identify those factors from the large PCNA interactome that function at active replication forks.
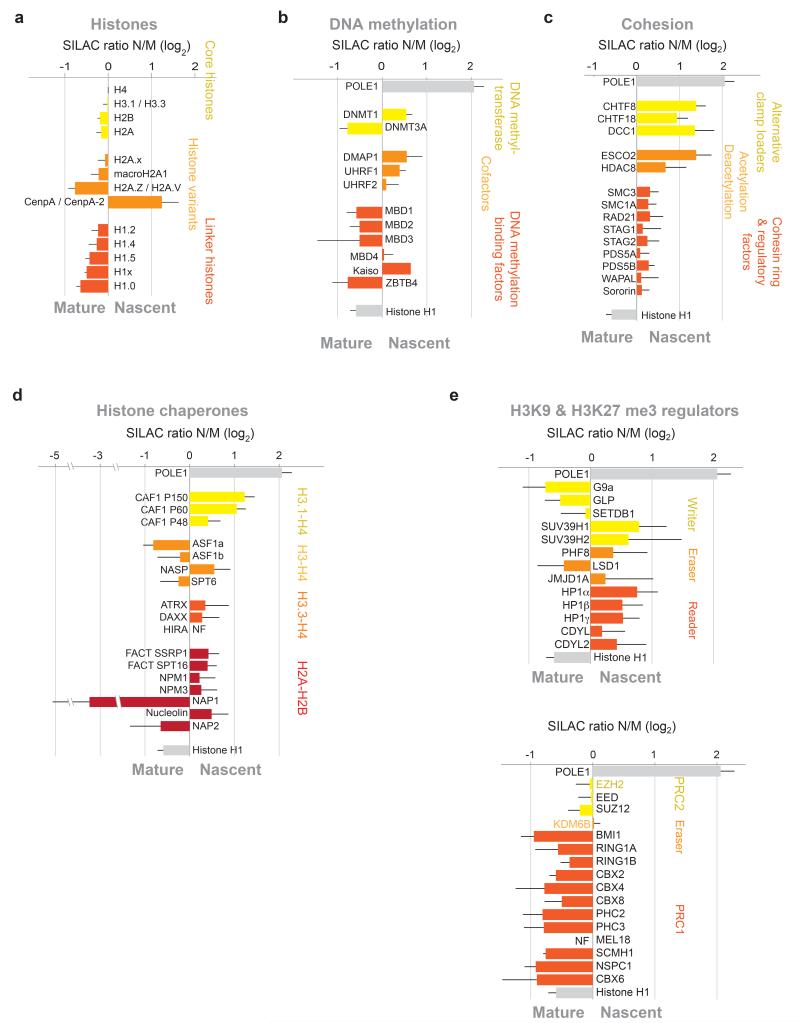
Chromatin assembly and maturation
We quantified 806 known chromatin components and transcriptional regulators in our analysis (Supplementary Table 1, Fig. 3), providing insight into their dynamic behaviour during chromatin replication. Although less sensitive, western blotting of selected factors generally mirrored SILAC enrichments (Supplementary Fig. 3d). As expected, we found chaperones like CAF-1 and FACT, DNMT1 with co-factors UHRF1 and DMAP1, and the cohesin acetyltransferase ESCO2 enriched in nascent chromatin (Fig. 3b, 3c and 3d). The Asf1a and Asf1b histone chaperone also play important roles during replication24, 25, yet they were not enriched in nascent chromatin (Fig. 3d). Likewise, previous studies could not identify these chaperones at replication sites by immunofluorescence25. This probably reflects their dynamic behaviour and additional replication-independent functions. Unexpectedly, the centromeric H3 variant CENP-A was highly enriched in nascent chromatin (Fig. 3a). However, we did not identify the CENP-A histone chaperone HJURP. Other interesting factors enriched in nascent chromatin include the H3K9me3 reader and writer HP1 and SUV39h1/2 (Fig. 3e), the NASP histone chaperone (Fig. 3d), the H3K4me3 demethylase KDM5D/C (Supplementary Fig. 3e), the deacetylases HDAC8 and SIR2L, components of the NuA4 (YEATS4, MORF4L2, and BRD8) and CREBBP acetyltransferases, RIF1 and several nuclear lamins (LAMIN-A/C, Lamin B1, B2) (Supplementary Table 1).
Figure 3. Chromatin assembly and maturation.
(a) Nascent chromatin enrichment of canonical and replacement histones. (b) Nascent chromatin enrichment of DNA methyl transferases, cofactors and methyl-binding proteins. (c) Nascent chromatin enrichment of the cohesin complex and regulatory factors. (d) Nascent chromatin enrichment of histone chaperones. (e) Nascent chromatin enrichment of H3K9 (top) and H3K27 (bottom) methylation writers, erasers and readers. Only core components of PRC1 and PRC2 are shown. All enrichments are presented as in Figure 1h.
The canonical histones showed a ratio close to 0 (Fig. 3a), consistent with their rapid assembly onto newly synthesized DNA and thus a similar occupancy on nascent and mature chromatin. This was also true for H2AX and MacroH2A1 (Fig. 3a), likely reflecting that they are transferred during replication and additional incorporation does not take place within 2 hours of maturation. A large number of other components also showed similar occupancy in nascent and mature chromatin, including cohesin rings (Fig. 3c), CTCF, the chromatin remodellers SMARCA1, 4 and 5, HDAC1 and 2, and the PRC2 complex (Fig. 3e, Supplementary Table 1). This probably reflects that these components, like canonical histones, are only transiently released during replication.
Most linker histone H1 isoforms and the replacement variant H2AZ were enriched in mature chromatin (Fig. 3a). This implies that incorporation of these histones is separated from replication (replication-independent), but commences within 2 hours after DNA synthesis. Several methyltransferases catalysing active methylation marks such as H3K4me3 and H3K36me2 were also enriched in mature chromatin (e.g. MMSET, ASH1, MLL1 and MLL2) (Supplementary Fig. 3e)26, correlating with RNA Pol II association (Supplementary Fig. 3f). Strikingly, the de novo enzyme DNMT3A and the PRC1 complex also preferentially occupied mature chromatin (Fig. 3b and 3e). Most methyl-binding proteins behaved like DNMT3 (Fig. 3b), as expected if their recruitment awaits restoration of methylation by DNMT1-UHRF1. Thus, late recruitment characterizes a broad group of factors involved in both transcriptional activation (MMSET, MLL1, H2AZ) as well as repression (H1, PRC2, DNMT3).
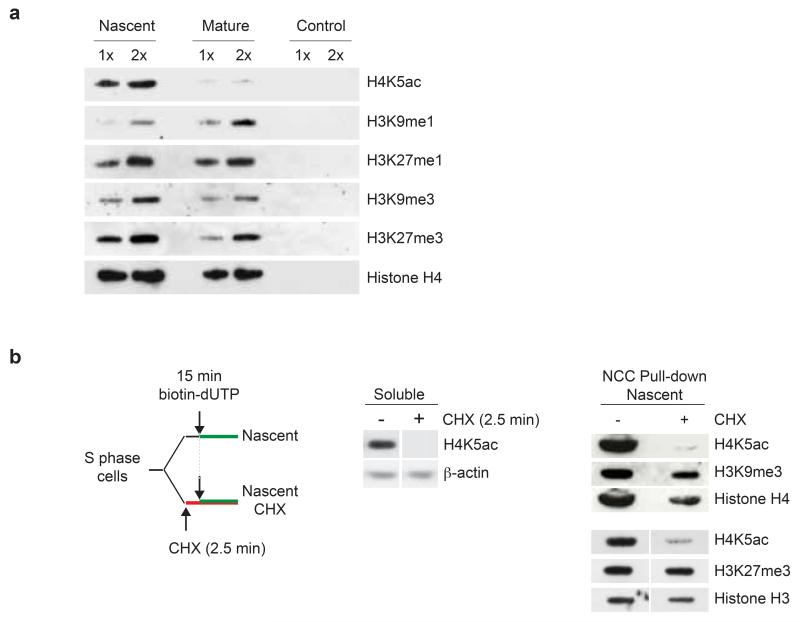
H3K9me3 and H3K27me3 are transferred with old histones
To explore the relationship between histone marks and the dynamics of the modifying enzymes, we followed H3K9me3 and H3K27me3 levels. In contrast to the rapid loss of H4K5K12diAc, neither H3K27me3 nor H3K9me3 levels changed significantly within the first two hours after replication (Fig. 4a). H3K9me1 levels were increased in mature chromatin, probably reflecting that a fraction of new histones are mono-methylated during this time-window. SILAC analysis of new histones has shown that establishment of H3K27me3 and H3K9me3 is slow, whereas mono-methylation is more rapid27, 28. Together with our NCC data, this predicts that H3K9me3 and H3K27me3 marks detected in nascent and mature chromatin originate from recycled old histones. To test this directly, we modified our NCC set-up to label DNA under conditions where new histone biosynthesis is repressed by cycloheximide (CHX). Short pre-treatment with CHX depleted the soluble pool of new histones marked by H4K5ac (Fig. 4b, left), thereby reducing their incorporation into nascent chromatin (Fig. 4b, right). However, H3K9me3 and H3K27me3 levels were not affected by the lack of new histone deposition (Fig. 4b, right), corroborating the long-standing model that histone marks are transferred with old histones onto newly synthesized DNA2, 3.
Figure 4. Histone mark dynamics during chromatin maturation.
(a) Histone marks analysed by western blot of NCC pull-downs of nascent and mature chromatin. 2x indicates the double amount of sample loaded in 1x. Antibody specificity was verified9. (b) Histone marks in nascent chromatin replicated in the presence of cycloheximide (CHX) to prevent new histone deposition. Left, experimental design. Middle, western blot of new histone H4 acetylated at K5 in the soluble fraction prior to biotin-dUTP labelling. Right, western blot of histone marks after NCC pull-down.
Predicting replication-coupled functions for uncharacterized proteins by in silico purification
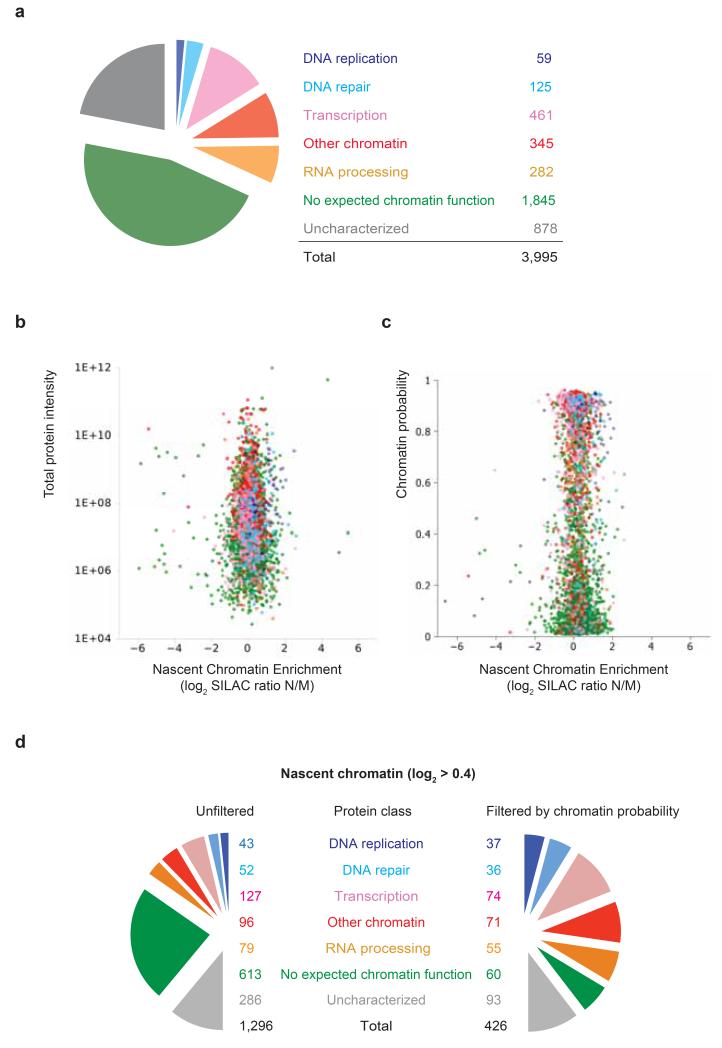
Our NCC-SILAC experiments quantified 3995 factors, manually distributed into 7 categories based on literature (Fig. 5a). Overall, more factors were enriched in nascent chromatin compared to mature, possibly due to its open and highly acetylated state. Importantly, DNA replication stands out as the most highly enriched category in nascent chromatin, suggesting a replication-coupled function of proteins that are similarly enriched (Fig. 5b and Supplementary Fig. 4). We found 878 functionally uncharacterized proteins as well as many factors with no expected chromatin function that for the most part are likely to be ‘hitchhikers’, considered biological/biochemical background13. Unexpectedly, some hitchhikers showed enrichment in nascent or mature chromatin (Supplementary Fig. 4), limiting the predictive value for functions of uncharacterized proteins. To add a second dimension to the data, such that functionally relevant proteins can be distinguished from background, we used a chromatin probability table for 7600 proteins (Kustatscher et al., in preparation). This is based on multi-classifier combinatorial proteomics, MCCP13, integrating many chromatin proteomics experiments unrelated to the current study but collectively indicating chromatin function. Applying chromatin probability as an in silico purification step to our data set, allowed to discriminate factors with a chromatin function from those with no expected chromatin function according to literature (Fig. 5c, Supplementary Table 1). Focusing on factors with nascent chromatin enrichment larger than the 40 most enriched core replication factors (SILAC log2 ratio >0.4), in silico purification remove 90 % of the factors with no expected chromatin function while keeping 90 % of the replication factors. A large number of uncharacterized proteins (60 %) were also removed, ultimately identifying 93 uncharacterized factors with high probability of having a function at replication forks or on newly replicated chromatin behind the fork (Supplementary Table 3).
Figure 5. Identification of novel proteins with a predicted function in chromatin replication.
(a) Pie-diagram illustrating the distribution of all proteins identified in NCC-SILAC into 7 functional classes (Supplementary Table 1). (b) Volcano plot showing the logarithmic ratio of protein intensities in the combined data set plotted against their median logarithmic ratio of heavy versus light peptides (nascent chromatin enrichment). (c) Plot of chromatin probability against nascent chromatin enrichment. (d) Barrel plot illustrating the distribution of proteins with a SILAC log2 ratio >0.4 before (left) and after (right) using chromatin probability to separate chromatin from non-chromatin proteins (Supplementary Table 3). Factors with a chromatin probability above 0.4 were considered chromatin, as this included roughly 90 % of the core replication factors (Fig. 1h).
Identification of novel chromatin replication factors
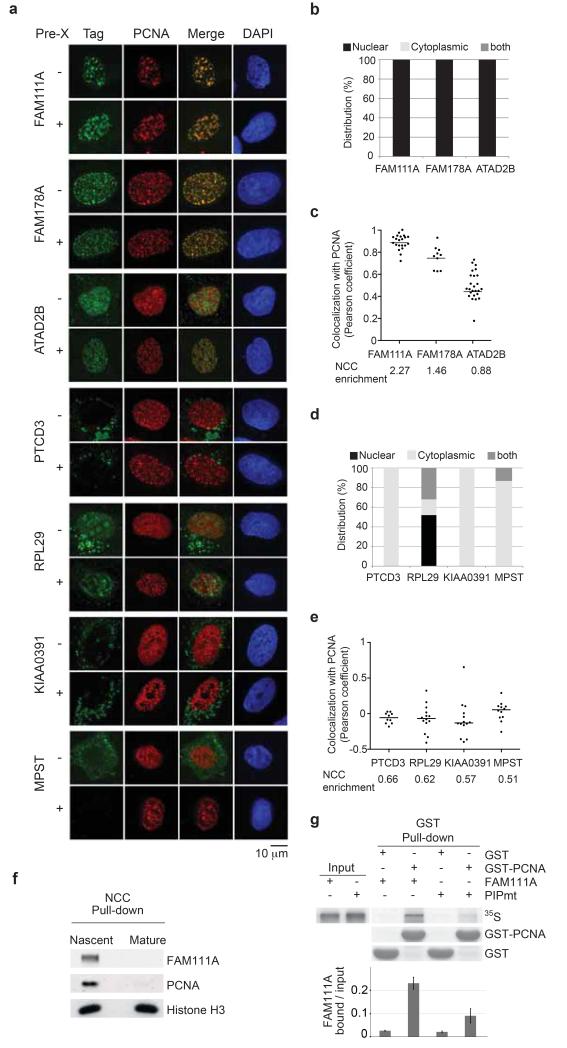
First, we tested the localization of seven uncharacterized proteins enriched on nascent chromatin, three with high chromatin probability and four with low probability (Kustatscher et al., in preparation). All three factors with a highly likely chromatin function, FAM111A, FAM178A and ATAD2B, showed nuclear localization, displayed a focal staining pattern and were resistant to Triton extraction (Fig. 6a-b and Supplementary Fig. 5a), as expected for chromatin bound proteins. In contrast, the four proteins with low chromatin probability were mainly cytoplasmic (Fig 6a-d and Supplementary Fig. 5c). FAM111A and FAM178A co-localized almost perfectly with PCNA, whereas ATAD2B showed partial co-localization consistent with the lower NCC enrichment (Fig. 6c and Supplementary Fig. 5b). We noted that both FAM111A and FAM178A contained putative PCNA interaction motifs, a PIP box and an APIM motive, respectively (Supplementary Fig. 6a). While mutation of the FAM178A APIM motif did not affect its localization (Supplementary Fig. 6b), disruption of the FAM111A PIP box led to diffuse pan-nuclear distribution (see below). We thus focused our attention on FAM111A and first confirmed the enrichment in nascent chromatin by western blot (Fig. 6f). Then we addressed PCNA binding by GST pull-down and found that FAM111A interacts directly with PCNA in PIP box dependent manner in vitro (Fig. 6g).
Figure 6. Localization of novel proteins with a predicted function in chromatin replication.
(a) Analysis of GFP- or FLAG-tagged proteins in U-2-OS cells stably expressing RFP-PCNA. Cells were fixed directly or after pre-extraction with Triton (Pre-X). Localization pattern (b, d) and co-localization with PCNA measured by Pearson coefficient in individual nuclei were scored after direct fixation (c, e) or Pre-X (Supplementary Figure 5a, 5c and 5b, 5d). Horizontal lines represent the median. (f) Western blot of NCC pull-downs. (g) In vitro analysis of PCNA binding using GST-PCNA to pull down in vitro translated 35S-labelled FAM111A wild type (WT) or PIP box mutant (PIPmt). The diagram shows the quantification of bound FAM111A relative to input with error bars representing the s.d. (n=3).
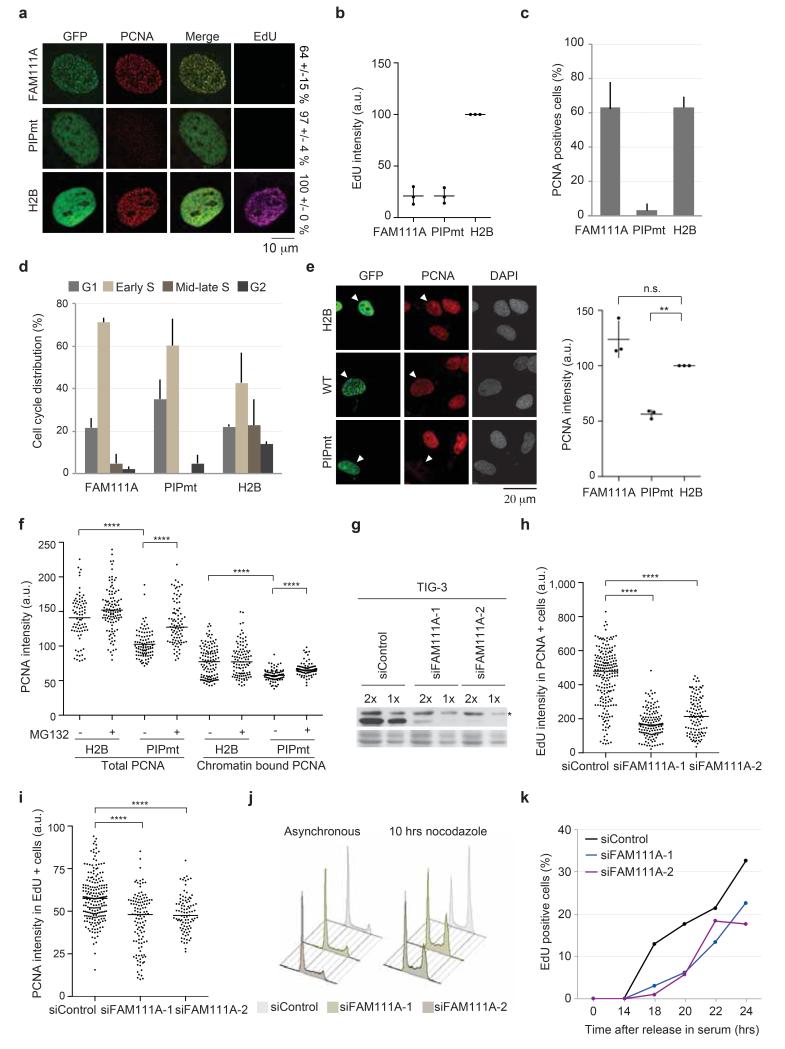
GFP-FAM111A co-localized with PCNA at replication sites, but at the same time impaired EdU incorporation and arrested cells in early S phase (Fig. 7a, 7b, Supplementary Fig. 6c). This probably reflects that PIP box-containing proteins upon overexpression can interfere with DNA replication by competing with other PCNA binding partners (i.e. DNA pol δ, DNA pol ε, FEN-1 and DNA ligase I)29-31. The FAM111A PIP box mutant (PIPmt) showed a pan-nuclear distribution (Fig. 7a), and, surprisingly, these cells were all negative for PCNA and EdU (Fig. 7b, 7c). This suggested that the mutant arrested cells outside S phase. We thus determined the cell cycle distribution based on MCM2 staining patterns and EdU labelling for one hour to detect slowly replicating cells32 (Supplementary Fig. 6d). Intriguingly, a substantial proportion of cells expressing the PIP mutant were in early S phase and incorporated low levels of EdU (Fig. 7d and Supplementary Fig. 6e), regardless that PCNA foci were hardly detectable (Fig. 7a, 7c, and Supplementary Fig. 6e). This suggested that the PIP mutant might affect PCNA loading and/or stability in a dominant negative manner. Quantitative analysis showed that the PIP mutant reduced the level of both chromatin-bound and total PCNA in the nucleus (Fig. 7e, 7f). Similar results were obtained in cells stably expressing RFP-PCNA from an exogenous promoter (Supplementary Fig. 6f), excluding indirect effects on transcription. We thus tested whether PCNA stability was affected by inhibiting the proteasome with MG132. This largely restored total PCNA levels, but only moderately increased loading of PCNA on chromatin (Fig. 7f).
Figure 7. FAM111A facilitates PCNA loading and S phase entry.
(a) Analysis of PCNA and EdU in Triton extracted U-2-OS cells. Localization patterns were quantified in transfected cells for 3 independent experiments. GFP-H2B was used as control. (b) Quantification of EdU intensities in transfected cells. The mean of 3 independent experiments with s.d. is shown, >200 cells scored per experiment. (c) Quantification of PCNA positive cells. Only transfected cells were scored. Error bars represent s.d. (n=3). The distribution of PCNA patterns is shown in Supplementary Figure 6c. (d) Cell cycle distribution based on combined analysis of MCM2 pattern and EdU incorporation in transfected cells. Supplementary Figure 6d shows representative patterns. Error bars represent s.d. (n=3), >75 cells counted per condition. (e) Analysis of total PCNA in transfected cells fixed without pre-extraction. (left) Representative images, arrowheads mark transfected cells. (right) Quantification, the mean of three independent experiments with s.d. is shown, (**) p = 0,022, (n.s.) non significant p = 0,13 (One sample t test). >200 cells scored per experiment. (f) Quantification of total and chromatin-bound PCNA in transfected cells treated or not with MG132. One representative experiment is shown. Horizontal lines represent the median, ****p < 0.0001 (unpaired t test, 77 < n). (g-k) FAM111A depletion in TIG-3 cells transfected with two independent siRNAs for 48 hours. (g) Western blot, 2x indicates the double amount of sample loaded in 1x. (*) unspecific band. Quantification of EdU incorporation (h) and chromatin-bound PCNA (i). Dot plots show EdU intensities in PCNA positive cells and PCNA intensities in EdU positive cells. Similar results were obtained in total cell populations and in U-2-OS cells (Supplementary Figure 6g-i). One representative experiment is shown. Horizontal lines represent the median, ****p < 0,0001 (unpaired t test, 108 < n). (j) Cell cycle profiles of cells treated without (left) or with (right) nocodazole for 10 hours. (k) S-phase entry scored by EdU incorporation after release from quiescence by re-stimulation with serum. One representative experiment is shown, >100 cells scored per time point. Statistics source data are available in Supplementary Table 4.
Next we used RNAi depletion to address directly whether FAM111A function is required for DNA replication. Depletion of FAM111A significantly impaired EdU incorporation (Fig. 7g, 7h, Supplementary Fig. 6g, 6h) and this was accompanied by reduced loading of PCNA onto chromatin (Fig. 7i, Supplementary Fig. 6i) in both primary TIG-3 primary fibroblasts and U-2-OS osteosarcoma cells. The cell cycle was not dramatically perturbed after 48 hours of FAM111A siRNA treatment (Fig. 7j). However, treatment with nocodazole to trap cells in mitosis showed that progression from G1 through S phase to G2/M was markedly delayed in the absence of FAM111A (Fig. 7j). To address G1/S transition directly, we released siRNA treated TIG-3 cells from quiescence and followed S phase entry by EdU incorporation. This revealed that FAM111A depleted cells enter S phase and start DNA replication substantially later than control cells (Fig. 7k).
DISCUSSION
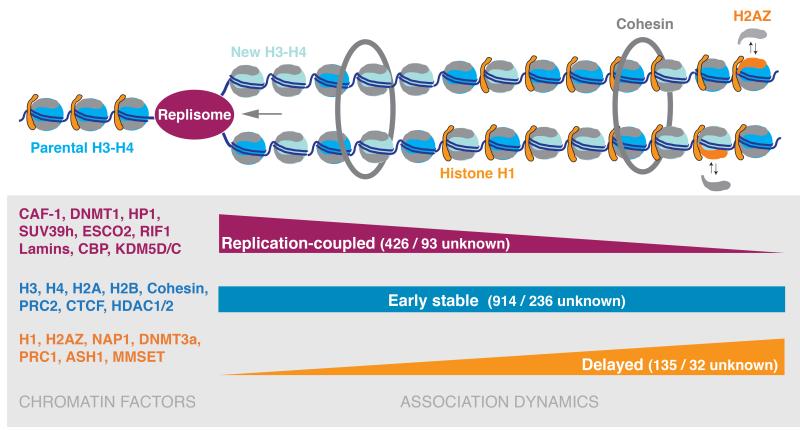
Here, we describe a strategy for profiling dynamics of the chromatin proteome and histone marks during replication. We provide association dynamics for 3995 factors during DNA replication and chromatin maturation, and predict a replication-coupled function for 93 uncharacterized proteins. Our analysis reveals that replication dramatically perturbs chromatin composition, with 561 chromatin factors changing association within the first hours post-replication (Fig. 8). This correlates with deacetylation of new histones, but surprisingly repressive marks like H3K9me3 and H3K27me3 remain largely unchanged. Overall, this large quantitative analysis provides a unique resource to understand epigenome maintenance in proliferating cells.
Figure 8. Chromatin dynamics during DNA replication.
Association dynamics of chromatin proteins during the first two hours of chromatin assembly and maturation determined by NCC-SILAC enrichment and chromatin probability (Supplementary Table 1). Selected factors are listed along with the total number of chromatin proteins identified and the functionally uncharacterized.
Two other methods for isolation of proteins on new DNA, iPOND and Dm-ChP, were recently described33, 34. These rely on EdU labeling of DNA and chemical ligation of biotin post-fixation (Click-iT). Introduction of biotin-dUTP requires a short hypotonic shift, but circumvents chemical ligation of biotin. Furthermore, NCC is developed for large-scale proteomics and thus optimized with respect to i) strong cross-linking to preserve larger chromatin-bound complexes, ii) isolation of nuclei to reduce contaminants, and iii) use of HeLa S3 spinner cultures and SILAC to quantify chromatin dynamics. Our analysis captures the first well-established steps in chromatin replication: DNA unwinding (CMG helicase), DNA synthesis (Pol α, β and ε), Okazaki fragment processing (DNA ligase I, FEN1), nucleosome assembly (CAF-1), maintenance DNA methylation (DNMT1-UHRF1) and establishment of sister chromatid cohesion (Esco2). The comprehensive identification of known fork components by NCC probably reflects our chromatin preparation method along with SILAC-based quantification of chromatin composition at two distinct maturation states.
NCC-SILAC should thus be a perfectly controlled approach to discover novel DNA replication and chromatin maturation factors. Surprisingly, many proteins enriched in nascent chromatin had well-established functions elsewhere in the cell. While some may function in replication, others likely constitute background. This is possible if the functional and structural differences between nascent and mature chromatin also impact on the background, in which case there is no perfect control. To pick the best candidates for follow-up studies, we thus took advantage of a chromatin-probability table for 7600 proteins established by proteomics of interphase chromatin (Kustatscher et al., in preparation). By this means, we shortlist 93 uncharacterized proteins that with high probability have a function at replication forks or in nascent chromatin. Corroborating this resource, three of the top candidates exclusively (FAM111A, FAM178A) or in part (ATAD2B) localize to replication sites. We thus anticipate that FAM111A and FAM178A primarily function in chromatin replication, while ATAD2B serve additional roles in chromatin. Indeed, FAM111A interacts directly with PCNA and promotes S phase entry and DNA synthesis. A direct function of FAM111A at replication forks might explain recent findings that SV40 large T antigen binds FAM111A to overcome host-range restriction35 and that variations in the chromosomal loci harbouring FAM111A (11q12) confer susceptibility to prostate cancer36. We speculate that FAM111A could play a role in replication initiation by affecting PCNA loading. Overexpression of the FAM111A PIP mutant compromises PCNA stability and chromatin-binding. Yet, stabilization of PCNA did not fully restore chromatin-binding (Fig. 7f), and overexpression of PCNA together with the PIP mutant could not rescue replication (data not shown). The FAM111A PIP mutant might thus interfere with PCNA loading or maintenance on chromatin in a dominant negative manner. Consistent with this, cells lacking FAM111A show low levels of chromatin-bound PCNA and impaired replication.
Our study defines the dynamic changes in chromatin composition during replication (Fig. 8). The challenge is now to identify recruitment mechanisms and uncover functional dependencies. In example, the delayed recruitment of linker histone H1 support the model that deacetylation of new histones pave the way for linker histone binding and compaction6, 37. PRC1 also promotes compaction38 and together with PRC2 constitute an important cellular memory system3. An in vitro study of SV40 replication proposed that PRC1 remains bound to chromatin during DNA replication39. Our work argues that a substantial part of PRC1 is recruited later during chromatin maturation. In contrast the PRC2 complex is present in both nascent and mature chromatin, consistent with rapid recruitment to nascent chromatin by parental histones carrying H3K27me340, 41. As H3K27me3 levels did not change, other features must contribute to delayed recruitment of PRC1.
The centromeric H3 variant CENP-A was enriched in nascent chromatin (Fig. 3a), despite that stable de novo CENP-A incorporation mainly occurs in early G142, 43. We envision that CENP-A either associates spuriously to accessible genomic regions such as nascent chromatin (this study) and sites of DNA repair44, or that structural changes in CENP-A nucleosomes after replication45 influence cross-linking efficiency. The histone variant H2AZ also decorates centric chromatin in addition to pericentric regions, promoters and gene bodies42. In contrast to CENP-A, H2AZ levels increase with chromatin maturation. This probably reflects dilution of H2AZ during replication46, followed by replication-independent incorporation of new H2AZ42.
NCC provides a tool to monitor transmission of histone modifications that complements current SILAC-based approaches to follow new and old histones27, 28. We took advantage of short cycloheximide treatment as a crude, but instant, approach to block new histone biosynthesis8, 47. Our data suggest that H3K27me3 and H3K9me3 in nascent chromatin largely reflect transmission with old histones, confirming current models of epigenetic memory2, 3. A recent study in drosophila embryos could not detect H3K4me3 and H3K27me3 on newly synthesized DNA and proposed that these marks were not inherited with parental histone48. While transmission of histone marks may differ in drosophila embryos and human cells, it is important to note that our results are entirely consistent with recent SILAC-based comparison of new and old histone in cycling human cells27, 28. Together these complementary studies support a model in which H3K9me3 and H3K27me3 are transferred with old histones and imposed on new histones with slow kinetics, while mono-methylation is more rapid.
NCC combined with quantitative mass spectrometry is a strong tool to address how the epigenetic framework is maintained in dividing cells. To understand cellular memory it is evident that we need to unravel how chromatin is replicated. The resource and technology we provide here should thus be relevant to questions about cell identity during development, proliferative exhaustion of aging cells and epigenetic alterations in cancer cells that experience uncontrolled proliferation. Given the wide use of chemotherapeutic drugs that target replication, NCC could also provide an avenue to drug design.
ONLINE METHODS
Cell lines
For SILAC experiment, HeLa S3 cells in spinners were grown in SILAC medium depleted for Arginine and Lysine (Thermo scientific RPMI 1640 Medium) and supplemented with dialyzed FBS (#26400-036 Invitrogen), MEM non-essential amino acid mix (#11140 Invitrogen), Glutamax (#35050-038 Invitrogen), 66 mg/L of Arginine and 100 mg/L of Lysine (SIGMA). Heavy medium was complemented with heavy Lysine and Arginine (Cambridge lab #CNLM-291-0.25, #CLM-2265-0). Cells were maintained for 8 to 9 divisions in heavy or light medium prior to NCC analysis.
Synchronization and drug treatment
Cells were synchronized at the G1/S border by single thymidine block (2 mM, 17 hours) and released into fresh media containing deoxycytidine (24 μM). TIG-3 cells were blocked in G2/M by Nocodazole treatment (100ng/ml, 10 hours) 42 hours after siRNA transfection. To follow S phase entry, TIG-3 cells were serum starved (0 % FBS) for 40 hours and released by replating in 10 % FBS. Cells were treated with cycloheximide (CHX, 200 μg/ml) for 2.5 minutes prior to biotin-dUTP labelling and with MG132 (5μM) for 6 hours.
Immunofluorescence
Cells grown on coverslips or cytospined onto glass slides were pre-extracted with CSK (10 mM PIPES pH7, 100 mM NaCl, 300 mM Sucrose, 3 mM MgCl2) 0.5% Triton for 5 minutes on ice to remove soluble proteins, and fixed with 4% formaldehyde for 7 minutes. For biotin detection, we used the streptavidin conjugated to Alexa Fluor 488 (Invitrogen #S32354). For EdU (5-ethynyl-2′-deoxyuridine) labelling, cells were incubated with 40 μM EdU (Invitrogen #A10044) for 10-15 minutes (Figure 7a, 7h and 7k) or 1 hour (Figure 7b, 7d). EdU was detected using Click-IT Alexa Fluor 488 azide (Invitrogen #A10266) and Click-IT cell reaction buffer kit (Invitrogen #C10269). Images were acquired using an Axiovert 200M confocal microscope equipped with LSM510 laser module (Zeiss), or a DeltaVision system, analysed and quantified with SoftWorKx 5.0.0 software (Applied Precision). For statistical analysis, data were processed in Prism.6 using t test when appropriate. For co-localization analysis, 3D images of single cells were acquired and deconvolved before co-localization was measured by the Pearson Coefficient using SoftWorKx.
Nascent Chromatin Capture
HeLa S3 cells growing in suspension were released from a single thymidine block for 3 hours. For SILAC analysis, 5 × 108 cells were used as starting material for heavy and light cultures. For biotin-dUTP labeling, cells were incubated for 5 minutes in a hypotonic buffer (50 mM KCl, 10 mM Hepes) containing biotin-dUTP and resuspended into fresh cell culture medium. Cells were fixed in 2% formaldehyde after 20 minutes (nascent chromatin) or chased for 2 hours in fresh medium prior to fixation (mature chromatin). Cross-linking was stopped after 15 minutes by adding glycine to a final concentration of 1 % and incubating 5 minutes at room temperature. Nuclei were mechanically isolated in a sucrose buffer (0.3 M Sucrose, 10 mM HEPES-NaOH pH 7.9, 1% Triton X-100, 2 mM MgOAc), and chromatin was solubilized by sonication in a Diagenode Bioruptor at 4 °C in sonication buffer (10 mM HEPES-NaOH pH 7.9, 100 mM NaCl, 2 mM EDTA pH 8, 1 mM EGTA pH 8, 0.2% SDS, 0.1% Sodium Sarkosyl, 1mM PMSF) (Bioruptor setting: High, 28 cycles of 30 sec sonication and 90 sec pause). Biotinylated chromatin fragments were purified on streptavidin-coated magnetic beads (MyC1 Streptavidin beads) by over night end-over-end rotation at 4 °C and 5 stringent washes (10 mM HEPES-NaOH pH 7.9, 100 mM NaCl, 2 mM EDTA pH 8, 1 mM EGTA pH 8, 0.1% SDS, 1 mM PMSF). For SILAC analysis, nascent and mature samples were mixed in the last wash. To release chromatin and reverse the cross-link, beads were boiled in LSB for 40 min at 100 °C, including a brief vortex and short spin every 10 min to prevent drying.
Proteomics
Proteins from the NCC purification were digested with trypsin. The resulting peptides were fractionated by offline SCX and analysed on an LTQ Orbitrap Velos with online UPLC. Initial data processing was done using MaxQuant50. We performed in-gel digestion51 on NCC purified proteins and fractionated the peptides using a PolyLC SCX column on Dionex UltiMate 3000. The resulting 30 fractions were desalted using StageTips52 and separated online on a Waters Acquity UPLC. Each fraction was run on a 2, 3 or 4 hours gradient according to estimated material from the offline UV trace; fractions of very low content were combined. We used a formic acid/acetonitrile buffer system, and gradients optimised for complex samples. Analysis was performed with an LTQ Orbitrap Velos (Thermo Fisher Scientific) using a nanoelectrospray ion source. Precursor scans were acquired with lock mass in the Orbitrap, and the top 20 ions, with dynamic exclusion, were selected for CID fragmentation and measurement in the LTQ. Initial data processing was done using MaxQuant v 1.2.2.550, using the final release of IPI. We used the default parameters, except that we adjusted minimum unique peptides to 1 and minimum ratio count to 1. We also use MaxQuant to combine our data with the chromatin probability data. We apply normalization, and derived statistics from the MaxQuant evidence file using in-house Perl scripts. Normalization is based on the ratio of H4, which we assume to be stable on chromatin from nascent to 2 hours mature. For each protein we represent the spread of data using the median absolute deviation (m.a.d.), which we observe to be more robust than the standard deviation of the mean (s.e.m.) for those proteins that have a great many quantitative measurements, and it is more in keeping with reporting the median ratio observed for each protein. It should be noted that the m.a.d. is estimated to be approximately 0.8 of the s.e.m.
Quantitation of histones by mass spectrometry
The large number of histone genes with highly conserved but not completely identical sequences makes it difficult to unambiguously assign peptides to one particular histone gene. In order to simplify data-processing of histone sequences, a simplified human histone database was created. This database contains only one representative sequence of each of the four replication-dependent core histones H2A, H2B, H3 and H4. The protein sequence most closely approaching the consensus sequence was chosen as representative sequence. In addition, this simplified histone database contains all non-canonical core histones (variants, isoforms of variants, but not pseudogenes) and all forms of the linker histone H1. Canonical, replication-dependent histones were distinguished from histone variants according to53 and, where necessary, based on Uniprot annotations and sequence comparisons. For histone quantitation, MS raw data from the NCC experiments where searched against this simplified human histone database using MaxQuant 1.2.2.5 software, omitting potential post-translational modifications from the search and using only unique peptides for quantitation. Each experiment was normalized to histone H4.
Chromatin probability
We used a chromatin probability table for 7600 proteins (Kustatscher et al., in preparation). This table is derived using multi-classifier combinatorial proteomics, MCCP13, to integrate many chromatin proteomics experiments unrelated to the current study but collectively indicating chromatin function as assessed by known chromatin proteins.
GST-pull down experiments
35S-labelled FAM111A WT and PIP mutant were produced using the TnT T7 Quick Coupled Transcription/Translation System (Promega) and incubated with recombinant GST-PCNA proteins in binding buffer (150 mM NaCl, 0.2% NP-40, 50 mM Tris, pH 7.6, 2 mM EDTA, 5% glycerol and phosphatase inhibitors) for 3 hours at 4°C. The reactions were washed 6 times in binding buffer containing 300 mM NaCl and analysed by autoradiography and western blot.
Transfection
siRNAs and plamids were introduced by Oligofectamine (Invitrogen) and Lipofectamin (Invitrogen), respectively, according to manufactures recommendation.
Statistical methods
Fig. 7e, 7h-j and Supplementary Fig. 1d, 6h, the values for each independent experiment are provided in Supplementary Table 4. The median is shown. An unpaired t test with Welch’s correction has been performed. The normality of the data has been tested by analyzing the histogram distribution in Prism.6, and by Q-Q plot in SAS (version 9.2 SAS institute, cary, n.c. USA). The difference of variance between two populations was measured in Prism.6. We used a Welch’s correction when the variances were not equal. p values are provided and defined in the legend of the figure. Fig. 7f, each value on the graph is the median from one experiment. Values for each independent experiment are provided in Supplementary Table 4. The mean is shown with the s.d. We have used a one sample t test to compare the two groups to the control in SAS. p values are provided and defined in the legend of the figure.
Primary antibodies, plasmids and siRNA
PCNA (Abcam ab29, clone PC10, 1/1000), FLAG (Sigma F7425, 1/2000), H4K12ac (Upstate Millipore 07-595, 1/1000), H4K5ac (Abcam ab51997, clone EP1000Y, 1/1000), Histone 3 (Abcam ab10799, clone mAbcam 10799, 1/2000), Histone H4 (Upstate Millipore 05-858, clone 62-141-13, 1/1000), Histone H2B (abcam ab1790, 1/1000), Smc3 (Abcam ab9263, 1/1000), Ctf18 (Bethyl A301-883A, 1/500), RPA/p70 (Abcam ab79398, clone EPR3472, 1/500), CAF1p15054 (1/500), CAF1p6054 (1/500), HP1γ (Abcam ab56978, (1/500), Dnmt1 (Abcam ab16632, 1/5000), H3K9me1 (Upstate Millipore 07-450, 1/1000), H3K9me3 (Upstate 07-442, 1/1000), H3K27me1 (Upstate Millipore 07-448, 1/500), H3K27me3 (CST 9756, 1/500), β-actin (Sigma A5441, clone AC-15, 1/25000), FAM111A (Sigma HPA040176, 1/500), MCM2 (BD Transduction laboratories 610701, clone 46/BM28, 1/1000). The FAM111A-tGFP (RG210012), FAM178A-FLAG (RC227146), PTCD3-FLAG (RC204119), RPL29-FLAG (RC203616), KIAA0391-FLAG (RC206805) and MPST-FLAG (RC202466) plasmids are from OriGene. The FAM111A PIP mutant was generated by site directed mutagenesis of Y24A and F25A, and FAM178A APIM mutant was generated by site directed mutagenesis of Y142A. ATAD2B-FLAG is a kind gift from K. Helin. siRNAs against FAM111A were GGAAGAUAACCACAUAUUUGGCAGG (siFAM111A-1, OriGene) and AGAGCUAAAUGCUUGAUUAGAAATG (siFAM111A-2, OriGene).
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank J. Dejardin for fruitful discussions, K. Helin for reagents, Ib J. Christensen for support on statistics, and C. Wu, W.C. Earnshaw and Z. Jasencakova for critical reading of this manuscript. The A.G. laboratory is supported by a European Research Council Starting Grant (ERC2011StG, no. 281,765), the Lundbeck Foundation, the Danish Cancer Society, the Danish National Research Foundation (DNRF82) and Medical Research Council, the Novo Nordisk Foundation and FP7 Marie Curie Actions ITN Nucleosome4D. The Wellcome Trust generously supported this work through a Senior Research Fellowship to J.R. (084229), two Wellcome Trust Centre Core Grants (077707, 092076) and an instrument grant (091020). CA was supported by fellowships from HFSP and the Danish Medical Research Council. GK was supported by a FEBS Long-Term fellowship.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Mechali M. Eukaryotic DNA replication origins: many choices for appropriate answers. Nat Rev Mol Cell Biol. 2010;11:728–738. doi: 10.1038/nrm2976. [DOI] [PubMed] [Google Scholar]
- 2.Alabert C, Groth A. Chromatin replication and epigenome maintenance. Nat Rev Mol Cell Biol. 2012;13:153–167. doi: 10.1038/nrm3288. [DOI] [PubMed] [Google Scholar]
- 3.Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet. 2010;11:285–296. doi: 10.1038/nrg2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Worcel A, Han S, Wong ML. Assembly of newly replicated chromatin. Cell. 1978;15:969–977. doi: 10.1016/0092-8674(78)90280-5. [DOI] [PubMed] [Google Scholar]
- 6.Annunziato AT, Seale RL. Histone deacetylation is required for the maturation of newly replicated chromatin. J Biol Chem. 1983;258:12675–12684. [PubMed] [Google Scholar]
- 7.DePamphilis ML, Wassarman PM. Replication of eukaryotic chromosomes: a close-up of the replication fork. Annu Rev Biochem. 1980;49:627–666. doi: 10.1146/annurev.bi.49.070180.003211. [DOI] [PubMed] [Google Scholar]
- 8.Annunziato AT. Assembling chromatin: The long and winding road. Biochim Biophys Acta. 2012;1819:196–210. doi: 10.1016/j.bbagrm.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 9.Sobel RE, Cook RG, Perry CA, Annunziato AT, Allis CD. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc Natl Acad Sci U S A. 1995;92:1237–1241. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sirbu BM, et al. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev. 2011;25:1320–1327. doi: 10.1101/gad.2053211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taddei A, Roche D, Sibarita JB, Turner BM, Almouzni G. Duplication and maintenance of heterochromatin domains. J Cell Biol. 1999;147:1153–1166. doi: 10.1083/jcb.147.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dejardin J, Kingston RE. Purification of proteins associated with specific genomic Loci. Cell. 2009;136:175–186. doi: 10.1016/j.cell.2008.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohta S, et al. The protein composition of mitotic chromosomes determined using multiclassifier combinatorial proteomics. Cell. 2010;142:810–821. doi: 10.1016/j.cell.2010.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maya-Mendoza A, Olivares-Chauvet P, Kohlmeier F, Jackson DA. Visualising chromosomal replication sites and replicons in mammalian cells. Methods. 2012 doi: 10.1016/j.ymeth.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 15.Koberna K, et al. Nuclear organization studied with the help of a hypotonic shift: its use permits hydrophilic molecules to enter into living cells. Chromosoma. 1999;108:325–335. doi: 10.1007/s004120050384. [DOI] [PubMed] [Google Scholar]
- 16.Ong SE, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 17.Maser RS, et al. Mre11 complex and DNA replication: linkage to E2F and sites of DNA synthesis. Mol Cell Biol. 2001;21:6006–6016. doi: 10.1128/MCB.21.17.6006-6016.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Betous R, et al. SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication. Genes Dev. 2012;26:151–162. doi: 10.1101/gad.178459.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duro E, et al. Identification of the MMS22L-TONSL complex that promotes homologous recombination. Mol Cell. 2010;40:632–644. doi: 10.1016/j.molcel.2010.10.023. [DOI] [PubMed] [Google Scholar]
- 20.Bubeck D, et al. PCNA directs type 2 RNase H activity on DNA replication and repair substrates. Nucleic Acids Res. 2011;39:3652–3666. doi: 10.1093/nar/gkq980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nielsen FC, Jager AC, Lutzen A, Bundgaard JR, Rasmussen LJ. Characterization of human exonuclease 1 in complex with mismatch repair proteins, subcellular localization and association with PCNA. Oncogene. 2004;23:1457–1468. doi: 10.1038/sj.onc.1207265. [DOI] [PubMed] [Google Scholar]
- 22.Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 23.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 24.Groth A, et al. Regulation of replication fork progression through histone supply and demand. Science. 2007;318:1928–1931. doi: 10.1126/science.1148992. [DOI] [PubMed] [Google Scholar]
- 25.Jasencakova Z, et al. Replication stress interferes with histone recycling and predeposition marking of new histones. Mol Cell. 2010;37:736–743. doi: 10.1016/j.molcel.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 26.Albert M, Helin K. Histone methyltransferases in cancer. Semin Cell Dev Biol. 2010;21:209–220. doi: 10.1016/j.semcdb.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 27.Scharf AN, Barth TK, Imhof A. Establishment of histone modifications after chromatin assembly. Nucleic Acids Res. 2009;37:5032–5040. doi: 10.1093/nar/gkp518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu M, Wang W, Chen S, Zhu B. A model for mitotic inheritance of histone lysine methylation. EMBO Rep. 2011;13:60–67. doi: 10.1038/embor.2011.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fridman Y, et al. Subtle alterations in PCNA-partner interactions severely impair DNA replication and repair. PLoS Biol. 2010;8:e1000507. doi: 10.1371/journal.pbio.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li R, Waga S, Hannon GJ, Beach D, Stillman B. Differential effects by the p21 CDK inhibitor on PCNA-dependent DNA replication and repair. Nature. 1994;371:534–537. doi: 10.1038/371534a0. [DOI] [PubMed] [Google Scholar]
- 31.Warbrick E, Lane DP, Glover DM, Cox LS. A small peptide inhibitor of DNA replication defines the site of interaction between the cyclin-dependent kinase inhibitor p21WAF1 and proliferating cell nuclear antigen. Curr Biol. 1995;5:275–282. doi: 10.1016/s0960-9822(95)00058-3. [DOI] [PubMed] [Google Scholar]
- 32.Masata M, Juda P, Raska O, Cardoso MC, Raska I. A fraction of MCM 2 proteins remain associated with replication foci during a major part of S phase. Folia Biol (Praha) 2011;57:3–11. doi: 10.14712/fb2011057010003. [DOI] [PubMed] [Google Scholar]
- 33.Sirbu BM, Couch FB, Cortez D. Monitoring the spatiotemporal dynamics of proteins at replication forks and in assembled chromatin using isolation of proteins on nascent DNA. Nat Protoc. 2012;7:594–605. doi: 10.1038/nprot.2012.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kliszczak AE, Rainey MD, Harhen B, Boisvert FM, Santocanale C. DNA mediated chromatin pull-down for the study of chromatin replication. Sci Rep. 2011;1:95. doi: 10.1038/srep00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fine DA, et al. Identification of FAM111A as an SV40 host range restriction and adenovirus helper factor. PLoS Pathog. 2012;8:e1002949. doi: 10.1371/journal.ppat.1002949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akamatsu S, et al. Common variants at 11q12, 10q26 and 3p11.2 are associated with prostate cancer susceptibility in Japanese. Nat Genet. 2012;44:426–429. doi: 10.1038/ng.1104. [DOI] [PubMed] [Google Scholar]
- 37.Perry CA, Annunziato AT. Histone acetylation reduces H1-mediated nucleosome interactions during chromatin assembly. Exp Cell Res. 1991;196:337–345. doi: 10.1016/0014-4827(91)90269-z. [DOI] [PubMed] [Google Scholar]
- 38.Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- 39.Francis NJ, Follmer NE, Simon MD, Aghia G, Butler JD. Polycomb proteins remain bound to chromatin and DNA during DNA replication in vitro. Cell. 2009;137:110–122. doi: 10.1016/j.cell.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hansen KH, et al. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- 41.Margueron R, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boyarchuk E, Montes de Oca R, Almouzni G. Cell cycle dynamics of histone variants at the centromere, a model for chromosomal landmarks. Curr Opin Cell Biol. 2011;23:266–276. doi: 10.1016/j.ceb.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 43.Jansen LE, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeitlin SG, et al. Double-strand DNA breaks recruit the centromeric histone CENP-A. Proc Natl Acad Sci U S A. 2009;106:15762–15767. doi: 10.1073/pnas.0908233106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bui M, et al. Cell-Cycle-Dependent Structural Transitions in the Human CENP-A Nucleosome In†Vivo. Cell. 2012;150:317–326. doi: 10.1016/j.cell.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nekrasov M, et al. Histone H2A.Z inheritance during the cell cycle and its impact on promoter organization and dynamics. Nat Struct Mol Biol. 2012;19:1076–1083. doi: 10.1038/nsmb.2424. [DOI] [PubMed] [Google Scholar]
- 47.Sariban E, Wu RS, Erickson LC, Bonner WM. Interrelationships of protein and DNA syntheses during replication of mammalian cells. Mol Cell Biol. 1985;5:1279–1286. doi: 10.1128/mcb.5.6.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petruk S, et al. TrxG and PcG Proteins but Not Methylated Histones Remain Associated with DNA through Replication. Cell. 2012;150:922–933. doi: 10.1016/j.cell.2012.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dimitrova DS, Todorov IT, Melendy T, Gilbert DM. Mcm2, but not RPA, is a component of the mammalian early G1-phase prereplication complex. J Cell Biol. 1999;146:709–722. doi: 10.1083/jcb.146.4.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
REFERENCES ONLINE METHODS
- 50.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 51.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 52.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrixassisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 53.Marzluff WF, Gongidi P, Woods KR, Jin J, Maltais LJ. The human and mouse replication-dependent histone genes. Genomics. 2002;80:487–498. [PubMed] [Google Scholar]
- 54.Green CM, Almouzni G. Local action of the chromatin assembly factor CAF-1 at sites of nucleotide excision repair in vivo. EMBO J. 2003;22:5163–5174. doi: 10.1093/emboj/cdg478. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.