Abstract
Methyl jasmonate (MeJA) elicitation is an effective strategy to induce and enhance synthesis of the anticancer agent paclitaxel (Taxol®) in Taxus cell suspension cultures; however, concurrent decreases in growth are often observed, which is problematic for large scale bioprocessing. Here, increased accumulation of paclitaxel in Taxus cuspidata suspension cultures with MeJA elicitation was accompanied by a concomitant decrease in cell growth, evident within the first three days post-elicitation. Both MeJA-elicited and mock-elicited cultures exhibited similar viability with no apoptosis up to day 16 and day 24 of the cell culture period, respectively, suggesting that growth repression is not attributable to cell death. Flow cytometric analyses demonstrated that MeJA perturbed cell cycle progression of asynchronously dividing Taxus cells. MeJA slowed down cell cycle progression, impaired the G1/S transition as observed by an increase in G0/G1 phase cells, and decreased the number of actively dividing cells. Through a combination of deep sequencing and gene expression analyses, the expression status of Taxus cell cycle-associated genes correlated with observations at the culture level. Results from this study provide valuable insight into the mechanisms governing MeJA perception and subsequent events leading to repression of Taxus cell growth.
Keywords: Cell cycle, next-generation sequencing, methyl jasmonate, paclitaxel, plant cell culture, Taxus
INTRODUCTION
Dedifferentiated plant cell suspension cultures provide a regulated environment independent of geographical and external environmental factors for the synthesis of plant-based secondary metabolites. Cell suspension culture offers a distinct advantage over tissue or organ culture as existing bioprocess technology developed for microbial and mammalian cells can be easily adapted to plant cells (Kieran, MacLoughlin and Malone 1997). The use of elicitors in plant cell suspension culture can both increase product yields and consequently decrease the long fermentation times, facilitating the use of plant cell culture technology in commercial applications. Jasmonic acid (JA) and its methyl ester, methyl jasmonate (MeJA) have been widely used as elicitors to induce secondary metabolite production in a variety of plant cell culture systems (Gundlach et al. 1992, Lijavetzky et al. 2008, Pauwels et al. 2008, Yazaki et al. 1997). In particular, jasmonates have been effective at enhancing production of the anticancer drug paclitaxel (Taxol®) in a variety of Taxus species and cell cultures (Bonfill et al. 2006, Ketchum et al. 1999, Yukimune et al. 1996). Paclitaxel is widely used for treatment of breast, ovarian and lung cancers as well as AIDS-related Kaposi’s sarcoma, and is being investigated for use in the treatment of neurological disorders and in post-surgery heart patients (Vongpaseuth and Roberts 2007). Paclitaxel titers of up to 900 mg/L have been achieved in industrial environments using a combination of MeJA elicitation and cell culture optimization strategies (Bringi et al. 2007).
Increased secondary metabolite accumulation upon MeJA elicitation is often accompanied with concurrent decreases in culture growth (Kim et al. 2005), Thanh et al. 2005, Zhang and Turner 2008, Sun et al. 2013). MeJA has been shown to broadly induce defense responses and secondary metabolism in plants (Farmer and Ryan 1990, Reymond and Farmer 1998, Seo et al. 2001), which diverts carbon resource allocation from primary metabolism (Logemann et al. 1995, Pauwels et al. 2009). Recent studies indicate that MeJA-mediated growth inhibition is associated with perturbations in mitochondrial membrane integrity along with decreases in the biosynthesis of ATP (Ruiz-May et al. 2011) and proteins related to energy metabolism (Cho et al. 2007).
At a mechanistic level, MeJA has demonstrated an inhibitory effect on growth at the level of the cell cycle (Pauwels et al. 2008, Swiatek et al. 2002). Most studies to understand the effect of jasmonates on the cell cycle have been done in angiosperms, such as Arabidopsis thaliana and tobacco BY-2 cell suspension cultures (Pauwels et al. 2008, Swiatek et al. 2002). Exogenously applied MeJA blocks the G1/S and G2/M transitions in the cell cycle of cultured tobacco BY-2 cells (Swiatek et al. 2002). Micromolar concentrations of MeJA added to A. thaliana suspension cultures repressed the activation of M phase genes, arresting cells in G2 phase (Pauwels et al. 2008). Genomic information and established protocols for synchronizing cell cultures (Kumagai-Sano et al. 2006, Menges et al. 2002) to understand cell cycle events are readily available for these plant species, facilitating mechanistic studies. In contrast, gymnosperms such as Taxus have not been as well studied with regard to cell cycle progression and the mechanism of MeJA-repressed growth.
While a number of studies have reported increased taxane biosynthetic pathway gene products upon MeJA elicitation (Jennewein et al. 2004, Nims et al. 2006, Patil et al. 2012, Li et al. 2012), there have been few reports regarding the role of MeJA on growth inhibition and cell cycle progression in Taxus cultures (Kim et al. 2005, Naill and Roberts, 2005a). In the present study we investigate the influence of MeJA on both cell growth and viability of Taxus cells in batch culture. The effect of MeJA on cell cycle progression was determined using asynchronous Taxus cuspidata cells. Actively dividing cells were quantified and cell cycle kinetics were determined by cumulative and pulse-labeling using 5-ethynyl-2’-deoxyuridine (EdU), a nucleoside analog of thymidine. Recently obtained 454 and Illumina transcriptome sequencing data for both MeJA-elicited and mock-elicited cultures were used to obtain the expression status of cell cycle-associated genes in the asynchronous T. cuspidata cultured cells. There is currently minimal sequence information on cell cycle regulated genes derived from this division of the plant kingdom (Li et al. 2012, Sun et al. 2013), and these studies provide the first insight into cell cycle control upon elicitation with MeJA. Because the mechanism of action of MeJA has not been investigated to date for gymnosperms such as Taxus, strategies to promote growth, while still enhancing secondary metabolite synthesis for bioprocessing, have not been identified or tested. The results here show that MeJA-induced growth repression in Taxus growth occurs at the level of cell cycle, providing important mechanistic information on the influence of MeJA on Taxus cell proliferation.
MATERIALS AND METHODS
Cell culture maintenance and MeJA elicitation
The T. cuspidata P93AF cell line was provided by the U.S. Plant Soil and Nutrition Laboratory (Ithaca, NY), and maintained in our laboratory on gyratory shakers as described previously (Kolewe et. al 2010). For elicitation, 40 μL of 100% methyl jasmonate (MeJA) was added to 460 µL of 95% (v/v) ethanol and 500 µL nanopure water. This solution was vortexed and then filtered through a 0.2 µm Gelman PVDF filter into a sterile container. This solution was then added to the cultures on day 7 post-transfer to yield a final concentration of 150 µM. The foam closures were covered with aluminum foil to prevent evaporation. Mock-elicited cultures were generated by using equivalent amounts of sterile water instead of MeJA. All chemicals were purchased from Sigma-Aldrich unless otherwise specified.
Biomass and taxane content measurements
A Multisizer 3™ Coulter counter equipped with a 2000 µm aperture (Beckman Coulter, Brea, CA, USA) was used to determine total biomass dry weight based on previously published correlations (Kolewe et al. 2010). For analysis, two × 2 mL samples of well-mixed culture broth (cells plus media) were taken from each flask. Taxane content in mock-elicited and MeJA-elicited cultures were analyzed at several time points post-elicitation using Ultra Performance Liquid Chromatography (UPLC), as described previously (Patil et al. 2012).
Viability analysis
Qualitative analysis of viability was performed by staining with fluorescein diacetate (FDA) and propidium iodide (PI). 500 µL of well-mixed culture was sampled, to which 10 µL of a 0.5 mg/mL FDA stock solution and 4 µL of a 1 mg/mL PI stock solution was added. After a 10 min incubation in the dark, supernatant was removed, and 500 µL of fresh Gamborg’s B5 medium was added. 100 µL of cell suspension was observed under a fluorescence microscope. A Zeiss Axiovert 200 inverted microscope fitted with a blue filter set (excitation at 450–490 nm and emission above 515 nm) for FDA fluorescence and a green filter set (excitation at 530–560 nm and emission above 580 nm) for PI fluorescence was used. FDA detects living cells, whereas PI detects non-viable cells. Note that esterases released from non-viable cells can result in background fluorescence.
DNA laddering assay
Total genomic DNA was extracted following the method of Dellaporta et al. (1983) with a slight modification. Fresh cells (0.2 g) were ground in liquid nitrogen with mortar and pestle. Ground cells were transferred to a sterilized Eppendorf tube and dissolved in 600 µL buffer (pH 8.0) consisting of NaCl (100 mM), Tris/HCl (50 mM), EDTA (25 mM), sodium dodecyl sulfate (1%, w/v), and β-mercaptoethanol (10 mM). The mixture was shaken vigorously through inversion and incubated in a water bath at 65 oC for 10 min. 250 µl of potassium acetate (5 M) was added to the mixture, which was incubated on ice for 30 min followed by centrifugation at 10,000 × g for 10 min at 4 oC. The supernatant was collected and mixed with an equal volume of isopropanol (approximately 600 µL). The precipitate formed was centrifuged, washed with 70% (v/v) ethanol, and redissolved in 200 µl of buffer (10 mM Tris/HCl, 5 mM EDTA, pH 8.0). Further precipitation was achieved by addition of 20 µL sodium acetate (3 M, pH 5.2), followed by 500 µL of 100% (v/v) ethanol and gentle inversion to completely mix the two phases. Precipitates were centrifuged, washed with 70% (v/v) ethanol and then dried at 25 °C. Pellets were resuspended in 40 µL buffer (10 mM Tris/HCL, 1 mM EDTA, pH 8.0). RNAse A (100 µg/mL) was added to digest RNA at 37 oC for 30 min. DNA concentrations were quantified using a Nanodrop 1000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). For analysis of DNA laddering, equal amounts of DNA were run on a 1.5 % (w/v) agarose gel stained with ethidium bromide (0.5 µg/mL), and observed under a UV transilluminator. 1 kbp and 100 bp DNA ladders (New England Biolabs) were used as molecular weight markers.
Isolation and fixation of intact nuclei
Intact nuclei were isolated from Taxus cells in Galbraith’s buffer [45 mM MgCl2, 30 mM sodium citrate, 20 mM 3-(N-morpholino)-propanesulfonic acid (MOPS), 0.3 % (w/v) Triton X-100, pH 7.0] as described previously (Patil et al. 2013, Gaurav et al. 2010, Galbraith et al. 1983). Post isolation, nuclei were fixed with 1% paraformaldehyde (dissolved in Galbraith’s buffer) at 4 oC for 30 min. Fixed nuclei were washed twice with Galbraith’s buffer by centrifuging at 700 × g for 4 min at 4 oC. After washing, nuclei were resuspended in 1 mL of Galbraith’s buffer and stored at 4 oC until further analysis.
Distribution of cells in different phases of the cell cycle
Triplicate samples of MeJA-elicited and mock-elicited cultures were sampled and nuclei were isolated at several time points post-elicitation as described above. 1 mL of the nuclei solution were aliquoted, and 50 µL of 1 mg/mL RNAse and 50 µL of 1 mg/mL PI were added. Samples were stained for at least 30 min on ice before flow cytometric analysis (see below).
EdU incorporation assay
MeJA-elicited and mock-elicited Taxus cell cultures were maintained in medium containing 10 µM EdU for the required incubation period (see cumulative and pulse labeling below). Nuclei were isolated from the EdU-labeled cultures, fixed and incubated with 250 µL EdU detection cocktail (Click-iT EdU Alexa Fluor 488 Flow Cytometry Assay kit, cat no: C35002, Invitrogen, Carlsbad, CA) at 25 °C for 30 min as per manufacturer’s protocol, with slight modifications. For one sample reaction (250 µL), the following amounts of kit components were used: 219 µL of 1X Click-iT Reaction buffer (Component G, diluted to 1X in Galbraith’s buffer), 5 µL Copper (II) sulfate solution (Component H, 100 mM aqueous CuSO4), 1.25 µL Alexa Fluor 488 azide (Component B) and 25 µL 1X buffer additive (component I, diluted to 1X in Galbraith’s buffer). Post incubation, 2 mL of Galbraith’s buffer were added to the samples as a wash; samples were centrifuged at 700 × g and 4 °C for 4 min, and resuspended in 0.5 mL Galbraith’s buffer for subsequent staining and analysis. 1 mg/mL of RNAse A was added followed by 1 mg/mL 7-aminoactinomycin D (7-AAD) (Invitrogen, Carlsbad, CA). Samples were incubated for 60 min at 25 °C before flow cytometric analysis (see below).
Pulse labeling of MeJA-elicited and mock-elicited cultures
MeJA-elicited and mock-elicited cultures on day 5 of the culture period were pulse-labeled for 4 hours with 10 µM EdU. Cells were collected by centrifugation at 800 × g for 5 min, and washed with conditioned medium. Conditioned medium was obtained by decanting settled MeJA-elicited and mock-elicited cell suspensions incubated without EdU under the same conditions. The wash procedure was repeated two more times to eliminate excess EdU. The volume was then adjusted with conditioned medium to that before washing, and cultures were continued in the absence of EdU. Samples of pulse-labeled cells were taken periodically over 48 hours and processed for EdU analysis. A series of bivariate DNA-EdU distributions at various times after pulse-labeling was obtained using flow cytometry (see below).
Cumulative labeling of MeJA-elicited and mock-elicited cultures
MeJA-elicited and mock-elicited cultures on day 7 of the culture period were incubated with 10 µM EdU. After EdU addition, nuclei were isolated and fixed every 24 hours for the following 5 days. Isolated nuclei were stained for EdU (Alexa Fluor 488) and DNA content (7-AAD) (see above) and analyzed via flow cytometry (see below).
Flow cytometry
For nuclei analysis, a Becton Dickinson (San Jose, CA) LSRII analytical flow cytometer equipped with an argon laser tuned to 488 nm with the standard filter set-up was used. A minimum of 5000 events were collected in the gated region of a forward scatter and side scatter plot corresponding to nuclei. The scatter plots were manually gated to exclude debris and doublets. For cell cycle analysis, forward scatter and side scatter were collected on a logarithmic scale, and PI fluorescence was collected on a linear scale. For cumulative-and pulse-EdU labeling analysis, a bivariate plot of DNA-EdU was obtained. Alexa Fluor 488 EdU intensity was detected between 515–545 nm. For detection of 7-AAD intensity (DNA content) the 663–677 nm emission range was used with the standard filter sets available on the LSRII. The boundary of EdU-labeled nuclei in biparametric plots was obtained by subtracting the background using a non-EdU treated culture.
Cell cycle-associated contig generation, annotation and expression analysis
RNA was isolated from T. cuspidata P93AF cells (MeJA-elicited and mock-elicited) every 24 hours post-elicitation over a time period spanning 22 days of the culture period. Equal amounts of total RNA from each culture were pooled from each time point for 454 sequencing. After rRNA depletion and fragmentation, a transcriptome library was generated by sequencing the pooled RNA sample on one full PicoTiterPlate (PTP) using the 454 Genome Sequencer FLX Titanium System™, following the manufacturer's instructions (Roche, Branford, CT). In addition, 50 bp paired end mRNA-sequencing libraries were prepared from both MeJA-elicited and mock-elicited T. cuspidata P93AF cells at 18 and 72 hour time points using the Illumina HiSeq 2000 platform (Illumina, Inc. San Diego, CA). Reads from both 454 and Illumina sequencing libraries were used in de novo assembly to generate contigs using CLC genomics workbench (CLC Bio, Aarhus, Denmark) by setting A, C, G, T voting method for conflict resolution. A total of 48,614 contigs were generated (>200 bp, avg. >100 reads/contig, >50X coverage) with N50 contig length of 873 bp. These contigs were annotated using Blast2GO default parameters (Conesa et al. 2005). Based on the Blast2GO annotation, 149 contigs representing known cell cycle-associated genes were identified (Online Resource 2). Paired end reads from each Illumina library were mapped onto the contigs using CLC Genomics Workbench software. Gene expression for both MeJA-elicited and mock-elicited P93AF cells at the 18 and 72 hour post-elicitation time points were calculated by using the RPKM (Reads per kb per million reads) method (Mortazavi et al. 2008). To identify differentially expressed genes, the proportions-based test was used between any two RNA-seq libraries under comparison with a p-value <0 .05 (Kal et al. 1999). To calculate the fold change between any two conditions, quantile normalization of the RPKM values was used.
RESULTS
MeJA represses cell growth without significant changes in necrosis and apoptosis
Inhibition of growth after MeJA elicitation has been observed in a variety of Taxus species and cell lines (Bonfill et al. 2006, Kim et al. 2004, Laskaris et al. 1999, Yukimune et al. 1996), as well as other plant cell culture systems (Goossens et al. 2003, Thanh et al. 2005). Although most reports indicate a decrease in Taxus cell growth upon MeJA elicitation, some data show no difference in cell growth between MeJA-elicited and non-elicited Taxus cell lines (Bonfill et al. 2007, Ketchum et al. 1999). Upon addition of MeJA on day 7 of the culture period, T. cuspidata P93AF cell cultures clearly demonstrate repressed growth (Fig. 1a) and increased taxane production (Fig. 1b) as compared to mock-elicited cultures.
Fig. 1.
Effect of MeJA elicitation on T. cuspidata P93AF cultures growth, taxane production and viability. (a) Biomass concentrations in MeJA-elicited cultures as compared to mock-elicited cultures. Reported values are the average of three biological replicates and error bars represent standard error of the mean (SEM). (b) Taxane levels as determined by UPLC in MeJA-elicited cultures. Note that mock-elicited cultures did not produce detectable levels of taxanes. Reported values are the average of three biological replicates and error bars represent SEM. (c, d) Viability of mock-elicited and MeJA-elicited cultures on day 16 (Row 1), day 21 (Row 2), and day 24 (Row 3) of the culture period. Columns indicate brightfield (Column 1), FDA-stained (Column 2) and PI-stained (Column 3) images. Cultures were either mock-elicited or elicited with 150 µM MeJA on day 7 of the culture period
Subsequently, we examined the viability of cultures using fluorescein diacetate (FDA) and propidium iodide (PI), to indicate both viable and non-viable cells, respectively. Eight time points were examined spanning 24 days of the culture period; representative data are shown in Fig. 1c and 1d. Both MeJA-elicited and mock-elicited cultures exhibited approximately 90% to 95% viability until day 16 of the culture period (9d post-elicitation) (Fig. 1c and 1d). At later time points, the FDA fluorescence intensity decreased in both MeJA-elicited and mock-elicited cultures, implying a decrease in metabolic activity for all cultures (Li et al. 2011). More PI fluorescent cells were observed in MeJA-elicited cultures as compared to mock-elicited cultures on days 21 and 24 of the culture period (14d and 17d post-elicitation) (Fig. 1c and 1d), indicating compromised cell membrane integrity and the beginning of cell necrosis in MeJA-elicited cultures. There was a time lag between evidence of reduced viability and a measurable decrease in biomass (dry weight), as has been observed with other Taxus cell lines (Kim et al. 2005, Naill and Roberts, 2005a).
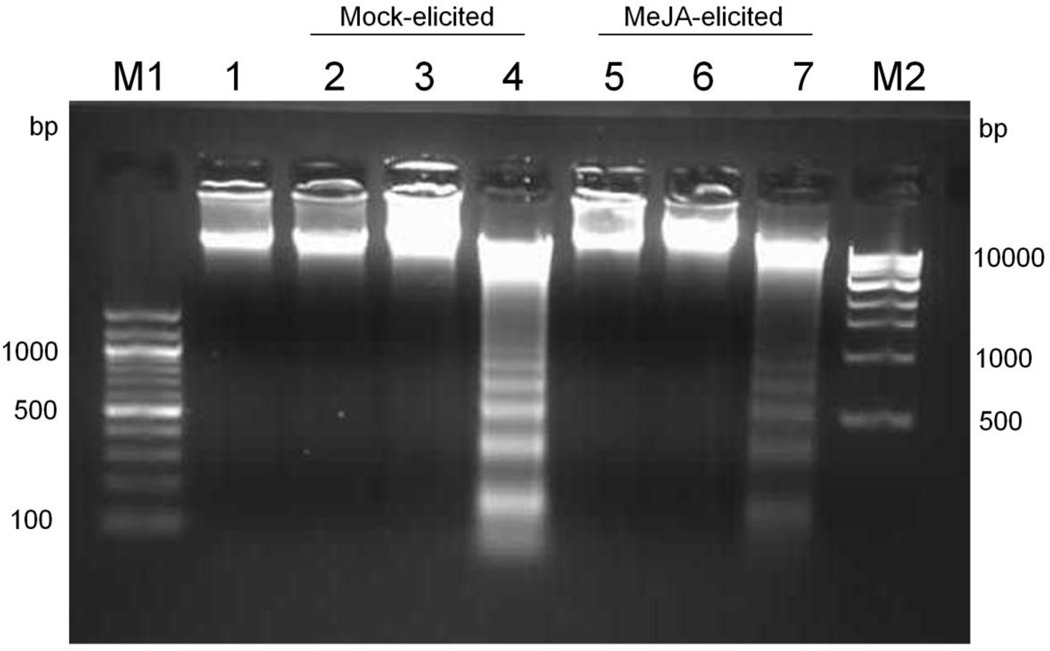
A hallmark feature of apoptotic cell death is the fragmentation of DNA into oligonucleosomal fragments of approximately 180–200 bp or multiples thereof, giving rise to a ladder during gel electrophoresis of genomic DNA (Ryerson and Heath, 1996, Yuan et al. 2002). Genomic DNA was isolated from MeJA-elicited and mock-elicited cultures on day 11, day 24 and day 30 of the culture period. A DNA laddering pattern was not observed until day 30 of the culture period (23d post-elicitation) in mock-elicited and MeJA-elicited cultures (Fig. 2), indicating that apoptosis is not a direct consequence of MeJA elicitation. Repression of cell growth thus occurs before significant necrosis and apoptosis begins in MeJA-elicited Taxus cultures, which necessitates further investigation into the cell cycle to understand the role of MeJA in growth inhibition.
Fig. 2.
Effect of MeJA elicitation on induction of oligonucleosomal fragmentation in cultured T. cuspidata P93AF cells. The agarose gel shows DNA extracted from mock-elicited and MeJA-elicited cultures. M1, 100 bp marker; lane 1, mock-elicited (day 7); lanes 2, 3 and 4, mock-elicited cultures on day 11, 24 and 30 of culture period, respectively; lanes 5, 6, and 7, MeJA-elicited cultures on day 11, 24 and 30 of the culture period, respectively; M2, 1 kbp marker. Cultures were either mock-elicited or elicited with 150 µM MeJA on day 7 of the culture period
MeJA causes a transient increase in G2 phase cells and a decrease in S phase cells, followed by an arrest at G0/G1 in asynchronous Taxus suspension cultures
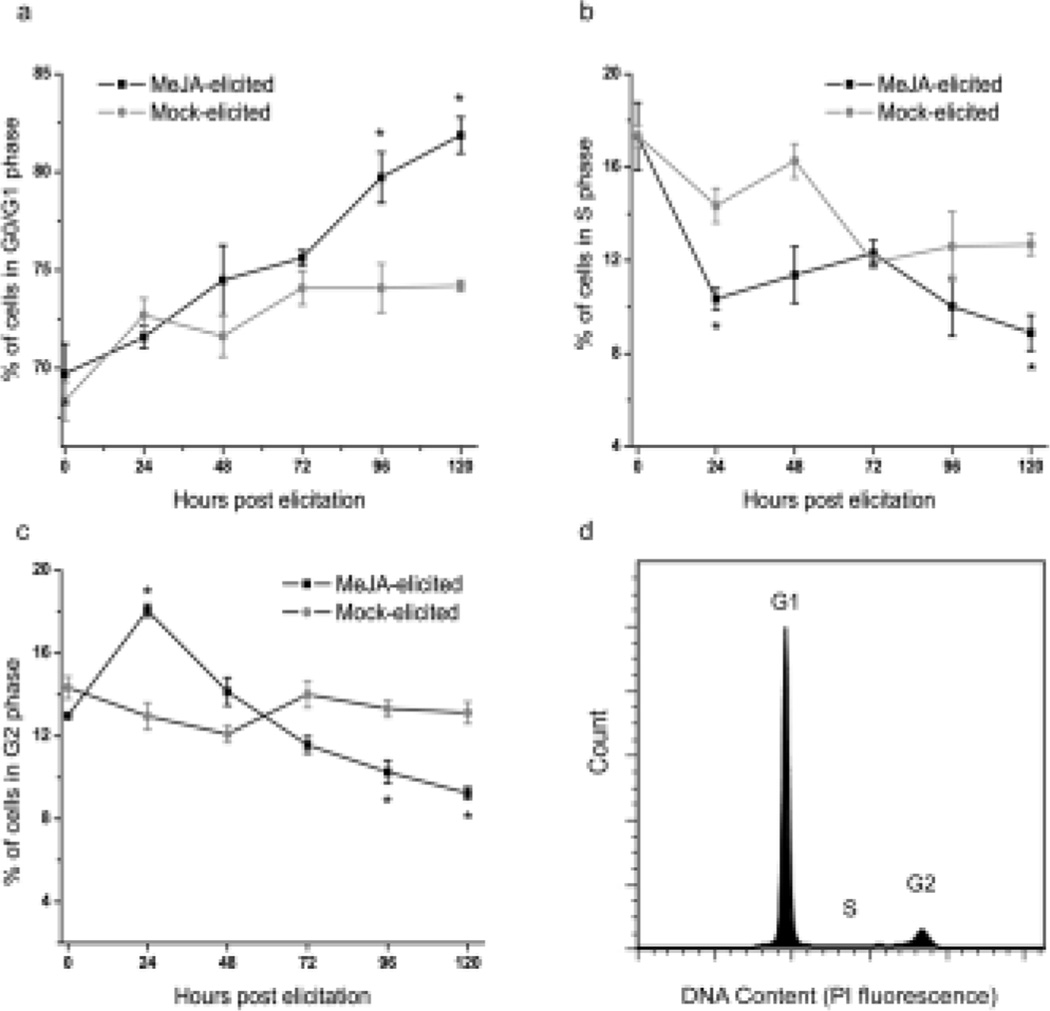
Nuclei were isolated from mock-elicited and MeJA-elicited cultures and stained with PI for flow cytometric-based DNA quantification. A flow cytometric DNA histogram of Taxus nuclei is shown in Fig. 3d for reference. All data presented were extracted from similar histograms. The percentage of cells in the different cell cycle phases were calculated using the Watson Pragmatic Model of FlowJo software (v 7.6, Tree Star, Inc.). The proportion of cells in each cell cycle phase in mock-elicited and MeJA-elicited cultures is shown in Fig. 3 (a–c). Within the first 24 hours post-elicitation, MeJA-elicited cultures had more cells in G2 phase (18% in MeJA-elicited, 13% in mock-elicited cultures) and fewer cells in S phase (10% in MeJA-elicited, 14.5% in mock-elicited cultures) when compared to mock-elicited cultures. This trend was confirmed by further analyzing cultures at shorter time increments before 24 hours post-elicitation (Online Resource 1). After 72 hours post-elicitation the percentage of cells in both G2 and S phases decreased and the distributions shifted towards a higher percentage of cells in G0/G1. An increased G0/G1 cell population post-MeJA-elicitation has also been observed with another Taxus cell suspension line (Naill and Roberts, 2005a).
Fig. 3.
Cell cycle distribution in MeJA-elicited and mock-elicited T. cuspidata P93AF cultures. The percentage of cells in (a) G0/G1, (b) S, and (c) G2/M phases is shown. (d) Cell cycle analysis using the Watson Pragmatic Model of the FlowJo (v7.6) software. RNAse A (50 µg/mL) treatment, followed by staining with propidium iodide (PI) (50 µg/mL) was performed to obtain DNA histograms. Reported values are the average of three biological replicates and error bars represent SEM. The asterisk (*) indicates a statistically significant difference (P < 0.05; paired Student's t test) between MeJA-elicited and mock-elicited conditions. Cultures were either mock-elicited or elicited with 150 µM MeJA on day 7 of the culture period
MeJA slows down the cell cycle
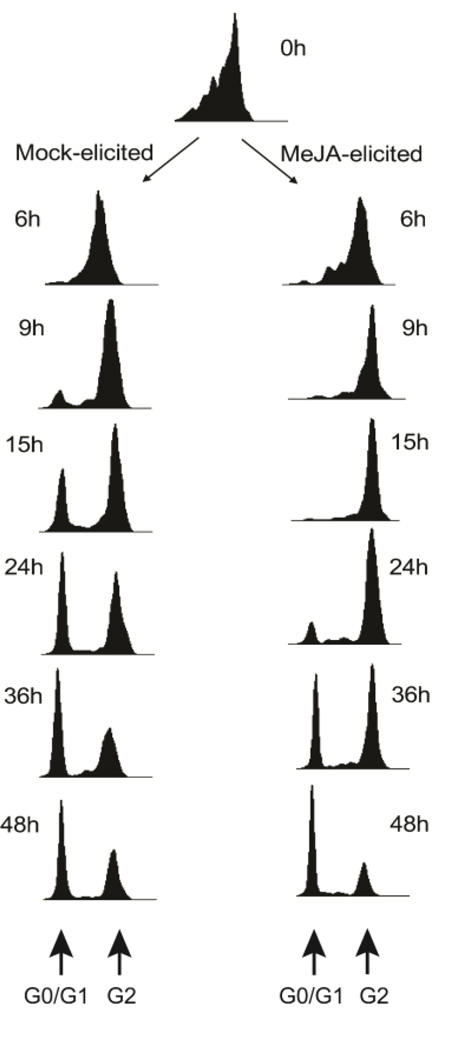
5-ethynyl-2’-deoxyuridine (EdU) pulse labeling was used to investigate the effect of MeJA on cell cycle kinetics. A four-hour EdU pulse was provided to both MeJA-elicited and mock-elicited cultures. As cells only in S-phase are able to incorporate EdU, labeled cells were quantified as they pass through different phases of the cell cycle (S to G2/M to G0/G1) using the bivariate DNA-EdU distributions obtained with flow cytometry. Approximately 10% EdU incorporation was used for pulse labeling, which allows enough cells to be labeled for accurate quantification using flow cytometry. A similar percentage of 5-bromo-2'-deoxyuridine (BrdU) incorporation was used successfully in pulse labeling studies of Solanum aviculare cells (Yanpaisan et al. 1998). The G0/G1 peak in mock-elicited cultures appeared 9 hours after the EdU pulse; whereas it took 24 hours after the EdU pulse to observe the G0/G1 peak in MeJA-elicited cultures (Fig. 4). This later appearance of the G0/G1 peak in MeJA-elicited cultures clearly demonstrates that MeJA repressed cell cycle progression in Taxus cultures. However, the appearance of the G0/G1 peak in MeJA-elicited cultures indicates that the cell cycle is not arrested at the G2/M transition.
Fig. 4.
Progression of EdU pulse labeled cells in mock-elicited and MeJA-elicited cultures. Only EdU positive cells were selected from bivariate histograms of EdU/DNA content. Cultures were either mock-elicited or elicited with 150 µM MeJA on day 7 of the culture period. Four hours later 10 µM EdU was added to all cultures
MeJA decreases the number of cycling cells
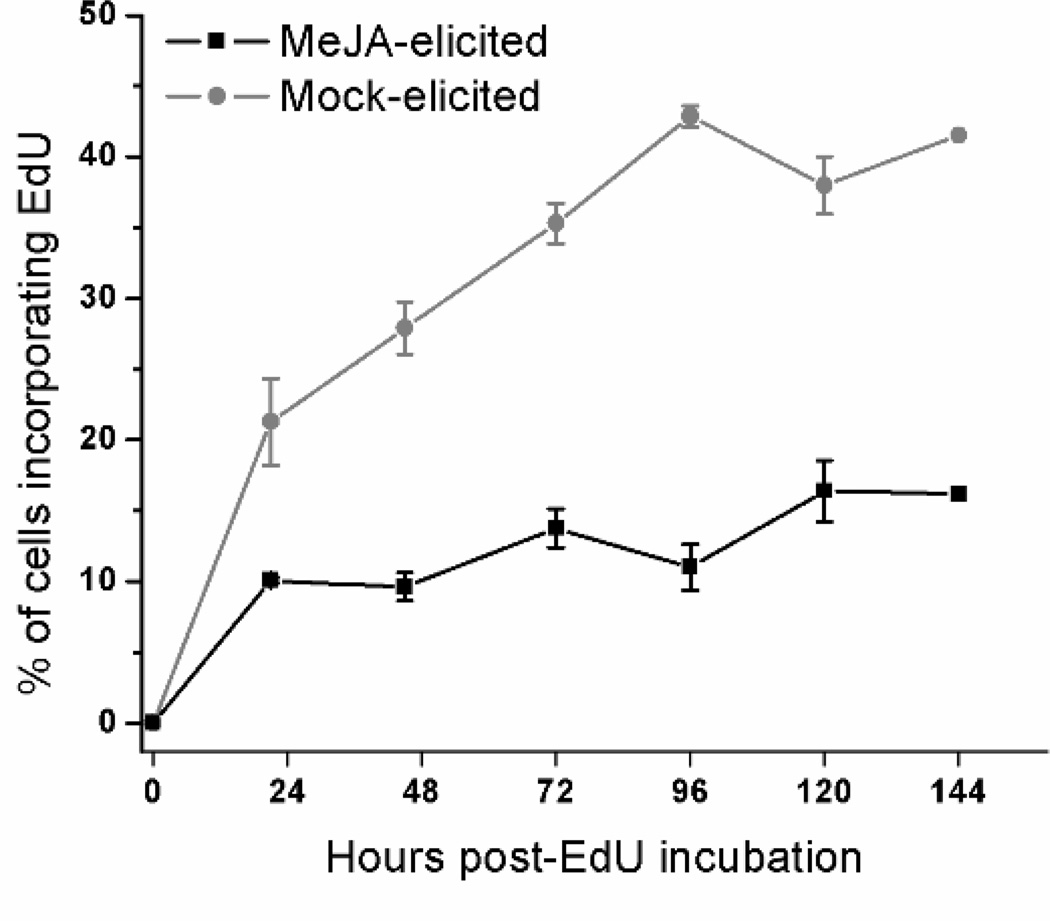
Cumulative EdU incorporation in MeJA-elicited and mock-elicited Taxus cultures is shown in Fig. 5. Five days after incubation with EdU, the number of cells incorporating EdU in mock-elicited cultures was approximately 45% of the total cell population, whereas in MeJA-elicited cultures it was only 12% of the total cell population. These data indicate that MeJA addition results in fewer cells participating in DNA synthesis, hence a lower number of actively cycling cells in culture. Repression of DNA synthesis and blockage of cells in G1 and G2 phases of the cell cycle upon MeJA-elicitation have also been observed in synchronized tobacco BY-2 cell cultures (Swiatek et al. 2002).
Fig. 5.
Total EdU incorporation in mock-elicited and MeJA-elicited cultures. Circles represent mock-elicited cultures. Squares represent MeJA-elicited cultures. Cultures were either mock-elicited or elicited with 150 µM MeJA on day 7 of the culture period. Four hours later 10 µM EdU was added to all cultures
MeJA represses a number of genes participating in cell cycle progression
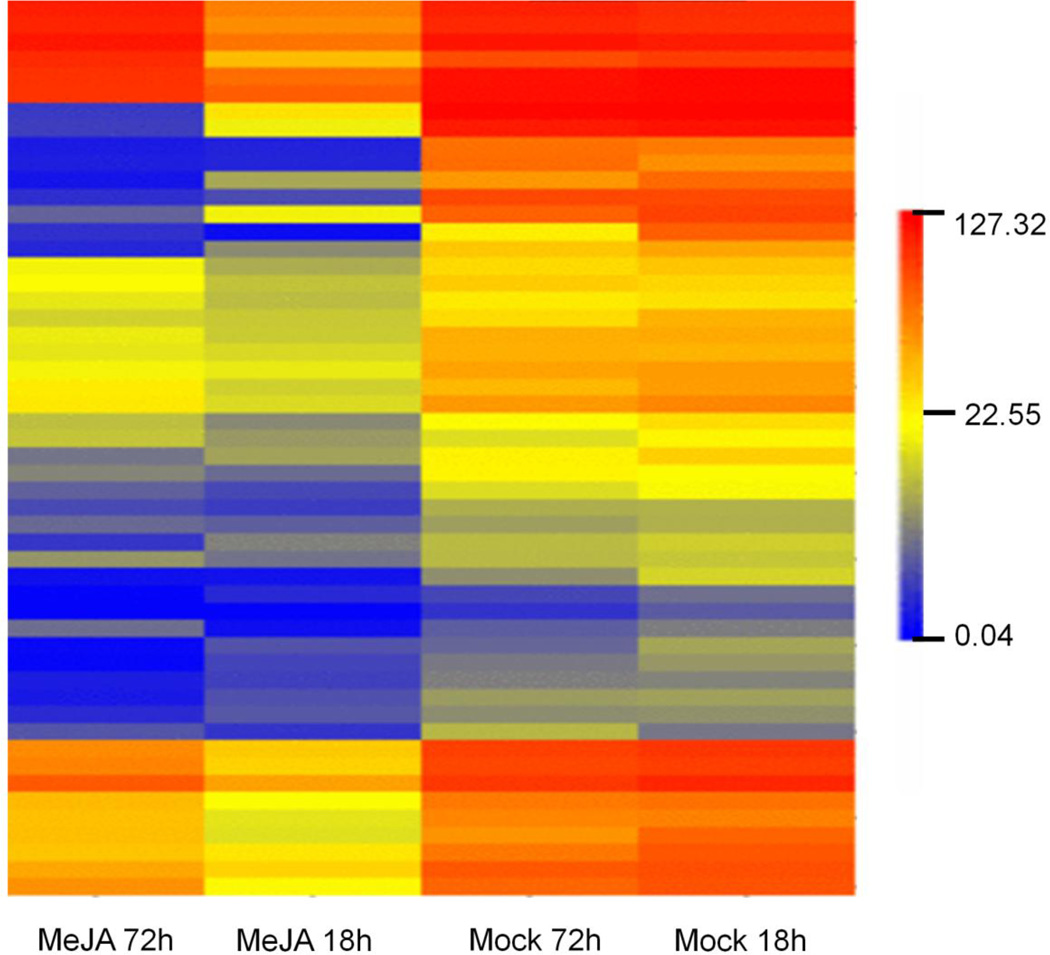
Using a combination of deep sequencing technologies and Blast2GO annotation, 149 cell cycle-associated contigs (referred to as cell cycle-associated genes from here onwards) were identified in the transcriptome of mRNA isolated from cultured Taxus cells (Online Resource 2). A comparison of gene expression between mock-elicited and MeJA-elicited cultures was done at 18 hours and 72 hours post-elicitation. At both 18 hours and 72 hours post-elicitation, none of the 149 cell cycle associated-genes were upregulated in MeJA-elicited cultures relative to the mock-elicited cultures. However, a total of 52 cell cycle genes were significantly downregulated (> 2-fold down regulation, P < 0.05) at 18 hours and 72 hours post-elicitation in MeJA-elicited cultures relative to mock-elicited cultures. Fig. 6 shows the hierarchical clustering of the 52 cell cycle genes that were downregulated in MeJA-elicited cultures. At 18 hours post-elicitation, 49 genes that were homologous to genes involved in the G2/M and G1/S transition in other plant species were downregulated in MeJA-elicited cultures (Table 1). In particular, the gene representing E2F target protein 1 (ETG1), which is a component of the replisome complex and needed for DNA replication (Takahashi et al. 2008), was drastically downregulated (∼160 fold). The transcription factor E2F, which in Arabidopsis governs expression of ETG1 and other cell cycle genes [about 70 target genes, (Vandepoele et al. 2005)] involved in the G1/S transition was also downregulated. Genes representing CDC6 (Castellano et al. 2001), CDC45 (Stevens et al. 2004), CDC48 and D-type cyclins (Meijer and Murray, 2000), whose expression typically peaks during G1 or early S phase, and some of the genes representing B-type cyclins and other G2 and M phase-specific genes that facilitate progression through mitosis were also downregulated.
Fig. 6.
Heat map showing expression patterns of significantly downregulated cell cycle related-genes in MeJA-elicited cultures as compared to mock-elicited cultures. Hierarchical clustering was performed using average linkage and Euclidean distance as a measurement of similarity. Changes in gene expression were calculated based on the RPKM values of corresponding genes
Table 1.
Contigs expressed in Taxus cell cultures and annotated as cell cycle-related genes downregulated in MeJA-elicited cultures relative to mock-elicited cultures at 18 hours post-elicitation.
| Contig No. | Sequence description | Fold Change (Normalized) |
|---|---|---|
| 140962 | E2F target protein 1 | −163.64 |
| 121259 | cell division control protein 6 homolog | −143.52 |
| 65828 | ccb22_orysj ame: G2 mitotic-specific cyclin-B2-2 | −36.41 |
| 12355 | probable G2 mitotic-specific cyclin | −33.23 |
| 10406 | cyclin dependent kinase regulator | −26.23 |
| 11366 | D2 4-type cyclin | −19.75 |
| 68975 | G2 mitotic-specific cyclin S13-6 | −14.18 |
| 61510 | D3-type cyclin | −6.19 |
| 119383 | cyclin dependent kinase B | −5.46 |
| 118076 | cell division control protein 45 homolog | −5.21 |
| 19091 | cell division cycle-associated 7-like isoform 2 | −5.03 |
| 68106 | B1-type cyclin dependent kinase | −5.00 |
| 66244 | cyclin-dependent kinases regulatory subunit | −4.41 |
| 141136 | cyclin A1 | −4.35 |
| 18492 | cyclin B 2 | −4.17 |
| 62294 | E2F protein | −3.49 |
| 142510 | antagonist of E2F–DP complex | −3.26 |
| 9782 | cyclin-dependent kinases regulatory subunit | −3.14 |
| 11005 | cell division cycle protein | −3.05 |
| 12010 | cyclin-dependent kinases regulatory subunit | −2.89 |
| 75242 | cyclin B3-1 | −2.88 |
| 108329 | cell division cycle protein | −2.81 |
| 57179 | cell division | −2.74 |
| 105427 | cell division cycle protein 48 homolog | −2.72 |
| 91147 | cell division cycle protein 48 homolog | −2.60 |
| 113240 | cell division control protein | −2.57 |
| 134490 | cell division cycle protein | −2.56 |
| 99133 | cell division cycle protein 48 homolog | −2.55 |
| 100939 | cell division cycle protein 48 homolog | −2.51 |
| 10476 | transcription factor E2F | −2.50 |
| 103306 | cell division cycle protein 48 homolog | −2.43 |
| 110709 | cell division cycle protein 48 homolog | −2.41 |
| 97765 | cell division control protein | −2.39 |
| 134513 | cell division cycle protein | −2.37 |
| 127620 | cell division cycle protein 48 homolog | −2.37 |
| 109918 | cell division cycle protein 48 homolog | −2.35 |
| 112279 | cell division cycle protein 48 homolog | −2.35 |
| 105698 | cell division cycle protein 48 homolog | −2.34 |
| 106161 | cell division control protein | −2.33 |
| 59549 | D2 4-type cyclin | −2.31 |
| 116949 | cell division cycle protein 48 homolog | −2.26 |
| 111055 | cell division cycle protein 48 homolog | −2.18 |
| 107757 | cell division cycle protein 48 homolog | −2.18 |
| 109405 | cell division cycle protein 48 homolog | −2.11 |
| 102800 | cell division cycle protein | −2.07 |
| 13227 | cyclin dependent kinase A | −2.05 |
| 109272 | cell division cycle protein 48 homolog | −2.03 |
| 18149 | cyclin-dependent kinase F-1 | −2.00 |
| 100185 | cell division cycle protein 48 homolog | −2.00 |
At 72 hours post-elicitation, about 20 cell cycle genes were significantly downregulated in MeJA-treated cultures (Table 2). Most of these genes (17 out of 20) were also downregulated at the 18 hour time point when compared to mock-elicited cultures. However, in contrast to the 18 hour time point, which had genes representing both the G1/S and G2/M transition, the majority of the downregulated genes at 72 hours were those whose expression typically peaks during G2 and M phase. For example, Cyclin A1, B1, B2, cyclin dependent kinase B (CDKB) and -other G2/M specific cyclins, whose transcripts are known to accumulate during G2 and M phases (Inze and De Veylder, 2006), were found to be the most downregulated (> 10-fold downregulation, Table 2). This result implies that in MeJA-elicited cultures, fewer cells are going through mitosis at the 72 hour time point as compared to mock-elicited cultures.
Table 2.
Contigs expressed in Taxus cell cultures and annotated as cell cycle-related genes downregulated in MeJA-elicited cultures relative to mock-elicited cultures at 72 hours post-elicitation.
| Contig No. | Sequence description | Fold change (normalized values) |
|---|---|---|
| 141136 | cyclin A1 | −49.32 |
| 119383 | cyclin dependent kinase B | −44.40 |
| 65828 | ccb22_orysj ame: G2 mitotic-specific cyclin-B2-2 | −42.36 |
| 12355 | probable G2 mitotic-specific cyclin | −40.96 |
| 142510 | antagonist of E2F–DP complex | −40.53 |
| 68975 | G2 mitotic-specific cyclin S13-6 | −32.15 |
| 68106 | B1-type cyclin dependent kinase | −27.65 |
| 118076 | cell division control protein 45 homolog | −23.96 |
| 66244 | cyclin-dependent kinases regulatory subunit | −23.03 |
| 11005 | cell division cycle protein | −20.99 |
| 10406 | cyclin dependent kinase regulator | −14.94 |
| 140962 | E2F target protein 1 | −12.52 |
| 75242 | cyclin B3-1 | −9.86 |
| 9782 | cyclin-dependent kinases regulatory subunit | −8.81 |
| 79802 | cell division cycle cofactor of APC complex | −6.64 |
| 10476 | transcription factor E2F | −5.79 |
| 81095 | anaphase-promoting complex subunit CDC20 | −5.69 |
| 61510 | D3-type cyclin | −3.68 |
| 12010 | cyclin-dependent kinases regulatory subunit | −3.51 |
| 125598 | cell division control protein | −3.25 |
| 19091 | cell division cycle-associated 7-like isoform 2 | −3.12 |
| 18492 | cyclin B 2 | −2.99 |
A comparison was made between the gene expression levels at 18 hours and 72 hours for both mock-elicited and MeJA-elicited cultures (18h mock-elicited vs. 72h mock-elicited and 18h MeJA-elicited vs 72h MeJA-elicited). There was no change in expression of cell cycle-related genes in the mock-elicited cultures. However, comparison between 18h MeJA-elicited and 72h MeJA-elicited cultures showed that genes that are usually expressed during G2 and M phase (e.g., cyclin dependent kinase B, cyclin A1, cyclin B2 and B3, etc.) were downregulated at 72 hours as compared to 18 hours (Table 3). Some G1 phase-specific genes (e.g., D-type cyclins and CDC 48 homologs) were upregulated at 72 hours as compared to 18 hours (Table 4).
Table 3.
Contigs expressed in Taxus cell cultures whose expression is downregulated 72 hours post-elicitation as compared to 18 hours post-elicitation in MeJA-elicited cultures.
| Contig No. | Sequence Description | Fold Change (normalized values) |
|---|---|---|
| 119383 | cyclin dependent kinase B | −17.90 |
| 141136 | cyclin A1 | −13.62 |
| 66244 | cyclin-dependent kinases regulatory subunit | −13.34 |
| 68975 | G2 mitotic-specific cyclin S13-6 | −8.09 |
| 68106 | B1-type cyclin dependent kinase | −7.96 |
| 75242 | cyclin B3-1 | −6.74 |
| 12355 | probable G2 mitotic-specific cyclin | −5.50 |
| 79802 | cell division cycle cofactor of APC complex | −4.97 |
| 65828 | ccb22_orysj ame: G2 mitotic-specific cyclin-B2-2 | −4.79 |
| 81095 | anaphase-promoting complex subunit CDC20 | −4.06 |
| 9782 | cyclin-dependent kinases regulatory subunit | −3.62 |
Table 4.
Contigs expressed in Taxus cell cultures whose expression is upregulated 72 hours post-elicitation as compared to 18 hours post-elicitation in MeJA-elicited cultures.
| Contig No. | Sequence Description | Fold Change (normalized values) |
|---|---|---|
| 11366 | D2 4-type cyclin | 16.25 |
| 11041 | cyclin-dependent protein | 3.50 |
| 59549 | D2 4-type cyclin | 2.55 |
| 107757 | cell division cycle protein 48 homolog | 2.19 |
| 100185 | cell division cycle protein 48 homolog | 2.18 |
| 112647 | cell division cycle protein 48 homolog | 2.15 |
| 109272 | cell division cycle protein 48 homolog | 2.13 |
| 145394 | cell division control protein | 2.12 |
| 113719 | cell division cycle protein 48 homolog | 2.04 |
| 92332 | cell division cycle protein 48 homolog | 2.04 |
| 101973 | cell division cycle protein 48 homolog | 2.04 |
| 107852 | cell division cycle protein 48 homolog | 2.01 |
Apart from the cell cycle-specific genes, genes representing the subclasses of core histones, H2A, H2B, H3 and H4, which are essential for cell proliferation and required for the packaging of DNA into chromatin (Gutierrez, 2009, Yi et al. 2006) were also repressed in MeJA-elicited cultures at both 18 and 72 hours (Online Resource 3 and Online Resource 4).
DISCUSSION
In the present study, we quantified the effect of MeJA, a widely used enhancer of plant secondary metabolism, on asynchronously dividing T. cuspidata cell cultures. Biomass measurements showed that MeJA repressed culture growth (Fig. 1a). There was a time lag between growth inhibition and cell death, indicating that MeJA-mediated growth inhibition was not due to necrosis and/or rupturing of cell membranes (Fig. 1c, d). Biotic (fungal derived) and abiotic (Cerium (Ce+4)) elicitors induce apoptotic cell death in Taxus cultures within a few hours to few days post-elicitation (Ge et al. 2002, Qiao et al. 2003, Yuan et al. 2002). Here, we observed DNA fragmentation, a hallmark of apoptotic cell death, only at a very late time point (day 30 of the culture period), implying that apoptosis is not directly linked to MeJA elicitation (Fig. 2). Similar results have been reported for T. cuspidata P991 and T. canadensis C093D cell lines upon MeJA elicitation (Kim et. al, 2005). One explanation for this anomalous behavior for MeJA is that MeJA elicitation in Taxus suspensions did not markedly increase the phosphatic acid (PA) levels, which is associated with regulating cell death response (Yang et al. 2008). Ce+4 caused both increased paclitaxel levels and increased PA levels, leading to cell death (Yang et al. 2008). Repression of growth without cell death and a delayed onset of apoptosis suggest that MeJA is repressing growth by altering cellular metabolism and/or affecting the cell cycle. We previously demonstrated that MeJA upregulates paclitaxel biosynthetic pathway genes (Lenka et al. 2012, Nims et al. 2006, Patil et al. 2012), and thus metabolism is shifted towards synthesis of paclitaxel. Here, the influence of MeJA on the Taxus cell cycle and the mechanism of growth inhibition have been elucidated.
MeJA addition to asynchronously dividing Taxus cultures resulted in four effects on the cell cycle, as revealed by flow cytometric analyses: i) transient increase in G2 phase cells, ii) transient decrease in S phase cells, iii) increase in G0/G1 phase cells at later stages post-elicitation, and iv) decreases in G2 and S phase cells at later stages post-elicitation (Fig. 3 and Online Resource 1). At a mechanistic level, the effect of jasmonates has been shown to be dependent on the phase of the cell cycle in synchronized tobacco BY-2 cells (Swiatek et al. 2002). When JA was applied before the G1/S transition it prevented DNA replication, keeping BY-2 cells in G1 phase. When JA was applied during S phase, it prevented cells from entering mitosis without directly affecting their DNA synthesis (Swiatek et al. 2002, Swiatek et al. 2004). Though our cultures were not synchronized, a similar effect was observed upon MeJA elicitation, with a drop in the percentage of cells in S phase and an increase in the percentage of cells in G2 phase immediately following elicitation. The decreased percentage of cells in G2 and S and increased percentage of cells in G0/G1 at 96 hours post-elicitation suggest that the cells were not permanently arrested in G2/M, but rather moving slowly. An EdU pulse label assay confirmed these data showing clearly that MeJA addition slowed progression through the cell cycle (Fig. 4).
The transcription of several cell cycle genes at 18 hours and 72 hours post-elicitation with MeJA also correlated with the flow cytometric data trends. Concomitant with the decrease in S phase cells (Fig. 3b), significant downregulation of expression of ETG1 and CDC6 genes at the 18 hour time point was observed. Expression of ETG1 and CDC6 genes has been shown to be necessary for the G1/S transition in Arabidopsis (Castellano et al. 2001, Takahashi et al. 2008). Genes representing CDC45 (Stevens et al. 2004) and CDC48 homologs (Feiler et al. 1995), which play a role in the G1/S transition were also downregulated. Plant cyclins are known to interact with cyclin-dependent kinases (CDKs), and these CDK-cyclin complexes regulate the key G1/S and G2/M transition points responsible for DNA replication and mitosis, respectively. Plant cyclins, especially the A and B types, show oscillatory behavior at the transcription level, where transcript levels peak during certain phases of the cell cycle. Generally, D-type cyclins are known to regulate the G1/S transition, A-type cyclins regulate the S/M transition, and B-type cyclins regulate both the G2/M transition and M phase control (Breyne and Zabeau, 2001, Inze and De Veylder, 2006); though a number of exceptions to these functional assignments have been reported (Inze and De Veylder, 2006, Kawamura et al. 2006). Genes representing all these three classes of cyclins were downregulated at the 18 hour time point in MeJA-elicited cultures relative to mock-elicited cultures. Also, cyclin-dependent kinase B (CDKB) genes, which are regulated at the transcript level and are necessary for the G2/M transition (Inze and De Veylder, 2006), were downregulated at 18 hours in MeJA-elicited cultures. The expression patterns of some of these cell cycle genes are thus consistent with the increase in G2 phase cells within 24 hours post-elicitation with MeJA (Fig. 3c). At 72 hours post elicitation, the majority of downregulated genes were those for which expression peaked during G2 and M phases (e.g., cyclin A1, CDKB, cyclin B2, etc.). Some of the genes that are involved in G1/S transition (e.g., ETG1, CDC45, etc.) were also downregulated at the 72 hour time point (Table 2), indicating that cell proliferation and progression of cells from G1 for division is hindered.
Comparison of cultures at 18 and 72 hours post-elicitation with MeJA shows that G2/M-specific cell cycle genes (e.g., G2-mitotic specific cyclin, cyclin A1, CDKB, etc.) are downregulated at 72 hours as compared to 18 hours (Table 3). This result implies that the downregulation of genes required to drive cells through G2 and M phases causes more cells to remain in G2 phase. Some G1 phase genes (e.g., cyclin D (d2–4 type cyclin) and CDC 48, Table 4) are also upregulated at 72 hours as compared to 18 hours post-elicitation, implying that more cells are present in G0/G1 phase at 72 hours relative to 18 hours in MeJA-elicited cultures. These results correlate well with flow cytometric data (Fig. 3), where both a decrease in G2 and S phase cells and an increase in G0/G1 phase cells were observed after 72 hours post-elicitation.
Moreover, along with the cell cycle-specific genes, histone-encoding genes were also downregulated upon MeJA elicitation. Histones are highly conserved across eukaryotic species and have been classified into four subcategories of core histones (H2A, H2B, H3, and H4) and linker histones (H1). For most species, each core histone protein is encoded by a multigene family (Piontkivska et al. 2002). Biotic and abiotic stresses can repress expression of histone genes in plants. For example, in cultured parsley cells, UV radiation and fungal elicitors repressed expression of several genes essential for cell cycle progression including histones H2A, H2B, H3, and H4 (Logemann et al. 1995), leading to growth inhibition. Similar results were observed here in MeJA-elicited T. cuspidata cultures, where all the genes representing histones were downregulated relative to mock-elicited cultures at both 18 and 72 hours post-elicitation. Thus, histone gene repression can be correlated to decreased cell division in Taxus, analogous to observations in parsley cells (Logemann et al. 1995).
The number of cells incorporating EdU was significantly lower in MeJA-elicited cultures when compared to mock-elicited cultures (Fig. 5). This result suggests that over time the number of actively dividing cells (i.e., synthesizing DNA) decreased in MeJA-elicited cultures. This result is supported by the increase in G0/G1 phase cells observed in MeJA-elicited cultures (Fig. 3a). One explanation is that cells remain in G0 phase and are specialized for accumulation of paclitaxel and potentially other secondary metabolites (Naill and Roberts, 2005a). Total metabolic activity inferred from total cellular protein content is relatively uniform in MeJA-elicited cultures (Naill and Roberts, 2005b), indicating that non-cycling cells are still metabolically active, but potentially redirect carbon flux away from primary metabolism towards secondary metabolism. Alkaloid accumulation increased in cultures of Solanum aviculare where cell cycle progression was inhibited using a cell cycle arrest agent, again suggesting that the metabolic flux may be directed towards secondary pathways in non-proliferating cells (Mak and Doran, 1993). Research to date in plant systems has been able to identify a significant G0 population in culture and presents indirect evidence to suggest a function of this population, but has not been able to explicitly correlate the non-cycling cell population to other metabolic information. A multi-parametric flow cytometry study to simultaneously analyze non-cycling cells (G0 phase cells) and paclitaxel-accumulating cells can reveal this relationship in the Taxus cell culture system, and warrants further investigation.
In summary, the MeJA-mediated repression of cell growth in Taxus cultures was shown to correlate with inhibition of cell cycle progression as evident both at the culture level through flow cytometric analyses and at the transcriptional level by repression of key cell cycle-associated genes. The newly annotated Taxus cell cycle-associated genes will provide an importance resource for future cell cycle studies of both Taxus and related gymnosperms. The cell cycle progression patterns in culture closely parallel the transcriptional regulation of cell cycle-associated genes in MeJA-elicited and mock-elicited Taxus cell cultures. The results from this study advance fundamental understanding of the mechanism of action of secondary metabolite elicitors such as MeJA on repression of plant cell division. This result is especially important for species such as Taxus, where most research has been focused on improving paclitaxel synthesis with less attention paid to the negative effect of MeJA on growth and implications on bioprocessing.
Supplementary Material
KEY MESSAGE.
Methyl jasmonate elicitation of Taxus cultures enhances paclitaxel accumulation but represses growth by inhibition of cell cycle progression. Growth repression is evident both at the culture level and transcriptional level.
ACKNOWLEDGEMENTS
This work was funded by National Institute of Health (grant number GM070852). We acknowledge the National Science Foundation-sponsored Institute for Cellular Engineering IGERT Program (DGE-0654128) for facilities and financial support. We thank Dr. Donna Gibson of the USDA, Agricultural Research Service, for the Taxus cell cultures. We also thank the UMass Amherst Flow Cytometry Facility and Prof. Shelly Peyton for the use of her Zeiss Axiovert 200 microscope.
Footnotes
AUTHOR CONTRIBUTION STATEMENT
R.A.P. designed the study, performed all the experiments, analyzed the data and wrote the manuscript. S.K.L performed the bioinformatic analysis. J.N and E.L.W helped supervise the experiment activities and contributed to designing the study. S.C.R. supervised the project, designed the study’s strategy, helped in data analysis and writing of the manuscript. All authors commented on the manuscript. All authors take full responsibility for the content of the paper.
CONFLICT OF INTEREST
The authors declare that they are no conflicts of interest.
REFERENCES
- Bonfill M, Bentebibel S, Moyano E, Palazón J, Cusidó R, Eibl R, Piñol M. Paclitaxel and baccatin III production induced by methyl jasmonate in free and immobilized cells of Taxus baccata. Biologia Plantarum. 2007;51:647–652. [Google Scholar]
- Bonfill M, Exposito O, Moyano E, Cusido RM, Palazon J, Pinol MT. Manipulation by culture mixing and elicitation of paclitaxel and baccatin III production in Taxus baccata suspension cultures. In Vitro Cell. Dev. Biol. Plant. 2006;42:422–426. [Google Scholar]
- Breyne P, Zabeau M. Genome-wide expression analysis of plant cell cycle modulated genes. Current Opinion in Plant Biology. 2001;4:136–142. doi: 10.1016/s1369-5266(00)00149-7. [DOI] [PubMed] [Google Scholar]
- Bringi V, Kadkade P, Prince CL, Roach B. Enhanced production of taxol and taxanes by cell cultures of Taxus species. U.S. Patent - 7264954 B1. 2007
- Castellano MM, del Pozo JC, Ramirez-Parra E, Brown S, Gutierrez C. Expression and stability of Arabidopsis CDC6 are associated with endoreplication. Plant Cell. 2001;13:2671–2686. doi: 10.1105/tpc.010329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho K, Agrawal GK, Shibato J, Jung YH, Kim YK, Nahm BH, et al. Survey of Differentially expressed proteins and genes in Jasmonic acid treated rice seedling shoot and root at the Proteomics and Transcriptomics levels. Journal of Proteome Research. 2007;6:3581–3603. doi: 10.1021/pr070358v. [DOI] [PubMed] [Google Scholar]
- Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: version II. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- Farmer EE, Ryan CA. Interplant communication - airborne methyl jasmonate induces synthesis of proteinase-inhibitors in plant leaves. Proc Natl Acad Sci U S A. 1990;87:7713–7716. doi: 10.1073/pnas.87.19.7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiler HS, Desprez T, Santoni V, Kronenberger J, Caboche M, Traas J. The higher-plant Arabidopsis thaliana encodes a functional Cdc48 homolog which is highly expressed in dividing and expanding cells. Embo Journal. 1995;14:5626–5637. doi: 10.1002/j.1460-2075.1995.tb00250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E. Rapid flow cytometric analysis of the Cell-cycle in intact plant-tissues. Science. 1983;220:1049–1051. doi: 10.1126/science.220.4601.1049. [DOI] [PubMed] [Google Scholar]
- Gaurav V, Kolewe ME, Roberts SC. Flow cytometric methods to investigate culture heterogeneities for plant metabolic engineering. Methods Mol Biol. 2010;643:243–262. doi: 10.1007/978-1-60761-723-5_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge ZQ, Yuan YJ, Wang YD, Ma ZY, Hu ZD. Ce4+-Induced apoptosis of Taxus cuspidata cells in suspension culture. Journal of Rare Earths. 2002;20:139–144. [Google Scholar]
- Goossens A, Hakkinen ST, Laakso I, Seppanen-Laakso T, Biondi S, De Sutter V, Lammertyn F, Nuutila AM, Soderlund H, Zabeau M, Inze D, Oksman-Caldentey KM. A functional genomics approach toward the understanding of secondary metabolism in plant cells. Proc Natl Acad Sci U S A. 2003;100:8595–8600. doi: 10.1073/pnas.1032967100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundlach H, Muller MJ, Kutchan TM, Zenk MH. Jasmonic acid is a signal transducer in elicitor induced plant cell cultures. Proc Natl Acad Sci U S A. 1992;89:2389–2393. doi: 10.1073/pnas.89.6.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez C. The Arabidopsis cell division cycle. Arabidopsis Book. 2009;7:e0120. doi: 10.1199/tab.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inze D, De Veylder L. Cell cycle regulation in plant development. Annual Review of Genetics. 2006;40:77–105. doi: 10.1146/annurev.genet.40.110405.090431. [DOI] [PubMed] [Google Scholar]
- Jennewein S, Wildung MR, Chau M, Walker K, Croteau R. Random sequencing of an induced Taxus cell cDNA library for identification of clones involved in Taxol biosynthesis. Proc Natl Acad Sci U S A. 2004;101:9149–9154. doi: 10.1073/pnas.0403009101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kal AJ, van Zonneveld AJ, Benes V, van den Berg M, Koerkamp MG, Albermann K, Strack N, Ruijter JM, Richter A, Dujon B, Ansorge W, Tabak HF. Dynamics of gene expression revealed by comparison of serial analysis of gene expression transcript profiles from yeast grown on two different carbon sources. Mol Biol Cell. 1999;10:1859–1872. doi: 10.1091/mbc.10.6.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura K, Murray JAH, Shinmyo A, Sekine M. Cell cycle regulated D3-type cyclins form active complexes with plant-specific B-type cyclin-dependent kinase in vitro. Plant Molecular Biology. 2006;61:311–327. doi: 10.1007/s11103-006-0014-y. [DOI] [PubMed] [Google Scholar]
- Ketchum REB, Gibson DM, Croteau RB, Shuler ML. The kinetics of taxoid accumulation in cell suspension cultures of Taxus following elicitation with methyl jasmonate. Biotechnol Bioeng. 1999;62:97–105. doi: 10.1002/(sici)1097-0290(19990105)62:1<97::aid-bit11>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Kieran PM, MacLoughlin PF, Malone DM. Plant cell suspension cultures: some engineering considerations. J Biotechnol. 1997;59:39–52. doi: 10.1016/s0168-1656(97)00163-6. [DOI] [PubMed] [Google Scholar]
- Kim BJ, Gibson DM, Shuler ML. Effect of subculture and elicitation on instability of Taxol production in Taxus sp suspension cultures. Biotechnol Progr. 2004;20:1666–1673. doi: 10.1021/bp034274c. [DOI] [PubMed] [Google Scholar]
- Kim BJ, Gibson DM, Shuler ML. Relationship of viability and apoptosis to taxol production in Taxus sp suspension cultures elicited with methyl jasmonate. Biotechnol Progr. 2005;21:700–707. doi: 10.1021/bp050016z. [DOI] [PubMed] [Google Scholar]
- Kolewe ME, Henson MA, Roberts SC. Characterization of aggregate size in Taxus suspension cell culture. Plant Cell Rep. 2010;29:485–494. doi: 10.1007/s00299-010-0837-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai-Sano F, Hayashi T, Sano T, Hasezawa S. Cell cycle synchronization of tobacco BY-2 cells. Nature Protocols. 2006;1:2621–2627. doi: 10.1038/nprot.2006.381. [DOI] [PubMed] [Google Scholar]
- Laskaris G, Bounkhay M, Theodoridis G, van der Heijden R, Verpoorte R, Jaziri M. Induction of geranylgeranyl diphosphate synthase activity and taxane accumulation in Taxus baccata cell cultures after elicitation by methyl jasmonate. Plant Science. 1999;147:1–8. [Google Scholar]
- Lenka SK, Boutaoui N, Paulose B, Vongpaseuth K, Normanly J, Roberts SC, Walker EL. Identification and expression analysis of methyl jasmonate responsive ESTs in paclitaxel producing Taxus cuspidata suspension culture cells. BMC Genomics. 2012;13:148. doi: 10.1186/1471-2164-13-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zhang P, Zhang M, Fu CH, Zhao CF, et al. Transcriptional profile of Taxus chinensis cells in response to methyl jasmonate. BMC genomics. 2012;13:295. doi: 10.1186/1471-2164-13-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ou D, Zheng L, Gan N, Song L. Applicability of the fluorescein diacetate assay for metabolic activity measurement of Microcystis aeruginosa (Chroococcales, Cyanobacteria) Phycological Research. 2011;59:200–207. [Google Scholar]
- Lijavetzky D, Almagro L, Belchi-Navarro S, Martinez-Zapater J, Bru R, Pedreno M. Synergistic effect of methyljasmonate and cyclodextrin on stilbene biosynthesis pathway gene expression and resveratrol production in Monastrell grapevine cell cultures. BMC Research Notes. 2008;1:132. doi: 10.1186/1756-0500-1-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logemann E, Wu SC, Schroder J, Schmelzer E, Somssich IE, Hahlbrock K. Gene activation by UV light, fungal elicitor or fungal infection in Petroselinum crispum is correlated with repression of cell cycle-related genes. Plant Journal. 1995;8:865–876. doi: 10.1046/j.1365-313x.1995.8060865.x. [DOI] [PubMed] [Google Scholar]
- Mak YX, Doran PM. Effect of Cell-cycle inhibition on synthesis of steroidal alkaloids by Solanum aviculare plant cells. Biotechnol Lett. 1993;15:1031–1034. [Google Scholar]
- Meijer M, Murray JAH. The role and regulation of D-type cyclins in the plant cell cycle. Plant Molecular Biology. 2000;43:621–633. doi: 10.1023/a:1006482115915. [DOI] [PubMed] [Google Scholar]
- Menges M, Hennig L, Gruissem W, Murray JAH. Cell Cycle-regulated Gene Expression in Arabidopsis . Journal of Biological Chemistry. 2002;277:41987–42002. doi: 10.1074/jbc.M207570200. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Naill MC, Roberts SC. Cell cycle analysis of Taxus suspension cultures at the single cell level as an indicator of culture heterogeneity. Biotechnol Bioeng. 2005a;90:491–500. doi: 10.1002/bit.20446. [DOI] [PubMed] [Google Scholar]
- Naill MC, Roberts SC. Flow cytometric analysis of protein content in Taxus protoplasts and single cells as compared to aggregated suspension cultures. Plant Cell Rep. 2005b;23:528–533. doi: 10.1007/s00299-004-0875-y. [DOI] [PubMed] [Google Scholar]
- Nims E, Dubois CP, Roberts SC, Walker EL. Expression profiling of genes involved in paclitaxel biosynthesis for targeted metabolic engineering. Metab Eng. 2006;8:385–394. doi: 10.1016/j.ymben.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Patil RA, Kolewe ME, Normanly J, Walker EL, Roberts SC. Contribution of taxane biosynthetic pathway gene expression to observed variability in paclitaxel accumulation in Taxus suspension cultures. Biotechnology Journal. 2012;7:418–427. doi: 10.1002/biot.201100183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil RA, Kolewe ME, Roberts SC. Cellular aggregation is a key parameter associated with long term variability in paclitaxel accumulation in Taxus suspension cultures. Plant Cell Tissue Organ Culture. 2013;112:303–310. doi: 10.1007/s11240-012-0237-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L, Inzé D, Goossens A. Jasmonate-inducible gene: what does it mean? Trends in Plant Science. 2009;14:87–91. doi: 10.1016/j.tplants.2008.11.005. [DOI] [PubMed] [Google Scholar]
- Pauwels L, Morreel K, Witte ED, Lammertyn F, Montagu MV, Boerjan W, Inzé D, Goossens A. Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc Natl Acad Sci U S A. 2008;105:1380–1385. doi: 10.1073/pnas.0711203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piontkivska H, Rooney AP, Nei M. Purifying selection and birth-and-death evolution in the histone H4 gene family. Mol Biol Evol. 2002;19:689–697. doi: 10.1093/oxfordjournals.molbev.a004127. [DOI] [PubMed] [Google Scholar]
- Qiao JJ, Yuan YJ, Zhao H, Wu JC, Zeng AP. Apoptotic cell death in suspension cultures of Taxus cuspidata co-treated with salicylic acid and hydrogen peroxide. Biotechnol Lett. 2003;25:387–390. doi: 10.1023/a:1022494213446. [DOI] [PubMed] [Google Scholar]
- Reymond P, Farmer EE. Jasmonate and salicylate as global signals for defense gene expression. Current Opinion in Plant Biology. 1998;1:404–411. doi: 10.1016/s1369-5266(98)80264-1. [DOI] [PubMed] [Google Scholar]
- Ruiz-May E, De-la-Peña C, Galaz-Ávalos RM, Lei Z, Watson BS, Sumner LW, Loyola-Vargas VM. Methyl jasmonate induces ATP biosynthesis deficiency and accumulation of proteins related to secondary metabolism in Catharanthus roseus (L.) G. hairy roots. Plant and Cell Physiology. 2011;52:1401–1421. doi: 10.1093/pcp/pcr086. [DOI] [PubMed] [Google Scholar]
- Ryerson DE, Heath MC. Cleavage of nuclear DNA into oligonucleosomal fragments during cell death induced by fungal infection or by abiotic treatments. Plant Cell. 1996;8:393–402. doi: 10.1105/tpc.8.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Song JT, Cheong JJ, Lee YH, Lee YW, Hwang I, Lee JS, Choi YD. Jasmonic acid carboxyl methyltransferase: A key enzyme for jasmonate-regulated plant responses. Proc Natl Acad Sci U S A. 2001;98:4788–4793. doi: 10.1073/pnas.081557298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens R, Grelon M, Vezon D, Oh JS, Meyer P, Perennes C, Domenichini S, Bergounioux C. A CDC45 homolog in Arabidopsis is essential for meiosis, as shown by RNA interference-induced gene silencing. Plant Cell. 2004;16:99–113. doi: 10.1105/tpc.016865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Yang Y, Xie F, Wen J-F, Wu J, Wilson IW, Tang Q, Liu H, Qiu D. Deep sequencing reveals transcriptome re-programming of Taxus × media cells to the elicitation with methyl jasmonate. PLoS ONE. 2013;8(4):1. doi: 10.1371/journal.pone.0062865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiatek A, Lenjou M, Van Bockstaele D, Inze D, Van Onckelen H. Differential Effect of Jasmonic Acid and Abscisic Acid on Cell Cycle Progression in Tobacco BY-2 Cells. Plant Physiology. 2002;128:201–211. [PMC free article] [PubMed] [Google Scholar]
- Swiatek A, Van Dongen W, Esmans EL, Van Onckelen H. Metabolic fate of jasmonates in tobacco bright yellow-2 cells. Plant Physiology. 2004;135:161–172. doi: 10.1104/pp.104.040501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Lammens T, Boudolf V, Maes S, Yoshizumi T, De Jaeger G, Witters E, Inze D, De Veylder L. The DNA replication checkpoint aids survival of plants deficient in the novel replisome factor ETG1. EMBO J. 2008;27:1840–1851. doi: 10.1038/emboj.2008.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanh NT, Murthy HN, Yu KW, Hahn EJ, Paek KY. Methyl jasmonate elicitation enhanced synthesis of ginsenoside by cell suspension cultures of Panax ginseng in 5-l balloon type bubble bioreactors. Appl Microbiol Biotechnol. 2005;67:197–201. doi: 10.1007/s00253-004-1759-3. [DOI] [PubMed] [Google Scholar]
- Vandepoele K, Vlieghe K, Florquin K, Hennig L, Beemster GTS, Gruissem W, Van De Peer Y, Inze D, De Veylder L. Genome-wide identification of potential plant E2F target genes. Plant Physiology. 2005;139:316–328. doi: 10.1104/pp.105.066290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vongpaseuth K, Roberts SC. Advancements in the understanding of Paclitaxel metabolism in tissue culture. Curr Pharm Biotechnol. 2007;8:219–236. doi: 10.2174/138920107781387393. [DOI] [PubMed] [Google Scholar]
- Yang S, Lu SH, Yuan YJ. Lipidomic analysis reveals differential defense responses of Taxus cuspidata cells to two elicitors, methyl jasmonate and cerium (Ce4+) Biochimica Et Biophysica Acta-Molecular and Cell Biology of Lipids. 2008;1781:123–134. doi: 10.1016/j.bbalip.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Yanpaisan W, King NJC, Doran PM. Analysis of cell cycle activity and population dynamics in heterogeneous plant cell suspensions using flow cytometry. Biotechnol Bioeng. 1998;58:515–528. doi: 10.1002/(sici)1097-0290(19980605)58:5<515::aid-bit8>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Yazaki K, Takeda K, Tabata M. Effects of methyl jasmonate on shikonin and dihydroechinofuran production in Lithospermum cell cultures. Plant and Cell Physiology. 1997;38:776–782. [Google Scholar]
- Yi H, Sardesai N, Fujinuma T, Chan CW, Veena , Gelvin SB. Constitutive expression exposes functional redundancy between the Arabidopsis histone H2A gene HTA1 and other H2A gene family members. Plant Cell. 2006;18:1575–1589. doi: 10.1105/tpc.105.039719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y-J, Li C, Hu Z-D, Wu J-C, Zeng A-P. Fungal elicitor-induced cell apoptosis in suspension cultures of Taxus chinensis var. mairei for taxol production. Process Biochemistry. 2002;38:193–198. [Google Scholar]
- Yukimune Y, Tabata H, Higashi Y, Hara Y. Methyl jasmonate-induced overproduction of paclitaxel and baccatin III in Taxus cell suspension cultures. Nat Biotechnol. 1996;14:1129–1132. doi: 10.1038/nbt0996-1129. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Turner JG. Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS ONE. 2008;3:e3699. doi: 10.1371/journal.pone.0003699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.