Abstract
Vesicle transport is intimately connected with key nuclear functions and transcriptional regulation. Here, children born with congenital genitourinary tract masculinization disorders were analyzed by array-Comparative Genomic Hybridization, which revealed the presence of de novo copy number gains on Xq28 encompassing the VAMP7 gene encoding a vesicle-trafficking protein. Humanized VAMP7 BAC transgenic mice displayed cryptorchidism, urethral defects, and hypospadias. Mutant mice exhibited reduced penile length, focal spermatogenic anomalies, diminished sperm motility, and subfertility. VAMP7 colocalized with estrogen receptor alpha (ESR1) in the presence of ligand. Elevated levels of VAMP7 markedly intensified ESR1 transcriptional activity by increasing ESR1 protein cellular content upon ligand stimulation and up-regulated the expression of estrogen-responsive genes including ATF3, CYR61, and CTGF, all of which are implicated in human hypospadias. Hence, increased gene dosage of the SNARE protein, VAMP7, enhances estrogen receptor action in male genitourinary tissues, affects the virilization of the reproductive tract, and results in genitourinary birth defects in humans.
INTRODUCTION
Estrogens are generally perceived to be inhibitory in male development or at least to antagonize androgen action1. Androgen action is essential for the acquisition of a normal male phenotype. Following the early steps of bipotential gonad formation and testis determination, androgens are required for the stabilization of the Wolffian duct system and its differentiation into the epididymides, vasa deferentia, and seminal vesicles. Their action is also critical for proper virilization of the external genitalia and migration of the testes through the inguinal canal into the scrotum2. The dependence of the masculinization of the reproductive system on androgen, however, renders this process inherently susceptible to destabilization by factors interfering with hormone synthesis, metabolism, or action. Such disturbances represent significant etiological determinants that potentially contribute to the high prevalence of human masculinization disorders, such as hypospadias (male urethral dysmorphogenesis) and cryptorchidism (failure of testicular descent).
Epidemiological studies provide links between inappropriate estrogen exposure and increased incidence of reproductive abnormalities in men (for review3,4). Similarly, reproductive tract lesions, such as cryptorchidism and penile defects, were present in male mice after pre- and neonatal exposure to the potent nonsteroidal synthetic estrogen, diethylstilbestrol5–8. Furthermore, transgenic male mice overexpressing the enzyme aromatase, necessary for the conversion of androgens to estrogens, display several genital abnormalities, including undescended testes, subtle effects on penile development, and spermatogenic arrest9.
The tight interrelationship between the androgens and estrogens working together to regulate the differentiation of the male reproductive system relies on the balance between androgen and estrogen action, rather than on the absolute circulating concentrations of these hormones. Any change in this homeostasis may disturb the internal milieu required for normal development and function of the male genital tract and may translate into a wide range of genitourinary disorders that can subsequently affect normal fertility.
RESULTS
A terminal Xq28 copy number gain encompassing the VAMP7 gene in children born with masculinization disorders
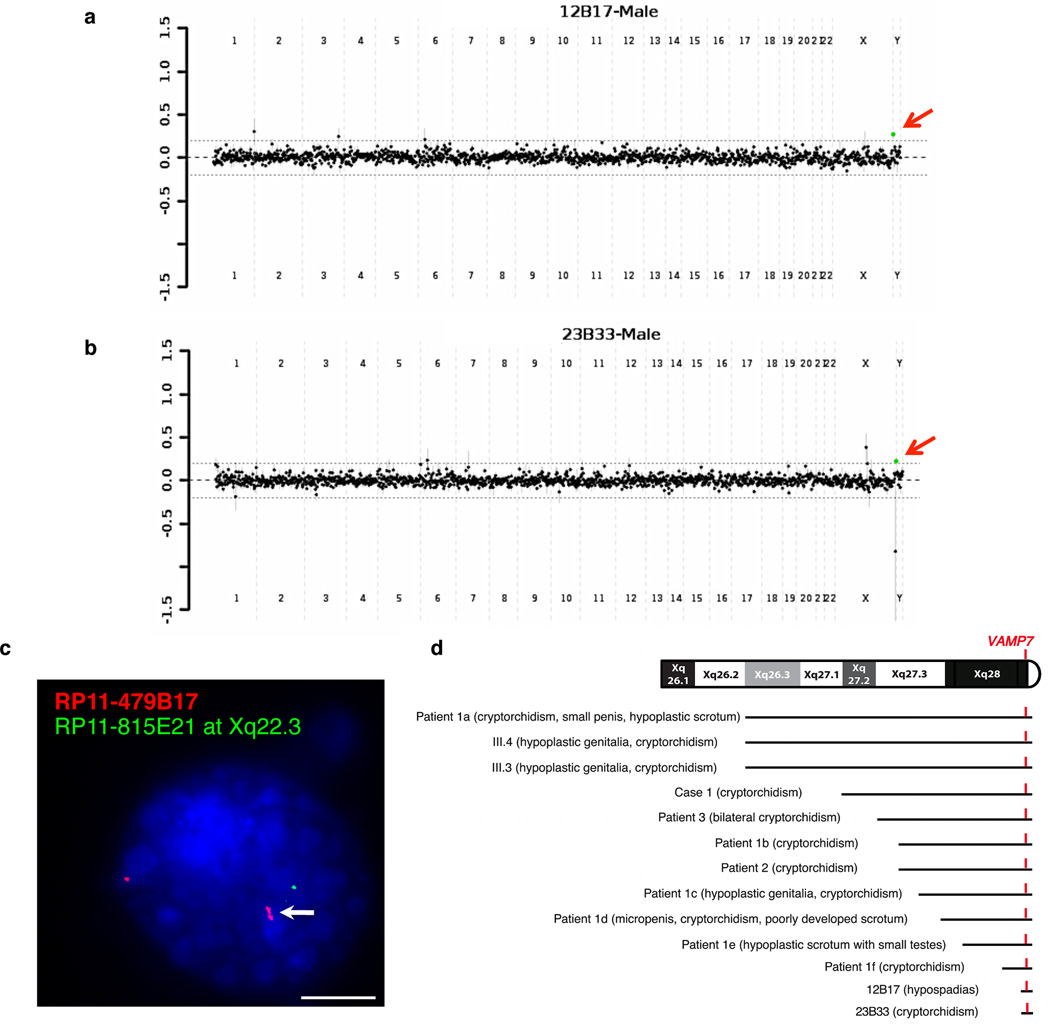
We used a clinically validated comparative genomic hybridization microarray platform to analyze DNA from 116 children presenting with idiopathic cases of 46, XY disorders of sexual development (DSD) including anomalies of testicular descent and defects of penile morphogenesis10. Structural DNA variation was highlighted as a potential underlying etiology for human disorders of sexual development, since frequent submicroscopic gains and losses of DNA segments were detected and strongly associated with defective urogenital traits10. Among the clinically significant imbalances identified, an identical copy number gain at the distal tip of the long arm of Xq28 was noted in two unrelated patients (Fig. 1a–b and Supplemental Fig. S1a–b). One child had bilateral cryptorchidism with a right inguinal testis and a left intra-abdominal gonad. The second child presented with midscrotal hypospadias with chordee and penoscrotal transposition. CGH array analysis of DNA from unaffected parents revealed the de novo occurrence of the terminal Xq28 gain (Supplemental Fig. S1c–d). This structural variant was secondarily confirmed by FISH analysis (Fig. 1c). When a two-tailed Fisher’s exact test was used, this copy number change was found to be more frequent in subjects presenting with congenital genitourinary defects (2 out of 116) than in control individuals without urogenital abnormalities (n = 8,951) run on the same array CGH platform (P = 2.2×10−3). While the Xq28 region had contiguous coverage on the microarray, the distal BAC clone RP11-479B17 was the only segment showing a copy number gain (Supplemental Fig. S1a–b). The maximal size of the defect exclusively included the pseudoautosomal region 2 (PAR2) while the minimal critical interval (RP11-479B17) solely encompassed VAMP7 (Supplemental Fig. S1e). The published literature reveals the presence of additional patients who presented with syndromic cryptorchidism and/or micropenis among their clinical features together with larger Xq28 terminal duplications that also encompassed VAMP7 (Fig. 1d and Supplemental Table 1).
Figure 1.
A terminal Xq28 gain encompassing VAMP7 in 46,XY children presenting with masculinization disorders of the urogenital tract. (a,b) Genomic hybridization profiles of unrelated 46,XY subjects with cryptorchidism or hypospadias compared with a gender-matched reference DNA. The mean log2 ratio (patient/reference) of intensity for each single probe is presented across individual chromosomes. Arrows indicate a terminal X copy gain (green). (c) Representative FISH hybridization of PHA-stimulated blood lymphocytes from Xq28 CGH array-detected patients using RP11-479B17 probe (red), specific to the pseudo-autosomal region 2, including Xq28 and Yq12 loci. RP11-815E21 at Xq22.3 (green) was used as a hybridization control. White arrow indicates copy number gain. Scale bar, 5 µm. (d) Literature review of patients with Xq28 duplication encompassing VAMP7 and presenting with defective virilization of the male reproductive tract. For references, see Supplemental Table 1.
On the basis of these observations, we hypothesized that Xq28 region and, more specifically, VAMP7 duplication play a role in human disorders of masculinization of the urogenital tract. VAMP7 is a highly conserved gene11 that encodes for a transmembrane protein belonging to the soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) family localized in late endosomes and lysosomes. VAMP7 is required for (i) heterotypic fusion of late endosomes with lysosomes and homotypic lysosomal fusion12–14, (ii) calcium-regulated lysosomal exocytosis15, and (iii) focal exocytosis of late endocytic vesicles during phagosome formation16,17. Quantitative PCR analysis of VAMP7 copy number in the genome of an additional cohort of 180 individuals presenting with isolated hypospadias (n = 83), cryptorchidism (n = 79), or both urogenital conditions (n = 18) identified one additional case of isolated inguinal cryptorchidism and one patient with glandular hypospadias and chordee. Both bore a VAMP7 copy gain (Supplemental Fig. S2a). A second independent replication study of primary cultures of human genital skin fibroblasts from 28 subjects with congenital genitourinary defects revealed 1 instance of mid-shaft hypospadias with a copy number gain of VAMP7 (3.6%) (Supplemental Fig. S2b), convincingly supporting the association of VAMP7 genomic gains with developmental disorders of the human genitourinary tract.
VAMP7 is expressed in the human and mouse genital tract
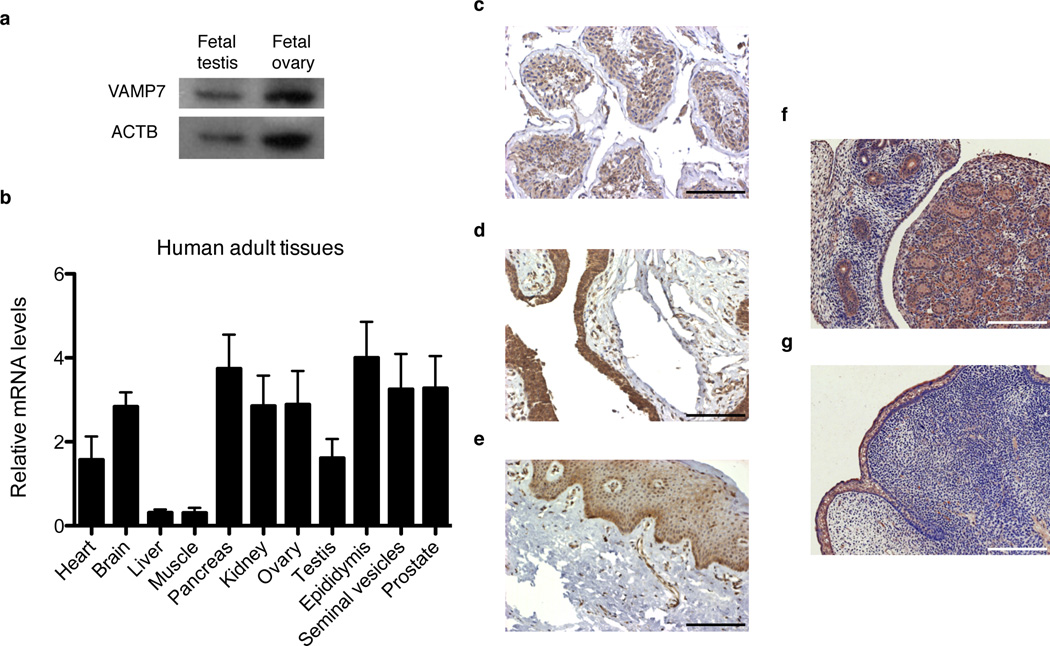
VAMP7 protein was detected in cytoplasmic lysates from human fetal testes and ovaries (Fig. 2a). At adult stages, VAMP7 mRNA levels were detected throughout the human male genital tract including the testes, epididymides, seminal vesicles, and prostatic and penile tissues (Fig. 2b). A punctuate pattern of VAMP7 staining was present in the Sertoli cells, as well as in the germ cells of the seminiferous tubules of human adult testes with histologically normal spermatogenesis (Fig. 2c). The cytoplasm of Leydig cells also contained VAMP7-positive immunoreactive staining (Fig. 2c). In the adult human external genitalia, VAMP7 was strongly expressed in the urethral epithelium (Fig. 2d). Moreover, the genital mesenchyme, including corpus cavernosa and the androgen-dependent penile spines of the preputial epithelium, showed significant levels of VAMP7 protein (Fig. 2e). Similarly, fetal testes and developing genital tubercles in mouse embryos at gestational day 16.5 were positively stained for VAMP7 (Fig. 2f–g). Thus, VAMP7 mRNA and protein are present in the human and murine genitourinary tract during development and at maturity.
Figure 2.
VAMP7 is present in human and murine male reproductive tissues. (a) Western-blot analysis of VAMP7 expression in human fetal testes and whole ovarian lysates. ACTB (β actin) was used a loading control. (b) qRT-PCR analysis of VAMP7 mRNA levels normalized to GAPDH in human adult tissues. n = 6 independent samples for each tissue. Data are presented as means ± s.e.m. (c–e) Immunohistological detection of VAMP7 in normal human adult testicular (c) and penile tissues including urethral (d) and preputial (e) epithelia. Scale bar, 125 µm. (f,g) Immunohistochemical detection of VAMP7 expression in fetal testis (f) and external genitalia (g) from wild-type male mice at embryonic stage E16.5. Scale bar, 125 µm.
Increased gene dosage of VAMP7 results in defects of testicular descent and morphogenesis of external genitalia in mice
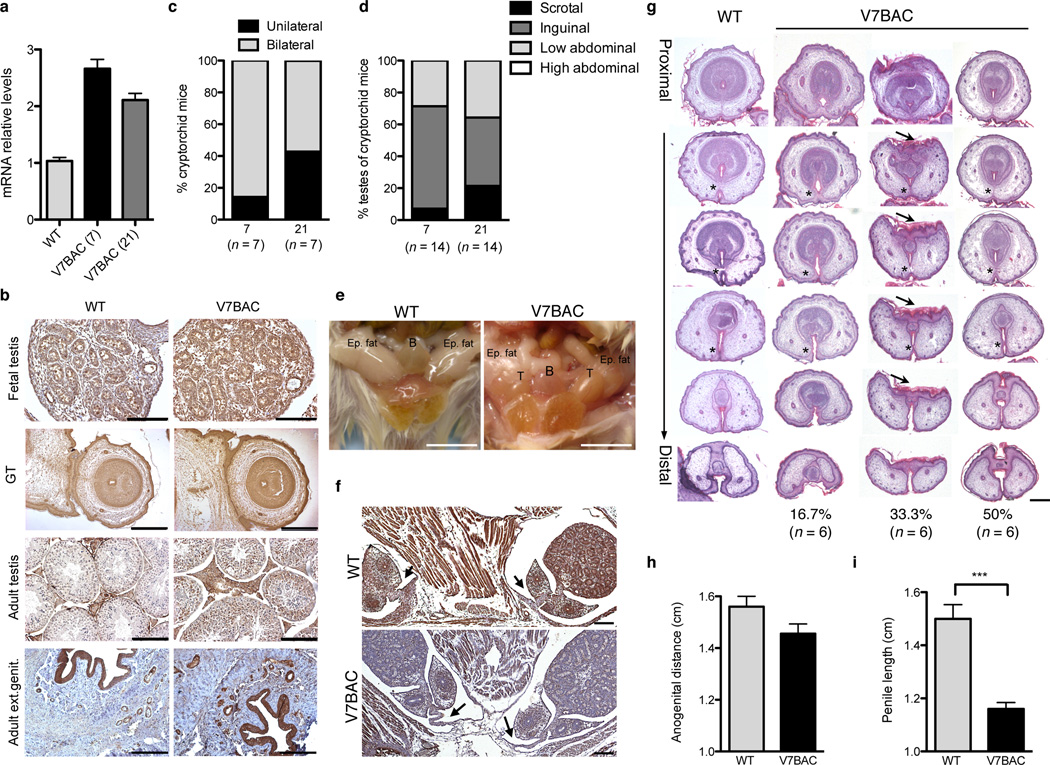
We sought to investigate in vivo the impact of genetically elevated VAMP7 levels on genitourinary development and reproductive physiology. A mouse model that expresses human VAMP7 under its endogenous regulatory sequences was produced to recapitulate the genetic gain observed in patients. Mutant mice were generated through microinjection of a 159-kilobase linearized bacterial artificial chromosome (BAC; clone RP11-479B17) including the human VAMP7 gene, into fertilized oocytes of FVB mice. Two independent VAMP7 founder lines, 7 and 21, were obtained. BAC transgenic mice were viable and had normal growth and lifespan. Western blot analysis using an antibody that specifically recognizes the human VAMP7 protein confirmed the expression of the transgene in the testes of mutant mice (Supplemental Fig. S3a). Using primers that recognize both human and mouse transcripts, we quantified mRNA levels of VAMP7 by qRT-PCR in the two lines (21 and 7) and found them to be only 2 to 3 times higher than the messenger levels of the endogenous Vamp7 in WT mice (Fig. 3a). Throughout the adult and fetal testes and external genitalia, the spatial expression pattern of the transgene mirrored that of the endogenous product in the wild-type animals and localized to cytoplasmic compartments as expected (Fig. 3b). A parallel study of the two transgenic lines controlled for potential insertion effects. Data from strain 7 are presented when identical outcomes were obtained for both lines (for line 21-see Supplemental Fig. S4).
Figure 3.
Mice overexpressing VAMP7 exhibit cryptorchidism and abnormal external genitalia. (a) qRT-PCR analysis of VAMP7 mRNA levels normalized to Gapdh in testis from WT (n = 4 animals) and humanized transgenic V7BAC mice (n = 5 mice for line 7 and n = 4 mice for line 21) using primers common to both mouse and human VAMP7 transcripts. Data are presented as means ± s.e.m. (b) VAMP7 immunohistological staining of fetal and adult testicular tissue and external genitalia from V7BAC transgenic and WT mice. Scale bars, 125 µm. For fetal genital tubercles, scale bar, 500 µm. (c)Distribution of unilateral and bilateral cryptorchidism in V7BAC transgenic mice harboring undescended testes. (n = 7 cryptorchid animals for each line). (d) Anatomical locations of cryptorchid testes in VAMP7 transgenic mice (n = 14 cryptorchid testes for each line). Distribution is presented in percentage. (e) Representative pictures of testis position in V7BAC and WT mice. B: bladder; T: testis; Epid. fat: epidydimal fat. Scale bar, 1 cm. (f) Desmin staining of testis and gubernaculum of V7BAC and WT male embryos at gestational age E18.5. Arrows indicate the gubernaculum cord. Scale bar, 500 µm. (g) Hematoxylin-eosin staining of cross-sections of genital tubercles from V7BAC male mouse embryos at gestational age E18.5 (n = 6) and WT littermates. Asterisks indicate normal and abnormal fusion of urethral folds or hypospadias, while arrows show abnormalities of the epithelial-lined prepuce housing the penis. The occurrence of penile defects was presented in percentage. Scale bar, 150 µm. (h) Measurement of anogenital distance in VAMP7 transgenic adult male mice (n = 9) and WT littermates (n = 5). Data are presented as means ± s.e.m. (i) Measurement of penile length in VAMP7 mutant adult male mice (n = 5) compared to WT animals (n = 7). Data are presented as means ± s.e.m. Mean differences were determined by unpaired, two-tailed Student’s t-test. *P < 0.05; **P < 0.01; ***P < 0.001.
Examination of individually-housed VAMP7 adult male mice revealed a high incidence of cryptorchid testes (87.5%; 7 out of 8 mice), either bilateral (85.7% for strain 7 and 57.2% for strain 21) or unilateral (14.3% for strain 7 and 42.8% for strain 21) (Fig. 3c). Testes were considered (i) “scrotal” in location if they had to be pulled out of the scrotal sac. (ii) “inguinal” when superior to the scrotum but inferior to the bladder at the inguinal ring, or (iii) “low abdominal” or “high abdominal” respectively when they were at, or superior to, the level of the bladder. The testicular location in uni- or bilaterally cryptorchid VAMP7 mutant mice was variable, predominantly inguinal in both strains (strain 7: 7.1% scrotal, 64.3% inguinal, and 28.6% low abdominal; strain 21: 21.4% scrotal, 42.9% inguinal, and 35.7% low abdominal) (Fig. 3d). Importantly, none of the cryptorchid gonads were located in a high intra-abdominal position (Fig. 3d–e), implying that the inguino-scrotal, androgen-dependent phase of testicular descent was defective in VAMP7 BAC mice. Examination of the gubernaculum, a mesenchymal tissue connecting the fetal testis to the developing scrotum, revealed a slightly feminized appearance with a thinner and elongated gubernacular cord connected to the lower cauda epididymis, as compared to the rudimentary gonadal ligament in wild-type male siblings (Fig. 3f). Interestingly, thin and elongated gubernaculum cords were seen previously in estrogen-treated mouse embryos18–21.
Histological analysis of VAMP7 mutant genital tubercles revealed variable penile defects (Fig. 3g). Half of the analyzed male transgenic embryos had normally developed genital tubercles with the ventral epithelial surfaces forming a closed urethral groove where epithelial surfaces fused to create the urethral seam and resulted in a true urethra. However, 16.7% (1 out of 6) of them exhibited hypospadias, a significant anomaly of the ventral urethral tube closure (Fig. 3g). Underdeveloped os penis, abnormalities of the epithelial-lined prepuce housing the penis, and a reduction in the thickness of the tunica albuginea capsule surrounding the corpora cavernosa were observed in 33.3% (2 out of 6) of VAMP7 BAC male embryos (Fig. 3g). Importantly, many of the penile defects in VAMP7 BAC animals resembled those observed following the prenatal or neonatal administration of estrogenic compounds in male mice6–8. Furthermore, anogenital distance tended to decrease in mutant VAMP7 male mice (Fig. 3h), but, more strikingly, penile length was significantly less in all adult transgenic animals than in their wild-type littermates (Fig. 3i).
Thus, increased levels of VAMP7 expression in the mouse male genitourinary tract mimicked the urogenital phenotypic traits observed in patients with VAMP7 copy number gain, i.e. cryptorchidism, hypospadias and micropenis. The VAMP7 BAC mouse models strongly suggest a causative link of VAMP7 duplication to congenital genitourinary defects in humans.
VAMP7 overexpression modestly alters androgen receptor action
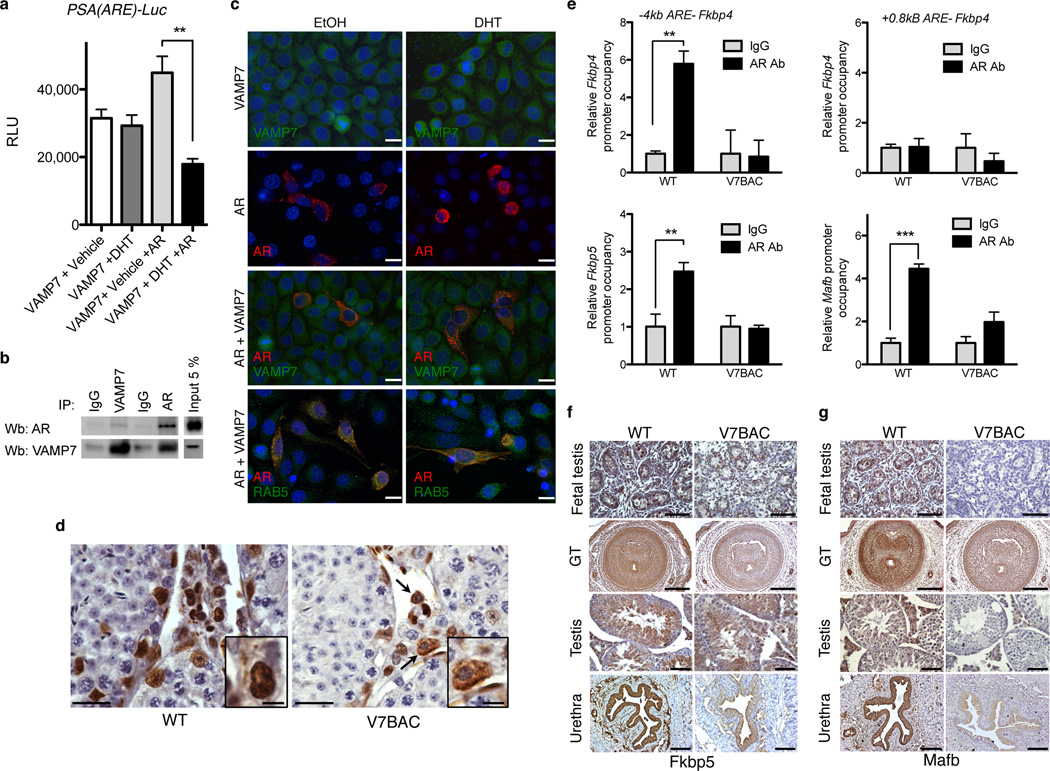
Since a VAMP7 genomic defect was present in males presenting with cryptorchidism, hypospadias, or micropenis, and since normal androgen action is crucial for penile development and testicular descent, we asked whether elevated levels of VAMP7 interfere with androgen action. To assess the impact of VAMP7 on androgen receptor (AR) transcriptional activity, we used a cell-culture-based assay using a firefly luciferase reporter gene driven by an androgen responsive element (ARE) of the PSA promoter. Interestingly, ligand-dependent AR transcriptional activity was suppressed by 60% with VAMP7 overexpression (Fig. 4a). This observed inhibition was not a result of a decrease of AR protein levels (Supplemental Fig. S3b). Physical co-presence of VAMP7 and AR in the same protein complexes was shown by co-immunoprecipitation assays, suggesting possible functional interactions between these two proteins (Fig. 4b). Since VAMP7 is involved in cellular sorting12,13,22, we questioned whether VAMP7 overexpression alters the subcellular localization of the androgen receptor at the time of ligand-dependent stimulation. Immunofluorescent staining after transient transfection of the androgen receptor and/or VAMP7 in the presence or absence of dihydrotestosterone revealed the co-localization of VAMP7 and AR in the cytoplasm upon DHT administration (Fig. 4c), implying a failure of efficient translocation of AR to the nucleus despite the presence of the ligand. Since VAMP7 localizes in the endosomes, we used RAB5A, a marker of early endosomes, to demonstrate a co-localization of AR and RAB5A in the presence of DHT (Fig. 4c). To examine the existence of a potential mis-localization of AR in vivo, we then analyzed the cellular distribution of AR in the testes of VAMP7 BAC mice, which express the transgene in ranges far below the high doses of our transfection conditions but similar to those in the human genomic gains. In vivo, AR was visible in the cytoplasm of few, but not all, positively stained cells of VAMP7 BAC testes (Fig. 4d), contrasting with its exclusive nuclear localization in the male gonads of wild type littermates (Fig. 4d). We concluded that the observed mosaic pattern of AR subcellular distribution in VAMP7 BAC testes reflects a partial interference of ligand-dependent shuttling of AR in vivo, likely due to VAMP7 overexpression.
Figure 4.
Elevated levels of VAMP7 modestly impair AR function. (a) Luciferase assays in HeLa cells co-transfected with VAMP7 or VAMP7 and AR and incubated in absence (EtOH) or in presence of 10−8 M dihydrotestosterone (DHT) for 24 h. n = 3 independent experiments for each condition. Data are presented as means ± s.e.m. One-way analysis of variance (ANOVA) with post hoc Bonferroni test was used for statistical analyses. *P < 0.05; **P < 0.01; ***P < 0.001. (b) Reciprocal co-immunoprecipitation of AR and VAMP7 following their co-transfection in HeLa cells.(c) Immunofluorescence staining of VAMP7, AR, and RAB5 in HeLa cells after transfection with AR, VAMP7, or both, upon stimulation with ethanol (EtOH) or 10−8 M dihydrotestosterone (DHT) for 24 h. Scale bar, 5 µm. (d) AR immunostaining of adult testis from V7BAC or WT mice. Arrows indicate cytoplasmic staining. Scale bar, 125 µm (Inset, 25 µm). (e) In vivo chromatin immunoprecipitation assays of testicular lysates from WT (n = 3) and V7BAC (n = 3) mice using IgG or AR antibody followed by qPCR of Fkbp5, Mafb and Fkbp4 promoters. Data are presented as means ± s.e.m. Two-way analysis of variance with post hoc Bonferroni test was used for statistical analyses. *P < 0.05; **P < 0.01; ***P < 0.001. (f,g) Immunostaining of Fkbp5 (f) and Mafb (g) in fetal and adult external genitalia and testis of WT and V7BAC mouse embryos (E18.5). For fetal tissues: scale bar, 250 µm. For adult urethra and testis: scale bar, 100 µm.
Under these conditions, we examined AR recruitment to its genomic occupancy sites in the presence of increased VAMP7 gene dosage. In vivo chromatin immunoprecipitation in testicular tissues of VAMP7 transgenic mice was performed using the androgen-responsive element of several genes involved in AR signaling, including Fkbp4, Mafb and Fkbp523–25. While a clear in vivo binding of AR was observed in wild-type testis, binding was significantly reduced in VAMP7 transgenic male gonads (Fig. 4e). Consistent with the observed AR occupancy, the immunoreactivity of the androgen-responsive genes, Fkbp5 and Mafb26,27 was lower in the fetal and adult genital tracts of VAMP7 BAC male mice than in those of wild-type littermates (Fig. 3f–g). The expression of other testicular androgen target genes such as Rhox5, Drd4, Eppin (Spinlw), Tubb3, and Cldn11 was defined. With the exception of a significant decrease of Cldn11 (encoding a tight junction protein at the blood-testis barrier), no drastic changes were observed in VAMP7 BAC testes compared to those in WT animals (Supplemental Fig. S3c). Thus, the impact of VAMP7 on AR transcriptional activity, although apparent when examined in an in vitro cellular context, is only partial in vivo. This is reflected by subtle alterations in expression of selective AR target genes. Accordingly, other signaling pathways were likely to contribute to the phenotypic anomalies observed in the VAMP7 BAC mice.
Elevated dosage of VAMP7 markedly enhances estrogen transcriptional action
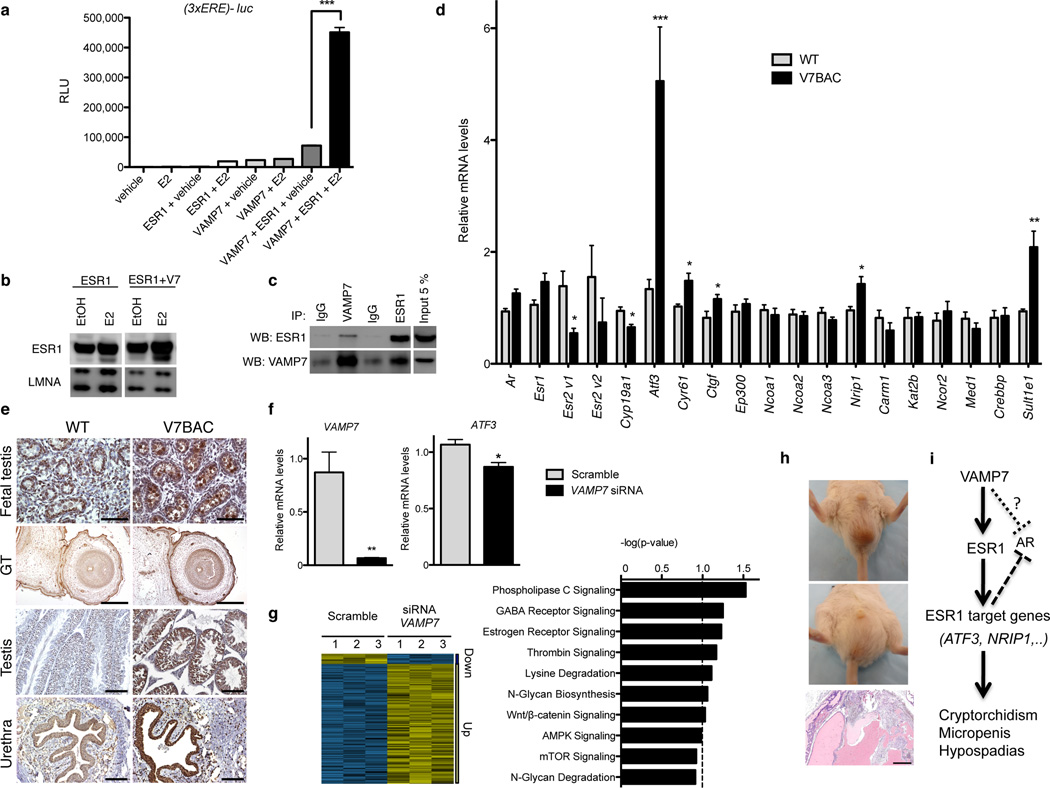
In vivo, the balance between androgen and estrogen receptor action, as opposed to androgen action alone, is an important factor for male reproductive tract development28. Consequently, further investigations focused on the impact of VAMP7 overexpression on the estrogen signaling. With a classic estrogen-responsive element driving the expression of a luciferase reporter, an unexpected, massive and largely significant, synergistic transcriptional effect was observed when VAMP7 was co-transfected with estrogen receptor alpha (ESR1) in the presence of estradiol (Fig. 5a). Under these conditions, VAMP7 increased the ligand-dependent protein concentration of ESR1 (Fig. 5b), perhaps through protein stabilization or through the utilization of posttranslational mechanisms, since endogenous ESR1 mRNA levels remained unaffected after a knockdown of VAMP7 in the human testicular embryonic carcinoma cell line NT2/D1 (Supplemental Fig. S3d), or after VAMP7 overexpression in the testes of our transgenic mice (Fig. 5). The potential for a functional interaction between VAMP7 and ESR1 was further substantiated by the fact that these two proteins were physically found in the same protein complexes (Fig. 5c) and co-localized in identical sub-cellular compartments (Supplemental Fig. S3e).
Figure 5.
VAMP7 enhances estrogen receptor transcriptional activity. (a) Luciferase assays following transfection with VAMP7 or ESR1 or VAMP7 and ESR1 in HeLa cells incubated in absence (EtOH) or presence of 17 beta-estradiol (10−8 M) for 24 h. n = 3 independent experiments for each condition. Data are presented as means ± s.e.m. One-way analysis of variance (ANOVA) with post hoc Bonferroni test was used for statistical analyses. *P < 0.05; **P < 0.01; ***P < 0.001. (b) Western Blot analysis of ESR1 and LMNA (lamin A/C) in nuclear protein extracts of HeLa cells co-transfected with ESR1 or ESR1 and VAMP7 in absence (EtOH) or presence of 17 beta-estradiol (10−8 M) for 24 h. (c) Reciprocal co-immunoprecipitation of ESR1 and VAMP7 following their co-transfection in HeLa cells. (d) qRT-PCR analysis of key genes of ESR1 signaling in testis from WT (n = 3) and V7BAC mice (line 7; n = 3). Data are expressed as mean ± s.e.m. Mean differences between WT and V7BAC were determined by unpaired, two-tailed Student’s t-test. *P < 0.05; **P < 0.01; ***P < 0.001. (e) ATF3 immunostaining of testis and external genitalia at fetal and adult stages of WT and V7BAC mice. For fetal tissues: scale bar, 250 µm. For adult urethra and testis: scale bar, 100 µm. (f) qRT-PCR of VAMP7 and ATF3 gene expression after incubation with non-targeting (scramble) or VAMP7 siRNA in NT2/D1. n = 3 independent experiments for each condition. Mean differences were determined by unpaired, two-tailed Student’s t-test. *P < 0.05; **P < 0.01; ***P < 0.001. (g) Left: Heatmap of testis gene expression profiles in NT2/D1 transiently transfected with non-targeting (scramble) or VAMP7-specific siRNA. Yellow and blue colors indicate increased and decreased expression, respectively, relative to scramble. Right: Ingenuity Pathway Analysis (IPA 8.5) top canonical pathways significantly altered (20% FDR) after VAMP7 knockdown in NT2/D1 cells. (i) Pictures of inguinal hernia in V7BAC males and hematoxylin-eosin staining of extensive granulomatous inflammatory response of entrapped tissues. Scale bar, 100 µm (j) Schematic representation of VAMP7 impact on estrogen receptor signaling and male phenotypic development.
To examine the in vivo functional interaction between VAMP7 and ESR1, we assessed gene expression of known players involved in the estradiol-mediated action in VAMP7 BAC mice. While expression levels were unchanged for the estrogen-dependent coregulators, including p160 family (Ncoa1, Ncoa2, Ncoa3), Crebbp, Kat2b, Med1, Ncor2, Carm1, and the nuclear receptors Ar and Esr1 (Fig. 5d and Supplemental Figure S4a), a significant up-regulation of mRNA levels of three estrogen-responsive genes, including activating transcription factor 3 (Atf3), connective tissue growth factor (Ctgf), and cysteine-rich angiogenic inducer 61 (Cyr61) was observed in the testes of the VAMP7 transgenic mice (Fig. 5d and Supplemental Figure S4a). These observations are of key importance, as ATF3, CTGF, and CYR61 are estrogen-responsive genes that are strongly up-regulated in hypospadias patients29–33. Moreover, variants of ATF3 are associated with hypospadias in humans34,35. Consistently, Atf3 protein levels were higher in fetal and adult testicular and penile tissues of VAMP7 transgenic animals, than in wild-type littermates’ tissues (Fig. 5e). Conversely, selective knockdown of VAMP7 in the human testicular embryonal carcinoma NT2/D1 cell line led to a significant decrease of ATF3 gene expression (Fig. 5f), confirming that VAMP7 is an important positive regulator of ATF3 gene expression. Concomitantly, the expression levels of additional estrogen responsive genes were altered in the testis of VAMP7 mutant mice compared to wild-type gonads. Indeed, Nrip1 mRNA levels, directly induced by estrogen receptor ction36, were higher in VAMP7 mutant mice than in WT littermates (Fig. 5d). The expression of Cyp19a1, which is down-regulated by estradiol in the testis37, was lower in the VAMP7 transgenic mouse testes than in WT gonads (Fig. 5d and Supplemental Figure S4a). Additionally, higher mRNA levels of estrogen sulfotransferase (Sult1e1) encoding an enzyme involved in the maintenance of the functional integrity of male-specific tissues against local estrogenic activity38, were observed in the testes of VAMP7-overexpressing mice than in WT animals (Fig. 5d and Supplemental Figure S4a). In line with these in vivo data, estrogen receptor signaling was among the top pathways affected following the acute VAMP7 knockdown in NT2/D1 cells, as revealed by the functional analysis of gene expression microarrays data by Ingenuity Pathway Analysis (Fig. 5g).
Finally, the occurrence of inguinal hernias in some VAMP7 BAC transgenic mice further supports our conclusions (Fig. 5h), since this phenotypic trait is observed with estrogen overstimulation in animal models, i.e. after administration of estrogens in male rodents39,40 and in aromatase overexpressing male mice9,41.
Collectively, increased gene dosage of VAMP7, a SNARE protein, positively impacts ESR1 transcriptional activity (Fig. 5i) and exerts a partial and inhibitory effect on AR action. ATF3, while positively induced by ESR1 action, acts as an AR corepressor by preventing its binding to target genes and inhibiting transcriptional activation42. This may explain the effects of VAMP7 gain on the expression of several androgen-responsive genes such as Cldn11, Fkbp5, and MafB. The dual differential action of VAMP7 may partially “estrogenize” males and subtly reduce AR action, leading to slightly feminized male reproductive development in VAMP7 BAC mice. This observation provides a molecular explanation for the clinical picture phenotype found in our cohort of human patients with genitourinary anomalies.
Focal disruption of spermatogenesis and reduced fertility in mice with elevated levels of VAMP7
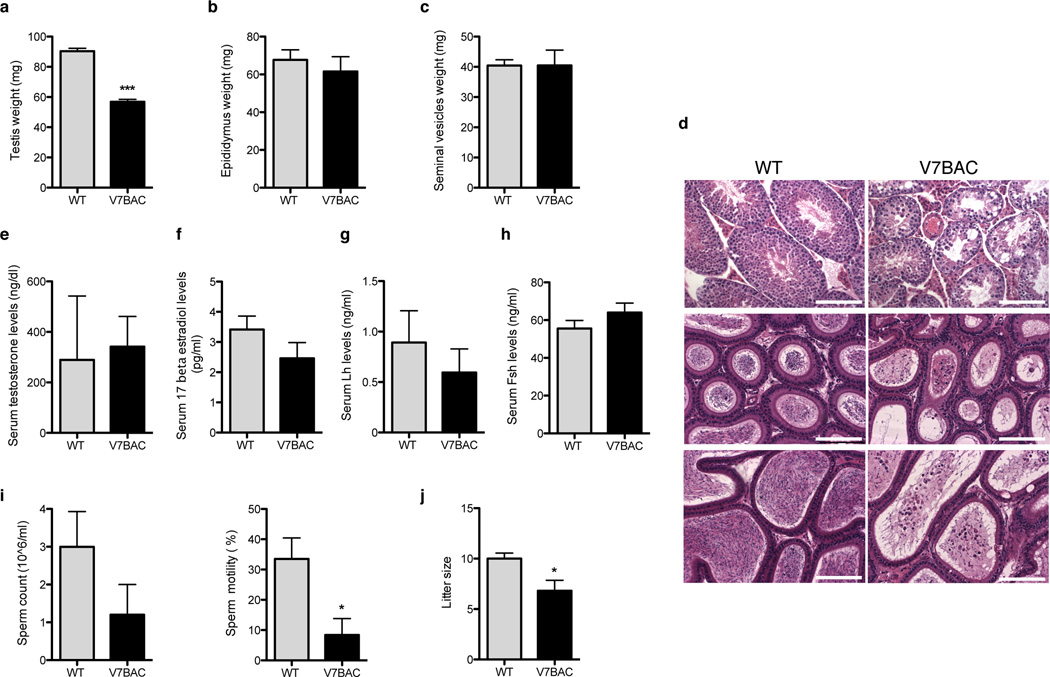
Since androgen and estrogen action are also important for spermatogenesis, we examined 6-month-old male mice with elevated levels of VAMP7. Transgenic animals had small testes and focal spermatogenic deficiency resulting in subfertility (Fig. 6 and Supplemental Fig. S4b–h). Testis weight was reduced to about 50% compared to that of wild-type scrotal testes (Fig. 6a). Epididymal and seminal vesicle weights were similar to those of wild-type controls (Fig. 6b–c and Supplemental Fig. S4c–d). The histological pattern of the adult mouse cryptorchid testes was less uniform, showing discrete foci of disrupted spermatogenesis. Arrest of spermatogenesis, sloughing of germ cells at various stages of maturation, and vacuolization of Sertoli cells were evident within the affected seminiferous tubules (Fig. 6d and Supplemental Fig. S4e). Large multinucleated round cells were often present in the lumen of the tubules (Fig. 6d and Supplemental Fig. S4e). Serum testosterone, 17β estradiol, Lh, and Fsh levels were not significantly different from those in wild-type males (Fig. 6e–h and Supplemental Fig. S4f). To quantify the functional consequences of the damage to the seminiferous epithelium, we measured sperm count and motility (Fig. 6i and Supplemental Fig. S4g). Although the number of caudal sperm was not significantly decreased, the total motility of sperm isolated from caudal epididymes of 6-month-old VAMP7 transgenic mice was reduced by 75% for strain 7 and 70% for strain 21, suggesting deficient epididymal function, perhaps due to altered hormone action (Fig. 6i and Supplemental Fig. S4g). Histological examination of VAMP7 BAC male reproductive tissues revealed morphological alterations of the epididymis in mutant mice, including vacuolization of the epididymal epithelium as well as the presence of numerous round cells in the lumen of caput and cauda V7BAC epididymides (Fig. 6d and Supplemental Fig. S4e). To assess whether the reduced motility of caudal sperm from the transgenic males was associated with reduced fertility, a controlled mating experiment was conducted in which four to five 6-week-old WT or transgenic males were mated with 8-week-old WT females. During a 4-month period, VAMP7 males sired a number of litters similar to those of the WT mice. However, the litter size in the transgenic group was significantly smaller (Fig. 6j and Supplemental Fig. S4h).
Figure 6.
Abnormal spermatogenesis and reduced motility and fertility in VAMP7 transgenic mice. (a) Weights of cryptorchid gonads (n = 7) from 6-month-old male V7BAC and scrotal testis (n = 8) from WT littermates. (b,c) Epididymis and seminal vesicles weights from 6-month-old male V7BAC (n = 8 mice) and WT (n = 6 animals). (d) Hematoxylin-eosin stained images of paraffin-embedded testis and caput and cauda epididymides sections from 6-month-old WT and V7BAC mice. Scale bar, 125 µm. (e–h) Serum testosterone, 17 beta-estradiol, Lh and Fsh hormone levels in 6-month-old male V7BAC and WT mice. (i) Epidydimal sperm count and motility in 6-month-old WT (n = 6) and V7BAC (n = 5) mice. (l) Litter size from WT (n = 12 litters) and V7BAC (n = 16 litters) male mice in a 4 month -continuous mating study. All data are expressed as mean ± s.e.m. Statistical significance was determined by unpaired, two-tailed Student’s t-test. *P < 0.05; **P < 0.01; ***P < 0.001.
In conclusion, VAMP7 transgenic male mice exhibited complex disturbances of reproductive development and function, characterized by multiple structural and functional abnormalities of the reproductive system. These changes may result from combined effects of a persistent intra-abdominal location of the testes, as well as from the prolonged imbalance of estrogen and androgen action, because several of the abnormalities observed resemble those described in mice with prenatal exposure to exogenous estrogens.
DISCUSSION
Copy-number variants account for significant human phenotypic variation and disease risk43. They exert their influence by modifying the expression of genes mapping within or close to the rearranged regions. A de novo copy number variant in 46,XY patients with mild masculinization defects suggested a causal link of VAMP7 gene duplication to the congenital genitourinary defects. Importantly, no copy number gain of VAMP7 was found in 8,951 control individuals, highlighting the high degree of significance for the VAMP7 copy number duplications and the strong association of VAMP7 gene dosage changes to human cryptorchidism, hypospadias, and micropenis (P value = 2.2×10−3; Exact Fisher test). We initially detected a gain of VAMP7 in 4 cases out of a total of 296 patients (i.e. 1.35%); then in a second replication study, we found 1 case in 28 distinct primary cultures of genital skin fibroblasts (3.6%). The incidence of VAMP7 gains represents the highest frequency reported to date for a genetic/genomic cause of genitourinary birth defects. Even previously identified pathogenetic mutations in genes such as INSL3, RXFP2, and AR exhibited lower frequencies ranging from 0.3 to 0.8% in patients with disorders of sexual differentiation44. Additionally, the Gene Expression Omnibus database repository of high throughput gene expression data (http://www.ncbi.nlm.nih.gov/geo/) shows that VAMP7 mRNA levels were significantly upregulated in human cryptorchid testes in two independent studies (GSE16191 and GSE25518)45,46. These data strongly substantiate our findings and support the fact that VAMP7 copy number change is a clinically significant factor in the etiology of human congenital genitourinary defects.
While our data supported a genomic basis of genitourinary birth defects, it also shed light on an unexpected molecular determinant impacting male phenotypic development. Under normal conditions, the vesicle trafficking protein, VAMP7, ensures proper membrane fusion and transport between various organelles12,13,22. This study shows that VAMP7 is a dosage-sensitive gene, as slight differences in the magnitude of the phenotypic defects were present between the two VAMP7 BAC transgenic lines. Indeed, line 7 mice, which had a higher level of VAMP7 expression than line 21, exhibited a higher incidence of bilateral cryptorchidism, as well as a slightly more marked decrease of testis weight and litter size, than line 21. Similarly, line 7 displayed a significant up-regulation of distinct mRNA levels (with Atf3, Sult1e1, and Cyr61 showing the most striking differences, and Ctgf and Nrip1 revealing modest impairment). Mice from strain 21 also displayed a significant up-regulation of Atf3 and Sult1e1 and a trend of increase of Cyr61 (P value = 0.09) while expression of Ctgf and Nrip1 genes was unchanged. Our observations provide evidence that higher protein levels of VAMP7 correlate with a more severe phenotype in mice.
Elevated levels of VAMP7 enhanced estrogen receptor-driven transcription of genes including Atf3, Ctgf, and Cyr61, all of which are up-regulated in patients with hypospadias29. The dual function in membrane sorting and transcriptional action was previously reported for a subset of factors involved in endocytosis and endosomal trafficking such as Huntingtin interacting protein 1 (HIP-1), Auxilin 2, Clathrin heavy chain, β-Arrestin 1 and 2, APPL1 and 2 (adaptor protein containing PH domain, PTB domain and leucine zipper motif 1 and 2) and CALM (clathrin assembly protein lymphoid myeloid)47–51. Several of these endocytic proteins exert AR coregulatory characteristics47–49, mainly through association with transcriptional factors and/or facilitation of chromatin modifications47,50,52. Our study reinforces the concept that vesicular transport and endosomal factors are able to perturb key nuclear functions and transcriptional regulation.
We demonstrated that elevated levels of VAMP7 can lead to an increase of ligand-dependent ESR1 protein content without affecting its mRNA levels and can result in a subsequent stimulation of transcriptional activity similar to the action of other previously reported proteins53. ESR1 stability and protein turnover are finely controlled by ubiquitination and proteosomal degradation to ensure efficient transcriptional activity of the nuclear receptor54–56. Recent evidence highlights the existence of cross talk between the ubiquitin-proteasome and autophagy-lysosome systems57. Perturbations in the flux through either pathway could affect the activity of the other system58. Since VAMP7 is a regulator of autophagosome formation17, it is tempting to speculate that one of the potential effects of increased VAMP7 might be the perturbation of the ubiquitin-proteasome pathway leading to abnormal processing of selective proteins including ESR1. Although beyond the scope of this present study, the exact underlying molecular mechanisms definitely merit further clarification.
In conclusion, genomic defects encompassing the gene encoding the vesicle SNARE, VAMP7, are present in a subset of human patients with congenital genitourinary disorders. Alterations of the gene dosage of this vesicular trafficking factor are responsible for abnormal human and murine male genitourinary tract development, mainly due to a marked enhancement of estrogen receptor transcriptional action. VAMP7 overexpression underlies the etiology of a subset of common disorders of male sexual differentiation.
ONLINE METHODS
Ethics statement, human subjects, and sample collection
This study was approved by the Institutional Review Board Committee at the Baylor College of Medicine (Houston, TX, USA). Probands affected with idiopathic cryptorchidism and/or hypospadias were enrolled through Texas Children’s Hospital and Ben Taub General Hospital, (Houston, TX, USA). Known causes of these birth defects, such as anomalies in the synthesis of testosterone or adrenal steroid hormones or exogenous modifiers, or karyotypic abnormalities were excluded etiologies after examination by pediatric urologists and/or neonatologists. Written informed assents were obtained for infant/child subjects and consents from their parents. Blood was collected from the children during surgery for correction of cryptorchidism or hypospadias. Parents provided saliva specimens for DNA isolation. Primary cultures of genital skin fibroblasts (GSF) isolated from the foreskin of male neonates at the time of circumcision or surgical correction were obtained from patients with hypospadias and/or cryptorchidism, after obtaining approval from the Baylor College of Medicine Institutional Review Board (Houston, TX, USA). GSF cell lines were maintained in MEM with 10% FBS and 1% penicillin and streptomycin.
CGH-based Microarray Analysis (CMA)
High molecular weight genomic DNA isolated from peripheral blood or saliva was submitted for chromosomal microarray analysis (CMA) to the Clinical Cytogenetics Laboratory at Baylor College of Medicine. CMA is a clinically-validated targeted CGH array that covers over 150 distinct human clinically relevant chromosomal loci (http://www.bcm.edu/geneticlabs/?pmid=16207). CMA V6 OLIGO microarray, a custom-targeted array manufactured by Agilent Technologies (Santa Clara, CA) using approximately 44,000 oligonucleotides to emulate the CMA V6 BAC arrays59 was used. CMA procedures and data analyses using an in-house analysis package for copy number analysis were used as described previously60. One unique DNA reference served as a control for CMA analysis and was from a pregnancy-proven fertile, gender-matched individual without any familial history of congenital genitourinary defects. The presence of imbalances was quantified by calculating the ratio of signal intensities of test DNA from patients compared with reference DNA extracted from a pregnancy-proven fertile, gender-matched individual with no familial history of congenital genitourinary defects. The log2 (Cy5/Cy3) scaling has the consequence of centering the ratios at approximately zero, making DNA copy number gains appear positive (positive shift of the probes signal).
FISH analysis
The BAC clone of interest (RP11-479B17) and a control probe RP11-815E21 (at Xq22.3) were grown in TB media with 20 mg ml−1 chloramphenicol. DNA was extracted from the BAC clones (Eppendorf Plasmid Mini Prep kit, Hamburg, Germany), directly labeled with fluorochrome conjugated-dUTP by nick translation (Vysis, Downer Grove, IL) according to the manufacturer’s instructions, and hybridized to PHA-stimulated patient lymphocyte cultures. Digital FISH images were captured using MacProbe version 4.4 software (Applied Imaging, San Jose, CA).
CNV Taqman assays
Copy Number Variant TaqMan® assays (Applied Biosystems) custom designed for VAMP7, were performed according to the manufacturer’s protocol using One Step Plus Real-Time PCR. TaqMan® Copy Number Reference Assay RNase P was used as the standard reference assay for copy number analysis. Relative quantitation analysis was done to estimate copy numbers for each sample by using the Copy-Caller-Software, v.1.0 (Applied Biosystems, Grand Island, NY, USA).
Cell culture and transfections
For transfection experiments, HeLa cells were grown in DME without phenol red, in the presence of 10% stripped fetal calf serum. Cells were plated at 70% confluency in 6-well plates for luciferase assays or 50% for immunofluorescence. Cells were transfected with several reporter-gene plasmids containing either the androgen responsive element of PSA promoter or a multimerized ERE sequence. These constructs were co-transfected as indicated in each experiment with different expression vectors encoding VAMP7 in pCMV6-XL5 vector (Origene, Rockville, MD) as well as PCMV-AR and PCR3.1-ESR1. Treatment with 10−8 M DHT or 17β estradiol was done 24 h after transfection, and reporter-gene levels were determined 24 hours after hormonal induction using Promega reagents.
For knock-down experiments, NT2/D1 (ATCC CRL-1973) cells were grown in a modified Dulbecco's medium supplemented with 10% fetal bovine serum and L-glutamine in an atmosphere of 5% CO2. Cells were plated in 6-well plates (1 × 105 per well) for RNA extraction. The next day, cells were transfected for 72 hours with nonspecific (Scramble) or siRNA for human VAMP7 (Dharmacon, Lafayette, CO, USA) according to the manufacturer’s procedure.
Immunofluorescence
Cells were rinsed twice in ice-cold phosphate-buffered saline (PBS) and fixed with a freshly prepared solution of 4% paraformaldehyde in PBS for 20 minutes and permeabilized for 10 minutes with 0.1% digitonin in PBS. After several rinses in ice-cold PBS, cells were incubated in 3% normal goat serum in PBS (blocking solution) for 30 minutes at room temperature, followed by an overnight incubation at 4°C with primary antibodies. Cells were extensively washed with PBS-0.1% digitonin and then incubated with Alexa-conjugated secondary antibodies (Molecular Probes) for 60 minutes at room temperature. Coverslips were mounted in a mounting medium. Images were captured with an API Delta Vision deconvolution microscope using SoftWorx software (Applied Precision, Issaquah, Washington, USA). Primary antibodies used were anti-AR (Sc-816, 1:200, Santa Cruz Biotechnology Inc.), anti-VAMP7 (NB100-91356, 1:1,000, Novus Biologicals), ESR1 (MA1-310, 1:200, Thermo-Fisher) and anti-RAB5 (#2143, 1:100, Cell Signaling).
Quantitative RT-PCR analysis
Total RNA was purified using the RNeasy Kit including an optional DNase I treatment according to the manufacturer's instructions (Qiagen). cDNA was prepared from 500 ng of total RNA by reverse transcription. Real-time PCR was performed with TaqMan PCR Master Mix on an ABI StepOnePlus Realtime PCR System (Applied Biosystems). PCR conditions were: 50°C for 2 min, 94°C for 2 min, followed by 40 cycles of 94°C for 15 s and 60°C for 30 s. For each experimental sample, the relative abundance value was normalized to the value derived from the endogenous control (GAPDH) of the same sample. Relative mRNA levels were quantified by the comparative DDCT method.
Generation of VAMP7 BAC transgenic mice
All described procedures were approved by the Institutional Animal Care and Use Committee of Baylor College of Medicine (Houston, TX, USA) and conducted in compliance with the Guide for the Care and Use of Laboratory Animals in a facility accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC). BAC (RP11-479B17) containing human VAMP7 gene was purified using the NucleoBond BAC 100 Kit (Clontech). The purified transgene DNA was introduced into the pronuclei of fertilized oocytes from the FVB/N mice through the service of Genetically Engineered Mouse Core Facility at the Baylor College of Medicine (http://cb-apps01.cellb.bcm.tmc.edu/gemcore/home.htm).
Fertility studies and semen evaluation
The fertility and fecundity of 6-week-old FVB male VAMP7 and their WT littermates were determined in a continuous mating study in which each male was mated with two FVB wild type females (6 weeks of age) for 4 months. The females were monitored for pregnancy during and after the mating periods, and the number of litters and offspring was recorded. Penile length was measured after exposing the penis up to the ischial arch and its stretched length measured from the tip of the glans penis to the midpoint of the ischial arch. The cauda epididymides of 3- to 8-month-old V7BAC and WT mice were collected in M2 medium. Cuts were made and sperm were allowed to swim out into the medium for 15 min at 37°C temperature. Motility was manually assessed using a hemocytometer.
Hormone assays
Mice under anesthesia were exsanguinated by closed cardiac puncture. Serum samples were collected and stored frozen at −20°C until further use. RIAs for LH and FSH were performed using National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) hormone assay kits according to the standard protocols of the Ligand Assay and Analysis Core Laboratory (University of Virginia, Charlottesville, VA, USA).
Histological analysis
Bouin’s fixed, paraffin-embedded testes were cut in cross and longitudinal sections and stained with hematoxylin and eosin. For immunohistochemistry analysis, slides of paraffin-embedded samples were deparaffinized and dehydrated. Antigen retrieval was performed by heat inactivation in 0.1 M sodium citrate for 30 min. VAMP7 (HPA036733, 1:25, Sigma Prestige or NB100-91356, 1:200, Novus Biologicals), AR (sc-816, 1:200, Santa Cruz), ATF3 (sc-188, 1:400, Santa Cruz), FKBP5 (AF4094, 1:200, R&D Systems), MAFB (ABE55, 1:200, Millipore), and desmin (NB110-56931, 1:500, Novus Biologicals) primary antibodies were used for immunodetection. The staining was performed using the avidin-biotin peroxidase system (ABC-peroxidase), and positive signals were visualized as brown precipitates using 3,3'-diaminobenzidine tetra-hydrochloride. Control staining was conducted by omission of the primary antibody. Hematoxylin was used for counterstaining.
Western blot and co-immunoprecipitation
20 µg of protein aliquots from cell lysates of testis from V7BAC transgenic mice were separated by 15% SDS-PAGE and transferred to nitrocellulose membranes. The membranes were probed with primary antibodies against VAMP7 (sc-166394, 1:100, Santa-Cruz or sc-67060, 1:50, Santa-Cruz), AR (sc-816, 1:1,000, Santa-Cruz), ESR1 (MA1-310, 1:200, Thermo-Fisher), LMNA (sc-20681, 1:200, Santa-Cruz), GAPDH (sc-32233, 1:200, Santa-Cruz), and ACTB (AC-40, 1:5,000, Sigma). HeLa cells were cotransfected with AR or ESR1 or VAMP7 or in combination. After 24 h of overexpression, cell lysates were prepared and immunoprecipitation was carried out using antibodies raised against AR (sc-816, Santa-Cruz) or ESR1 (MA5-13065, Thermo-Fisher) or VAMP7 (sc-67060, Santa-Cruz) or control IgG antibody. The samples were later subjected to western blotting with the indicated antibodies. Input was 5% of the lysates.
Chromatin immunoprecipitation
In vivo ChIP was performed on single-cell collagenase-treated testis from VAMP7 mutant mice. Formaldehyde (1%) was added to produce cross-linking during 10 minutes at room temperature. The rest of the ChIP procedure was performed using the EZ ChIP kit (Millipore) following the manufacturer’s protocol. AR antibody was from Santa-Cruz (sc-816, Santa-Cruz). qPCR for ChIP was performed using the SYBR-Green technology (Applied Biosystems). Results were normalized to input in each case. Primer sequences are available upon request.
Gene expression microarray hybridization and analysis
NT2/D1 (ATCC CRL-1973) cells were grown in a modified Dulbecco's medium supplemented with 10% fetal bovine serum and L-glutamine in an atmosphere of 5% CO2. Cells were plated in 6-well plates (1 × 105/well) for RNA extraction. The next day, cells were transfected for 72 hours with non-specific (scramble) or siRNA for human VAMP7 (Dharmacon, Lafayette, CO, USA) according to the manufacturer’s procedure. Three samples treated with siRNA targeting VAMP7 in NT2 cells were compared with three samples of cells treated with scrambled siRNA using Affymetrix Human U133 2.0 Plus Array which contains 1,300,000 unique oligonucleotides 47,000 transcripts and variants which represent about 39,000 of the best characterized human genes. The probe set design was based on databases: GenBank, dbEST, RefSeq. Microarray results were normalized using Robust Multichip Average (RMA). Statistical analysis approach of SAM (Significance Analysis of Microarray) was applied using Multiple Experiment Viewer (MeV). Data have been deposited in the GEO database under the accession number: GSE56102. We identified a list of about 600 genes that are significantly altered at a 20% False Discovery Rate (FDR) when VAMP7 was knocked down in NT2/D1 cells. This list was then analyzed using Ingenuity Pathway Analysis (IPA) to identify pathways and biological functions involved.
Accession number
Gene expression microarray data have been deposited in the GEO database. Accession number: GSE56102.
Statistical analysis
Samples and animals were evaluated in a randomized manner by six investigators (M.T.L.; S.H.; J.F.L.; B.Z; K.R. and J.A.) who were blinded to the treatment conditions. No statistical method was used to predetermine sample size for animal or cell studies. No inclusion/exclusion criteria were defined for samples and animals. At least 3 independent experiments were performed for mouse tissues and cell lines and included in the statistical analyses, as reported in all figure legends. Each replicate represents an independent experiment. Data are presented as means ± s.e.m. One- or two-way analysis of variance (ANOVA) with post hoc Bonferroni test was run for all statistical analyses except where a Student's t-test was used. Significance threshold was set at P = 5.0 × 10−2. To analyze the frequency of de novo Xq28 copy number change in affected GU patients compared to unaffected GU individuals (non-GU controls), two-tailed Fisher's exact test was performed and statistical significance determined using SPSS software.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank all the referring physicians and especially Edmond T. Gonzales, Lars Cisek, Nicolette Janzen, David R. Roth, and Eric Jones, the affected individuals, and their family members who participated in this study. We thank Marco Marcelli (Baylor College of Medicine, Houston, TX, USA) for providing the foreskin fibroblasts for analysis. We thank Lisa White, Chad Shaw and Francesco De Mayo for their assistance. This study was supported in part by US National Institutes of Health grant K12 DK0083014 (KURe to D.J.L.) and R01DK078121 from The National Institute of Diabetes and Digestive and Kidney Diseases (to D.J.L.).
Footnotes
AUTHOR CONTRIBUTIONS
Both senior authors M.T.L. and D.J.L. conceived and supervised the study, conducted experiments, analyzed data, wrote and revised the manuscript. S.H. and J.F.L. performed experiments and analyzed data. B.Z., K.R., J.A. and A.S. performed experiments. S.W.C. conducted the human studies.
COMPETING INTERESTS STATEMENTS
The authors declare that they have no competing financial interests.
REFERENCES
- 1.Sharpe RM, Skakkebaek NE. Are oestrogens involved in falling sperm counts and disorders of the male reproductive tract? Lancet. 1993;341:1392–1395. doi: 10.1016/0140-6736(93)90953-e. [DOI] [PubMed] [Google Scholar]
- 2.Wilhelm D, Koopman P. The makings of maleness: towards an integrated view of male sexual development. Nat Rev Genet. 2006;7:620–631. doi: 10.1038/nrg1903. [DOI] [PubMed] [Google Scholar]
- 3.Vidaeff AC, Sever LE. In utero exposure to environmental estrogens and male reproductive health: a systematic review of biological and epidemiologic evidence. Reprod Toxicol. 2005;20:5–20. doi: 10.1016/j.reprotox.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 4.Storgaard L, Bonde JP, Olsen J. Male reproductive disorders in humans and prenatal indicators of estrogen exposure. A review of published epidemiological studies. Reprod Toxicol. 2006;21:4–15. doi: 10.1016/j.reprotox.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 5.McLachlan JA, Newbold RR, Bullock B. Reproductive tract lesions in male mice exposed prenatally to diethylstilbestrol. Science. 1975;190:991–992. doi: 10.1126/science.242076. [DOI] [PubMed] [Google Scholar]
- 6.Goyal HO, et al. Estrogen receptor alpha mediates estrogen-inducible abnormalities in the developing penis. Reproduction. 2007;133:1057–1067. doi: 10.1530/REP-06-0326. [DOI] [PubMed] [Google Scholar]
- 7.Goyal HO, Braden TD, Williams CS, Williams JW. Estrogen-induced developmental disorders of the rat penis involve both estrogen receptor (ESR)- and androgen receptor (AR)-mediated pathways. Biol Reprod. 2009;81:507–516. doi: 10.1095/biolreprod.108.071951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simon L, et al. Exposure of neonatal rats to anti-androgens induces penile mal-developments and infertility comparable to those induced by oestrogens. Int J Androl. 2012;35:364–376. doi: 10.1111/j.1365-2605.2011.01232.x. [DOI] [PubMed] [Google Scholar]
- 9.Li X, et al. Altered structure and function of reproductive organs in transgenic male mice overexpressing human aromatase. Endocrinology. 2001;142:2435–2442. doi: 10.1210/endo.142.6.8211. [DOI] [PubMed] [Google Scholar]
- 10.Tannour-Louet M, et al. Identification of de novo copy number variants associated with human disorders of sexual development. PLoS One. 2010;5:e15392. doi: 10.1371/journal.pone.0015392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kloepper TH, Kienle CN, Fasshauer D. An elaborate classification of SNARE proteins sheds light on the conservation of the eukaryotic endomembrane system. Mol Biol Cell. 2007;18:3463–3471. doi: 10.1091/mbc.E07-03-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pryor PR, et al. Molecular basis for the sorting of the SNARE VAMP7 into endocytic clathrin-coated vesicles by the ArfGAP Hrb. Cell. 2008;134:817–827. doi: 10.1016/j.cell.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pryor PR, et al. Combinatorial SNARE complexes with VAMP7 or VAMP8 define different late endocytic fusion events. EMBO Rep. 2004;5:590–595. doi: 10.1038/sj.embor.7400150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Advani RJ, et al. VAMP-7 mediates vesicular transport from endosomes to lysosomes. J Cell Biol. 1999;146:765–776. doi: 10.1083/jcb.146.4.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rao SK, Huynh C, Proux-Gillardeaux V, Galli T, Andrews NW. Identification of SNAREs involved in synaptotagmin VII-regulated lysosomal exocytosis. J Biol Chem. 2004;279:20471–20479. doi: 10.1074/jbc.M400798200. [DOI] [PubMed] [Google Scholar]
- 16.Fader CM, Sanchez DG, Mestre MB, Colombo MI. TI-VAMP/VAMP7 and VAMP3/cellubrevin: two v-SNARE proteins involved in specific steps of the autophagy/multivesicular body pathways. Biochim Biophys Acta. 2009;1793:1901–1916. doi: 10.1016/j.bbamcr.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 17.Moreau K, Ravikumar B, Renna M, Puri C, Rubinsztein DC. Autophagosome precursor maturation requires homotypic fusion. Cell. 2011;146:303–317. doi: 10.1016/j.cell.2011.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shono T, et al. Scanning electron microscopy shows inhibited gubernacular development in relation to undescended testes in oestrogen-treated mice. Int J Androl. 1996;19:263–270. doi: 10.1111/j.1365-2605.1996.tb00474.x. [DOI] [PubMed] [Google Scholar]
- 19.Nef S, Shipman T, Parada LF. A molecular basis for estrogen-induced cryptorchidism. Dev Biol. 2000;224:354–361. doi: 10.1006/dbio.2000.9785. [DOI] [PubMed] [Google Scholar]
- 20.Jiang XW, Li JH, Huang TH, Deng WD. Effect of prenatal exposure to diethylstilbestrol on gubernacular development in fetal male mice. Asian J Androl. 2004;6:325–329. [PubMed] [Google Scholar]
- 21.Cederroth CR, et al. Estrogen receptor alpha is a major contributor to estrogen-mediated fetal testis dysgenesis and cryptorchidism. Endocrinology. 2007;148:5507–5519. doi: 10.1210/en.2007-0689. [DOI] [PubMed] [Google Scholar]
- 22.Kent HM, et al. Structural Basis of the Intracellular Sorting of the SNARE VAMP7 by the AP3 Adaptor Complex. Dev Cell. 2012 doi: 10.1016/j.devcel.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Magee JA, Chang LW, Stormo GD, Milbrandt J. Direct, androgen receptor-mediated regulation of the FKBP5 gene via a distal enhancer element. Endocrinology. 2006;147:590–598. doi: 10.1210/en.2005-1001. [DOI] [PubMed] [Google Scholar]
- 24.Makkonen H, Kauhanen M, Paakinaho V, Jaaskelainen T, Palvimo JJ. Long-range activation of FKBP51 transcription by the androgen receptor via distal intronic enhancers. Nucleic Acids Res. 2009;37:4135–4148. doi: 10.1093/nar/gkp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen H, et al. Fkbp52 regulates androgen receptor transactivation activity and male urethra morphogenesis. J Biol Chem. 2010;285:27776–27784. doi: 10.1074/jbc.M110.156091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishida H, et al. Gene expression analyses on embryonic external genitalia: identification of regulatory genes possibly involved in masculinization processes. Congenit Anom (Kyoto) 2008;48:63–67. doi: 10.1111/j.1741-4520.2008.00180.x. [DOI] [PubMed] [Google Scholar]
- 27.Miyagawa S, et al. The role of sonic hedgehog-Gli2 pathway in the masculinization of external genitalia. Endocrinology. 2011;152:2894–2903. doi: 10.1210/en.2011-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rivas A, Fisher JS, McKinnell C, Atanassova N, Sharpe RM. Induction of reproductive tract developmental abnormalities in the male rat by lowering androgen production or action in combination with a low dose of diethylstilbestrol: evidence for importance of the androgen-estrogen balance. Endocrinology. 2002;143:4797–4808. doi: 10.1210/en.2002-220531. [DOI] [PubMed] [Google Scholar]
- 29.Wang Z, et al. Up-regulation of estrogen responsive genes in hypospadias: microarray analysis. J Urol. 2007;177:1939–1946. doi: 10.1016/j.juro.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 30.Liu B, et al. Estradiol upregulates activating transcription factor 3, a candidate gene in the etiology of hypospadias. Pediatr Dev Pathol. 2007;10:446–454. doi: 10.2350/06-04-0079.1. [DOI] [PubMed] [Google Scholar]
- 31.Liu B, et al. Activating transcription factor 3 is up-regulated in patients with hypospadias. Pediatr Res. 2005;58:1280–1283. doi: 10.1203/01.pdr.0000187796.28007.2d. [DOI] [PubMed] [Google Scholar]
- 32.Liu B, Agras K, Willingham E, Vilela ML, Baskin LS. Activating transcription factor 3 is estrogen-responsive in utero and upregulated during sexual differentiation. Horm Res. 2006;65:217–222. doi: 10.1159/000092402. [DOI] [PubMed] [Google Scholar]
- 33.Gurbuz C, et al. Is activating transcription factor 3 up-regulated in patients with hypospadias? Korean J Urol. 2010;51:561–564. doi: 10.4111/kju.2010.51.8.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalfa N, et al. Genomic variants of ATF3 in patients with hypospadias. J Urol. 2008;180:2183–2188. doi: 10.1016/j.juro.2008.07.066. discussion 2188. [DOI] [PubMed] [Google Scholar]
- 35.Beleza-Meireles A, et al. Activating transcription factor 3: a hormone responsive gene in the etiology of hypospadias. Eur J Endocrinol. 2008;158:729–739. doi: 10.1530/EJE-07-0793. [DOI] [PubMed] [Google Scholar]
- 36.Thenot S, Charpin M, Bonnet S, Cavailles V. Estrogen receptor cofactors expression in breast and endometrial human cancer cells. Mol Cell Endocrinol. 1999;156:85–93. doi: 10.1016/s0303-7207(99)00139-2. [DOI] [PubMed] [Google Scholar]
- 37.Lambard S, et al. Aromatase in testis: expression and role in male reproduction. J Steroid Biochem Mol Biol. 2005;95:63–69. doi: 10.1016/j.jsbmb.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 38.Song WC, Qian Y, Sun X, Negishi M. Cellular localization and regulation of expression of testicular estrogen sulfotransferase. Endocrinology. 1997;138:5006–5012. doi: 10.1210/endo.138.11.5512. [DOI] [PubMed] [Google Scholar]
- 39.Hazary S, Gardner WU. Influence of sex hormones on abdominal musculature and the formation of inguinal and scrotal hernias in mice. Anat Rec. 1960;136:437–443. doi: 10.1002/ar.1091360402. [DOI] [PubMed] [Google Scholar]
- 40.Reinert RB, et al. Tamoxifen-Induced Cre-loxP Recombination Is Prolonged in Pancreatic Islets of Adult Mice. PLoS One. 2012;7:e33529. doi: 10.1371/journal.pone.0033529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li X, Rahman N. Impact of androgen/estrogen ratio: lessons learned from the aromatase over-expression mice. Gen Comp Endocrinol. 2008;159:1–9. doi: 10.1016/j.ygcen.2008.07.025. [DOI] [PubMed] [Google Scholar]
- 42.Wang H, et al. The stress response mediator ATF3 represses androgen signaling by binding the androgen receptor. Mol Cell Biol. 2012;32:3190–3202. doi: 10.1128/MCB.00159-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Girirajan S, Campbell CD, Eichler EE. Human copy number variation and complex genetic disease. Annu Rev Genet. 2011;45:203–226. doi: 10.1146/annurev-genet-102209-163544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ferlin A, et al. Genetic alterations associated with cryptorchidism. JAMA. 2008;300:2271–2276. doi: 10.1001/jama.2008.668. [DOI] [PubMed] [Google Scholar]
- 45.Hadziselimovic F, et al. EGR4 is a master gene responsible for fertility in cryptorchidism. Sex Dev. 2009;3:253–263. doi: 10.1159/000249147. [DOI] [PubMed] [Google Scholar]
- 46.Hadziselimovic F, Hadziselimovic NO, Demougin P, Oakeley EJ. Testicular gene expression in cryptorchid boys at risk of azoospermia. Sex Dev. 2011;5:49–59. doi: 10.1159/000323955. [DOI] [PubMed] [Google Scholar]
- 47.Mills IG, et al. Huntingtin interacting protein 1 modulates the transcriptional activity of nuclear hormone receptors. J Cell Biol. 2005;170:191–200. doi: 10.1083/jcb.200503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ray MR, et al. Cyclin G-associated kinase: a novel androgen receptor-interacting transcriptional coactivator that is overexpressed in hormone refractory prostate cancer. Int J Cancer. 2006;118:1108–1119. doi: 10.1002/ijc.21469. [DOI] [PubMed] [Google Scholar]
- 49.Jasavala R, et al. Identification of putative androgen receptor interaction protein modules: cytoskeleton and endosomes modulate androgen receptor signaling in prostate cancer cells. Mol Cell Proteomics. 2007;6:252–271. doi: 10.1074/mcp.M600169-MCP200. [DOI] [PubMed] [Google Scholar]
- 50.Kang J, et al. A nuclear function of beta-arrestin1 in GPCR signaling: regulation of histone acetylation and gene transcription. Cell. 2005;123:833–847. doi: 10.1016/j.cell.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 51.Yang L, et al. APPL suppresses androgen receptor transactivation via potentiating Akt activity. J Biol Chem. 2003;278:16820–16827. doi: 10.1074/jbc.M213163200. [DOI] [PubMed] [Google Scholar]
- 52.Enari M, Ohmori K, Kitabayashi I, Taya Y. Requirement of clathrin heavy chain for p53-mediated transcription. Genes Dev. 2006;20:1087–1099. doi: 10.1101/gad.1381906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wei X, Xu H, Kufe D. MUC1 oncoprotein stabilizes and activates estrogen receptor alpha. Mol Cell. 2006;21:295–305. doi: 10.1016/j.molcel.2005.11.030. [DOI] [PubMed] [Google Scholar]
- 54.Reid G, et al. Cyclic, proteasome-mediated turnover of unliganded and liganded ERalpha on responsive promoters is an integral feature of estrogen signaling. Mol Cell. 2003;11:695–707. doi: 10.1016/s1097-2765(03)00090-x. [DOI] [PubMed] [Google Scholar]
- 55.Lonard DM, Nawaz Z, Smith CL, O'Malley BW. The 26S proteasome is required for estrogen receptor-alpha and coactivator turnover and for efficient estrogen receptor-alpha transactivation. Mol Cell. 2000;5:939–948. doi: 10.1016/s1097-2765(00)80259-2. [DOI] [PubMed] [Google Scholar]
- 56.Nawaz Z, Lonard DM, Dennis AP, Smith CL, O'Malley BW. Proteasome-dependent degradation of the human estrogen receptor. Proc Natl Acad Sci U S A. 1999;96:1858–1862. doi: 10.1073/pnas.96.5.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Korolchuk VI, Menzies FM, Rubinsztein DC. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett. 2010;584:1393–1398. doi: 10.1016/j.febslet.2009.12.047. [DOI] [PubMed] [Google Scholar]
- 58.Korolchuk VI, Mansilla A, Menzies FM, Rubinsztein DC. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell. 2009;33:517–527. doi: 10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
REFERENCES TO METHODS
- 59.Ou Z, et al. Branchiootorenal syndrome and oculoauriculovertebral spectrum features associated with duplication of SIX1, SIX6, and OTX2 resulting from a complex chromosomal rearrangement. Am J Med Genet A. 2008;146A:2480–2489. doi: 10.1002/ajmg.a.32398. [DOI] [PubMed] [Google Scholar]
- 60.Ou Z, et al. Bacterial artificial chromosome-emulation oligonucleotide arrays for targeted clinical array-comparative genomic hybridization analyses. Genet Med. 2008;10:278–289. doi: 10.1097/GIM.0b013e31816b4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.