Abstract
A polymorphism in the UL42 – UL43 region of the human cytomegalovirus genome has been characterized by nucleotide sequence analysis, revealing a 929-bp insertion following nt 54,612 relative to the published strain AD169-UK genome sequence (M. S. Chee et al., 1990, Curr. Top. Microbiol. Immunol. 154, 125 – 170). Although AD169-UK exhibited polymorphism in this genomic region, other CMV strains (Towne, Toledo, and AD169-ATCC) carried only the newly characterized longer form. The additional sequence altered the assignment of UL42 and UL43 open reading frames. UL42 decreased in size from 157 to 125 codons, retaining 76 of the previously reported carboxyl terminal codons, and UL43 increased in size from 187 to 423 codons, retaining 185 of the previously reported amino terminal codons. This additional sequence makes UL43 a more conserved betaherpesvirus US22 family member. Only AD169-UK exhibited restriction fragment length polymorphism in this region, suggesting that a deletion occurred during the propagation of this strain in cell culture. The additional sequence should be considered a bona fide part of the cytomegalovirus genome and the AD169 genome size should be corrected to 230,283 bp.
Introduction
Human cytomegalovirus (CMV) is a ubiquitous pathogen causing a variety of diseases primarily in immuno-compromised hosts and in the fetus (Alford and Britt, 1995). This virus exhibits a complex genome organization (Mocarski, 1995) and carries a linear double stranded DNA genome 229,354 bp in size based on the nucleotide sequence analysis of a set of HindIII clones derived from the AD169 genome (Oram et al., 1982) and limited analysis of cosmid clones (Fleckenstein et al., 1982) across HindIII sites (Chee et al., 1990) (EMBL Accession No. X17403). AD169 was isolated from cultured tissue taken from adenoids of a 7-year-old girl (Rowe et al., 1956); however, early passage samples of this widely used strain or descriptions of early passage history are not available. The AD169 genome sequence has been the basis for studies on CMV (Mocarski, 1995) despite the fact that it had been derived laboratory-propagated strain. Single nucleotide corrections to the original AD169 genome sequence have been reported in the UL102 (Smith and Pari, 1995) and US28 (Neote et al., 1993) genes. Recently, the genomes of strains AD169 and Towne have both been shown to contain sequence rearrangements and to be much less complex than low-passage strains (Cha et al., 1996).
We had previously observed very marked differences in the ability of CMV strains Towne, Toledo, and AD169 to replicate efficiently in thymic medullary epithelial cells in human thymus– liver implants carried by SCID-hu mice. We noted differences in the replication efficiency of the AD169 strain from two sources (ATCC vs UK) (Brown et al., 1995). We noticed that strain AD169-UK, which replicated several orders of magnitude better than AD169-ATCC, exhibited a restriction fragment length polymorphism within the HindIII M region of the viral genome that had been noted in one earlier analysis (Oram et al., 1982) but had not been noticed by others (Fleckenstein et al., 1982; Spector et al., 1982; Weststrate et al., 1983). Although two species of HindIII M fragment were observed in the work by Oram and colleagues, the smaller and more abundant fragment was cloned as representative of the viral genome (Oram et al., 1982) and was subsequently sequenced as a part of the strain AD169 genome analysis which led to the assignment of ORFs UL41 through UL48 (Chee et al., 1990).
Materials and Methods
Virus and cells
CMV strain AD169-ATCC, obtained from the American Type Culture Collection (ATCC VR 538), AD169-UK, obtained from H. Browne (Cambridge, United Kingdom), Toledo, obtained from S. Plotkin (Pasteur Merieux, Lyon, France), and Towne, passage 134, obtained from S. Plotkin were grown in low passage (4 – 20) human foreskin fibroblasts (HFs) using Dulbecco's minimal essential medium (Gibco-BRL) supplemented with 10% NuSerum (Collaborative Biomedical), amino acids, and antibiotics as previously described (Brown et al., 1995). Viral DNA was prepared as previously described (Spaete and Mocarski, 1985).
Plasmids and sequence analysis
Cosmids pCM1017 and pCM1049 have been described (Fleckenstein et al., 1982). pON2201 carries the HindIII M fragment from cosmid pCM1049 (Fleckenstein et al., 1982) cloned into pGEM3Zf/(Promega). pON2202 carries sequence between the HindIII Z/M site (nt 54,144) and a HpaI site (nt 54,813; codon 118 of UL43) within HindIII M and was prepared by deleting sequences from the HpaI site through the HindIII M/F site (nt 64,521) of pON2201. pON2128 carries a PstI fragment representing sequences at the left end of HindIII M between nt 54,767 and 60,045 of AD169 in pGEM3Zf/.
Nucleotide sequence of pON2202 was determined on both strands by automated dideoxy sequence analysis (Lagenaur et al., 1994; Sanger et al., 1977) at the Stanford PAN facility using the following primers: 5′ CAGGAAACAGCTATGACC 3′, 5′ GGCGACGGCGAACTAAC 3′, 5′ CTGGCTCCCGTTCAAGAC 3′, 5′ GGCAAACGACGGCTT-TTC 3′, 5′ CGCAGCACAGGAACCTCT 3′, 5′ TGTAAAACGACGGCCAGT 3′, 5′ CGTGGTGGAATCGCTGTG 3′, 5′ TGGTGAACGAGAGCGGCG 3′, 5′ CACTGCCACCGACATGGA 3′, and 5′ CGGGGACGCAAGCATCAT 3′. All alignments were performed with the MCB Search Launcher program (Human Genome Center, Baylor College of Medicine) and assembled using SeqVu 1.0.1 (Garvan Institute of Medical Research, Sydney, Australia).
Results and Discussion
We compared the HindIII digestion patterns of AD169 DNA from virus preparations at different passage levels and from two different sources. Figure 1 shows a comparison of HindIII digests of AD169-UK and AD169-ATCC DNA from different lots of virus stock (2, 4, 16, and 22). The HindIII M region was detected by hybridization with a specific probe, pON2128 (Fig. 1A). The HindIII digests were also hybridized with a pCM1017 probe to detect HindIII fragments Y, N, J, Z, and M (Fleckenstein et al., 1982) (data not shown). AD169-UK DNA contained two HindIII fragments spanning this region, one approximately 10 kbp which has been denoted HindIII M (Oram et al., 1982) and the other approximately 11 kbp, which we denote here as modified HindIII M (Mm). All lots of AD169-ATCC that were analyzed contained only the 11-kbp HindIII Mm fragment. Serial propagation of AD169-UK appears to have resulted in a deletion not observed in AD169-ATCC or in other strains of virus.
Fig. 1.
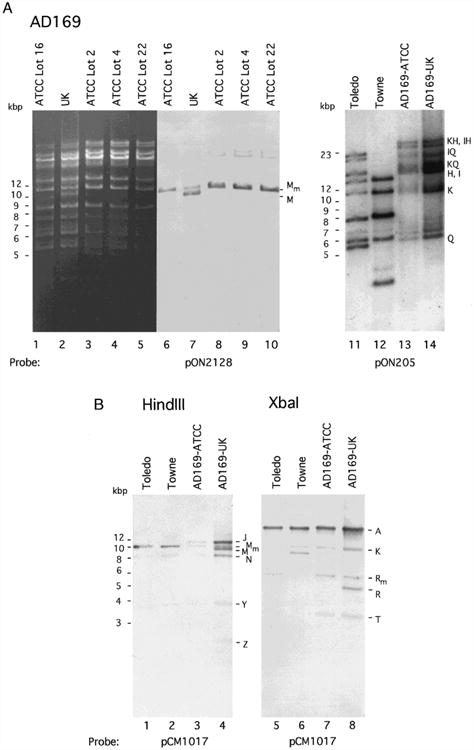
Size variation in the HindIII M region of CMV strain AD169-UK compared to AD169-ATCC, Towne, and Toledo. (A) Electrophoretically separated (0.5% agarose gel) HindIII-digested DNAs from AD169-ATCC (Lot 16), AD169-UK, AD169-ATCC (Lot 2), AD169-ATCC (Lot 4), and AD169-ATCC (Lot 22) were visualized by staining with ethidium bromide (lanes 1 – 5) and by subsequent DNA blot hybridization (lanes 5 – 10) with a fluorescein-11-dUTP (Amersham) random-primed labeled pON2128 probe detected by ECL (Amersham). Lots 2 and 4 of AD169 are the earliest stocks grown from material that was deposited with ATCC by the Rowe laboratory. Lot 16 represents material obtained from ATCC by our laboratory in 1987, and Lot 22, which has sustained 80 to 90 passages in culture, is a more recent (1995) preparation. Although early passages of AD169-UK were not examined, the strain we obtained from H. Browne (Cambridge) generated a pattern previously reported (Oram et al., 1982). This strain had been obtained from the Rowe laboratory in 1960 in its 14th passage and was passaged 54 times by Stern and colleagues (Elek and Stern, 1974) and was provided to J. Oram and H. Browne by J. Booth (St. Georges Hospital Medical School, London). (B) Electrophoretically separated (0.7% agarose gel) HindIII-digested (lanes 1 – 4) or XbaI-digested (lanes 5 – 8) strain Toledo, Towne, AD169-ATCC (Lot 16), and AD169-UK DNA fragments hybridized with a fluorescein-11-dUTP random-primed labeled pCM1017 probe (Fleckenstein et al., 1982) and detected by ECL. Note that Towne lacks the XbaI R/T site present at nt 56,067 in the AD169 genome and therefore exhibits a 9.5-kbp fragment instead of XbaI fragments Rm (6 kbp) and T (3.5 kbp) that are observed in the other strains. Size markers are given on the left and AD169 genome restriction fragment names (with subscript m indicating the modified fragments identified here) are indicated to the right of each set of lanes.
We next compared the organization of this region in AD169-UK with other laboratory-propagated (Towne, passage 134) as well as low passage level (Toledo, passage 12) strains of CMV (Plotkin et al., 1989) and found that these strains contained only the HindIII Mm fragment. Cosmids pCM1017, used as a probe here, and pCM1049, used to clone HindIII Mm, were derived from AD169 (Fleckenstein et al., 1982) and also contained only the HindIII Mm fragment (data not shown). Toledo and AD169-ATCC were also shown to contain an XbaI fragment, denoted XbaI Rm, that was approximately 1 kbp larger in size than the major XbaI R fragment in AD169-UK (Oram et al., 1982). This localized the region of polymorphism to between the HindIII site at nt 54,144 and the XbaI site at nt 56,067 on the strain AD169 genome. The Towne genome has a single restriction fragment representing a fusion of AD169 XbaI R and T (LaFemina and Hayward, 1980) which precludes direct comparison to Toledo or AD169. In order to further map the region of additional sequence, HindIII plus XbaI, HindIII plus EcoRI (nt 55,904), and HindIII plus HpaI (nt 54,813) digests of Toledo, Towne, AD169-ATCC, and AD169-UK DNA were blotted and hybridized with the pCM1017 probe (data not shown). The additional sequence mapped to the leftmost portion of HindIII Mm within a region that should have exhibited a 669-bp HindIII– HpaI fragment based on the published sequence. Instead, this fragment was approximately 1.5 kbp in size and was conserved in all strains tested. Thus, the region of variability mapped to the UL42 – UL43 region of the AD169 genome (nt 54144 to 54813) as shown in Fig. 2A. AD169-UK was the only strain exhibiting both short and long fragments.
Fig. 2.
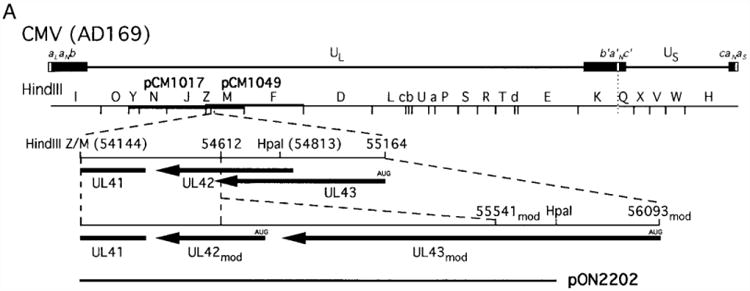

Nucleotide sequence arrangement and predicted aa sequence of UL42mod and UL43mod. (A) The top line shows the CMV strain AD169 genome with the thickened blocks representing repeated sequences (Mocarski, 1995). A HindIII map of the prototype genome is shown with the region carried by pCM1017 (nt 25921 to 64521) and the region carried by pCM1049 (nt 51805 to 84864) depicted by thickened lines below and above the HindIII map, respectively (Fleckenstein et al., 1982). The expanded region shows the published ORF arrangement of the region between nt 54,144 and 55,164 on the AD169 genome (Chee et al., 1990) and the modified region containing a 929-bp insert at nt 54,612 that alters the predicted size and aa sequence of UL42 and UL43. The plasmid clone pON2202, which carries 1599 bp of sequence aligning with a HindIII site at 54,144 and a HpaI site at 54,813 within UL43 (Chee et al., 1990). (B) Deduced aa sequence of UL43mod and alignment with aa 136 to 597 of murine CMV M43 (Rawlinson et al., 1996). Areas with US22 family motifs (I – IV) are shown above. Identical aa are boxed, conserved aa are shaded, and gaps are indicated by a dash. Identity across all four UL43 homologs (UL43mod, M43, U25 of HHV-6, and U25 of HHV-7) are denoted by an asterisk and identity in three of the four homologs along with a conservative substitution in the fourth are denoted by a dot below the aa. (C) Deduced aa sequence of UL42mod and alignment with murine CMV M42 (Rawlinson et al., 1996). The GenBank Accession No. for the 1602-bp region (including the complete HindIII and HpaI sites) is AF01963.
To further evaluate the arrangement of sequences in the HindIII M region, a plasmid clone, pON2202, was subjected to nucleotide sequence analysis with oligonucleotide primers indicated in Fig. 2 and an automated sequencer (Lagenaur et al., 1994; Sanger et al., 1977). nstead of the expected 669-bp based on published sequence, an insertion of 929 bp was found at nt 54,612, resulting in a total fragment size of 1599 bp (GenBank Accession No. AF01963).
The presence of this region in all CMV strains surveyed reinforced the notion that the longer fragment represented an authentic segment of CMV genome. The sequence was found to alter previously predicted ORFs, UL42 and UL43 (Chee et al., 1990). Modified UL43 (UL43mod) shared the amino terminal 185 codons with previously published UL43 (Chee et al., 1990), but had an additional 238 carboxyl-terminal codons. Thus, a 423-aa protein would be encoded by UL43mod. UL43mod is a much more convincing US22 family member that includes a complete set of conserved motifs (Chee et al., 1990; Efstathiou et al., 1992; Kouzarides et al., 1988; Nicholas and Martin, 1994), as shown in Fig. 2B. Modified UL42 (UL42mod) is 125 codons in size, sharing 76 carboxyl terminal codons with previously published UL42 (Chee et al., 1990) and is predicted to be a transmembrane protein due to the previously identified hydrophobic run near the carboxyl terminus. UL42mod starts at a strong consensus initiation codon and is more homologous (17% identity, 61% similar) to M42 of murine CMV (Rawlinson et al., 1996) than previously identified UL42, as depicted in Fig. 2C. A BLASTP search (Altschul et al., 1990) revealed no close relatives of UL42mod in the Swissprot database or in any other herpesvirus. Although UL42mod and UL43mod appeared to be the most likely protein-coding sequences in this region based on their size and the presence of initiation codons, other ORFs ranging in size from 33 to 205 codons (all from initiation codons) were also found in this region. These additional ORFs were not similar to any of the ORFs in murine CMV, HHV-6 or HHV-7, and were not considered further. The additional sequence also revealed a 63-bp region between the stop codon of UL43 and the initiation codon of UL42.
UL43mod shows much greater similarity to betaherpesvirus US22 family members (Chee et al., 1990; Efstathiou et al., 1992; Kouzarides et al., 1988; Nicholas and Martin, 1994) and includes not only a US22 family motif I (GxxoxoxWP; where “x” is any aa and “o” is a hydrophobic aa) previously described for UL43, but also complete motifs II (ooCCxxxLxxoG), III, and IV (Fig. 2B). A BLASTP search of the Swissprot database revealed UL43mod similarity with M43 (Rawlinson et al., 1996) in the murine CMV genome (24% identity and 58% homology over the entire 423 aa) as well as U25 ORFs of HHV-6 and HHV-7 (Fig. 2B). These UL43mod homologs align throughout their carboxyl termini but have divergent amino termini. M43 shows similarity to the predicted amino terminus of UL43mod beginning at M43 aa 140. UL43mod is similar to M43 over its entire length. Similarity to other US22 family members begins at motif I (UL43mod aa 142), which represents the amino terminus of many US22 family members including U25 of HHV-6 or HHV-7 (Gompels et al., 1995; Nicholas, 1996; Nicholas and Martin, 1994). Like other members of the human CMV US22 family members, UL43mod is more similar to US22 family members UL24, UL29, and UL36. After UL region family members, UL43mod appears more similar to TRS1/IRS1 than to other Us region family members. Similarity to Us region members is restricted to a small number of amino acids (aa) within the conserved motifs (Chee et al., 1990; Efstathiou et al., 1992; Kouzarides et al., 1988; Nicholas and Martin, 1994). Unlike other US22 family members, UL43mod exhibits a striking cluster of basic aa at the extreme carboxyl terminus (8 of the 13 carboxyl terminal residues are K or R), a characteristic that is also present in M43 (Fig. 2B). Like many US22 family members, UL43mod exhibits several consensus N-linked glycosylation sites (NxT/S) located at aa 26, 118, 299, 345 but has no hydrophobic stretch sufficiently long to span a membrane. UL43mod and M43 contain conserved cysteines at aa 23, 180, 181, 383, 404, 410. The UL43mod homolog U25 of HHV-6 has been shown to be a transactivator of gene expression (Nicholas and Martin, 1994), a characteristic that was first noted for other US22 family members (Cardin et al., 1993; Colberg-Poley et al., 1992; Geng et al., 1992; Stasiak and Mocarski, 1992).
This report has identified additional nucleotide sequence in the UL42 – UL43 region of the CMV genome. The additional sequence is present in all AD169 strain variants and was apparently overlooked during the selection of plasmid clones for sequencing in previous work. The overall identity of all other regions of the AD169-ATCC and AD169-UK genomes can be deduced from previous restriction enzyme mapping (Fleckenstein et al., 1982; Greenaway et al., 1982; Oram et al., 1982; Spector et al., 1982; Weststrate et al., 1980, 1983) as well as the absolute identity of many individual genes that have been sequenced (Boshart et al., 1985; Heilbronn et al., 1987; Jahn et al., 1987; Mach et al., 1986; Ruger et al., 1987) and compared to the complete genome sequence (Chee et al., 1990). Direct comparison of AD169-UK and AD169-ATCC restriction digests in our hands has not revealed any other differences (Fig. 1A and data not shown). We believe that strain AD169-UK may have developed a deletion in the UL42 – UL43 region during serial passage which was performed in a manner to attenuate virulence (Elek and Stern, 1974); however, the presence of the deletion does not correlate with reduced growth in thymic epithelial cells in SCID-hu mice (Brown et al., 1995). When AD169-UK was subjected to additional plaque purification, the two variants were able to grow independently and to similar levels, indicating that the 929-bp sequence is dispensable for growth in cultured fibroblasts (Dargan et al., 1997). Defective CMV genomes are known to be generated by purposeful high multiplicity passage (Stinski et al., 1979) and a similar process may lead to the loss of regions of the CMV genome that are dispensable for growth in cultured fibroblasts. Limited homology of a similar nature has been noticed at the borders of herpes simplex virus defective genomes which arise by a recombination mechanism during high multiplicity passage (Mocarski et al., 1985). The flanking regions matched the published sequence (Chee et al., 1990) for this region with one copy of the sequence 5′ GCAG 3′ at both ends of the insertion. It is possible that the 4-bp repeat (GCAG) bracketing the region played some role in recombination because one copy of this tetranucleotide remained in place following deletion. Although the AD169-UK strain is polymorphic, other strains, including the AD169 strain from ATCC lack evidence of polymorphism in this region. We propose that the additional 929 bp and ORFs UL42mod and UL43mod should be considered bona fide constituents of the CMV genome and the estimated size of the strain AD169 genome should be increased to 230,283 bp.
Acknowledgments
We thank Shinya Watanabe for helpful comments on the manuscript and acknowledge Maria Kirichenko for assistance with virus and cell culture. J.M.B. was supported by U. S. Public Health Service Grant K11 AI01197, M.N.P. was supported by U. S. Public Health Service National Research Service Award AI09008, C.S.T. was supported by U. S. Public Health Service Training Grant GM07276. This work was supported by U. S. Public Health Service Research Grant AI30363.
References
- Alford CA, Britt WJ. Cytomegalovirus. In: Fields BN, Knipe DM, Howley PM, editors. Fields Virology. Lippincott-Raven; New York: 1995. pp. 2493–2534. [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Boshart M, Weber F, Jahn G, Dorsch-Hasler K, Fleckenstein B, Schaffner W. A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell. 1985;41:521–530. doi: 10.1016/s0092-8674(85)80025-8. [DOI] [PubMed] [Google Scholar]
- Brown JM, Kaneshima H, Mocarski ES. Dramatic interstrain differences in the replication of human cytomegalovirus in SCID-hu mice. J Infect Dis. 1995;171:1599–1603. doi: 10.1093/infdis/171.6.1599. [DOI] [PubMed] [Google Scholar]
- Cardin RD, Boname JM, Abenes GB, Jennings SA, Mocarski ES. Reactivation of murine cytomegalovirus from latency. In: Plotkin S, Michelson S, editors. Multidisciplinary Approaches to Understanding Cytomegalovirus Disease. Elsevier; Amsterdam: 1993. pp. 101–110. [Google Scholar]
- Cha TA, Tom E, Kemble GW, Duke GM, Mocarski ES, Spaete RR. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J Virol. 1996;70:78–83. doi: 10.1128/jvi.70.1.78-83.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee MS, Bankier AT, Beck S, Bohni R, Brown CM, Cerny R, Horsnell T, Hutchison CAI, Kouzarides T, Martignetti JA, Preddie E, Satchwell SC, Tomlinson P, Weston KM, Barrell BG. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–170. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- Colberg-Poley AM, Santomenna LD, Harlow PP, Benfield PA, Tenney DJ. Human cytomegalovirus US3 and UL36 – 38 immediate– early proteins regulate gene expression. J Virol. 1992;66:95–105. doi: 10.1128/jvi.66.1.95-105.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dargan DJ, Jamieson FE, Maclean J, Dolan A, Addison C, McGeoch DJ. The published DNA sequence of human cytomegalovirus strain AD169 lacks 929 base pairs affecting genes UL42 and UL43. J Virol. 1997 doi: 10.1128/jvi.71.12.9833-9836.1997. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efstathiou S, Lawrence GL, Brown CM, Barrell BG. Identification of homologues to the human cytomegalovirus US22 gene family in human herpesvirus 6. J Gen Virol. 1992;73:1661–1671. doi: 10.1099/0022-1317-73-7-1661. [DOI] [PubMed] [Google Scholar]
- Elek SD, Stern H. Development of a vaccine against mental retardation caused by cytomegalovirus infection in utero. Lancet. 1974;1:1–5. doi: 10.1016/s0140-6736(74)92997-3. [DOI] [PubMed] [Google Scholar]
- Fleckenstein B, Muller I, Collins J. Cloning of the complete human cytomegalovirus genome in cosmids. Gene. 1982;18:39–46. doi: 10.1016/0378-1119(82)90054-3. [DOI] [PubMed] [Google Scholar]
- Geng YQ, Chandran B, Josephs SF, Wood C. Identification and characterization of a human herpesvirus 6 gene segment that trans activates the human immunodeficiency virus type 1 promoter. J Virol. 1992;66:1564–1570. doi: 10.1128/jvi.66.3.1564-1570.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompels UA, Nicholas J, Lawrence G, Jones M, Thomson BJ, Martin ME, Efstathiou S, Craxton M, Macaulay HA. The DNA sequence of human herpesvirus-6: Structure, coding content, and genome evolution. Virology. 1995;209:29–51. doi: 10.1006/viro.1995.1228. [DOI] [PubMed] [Google Scholar]
- Greenaway PJ, Oram JD, Downing RG, Patel K. Human cytomegalovirus DNA: BamHI, EcoRI and PstI restriction endo-nuclease cleavage maps. Gene. 1982;18:355–360. doi: 10.1016/0378-1119(82)90174-3. [DOI] [PubMed] [Google Scholar]
- Heilbronn R, Jahn G, Burkle A, Freese UK, Fleckenstein B, zur Hausen H. Genomic localization, sequence analysis, and transcription of the putative human cytomegalovirus DNA polymer-ase gene. J Virol. 1987;61:119–124. doi: 10.1128/jvi.61.1.119-124.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn G, Kouzarides T, Mach M, Scholl BC, Plachter B, Traupe B, Preddie E, Satchwell SC, Fleckenstein B, Barrell BG. Map position and nucleotide sequence of the gene for the large structural phosphoprotein of human cytomegalovirus. J Virol. 1987;61:1358–1367. doi: 10.1128/jvi.61.5.1358-1367.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T, Bankier AT, Satchwell SC, Preddy E, Barrell BG. An immediate early gene of human cytomegalovirus encodes a potential membrane glycoprotein. Virology. 1988;165:151–164. doi: 10.1016/0042-6822(88)90668-x. [DOI] [PubMed] [Google Scholar]
- LaFemina RL, Hayward GS. Structural organization of the DNA molecules from human cytomegalovirus. In: Fields BN, Jaenisch R, editors. Animal Virus Genetics. Academic Press; Orlando: 1980. pp. 39–55. [Google Scholar]
- Lagenaur LA, Manning WC, Vieira J, Martens CL, Mocarski ES. Structure and function of the murine cytomegalovirus sgg1 gene: A determinant of viral growth in salivary gland acinar cells. J Virol. 1994;68:7717–7727. doi: 10.1128/jvi.68.12.7717-7727.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach M, Utz U, Fleckenstein B. Mapping of the major glycoprotein gene of human cytomegalovirus. J Gen Virol. 1986;67:1461–1467. doi: 10.1099/0022-1317-67-7-1461. [DOI] [PubMed] [Google Scholar]
- Mocarski ES. Cytomegaloviruses and their replication. In: Fields BN, Knipe DM, Howley PM, editors. Fields Virology. 3rd. Lippincott-Raven; New York: 1995. pp. 2447–2492. [Google Scholar]
- Mocarski ES, Deiss LP, Frenkel N. Nucleotide sequence and structural features of a novel US-a junction present in a defective herpes simplex virus genome. J Virol. 1985;55:140–146. doi: 10.1128/jvi.55.1.140-146.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neote K, DiGregorio D, Mak JY, Horuk R, Schall TJ. Molecular cloning, functional expression, and signaling characteristics of a C– C chemokine receptor. Cell. 1993;72:415–425. doi: 10.1016/0092-8674(93)90118-a. [DOI] [PubMed] [Google Scholar]
- Nicholas J. Determination and analysis of the complete nucleotide sequence of human herpesvirus. J Virol. 1996;70:5975–5989. doi: 10.1128/jvi.70.9.5975-5989.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas J, Martin ME. Nucleotide sequence analysis of a 38.5-kilobase-pair region of the genome of human herpesvirus 6 encoding human cytomegalovirus immediate– early gene homologs and transactivating functions. J Virol. 1994;68:597–610. doi: 10.1128/jvi.68.2.597-610.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oram JD, Downing RG, Akrigg A, Dollery AA, Duggleby CJ, Wilkinson GW, Greenaway PJ. Use of recombinant plasmids to investigate the structure of the human cytomegalovirus genome. J Gen Virol. 1982;59:111–129. doi: 10.1099/0022-1317-59-1-111. [DOI] [PubMed] [Google Scholar]
- Plotkin SA, Starr SE, Friedman HM, Gonczol E, Weibel RE. Protective effects of Towne cytomegalovirus vaccine against low-passage cytomegalovirus administered as a challenge. J Infect Dis. 1989;159:860–865. doi: 10.1093/infdis/159.5.860. [DOI] [PubMed] [Google Scholar]
- Rawlinson WD, Farrell HE, Barrell BG. Analysis of the complete DNA sequence of murine cytomegalovirus. J Virol. 1996;70:8833–8849. doi: 10.1128/jvi.70.12.8833-8849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe WP, Hartley JW, Waterman S, Turner HC, Huebner RJ. Cytopathogenic agent resembling human salivary gland virus recovered from tissue cultures of human adenoids. Proc Soc Exp Biol Med. 1956;92:418–424. [PubMed] [Google Scholar]
- Ruger B, Klages S, Walla B, Albrecht J, Fleckenstein B, Tomlinson P, Barrell B. Primary structure and transcription of the genes coding for the two virion phosphoproteins pp65 and pp71 of human cytomegalovirus. J Virol. 1987;61:446–453. doi: 10.1128/jvi.61.2.446-453.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JA, Pari GS. Human cytomegalovirus UL102 gene. J Virol. 1995;69:1734–1740. doi: 10.1128/jvi.69.3.1734-1740.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaete RR, Mocarski ES. The a sequence of the cytomegalovirus genome functions as a cleavage/packaging signal for herpes simplex virus defective genomes. J Virol. 1985;54:817–824. doi: 10.1128/jvi.54.3.817-824.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DH, Hock L, Tamashiro JC. Cleavage maps for human cytomegalovirus DNA strain AD169 for restriction endonucleases EcoRI, BgIII, and HindIII. J Virol. 1982;42:558–582. doi: 10.1128/jvi.42.2.558-582.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasiak PC, Mocarski ES. Transactivation of the cytomegalovirus ICP36 gene promoter requires the a gene product TRS1 in addition to IE1 and IE2. J Virol. 1992;66:1050–1058. doi: 10.1128/jvi.66.2.1050-1058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinski MF, Mocarski ES, Thomsen DR. DNA of human cytomegalovirus: Size heterogeneity and defectiveness resulting from serial undiluted passage. J Virol. 1979;31:231–239. doi: 10.1128/jvi.31.1.231-239.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weststrate MW, Geelen JL, van der Noordaa J. Human cytomegalovirus DNA: Physical maps for restriction endonucleases BgIII, HindIII and XbaI. J Gen Virol. 1980;49:1–21. doi: 10.1099/0022-1317-49-1-1. [DOI] [PubMed] [Google Scholar]
- Weststrate MW, Geelen JL, Wertheim PM, van der Noordaa J. Comparison of the physical maps of the DNAs of two cytomegalovirus strains. J Gen Virol. 1983;64:47–55. doi: 10.1099/0022-1317-64-1-47. [DOI] [PubMed] [Google Scholar]


