Abstract
Three canine parainfluenza viruses type 5 (CPIV-5) were isolated from lung tissues of 3 Korean dogs with mild pneumonia between 2008 and 2009. The isolates were fully sequenced and compared with published reference sequences. The size of the genome was 15 246 nucleotides long and no remarkable differences were found when compared with previously published reference sequences. In phylogenetic analysis based on the F and P genes, parainfluenza virus 5 (PIV-5) strains were divided into at least 3 subgroups. Three CPIV-5 strains were clustered with CPIV-5 T1, H22 and 78524 strains. All PIV-5 strains were independent of the host species, geographical distribution, and the isolated period.
Résumé
Trois virus parainfluenza canins de type 5 (CPIV-5) furent isolés entre 2008 et 2009 à partir de tissu pulmonaire de trois chiens coréens souffrant de pneumonie légère. Les isolats furent complètement séquencés et comparés à des séquences de référence publiées. La taille du génome était de 15 246 nucléotides de longueur et aucune différence notable ne fut trouvée lorsque comparé avec les séquences de référence. Lors d’analyses phylogénétiques basées sur les gènes F et P, les souches de virus parainfluenza de type 5 (PIV-5) furent séparées en au moins trois sous-groupes. Les trois souches de CPIV-5 étaient regroupées avec les souches de CPIV-5 T1, H22 et 78524. Toutes les souches de PIV-5 étaient indépendantes de l’espèce hôte, de la distribution géographique et de la période à laquelle elles furent isolées.
(Traduit par Docteur Serge Messier)
Parainfluenza virus type 5 (PIV-5) is an enveloped, non-segmented, negative-sense RNA virus belonging to the genus Rubulavirus, family Paramyxoviridae. Other viruses in this group include the distantly genetically and antigenetically related human parainfluenza virus (HPIV types 2 and 4) and mumps virus (1). The name and host range of PIV-5 have been confused for decades because the virus was first found in cultures of rhesus and cynomolgus monkey kidney cells. Recently, the application of sequencing technology to simian virus 5-like viruses from different hosts revealed a striking lack of sequence variation at both the nucleotide (nt) and amino acid levels among the viruses. However, the potential for confusion remains because canine parainfluenza virus 5 (CPIV-5) is also called canine parainfluenza virus type 2 (CPIV-2), perhaps based on its relationship to HPIV-2, which causes respiratory disease in humans (1–3).
Canine parainfluenza virus 5 is pleomorphic virion comprising a nucleocapsid surrounded by a lipid envelope (1). A unique differentiating feature of CPIV-5 is the presence of a seventh gene, the SH gene, that is located between the fusion (F) and hemagglutinin-neuraminidase (HN) genes on the gene map (1).
Canine parainfluenza virus 5 has been recognized as an infectious cofactor in the canine respiratory disease complex or canine infectious tracheobronchitis [CITB; commonly known as canine infectious respiratory disease (CIRD)]. Other respiratory pathogens have been associated with the development of CITB, including Bordetella bronchiseptica and canine adenovirus type 2 (4,5). The clinical signs of CITB include pyrexia, mucosal nasal discharge, pharyngitis, and tonsillitis. Canine infectious tracheobronchitis is a major health problem for dogs worldwide (6).
A few PIV-5 were fully sequenced and analyzed (7). In this study, 3 CPIV-5 strains (08-1990, 1168-1, and D277) were identified from the lungs of mixed-breed dogs presenting with signs of pneumonia between 2008 and 2009, and the strains were characterized using genome sequencing and phylogenetic analysis.
To exclude PIV-5 contamination from cell origin, total RNA was purified directly from clinical samples and amplified DNA was sequenced in this study. Thirty-one primer sets were designed to amplify overlapping regions using the complete genome alignments of the PIV-5 (NC006430 and AF052755) sequences from GenBank. Total RNA was extracted directly from lung samples using the RNeasy mini kit (QIAGEN, Valencia, California, USA), according to the manufacturer’s directions. The amplified DNA fragment was purified using an agarose gel DNA extraction kit (iNtRON, Kyungki-Do, Korea) and subcloned into the pGEM-T vector (Promega, Madison, Wisconsin, USA), according to the manufacturer’s instructions. Automated nt sequencing was done on an ABI 3130XL genetic analyzer (Applied Biosystems, Foster City, California, USA) using a sequencing kit (Big Dye Terminator Cycle Sequencing Kit; Applied Biosystems). All nt positions were confirmed by 3 or more independent sequencing runs in both directions. The leader and trailer sequences were obtained using the 5′–3′-RACE Kit (Invitrogen, Carlsbad, California, USA), according to the manufacturer’s instructions. The complete genome was assembled and compiled from overlapping sequences of the CPIV-5 strains.
The nt and putative amino acid sequence alignments were created using computer software (BioEdit; Ibis Biosciences, Carlsbad, California, USA). The complete genomes of the 3 CPIV-5 strains (08-1990, 1168-1, and D277) have been deposited in GenBank under accession nos. KC237063 to KC237065.
The complete genome sequences of the CPIV-5 strains were compared to that of PIV-5, W3A, (NC006430) at both the nucleotide and amino acid levels. In addition, the complete genome sequences of the CPIV-5 strains were aligned with genus Rubulavirus sequences obtained from GenBank, using computer software (BioEdit). A phylogenetic analysis was conducted using computer software [BioEdit and Molecular Evolutionary Genetics Analysis (MEGA) 4.0] with bootstrap values calculated from 1000 replicates (8). The neighbor-joining phylogenetic algorithm was used to construct the trees.
The complete genomes of the 3 strains were sequenced and the genome length was 15 246 nucleotide, identical to the PIV-5 strain genome. No deletions or insertions were found in the complete sequences of the 3 strains. The sequences of the complete genomes of 3 CPIV-5 strains and the reported PIV-5 strain, W3A, (NC006430) showed 96.7% to 98.8% identity at the nt level and 96.2% to 98.6% at the amino acid level (Table I). The nt and amino acid similarities between the 1168-1 strain and PIV-5 were 98.8% and 98.6, respectively. The other 2 strains varied considerably from the PIV-5 strain in all of the non-coding and coding regions of the genome. The nucleotide identities for coding regions observed between the 3 CPIV-5 strains and PIV-5 were in the range of 83.7% to 99.2% and the amino identities between the 3 CPIV-5 strains and PIV-5 were in the range of 72.7% to 99.5%. The amino acid similarities of the SH gene region between the 3 CPIV-5 and PIV-5 varied markedly from 72.7% to 93.1%.
Table I.
Nucleotide and putative amino acid identities of PIV-5 (NC006430) with three CPIV-5 isolates (08-1990, 1168-1, and D277)
| Nucleotide identity (%) | Amino acid identity (%) | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Gene region (NC006430) | 08-1990 | 1168-1 | D277 | 08-1990 | 1168-1 | D277 |
| Leader UTR (nt 151) | 97.3 | 100 | 98.0 | — | — | — |
| Nucleoprotein(N) (nt 1530) | 97.1 | 99.1 | 97.0 | 98.4 | 99.4 | 98.4 |
| N-V/P UTR (nt 169) | 83.9 | 94.0 | 83.9 | — | — | — |
| V protein (V) (nt 670) | 97.4 | 98.9 | 97.3 | 97.7 | 99.5 | 97.7 |
| Phosphoprotein (P) (nt 1176) | 97.1 | 99.2 | 97.1 | 97.4 | 99.4 | 97.6 |
| P-M UTR (nt 114) | 88.4 | 98.2 | 90.2 | — | — | — |
| Matrix (M) protein (nt 1134) | 96.2 | 98.4 | 96.2 | 96.8 | 97.8 | 96.4 |
| M-F UTR (nt 255) | 93.3 | 97.6 | 93.3 | — | — | — |
| Fusion (F) protein (nt 1587) | 96.1 | 98.8 | 95.9 | 96.3 | 98.7 | |
| F-SH UTR (nt 187) | 93.3 | 98.6 | 93.3 | — | — | — |
| SH (nt 135) | 84.4 | 97.0 | 83.7 | 72.7 | 93.1 | 72.7 |
| SH-HN UTR (nt 146) | 94.5 | 100 | 93.8 | — | — | — |
| Hemagglutinin-neuraminidase (HN) (nt 1698) | 96.7 | 98.5 | 96.7 | 96.2 | 97.8 | 96.2 |
| HN-L UTR (nt 132) | 91.6 | 99.2 | 93.1 | — | — | — |
| Large (L) protein (nt 6768) | 97.9 | 99.1 | 97.9 | 98.9 | 99.4 | 99.0 |
| Trailer UTR (nt 65) | 96.9 | 96.9 | 95.3 | — | — | — |
| Complete genome (nt 15246) | 96.7 | 98.8 | 96.7 | 96.2 | 98.6 | 96.2 |
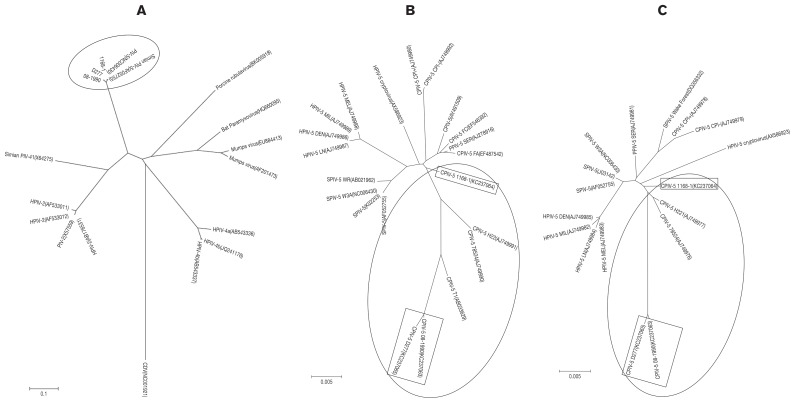
The nt sequence alignments of the complete genomes were used in the phylogenetic analysis. The derived phylogenetic tree showed that the 3 CPIV-5 strains clustered with the human and simian PIV-5 strains. The 3 CPIV-5 strains formed a lineage that was distinct from the PIV-2 and PIV-4 strains in the phylogenetic trees (Figure 1A). This supports the notion that all PIV-5 strains are genetically homologous independent of the host, as reported elsewhere (9). In addition, phylogenetic analysis based on the V/P and F genes were performed (Figures 1B, 1C). The PIV-5 strains were divided into at least 3 subgroups. Three CPIV-5 strains were clustered with CPIV-5 T1, H22, and 78524 strains.
Figure 1.
Phylogenetic analysis of parainfluenza virus type 5 strains based on the nucleotide sequences of the complete genome sequence (A), V/P gene (B), and F gene (C). The nucleotide sequences of various parainfluenza virus strains were aligned using BioEdit and MEGA 4. One-thousand bootstrap replicates were subjected to nucleotide sequence distance and neighbor-joining analyses, and the consensus phylogenetic tree is shown. 1A: canine distemper virus (NC001921) was used as an outgroup.
In the present study, the complete genomes of 3 CPIV-5 strains identified in Korea were sequenced, and found to be similar to that of the PIV-5 genome. Only a few CPIV-5 partial sequences have been reported (9,10) and a partial comparison of PIV-5 genomes in different species revealed a lack of sequence variation at both the nucleotide and amino acid levels (0 to 3%). In our study, all PIV-5 strains were independent of host species, geographical distribution, and the isolated period (9).
Interestingly, the amino acid similarities of the SH gene region between three CPIV-5 strains and PIV-5 differed markedly. These data support that the SH gene region may potentially be a genetic marker for the differentiation of PIV-5 from CPIV-5 (11). However, further genetic analyses are required to elucidate the precise relationship between species and genetic variation.
Acknowledgment
This research was supported by a grant from the Animal and Plant Quarantine Agency, Anyang, Republic of Korea.
References
- 1.Karron RA, Collins PL. Field Virology. 5th ed. Philadelphia, Pennsylvania: Lippincott, Williams & Wilkins; 2007. pp. 1497–1526. [Google Scholar]
- 2.Chanock RM, Johnson KM, Cook MK, Wong DC, Vargosko A. The hemadsorption technique, with special reference to the problem of naturally occurring simian parainfluenza virus. Am Rev Respir Dis. 1961;83:125–129. [Google Scholar]
- 3.Murphy FA, Gibbs EPJ, Horzinek MC, Studdert MJ. Veterinary Virology. 3rd ed. San Diego, USA: Academic Press; 1999. p. 148. [Google Scholar]
- 4.Dambro NN, Grad R, Witten ML, et al. Bronchoalveolar lavage fluid cytology reflects airway inflammation in beagle puppies with acute bronchiolitis. Pediat Pulmonol. 1992;12:213–220. doi: 10.1002/ppul.1950120404. [DOI] [PubMed] [Google Scholar]
- 5.Quan SF, Witten ML, Dambro NN, Lemen RJ. Canine parainfluenza type 2 and Bordetella bronchiseptica infection produces increased bronchoalveolar lavage thromboxane concentrations in beagle puppies. Prostaglandins Leukot Essent Fatty Acids. 1991;44:171–175. doi: 10.1016/0952-3278(91)90052-7. [DOI] [PubMed] [Google Scholar]
- 6.Ford RB. Infectious diseases of the dog and cat. 3rd ed. Philadelphia, Pennsylvania: Elsevier; 2006. pp. 54–61. [Google Scholar]
- 7.He B, Paterson RG, Ward CD, Lamb RA. Recovery of infectious SV5 from cloned DNA and expression of a foreign gene. Virol. 1997;237:249–260. doi: 10.1006/viro.1997.8801. [DOI] [PubMed] [Google Scholar]
- 8.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 9.Chatziandreou N, Stock N, Young D, et al. Relationships and host range of human, canine, simian and porcine isolates of simian virus 5 (parainfluenza virus 5) J Gen Virol. 2004;85:3007–3016. doi: 10.1099/vir.0.80200-0. [DOI] [PubMed] [Google Scholar]
- 10.Baty DU, Southern JA, Randall RE. Sequence comparison between the haemagglutinin-neuraminidase genes of simian, canine and human isolates of simian virus 5. J Gen Virol. 1991;72:3103–3107. doi: 10.1099/0022-1317-72-12-3103. [DOI] [PubMed] [Google Scholar]
- 11.Takeuchi K, Tanabayashi K, Hishiyama M, Yamada A, Sugiura A. Variation of nucleotide sequences and transcription of the SH gene among mumps virus strains. Virology. 1991;181:364–366. doi: 10.1016/0042-6822(91)90504-5. [DOI] [PubMed] [Google Scholar]