Abstract
The conserved transcription coactivator SAGA is comprised of several modules that are involved in activator binding, TBP binding, histone acetylation (HAT) and deubiquitination (DUB). Crosslinking and mass spectrometry, together with genetic and biochemical analyses, were used to determine the molecular architecture of the SAGA-TBP complex. We find that the SAGA Taf and Taf-like subunits form a TFIID-like core complex at the center of SAGA that makes extensive interactions with all other SAGA modules. SAGA-TBP binding involves a network of interactions between subunits Spt3, Spt8, Spt20, and Spt7. The HAT and DUB modules are in close proximity, and the DUB module modestly stimulates HAT function. The large activator-binding subunit Tra1 primarily connects to the TFIID-like core via its FAT domain. These combined results were used to derive a model for the arrangement of the SAGA subunits and its interactions with TBP. Our results provide new insight into SAGA function in gene regulation, its structural similarity with TFIID, and functional interactions between the SAGA modules.
Keywords: coactivator, gene regulation, proteomics, transcription
Introduction
Yeast SAGA and the closely related mammalian orthologs STAGA, PCAF, and TFTC are broadly conserved coactivator complexes that regulate the transcription of many inducible and developmentally regulated genes (Rodriguez-Navarro, 2009; Samara & Wolberger, 2011; Helmlinger, 2012; Weake & Workman, 2012). In yeast, SAGA regulates ˜ 10% of genes, with this gene set being highly enriched for genes induced by environmental stress and for genes with promoters containing a match to the consensus TATA element (Basehoar et al, 2004; Huisinga & Pugh, 2004). SAGA and related complexes are multifunctional, containing at least five separate activities: nucleosomal histone H3 acetyltransferase (HAT), histone H2B deubiquitinase (DUB), TBP binding, nucleosome binding, and activator binding. However, not all SAGA activities are required for transcription of every SAGA-dependent gene. For example, less than half of SAGA-dependent genes require the HAT activity for normal expression (Lee et al, 2000; Huisinga & Pugh, 2004). Similarly, the TBP-binding function of SAGA is essential for TBP recruitment at some genes, while at other genes, SAGA functions at steps after formation of the complete RNA Pol II preinitiation complex (Larschan & Winston, 2001; Bhaumik & Green, 2002; Mohibullah & Hahn, 2008; Chen et al, 2012). These diverse activities allow SAGA to modulate transcription at many promoters with different coactivator requirements by regulating steps encompassing chromatin modification, stimulation of PIC assembly, and enhancing steps in early transcription elongation.
SAGA from Saccharomyces cerevisiae is a 1.8 MDa complex comprised of 19–20 subunits with its various activities separated into distinct structural modules. The activity and specificity of the HAT and DUB catalytic functions are dictated by non-catalytic subunits within each module. For example, Ubp8, the subunit containing DUB activity, is inactive unless associated with the three other subunits of the DUB module (Köhler et al, 2008; Lang et al, 2011). The X-ray structure of this module shows a complex intertwining of the DUB module subunits and provides an explanation for the interdependence of the subunits in DUB function (Köhler et al, 2010; Samara et al, 2010). Similarly, the substrate specificity of Gcn5 HAT activity is altered by its associated subunits, Ada2, Ada3, and Sgf29, to acetylate multiple residues within the nucleosome histone H3 tail (Grant et al, 1999; Balasubramanian et al, 2002; Bian et al, 2011).
Other groups of subunits also contribute to the disparate functions of SAGA. Both genetic and biochemical experiments have shown that the SAGA subunits Spt3 and Spt8 contact TBP (Eisenmann et al, 1992; Warfield et al, 2004; Sermwittayawong & Tan, 2006; Laprade et al, 2007; Mohibullah & Hahn, 2008); however, the mechanism of these interactions and whether both subunits can simultaneously bind TBP are unclear. SAGA shares a set of subunits with the coactivator TFIID, which likely play a structural role in SAGA architecture (Grant et al, 1998). Within TFIID, two copies each of the histone fold pair-containing subunits Taf6-Taf9 and Taf4-Taf12, together with two copies of Taf5, form a symmetric complex (Bieniossek et al, 2013). This complex is bound by the Taf8-Taf10 heterodimer, generating an asymmetric core structure upon which the other TFIID-specific Tafs assemble. Likewise, SAGA contains a similar set of Tafs but with Taf4 and Taf8 replaced by the histone fold domain containing SAGA-specific subunits Ada1 and Spt7 (Gangloff et al, 2000, 2001).
Because of its complex composition, we know relatively little about the molecular interactions between SAGA subunits, the overall arrangement of the different modules, and whether the modules functionally interact. For example, purification of SAGA via a tag on subunit Spt7 in strains with deletions of other SAGA subunits revealed complex interactions among the SAGA subunits (Wu & Winston, 2002). Similarly, purification of SAGA from strains containing TAF12 conditional mutations showed defects in association of subunits from different SAGA modules (Durso et al, 2001). A systematic mass spectrometry analysis of SAGA purified from strains lacking individual non-essential SAGA subunits led to a model for the composition of each module and organization of the complex (Lee et al, 2011). However, this model differs in the proposed relative locations of the Tafs, Spt, and Tra1 subunits derived from an earlier model based on EM and immune localization of several subunits (Wu et al, 2004). Together, these initial findings suggest a series of complex interactions between different SAGA subunits and modules, perhaps somewhat analogous to the complex interactions between subunits of the DUB module.
To understand how the diverse functions of SAGA are coordinated to modulate gene expression, it is important to determine the overall architecture of SAGA, the arrangement of the different modules, and whether these modules interact. To approach this problem, we have used a combination of crosslinking and mass spectrometry, together with genetic and biochemical analysis, to determine molecular interactions between the SAGA subunits and its modules. Based on this work, we derive a new model for SAGA architecture with the TFIID-like Taf complex at the center of SAGA. We also find functional interactions between the two enzymatic modules and new insight into the mechanisms of SAGA-TBP binding.
Results
Formation of the SAGA-TBP complex
To determine the molecular architecture of the SAGA complex and its interactions with TBP, we first purified yeast SAGA using a TAP-tag at the C-terminus of subunit Spt7. This approach selectively purifies SAGA and not the related SLIK complex, which lacks the C-terminus of Spt7 (Wu & Winston, 2002). Purified SAGA was visualized on a silver-stained protein gel (Fig 1A, lane 3). Mass spectrometry analysis of this preparation identified all known SAGA subunits with the exception of Chd1 (Supplementary Table S1), a chromatin remodeler that reportedly associates with SAGA (Pray-Grant et al, 2005). In agreement with most previous findings, TBP did not copurify with SAGA, indicating a weak association. As a first attempt to generate the SAGA-TBP complex, we purified SAGA from a strain with the spt3-401 allele, containing the Spt3 mutation E240K, which was suggested to bind TBP more stably compared to wild-type SAGA (Laprade et al, 2007). Under our purification conditions, however, this SAGA derivative was not stably associated with TBP (Fig 1A, lane 5). As an alternative approach for finding conditions to generate the SAGA-TBP complex, purified SAGA was immobilized to calmodulin beads via the calmodulin-binding tag on Spt7. Immobilized SAGA was incubated with recombinant TBP for 90 min, washed, and then eluted from the beads using EGTA. Both wild-type and the Spt3-401 SAGA derivative specifically bound TBP (Fig 1A, lanes 4, 6, 7). Quantitation of the TBP/Ada1 ratio for the two preparations showed that the Spt3-401 SAGA bound about fourfold more TBP compared to wild-type SAGA (Fig 1B). Since wild-type SAGA bound TBP under these conditions, TBP and wild-type SAGA were combined in a 1:1.2 molar ratio and used as a substrate for subsequent crosslinking experiments.
Figure 1. TAP purified SAGA complex binds TBP.
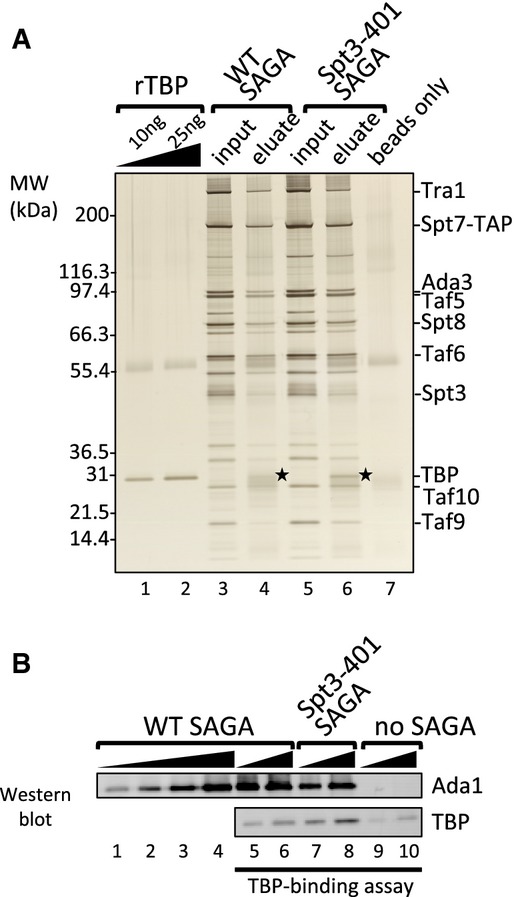
- A silver-stained protein gel showing the TBP-binding assay of wild-type and Spt3-401 (E240K) SAGA complexes. 3 μg SAGA was immobilized to calmodulin beads and incubated with 0.5 μg recombinant TBP (rTBP). EGTA eluted samples were analyzed for TBP binding. Stars denote bound TBP.
- Western blot analysis of the above TBP-binding assay probing for Ada1 and TBP.
Source data are available online for this figure.
Chemical crosslinking and mass spectrometry of SAGA
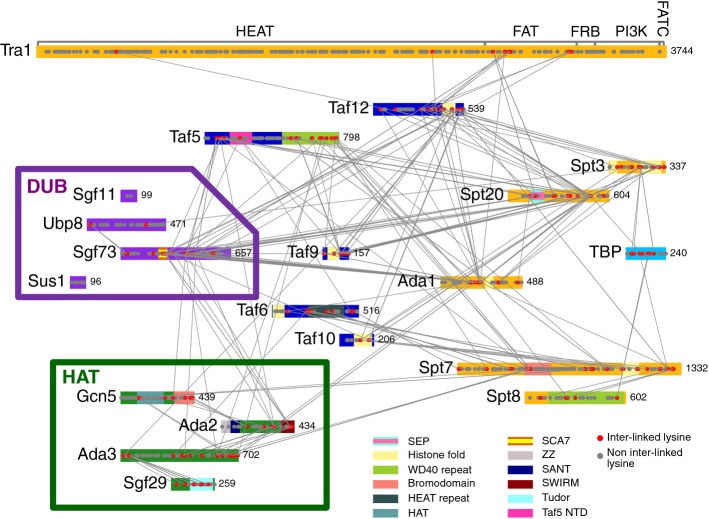
We next used a chemical crosslinking and mass spectrometry approach (Chen et al, 2010; Müller & Sinz, 2012) to map protein–protein interactions within SAGA. The SAGA-TBP complex was crosslinked using the bifunctional crosslinking reagent BS3, digested with trypsin, and the peptides analyzed by mass spectrometry. The BS3 crosslinker reacts with accessible primary amine groups found in lysine side chains and at the N-termini of polypeptides (Supplementary Fig S1A). Each SAGA subunit, except Sgf11, contains many lysine residues that are distributed throughout the polypeptides (Fig 2, Supplementary Fig S2), making SAGA a good substrate for this crosslinking strategy. A database search algorithm pLink (Yang et al, 2012) was used to identify the crosslinked peptides, and the identifications were then used to assemble site-specific linkage maps of all the crosslinked residues between and within the SAGA subunits (Fig 2, Supplementary Fig S2).
Figure 2. BS3 intermolecular crosslinking map of SAGA-TBP complex.
SAGA subunits are color-coded according to the different modules they belong to. Domains are highlighted as indicated based on previous studies or HHpred predictions (with > 95% probability score). Grey and red dots represent lysine residues (not crosslinked and crosslinked, respectively), while grey lines connecting red dots denote intermolecular crosslinked lysine pairs. SEP: Shp1, Eyc and p47; SCA7: spinocerebellar ataxia type 7; ZZ: zinc-binding domain; SANT: SWI3, ADA2, N-CoR and TFIIIB” DNA-binding domain; SWIRM: SWI3, RSC8 and MOIRA; Taf5 NTD: Taf5 N-terminal domain; FAT: FRAP, ATM and TRRAP; FRB: FKBP12 rapamycin binding; FATC: FAT C-terminal.
SAGA-TBP was crosslinked with either 2 or 5 mM BS3, and the reactions were independently analyzed by mass spectrometry for protein crosslinks. Results from both experiments were combined. Altogether, we identified 199 intermolecular crosslinks (between two different subunits) and 240 intramolecular crosslinks (within a single subunit) (Fig 2, Supplementary Figs S1C and S2, and Supplementary Table S2). To validate our crosslinking approach, we mapped the crosslinked lysine pairs within known structured domains of SAGA and TBP. BS3 has a theoretical maximum Cα-Cα crosslinking distance of ˜30 Å between two lysine residues (Kalkhof & Sinz, 2008; Mädler et al, 2009; Chen et al, 2010; Müller & Sinz, 2012; Merkley et al, 2014). Within domains for which high-resolution structural information is available, 17 out of the 19 crosslinks were within the theoretical crosslinking distance for BS3 (Supplementary Table S3).
A TFIID-like core complex at the center of SAGA
SAGA and TFIID share a subset of Tafs that are important for the function of both complexes. A recent study reported the cryo-EM structure of a human TFIID subcomplex, comprised of two copies each of Tafs 4, 5, 6, 9, and 12 (Bieniossek et al, 2013). Addition of the Taf8-Taf10 heterodimer to this complex generates an asymmetric TFIID core complex that acts as a scaffold to assemble the remaining TFIID subunits. All of the above-mentioned Tafs are also subunits of SAGA, with the exceptions of Taf4 and Taf8. The SAGA subunit Ada1 has a histone fold domain homologous to that of Taf4 and forms a histone fold pair with Taf12 (Gangloff et al, 2000), while Spt7 has a histone fold domain homologous to Taf8 and pairs with Taf10 (Gangloff et al, 2001). Indeed, we observed crosslinks between Taf12 and Ada1, and between Taf10 and Spt7, close to the histone fold domains (Fig 2), indicating that these proteins likely form heterodimers through histone fold pairing within the SAGA complex. We also observed crosslinks between all of the other Taf subunits except for Taf6 and Taf9, which are known to form a histone fold pair within TFIID (Bieniossek et al, 2013). Together with previous findings on the TFIID core complex, our combined results suggest that SAGA contains a TFIID-like core. As described in more detail below, this TFIID-like core complex crosslinks to all other SAGA modules (Fig 3A). Thus, similar to its role in TFIID, this TFIID-like core is likely positioned at the center of SAGA where it functions to nucleate assembly of other SAGA modules.
Figure 3. Crosslinking network of the SAGA complex.
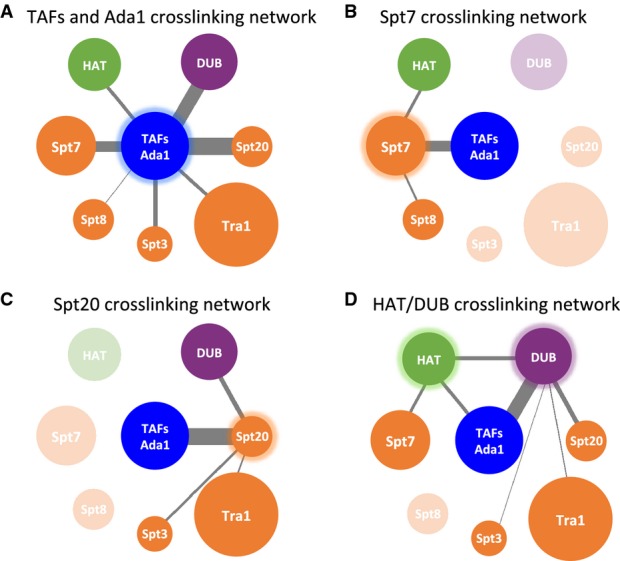
A–D Crosslinking network for (A) TAFs/Ada1, (B) Spt7, (C) Spt20, (D) HAT, and DUB modules. SAGA subunits or modules are represented as spheres and are color-coded similar to Fig 2. Grey lines connect crosslinked SAGA components, with the width of the lines proportional to the number of intermolecular crosslinks. Transparent spheres in (B), (C), and (D) denote the subunits or modules that do not crosslink to the highlighted components.
Subunits Spt7 and Spt20 crosslink to distinct SAGA modules
The SAGA subunits Spt7, Spt20, and Ada1 were initially proposed to be components of a core module (Wu & Winston, 2002), as mutation of any of these subunits disrupted conventional purification of SAGA (Sterner et al, 1999). This view was supported by systematic deletion analysis, suggesting that all Spt subunits were part of a common module (Lee et al, 2011). In contrast, mapping Spt7 and Spt20 within the SAGA EM structure led to an alternative view, suggesting that these two subunits are positioned at different ends of a large structural module (Wu et al, 2004). Consistent with this latter view, our crosslinking results suggest distinct roles for these two subunits in SAGA assembly as Spt7 and Spt20 crosslink to different sets of subunits but do not crosslink to each other.
We found that Spt7, part of the TFIID-like module, crosslinks to Spt8, one of the TBP-binding subunits (Fig 3B). This interaction occurs primarily between the C-terminus of Spt7 and WD40 repeats at the center of Spt8. This is in perfect agreement with previous biochemical results, which found that the 152 C-terminal residues of Spt7 are required for Spt8 binding to SAGA (Wu & Winston, 2002). Spt7 also crosslinks to Gcn5 and Ada3, showing close positioning of Spt7 to the HAT module. In contrast, Spt20 crosslinks to neither Spt7, Spt8, nor the HAT module (Fig 3C). Instead, we observe extensive crosslinking between Spt20 and the TFIID-like core domain, the DUB module, and the TBP-binding subunit Spt3.
Tra1, an activator-binding subunit of both SAGA and the coactivator NuA4, crosslinks to Spt20 at several sites within its FAT domain. The FAT domain also crosslinks to TAF12, in a region of Taf12 that also crosslinks to Spt20. Thus, Tra1 seems to primarily associate with SAGA via interactions between its FAT domain and segments of Taf12 and Spt20 that are in close proximity (Figs2 and 3). While the sparse Tra1-SAGA crosslinking outside of the FAT domain does not rule out other Tra1 interactions, our observation that the major Tra1-SAGA crosslinking is localized within the relatively small Tra1 FAT domain, together with our findings that the FAT domain crosslinks with two closely positioned SAGA subunits, suggests a peripheral position for the large Tra1 subunit within the SAGA complex, in agreement with a previous model (Wu et al, 2004).
A network of interactions between TBP and four SAGA subunits
Crosslinking of the SAGA-TBP complex revealed four subunits positioned close to TBP: Spt3, 8, 7, and 20 (Figs2 and 4A). While both genetic and biochemical studies have implicated Spt3 and Spt8 in TBP binding, Spt8 is the only subunit known to bind to TBP in the absence of other SAGA components (Warfield et al, 2004; Sermwittayawong & Tan, 2006). While the crosslinks we observe between Spt3, Spt8, and TBP are consistent with previous findings, the additional crosslinks of TBP with Spt20 and Spt7 suggest that these subunits may function to facilitate TBP binding. The lysine residues of TBP that crosslink to Spt3 and Spt8 are located on opposite ends of the TBP surface (Fig 4B). Previous studies using the photocrosslinker BPA (ρ-benzoyl-phenylalanine) positioned on TBP identified two TBP surfaces that are close to Spt3 and Spt8 in the preinitiation complex (Mohibullah & Hahn, 2008). Consistent with this, we found that the TBP lysine residues that crosslink to Spt3 or Spt8 are close to the previously identified BPA crosslinks, further supporting the existence of two separate SAGA binding interfaces on TBP. The region of TBP that crosslinks to Spt3 also agrees with the location of mutations in TBP that suppress Spt3 mutations (Fig 4B) (Eisenmann et al, 1992; Laprade et al, 2007).
Figure 4. Two distinct surface regions on TBP that bind Spt3 and Spt8.
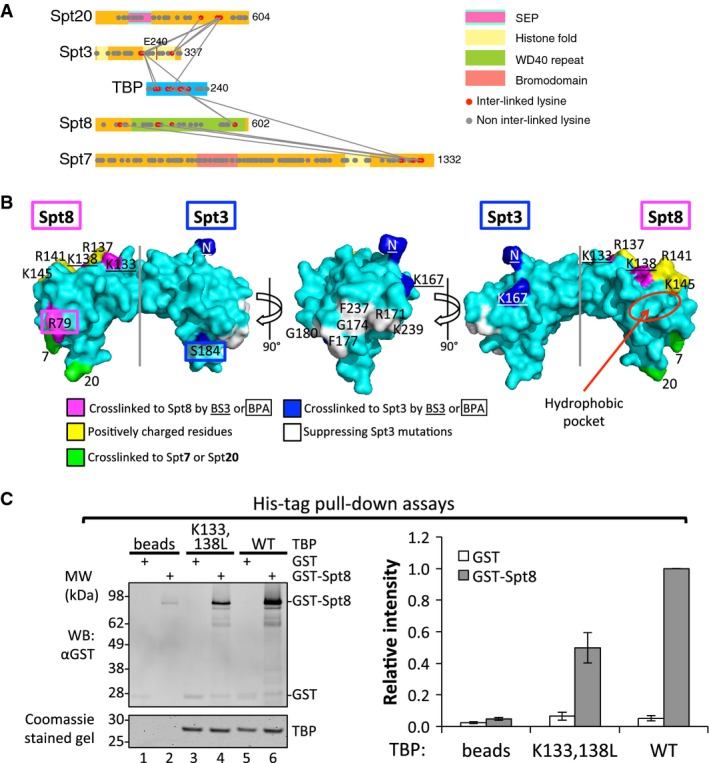
- A mini-crosslinking map between TBP and SAGA subunits.
- Surface representation of TBP (PDB ID 1TBP). TBP residues that crosslink to Spt8 and Spt3 are colored magenta and blue, respectively, with BS3 crosslinked residues underlined and BPA crosslinked residues boxed. Residues crosslinked to Spt7 (7) and Spt20 (20) are colored green. A conserved positively charged groove on the TBP surface is colored yellow. A hydrophobic pocket on TBP is shown as a red circle. Residues on the TBP surface that have been shown to suppress Spt3 mutations are colored white (Eisenmann et al, 1992; Laprade et al, 2007).
- Representative Western blot and Coomassie-stained gel of 6His-TBP pull-down experiments are shown on the left. 10% eluates were loaded each lane. Relative band intensity for GST or GST-Spt8 is shown as bar graph on the right. Intensity of GST-Spt8 bound with WT TBP (lane 6 on the left) was set to 1. Error bars represent standard deviation from three independent experiments.
Source data are available online for this figure.
Spt3 contains both halves of an interacting histone fold pair, located at the N and C-terminal ends of the protein (Birck et al, 1998). The spt3-401 allele (E240K), a suppressor of a TBP mutation in the Spt3-binding surface, lies within the C-terminal histone fold and implicates this region in TBP binding (Fig 4A) (Laprade et al, 2007). Consistent with this, we find that Spt3 lysine residue 190, located at one end of the C-terminal histone fold, crosslinks to the C-terminal half of TBP. Spt3 K190 also crosslinks to two residues within the disordered and non-conserved TBP N-terminal region. This latter interaction is probably not functionally important, as the N-terminal region of TBP can be deleted with no obvious phenotype (Reddy & Hahn, 1991). Interestingly, the C-terminal region of Spt20 crosslinks to both ends of the Spt3 C-terminal histone fold and to the opposite side of TBP near the Spt8-interacting surface (Fig 4A and B). Since Spt3 alone cannot stably bind TBP, our findings suggest that Spt20 may play a complex role in SAGA-TBP binding by modulating the Spt3-TBP interaction and possibly by direct interactions with TBP.
TBP residues K133 and K138 crosslink to the Spt8 WD40 repeats and are located in a conserved and highly positively charged groove on the convex surface of TBP (Fig 4B). These two lysine residues, together with K145 on TBP helix 2, are essential for interactions within Pol I, Pol II, and Pol III preinitiation complexes. For example, Mot1, the Taf1 TAND2 domain, and TFIIA all interact with this positively charged groove (Buratowski & Zhou, 1992; Kim & Roeder, 1994; Cui et al, 2011; Anandapadamanaban et al, 2013). Both the Taf1 TAND2 domain and Spt8 compete with TFIIA for binding TBP (Kokubo et al, 1998; Bagby et al, 2000; Warfield et al, 2004), suggesting that Spt8 might also bind TBP via the positively charged groove. To test this model, we introduced a double leucine substitution to TBP (K133, 138L) that is defective in forming the TBP-Mot1-DNA complex (Auble & Hahn, 1993) and in interacting with TFIIA (Buratowski & Zhou, 1992). These two lysine residues are both crosslinked to Spt8 by BS3 (Fig 4A and B). To test the effect of these TBP mutations on Spt8 binding, we performed a pull-down assay where His-tagged wild-type or K133,138L TBP were immobilized on Ni Sepharose resin and then incubated with GST or GST-tagged Spt8 (Fig 4C). Our results showed that the K133,138L mutation reduced TBP-Spt8 binding by twofold (Fig 4C). Taken together, our crosslinking and biochemical analyses suggest that Spt8 physically contacts the positively charged groove on TBP and that this interaction contributes to TBP-Spt8 binding.
Finally, we found that the C-terminal region of Spt7 crosslinks to both Spt8 and TBP (Fig 4A), suggesting that this region of Spt7 may be directly involved in TBP binding. Although recombinant Spt8 alone can bind TBP (Fig 4C), Spt7 may modulate this interaction. Interestingly, both Spt7 and Spt20 crosslink to nearby positions on the TBP N-terminal stirrup where they could potentially interact (Fig 4B). Thus, our crosslinking analysis suggests that Spt3, Spt20, Spt8, and Spt7 have a complex network of interactions with TBP.
A network of interactions linking SAGA with the HAT and DUB modules
Both the HAT and DUB modules function to enhance transcription (Weake & Workman, 2010). Recruitment of SAGA to promoters allows targeted acetylation of the H3 tail by Gcn5, promoting the binding of other regulatory factors and generating a permissive environment for chromatin remodeling. The DUB module removes ubiquitin from H2B, a histone modification that transiently increases at the 5′ end of transcribed regions upon gene induction and is linked to transcription elongation. Ubiquitin removal allows subsequent recruitment of the CTD Ser2 kinase CTK1 to coding regions (Wyce et al, 2007), promoting efficient transcription elongation and the placement of additional downstream chromatin marks. A mutant Poly Q-containing form of the DUB module subunit Ataxin7 (the human ortholog of yeast Sgf73) was shown to inhibit HAT function (McMahon et al, 2005; Palhan et al, 2005), and Gcn5 can enhance the stability of the DUB module in the related mammalian complex STAGA (Atanassov et al, 2009).
Our crosslinking data showed a surprising network of interactions between the HAT, DUB and TFIID-like core modules (Figs2 and 3D, and Supplementary Fig S3). Within the DUB module, the C-terminal region of Sgf73 makes extensive crosslinks with the TFIID-like core, Spt20, and the HAT module subunits Ada2 and Ada3. This agrees with previous results showing that Sgf73 is responsible for linking the DUB module to SAGA (Köhler et al, 2008; Lee et al, 2009). The HAT module subunit Ada3 crosslinks to the TFIID-like core through Taf5, Taf6, Spt7, and Ada1. Together, our results demonstrate that these two enzymatic modules are positioned near each other and raise the possibility that they functionally interact.
We next determined the regions of Sgf73, Ada2, and Ada3 required for integrity of each module and their association with SAGA using the crosslinks as a guide to test specific regions of these subunits. Internal deletions were designed within subunits Sgf73, Ada2, and Ada3, and complex integrity was monitored by immunoprecipitation (IP) experiments (Fig 5). Within Sgf73, mutations were generated between residues 196–575 in segments that crosslink to Spt20, Ada2, Ada3, or the TFIID-like core (Fig 5A and D, and Supplementary Fig S3A and B). An N-terminal Sgf73 deletion (Δ2–104) that was shown to block association of the other three DUB subunits (Köhler et al, 2008) was also included in the analysis. Whole cell extracts containing the Sgf73 derivatives were assayed by IP using either a 3× Flag tag at the C-terminus of Sgf73 or via the Spt7 TAP-tag (Fig 5A and D). Our results showed that deletion of Sgf73 residues 196–314 (Δ1; encompassing the nucleosome-binding SCA7 domain) or residues at the C-terminus (401–479 and 535–575; Δ2B and Δ3) had no effect on integrity of SAGA or the DUB module (Fig 5D, lanes 3, 6, 7). In contrast, deletion of the Sgf73 N-terminus (Δ4) caused dissociation of Ubp8 as expected (Fig 5D, lane 10). Deletion of Sgf73 residues 350–400 (Δ2A) had the most severe effect, resulting in release of Sgf73 and the DUB module from SAGA, as Sgf73 failed to copurify with the TFIID-like core and the HAT modules (Fig 5D, lane 5). In a yeast growth assay, this latter mutant had a slow growth rate similar to a strain with a complete deletion of SGF73 (Supplementary Fig S4A). While this important segment overlaps with the region of Sgf73 that crosslinks to the HAT module, Spt20, and the TFIID-like core, deletion of adjacent regions in Sgf73 that also crosslink to these subunits had no effect on DUB-SAGA association. We speculate that these adjacent regions of Sgf73 are in close proximity to other SAGA subunits but do not make direct protein–protein contacts.
Figure 5. Interactions linking SAGA with the HAT and DUB modules.
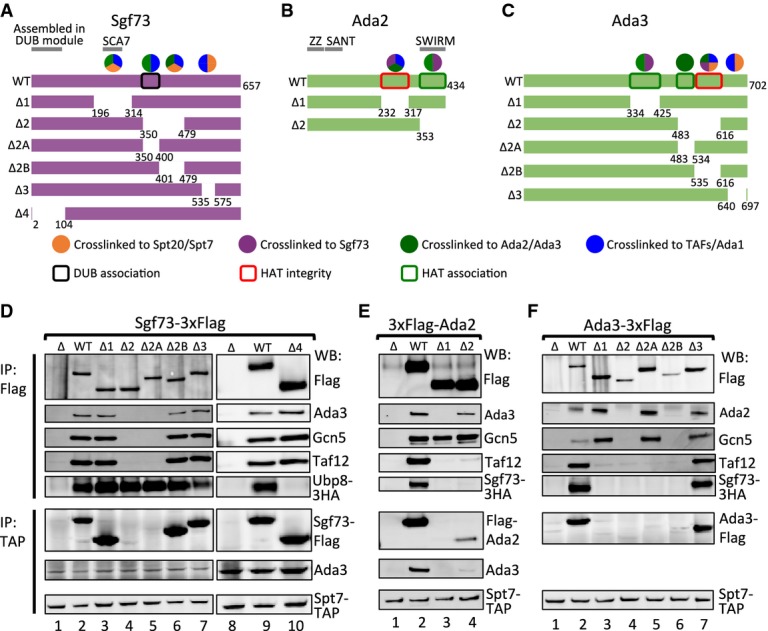
A–C Deletion schematics for (A) Sgf73, (B) Ada2, and (C) Ada3. Numbers indicate deleted residues. Sgf73 is colored purple, and Ada2 and Ada3 are colored green. Colored circles above wild-type (WT) constructs denote the crosslinking partners to corresponding regions: orange, Spt7/Spt20; purple, Sgf73; dark green, Ada2/Ada3; blue, Tafs/Ada1. Sgf73 and Ada3 constructs have a 3× Flag epitope tag at the C-terminus; Ada2 constructs have a 3× Flag epitope tag at the N-terminus. Boxes summarize the IP experiments in the lower panel: black, required for the association of the DUB module; red, required for the integrity of the HAT module; dark green, required for the association of the HAT module.
D–F IP experiments for (D) Sgf73 mutants, (E) Ada2 mutants and (F) Ada3 mutants are shown. Upper panels: anti-Flag IP; lower panels: anti-Spt7-TAP IP. Proteins are visualized in Western blot (WB) with corresponding antibodies labeled on the right of the blots.
Source data are available online for this figure.
Using the same strategy, we designed internal deletions surrounding the crosslinked lysine residues within HAT subunits Ada2 and Ada3 (Fig 5B and C). For Ada2, the IP assays of these mutant proteins agreed with the crosslinks and showed that the regions required for association with Ada3 and the TFIID-like core precisely overlap. Ada2 deletion Δ1, removing residues 232–317, retained association with Gcn5 but was released from its interactions with Ada3 and the TFIID-like core (Fig 5E, lane 3). This strain has a slow growth phenotype, equivalent to deletion of the ADA2 gene (Supplementary Fig S4B). Deletion of Ada2 residues 353–434 (Δ2) retained association with other HAT module subunits, but its binding to the TFIID-like core was diminished (Fig 5E, lane 4). This mutation has only a slight slow growth phenotype (Supplementary Fig S4B), suggesting that the HAT-SAGA interaction is not completely disrupted in vivo. Within Ada3, deletion of residues 334–425 (Δ1) and 483–534 (Δ2A) showed similar biochemical defects with an intact HAT module that was released from the TFIID-like core (Fig 5F, lanes 3, 5). In contrast, deletion of Ada3 residues 535–616 (Δ2B) disrupted the entire HAT module (Fig 5F, lane 6). All these mutant strains showed a slow growth phenotype (Supplementary Fig S4C). Interestingly, Ada3 deletion Δ3 (residues 640–697) was not affected for SAGA or HAT stability (Fig 5F, lane 7) but still showed a slow growth phenotype equivalent to deletion of the ADA3 gene (Supplementary Fig S4C). This suggests that the Ada3 C-terminus may play a role in modulating Gcn5 activity, perhaps related to its interactions with the TFIID-like core and Spt7.
While the IP assays did not reveal SAGA-HAT association defects resulting from mutations within the DUB module or vice versa, the fact that these two modules are in close proximity raised the possibility of a functional interaction. To examine whether one of these enzymatic modules could affect the other, we first measured subunit association in the mutant strains using a more stringent assay. Three mutations were selected based on the above IP experiments: (i) Sgf73 Δ4, which results in specific dissociation of DUB module subunits but leaves Sgf73-SAGA association intact, (ii) Sgf73 Δ2A, which results in release of Sgf73 and its associated DUB module subunits from SAGA, and (iii) Ada3 Δ2A, which results in release of the SAGA-HAT module.
SAGA was TAP-tag purified from each of these mutant strains, and Fig 6A shows a representative silver-stained gel of the preparations. The purified complexes were digested in-solution with trypsin and analyzed by mass spectrometry. The iBAQ (intensity-based absolute quantification) values (Schwanhäusser et al, 2011) were calculated using MaxQuant (Cox & Mann, 2008) to determine the relative abundance of each SAGA subunit in the purified complexes. The iBAQ value for each individual subunit was then normalized to the value of Spt7, because all complexes were purified through the tagged Spt7 subunit (Fig 6B, Supplementary Table S4A).
Figure 6. DUB module affects the nucleosomal HAT activity.
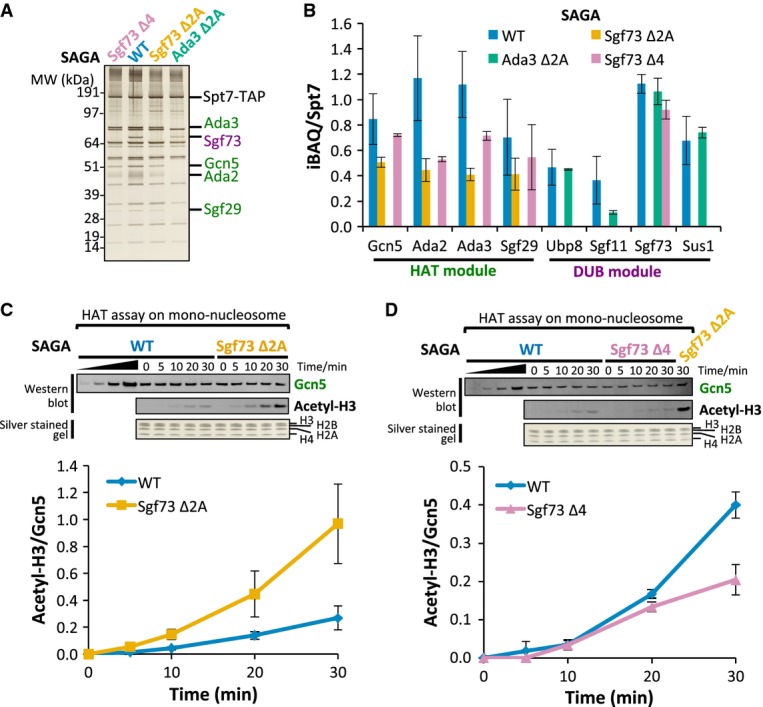
- A silver-stained protein gel showing the purification of Sgf73 Δ4, wild-type (WT), Sgf73 Δ2A, and Ada3 Δ2A SAGA complexes. DUB subunits are colored purple; HAT subunits are colored green; Sgf73 Δ4 complex is colored pink; WT complex is colored blue; Sgf73 Δ2A complex is colored orange; Ada3 Δ2A complex is colored light green.
- iBAQ analysis of WT, Sgf73 Δ2A, Ada3 Δ2A, and Sgf73 Δ4 SAGA complexes. Only iBAQ values normalized to Spt7 for DUB and HAT subunits are shown. See Supplementary Fig S5A and Supplementary Table S4A for iBAQ values for other SAGA subunits. Error bars represent standard deviation from 4 replicates for WT complex, 2 replicates for all three mutant complexes. Color scheme is the same as in (A).
- WT and Sgf73 Δ2A SAGA complexes were analyzed in an in vitro HAT assay using mono-nucleosome as substrate. Equivalent amounts of Gcn5 and all 4 histones were present at each time point as visualized by Western blot and silver staining (upper panel). Reactions were monitored by Western blot using antibody against acetylated H3K9 (Acetyl-H3). The acetylation signal was normalized to the amount of Gcn5 in the reaction at each time point (lower panel). Error bars represent standard deviation from 4 experiments (two biological replicates with two technical repeats). Color scheme is the same as in (A).
- Same as in (C), but comparing HAT activity between WT and Sgf73 Δ4 SAGA complexes. Sample from Sgf73 Δ2A complex at 30 min time point was also loaded as a control. Error bars represent standard deviation from 2 biological replicates.
Source data are available online for this figure.
For Sgf73 Δ4, the MS analysis confirmed that SAGA lacked the DUB subunits Ubp8, Sgf11, and Sus1, while retaining wild-type levels of Sgf73 (Fig 6B). As expected from the IP results, the more severe mutation Sgf73 Δ2A caused dissociation of all DUB associated subunits including Sgf73 (Fig 6B). In contrast, mutation of the HAT module subunit Ada3 (Δ2A) had no effect on the stability of the DUB module apart from a surprising 70% decrease in association of Sgf11 (Fig 6B). Finally, we found that both Sgf73 mutations affected the stability of the HAT module, consistent with crosslinks observed between Sgf73 and HAT subunits. SAGA purified from the mutants Sgf73 Δ2A and Δ4 showed a consistent 15–63% decrease in the amount of the associated HAT subunits (Fig 6B). We obtained similar results by analyzing the same data set and calculating the adjusted normalized spectral abundance factor (adjNSAF) using Abacus (Fermin et al, 2011) (Supplementary Table S4B). Together, our analysis of subunit composition in the HAT and DUB mutants suggests that the DUB module plays a modest role in stabilizing association of the HAT module with SAGA.
Mutations in the DUB module subunit Sgf73 affect SAGA nucleosomal HAT activity
Since the above analysis showed an interaction between the DUB and HAT modules, we tested whether mutations in Sfg73 affect HAT function using both recombinant histone H3 and mono-nucleosome substrates. The amounts of purified wild-type and mutant SAGA added to the HAT assays were normalized based on the amounts of added Gcn5. Histone H3 acetylation was monitored by Western blot assay using an antibody against acetylated H3K9. Unexpectedly, we observed that the two Sgf73 mutations have opposite effects on HAT activity.
SAGA purified from the more severe Sgf73 Δ2A mutation had HAT activity equivalent to wild-type SAGA using the histone H3 substrate, but showed about fourfold higher acetylation of the mono-nucleosome substrate after 30 min of incubation (Fig 6C, Supplementary Fig S5C). In contrast, SAGA purified from the less severe Sgf73 Δ4 mutation shows about twofold lower activity compared to wild-type SAGA on both the H3 and mono-nucleosome substrates (Fig 6D, Supplementary Fig S5C). Since this latter mutation only affects stability of the DUB module, it suggests that the DUB modestly enhances enzymatic activity of the HAT module. Because Sgf73 makes numerous protein–protein interactions with other SAGA modules, complete dissociation of Sgf73 in the Δ2A mutant may indirectly affect SAGA-HAT activity due to changes in other SAGA-HAT interactions. Our combined analysis of SAGA indicates a close proximity between the DUB and HAT modules and that the DUB module modestly enhances the enzymatic activity of the HAT module.
Discussion
SAGA is a multifunctional transcription coactivator that preferentially acts at inducible promoters where its multiple activities stimulate steps during transcription initiation and early elongation. The complexity of SAGA has led to several different models for the organization of its subunits and the architecture of the complex. To resolve these differences, we used an integrated approach to define molecular interactions between subunits and to determine how the different SAGA modules fit into the complete complex. Our work has led to a model for SAGA architecture, identification of molecular interactions between the different modules and new insight into the mechanisms and regulation of SAGA-TBP binding.
Previous studies revealed that SAGA contains a subset of the Taf subunits from the coactivator TFIID and two SAGA-specific subunits (Ada1 and Spt7) with Taf-like histone fold domains. Within TFIID, this set of Taf proteins forms an asymmetric core complex at the center of TFIID, which acts as a scaffold for assembly of the other Tafs. Our results are consistent with an analogous structure at the center of SAGA. First, the SAGA Taf and Taf-like subunits crosslink to each other as expected from an analogous complex within SAGA. Second, previous studies showed disruption of the SAGA complex upon inactivation of the SAGA Taf and Taf-like proteins. Our crosslinking data suggest that these subunits are at the center of SAGA, as they are a unique group of subunits that crosslink to all other SAGA modules. Within the TFIID-like core, SAGA-specific regions within Spt7 and Ada1 outside of their histone fold domains play an important role in bridging other structural modules with the TFIID-like core.
A Model for the architecture of SAGA
Figure7 (see also Supplementary Movie S1) shows a model for the architecture of SAGA with the TFIID-like structure at the center. Based on the proposed arrangement of the TFIID subunits, the central core of the SAGA model contains two copies each of the histone fold containing pairs Taf6-Taf9, and Ada1-Taf12 together with two copies of Taf5. This core structure contains extra SAGA-specific density (not shown) since Ada1 has over 170 residues both N- and C-terminal to its histone fold domain that are involved in interactions with other SAGA subunits. One copy of Spt7-Taf10 is predicted to bind this complex, analogous to the binding of Taf8-Taf10 in TFIID. Spt7 has a broad interface with this structure, as it crosslinks to the widely separated Taf5 N and C-terminal domains. Taf10, which requires only a 91-residue segment containing the histone fold domain for function (Kirschner et al, 2002), crosslinks to the histone fold domain of Taf12 in the lower half of the core.
Figure 7. Model for the molecular architecture of the SAGA complex.
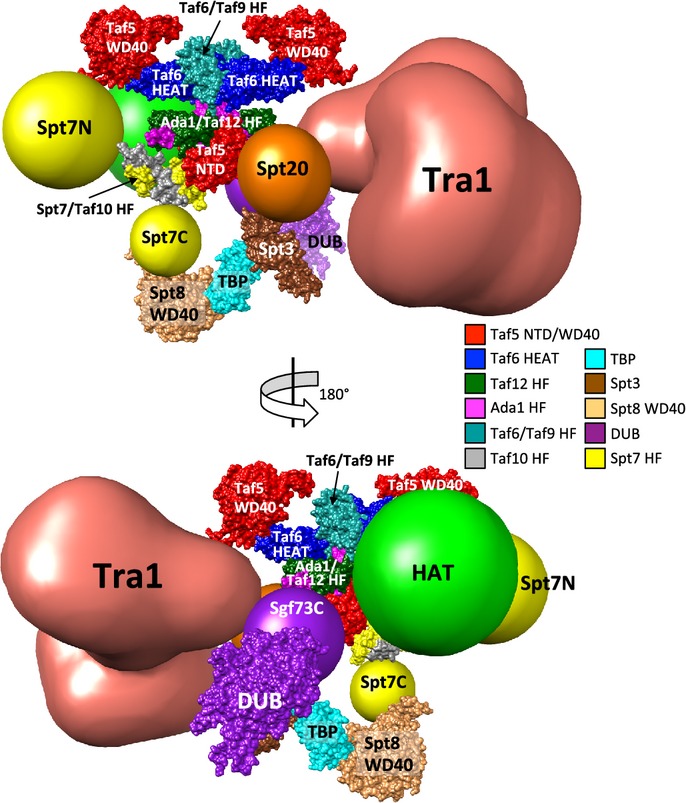
SAGA subunits were positioned around the TFIID core EM model (see Discussion section for details). Homology models of yeast Taf proteins and Ada1 were superimposed onto the coordinates kindly provided by Patrick Schultz (Bieniossek et al, 2013) and color-coded as indicated. Spt7/Taf10 HF (Histone Fold), Spt3, Spt8 WD40 are homology models (see Materials and Methods for details). Crystal structures used: TBP, 1TBP; DUB, 3M99; Taf5 NTD (N-terminal domain), 2J49. HF, histone fold domain; Spt7N/C, Spt7 N-terminal/C-terminal region; Sgf73C, Sgf73 C-terminal region. Images were prepared using UCSF Chimera (Pettersen et al, 2004).
Our crosslinking also reveals the interactions this TFIID-like core makes with the other SAGA modules and more peripheral subunits. For example, in addition to its crosslinks with the Taf5 N-terminal domain, the C-terminus of Spt7 also crosslinks to Spt8 WD40 repeats, likely positioning Spt8 in the lower half of the TFIID-like core. Sgf73 links the DUB module to the TFIID-like core, demonstrated by its extensive crosslinks with the histone fold domains of Ada1 and Taf12 as well as the Taf5 N-terminus. These crosslinks position the DUB module on the lower half of the TFIID-like core. Spt20, which crosslinks to the DUB module, Tra1, and Spt3, displays numerous crosslinks with the histone fold domains of Ada1, Taf9, Taf10, Taf12, and the N-terminal domain of Taf5. These results locate Spt20 in the center of the TFIID-like complex and explain how Spt20 and Sgf73 can extensively crosslink. Ada3, the HAT module subunit that links this module to SAGA, crosslinks to the histone fold domains of Ada1, and Taf12, the WD40 repeats in Taf5 and near the HEAT repeat of Taf6. This suggests that the HAT module is situated in the central and upper half of the TFIID-like core and in a position to crosslink with the C-terminus of Sgf73. The TBP-binding subunit, Spt3, crosslinks extensively to the same region of Spt20 that makes extensive crosslinks to the Taf12 histone fold domain and the Taf5 N-terminus. This maps Spt3 to the lower half of the TFIID-like complex, and potentially situated so that both Spt8 and Spt3 can simultaneously interact with TBP.
Wu et al reported an EM model of the SAGA complex, in which they mapped the positions of several SAGA subunits through immune labeling and electron microscopic analysis (Wu et al, 2004). Most of these mapping studies are in good agreement with our crosslinking results, with TAFs in a central position, Spt20 connecting with Spt3, and Spt7 close to Gcn5. The only discrepancy is their mapping of Tra1 and Spt20 at opposite ends of SAGA, in contrast to our crosslinking, which shows Tra1 and Spt20 in close proximity. We reason that this difference is likely due to the extended structure of Tra1, as Wu et al used Myc-antibody to probe the N-terminus of Tra1, and we observed Spt20 crosslinks near the Tra1 C-terminus (Fig 2). Our model is also distinct from another model, based on combinatorial depletion analysis, which linked all Spt subunits, Ada1 and Tra1 in a single group and positioned the Taf subunits in a more peripheral part of the protein–protein interaction network (Lee et al, 2011).
A surprisingly small interface between Tra1 and SAGA
Crosslinks between SAGA and the large activator-binding subunit Tra1 are primarily between the Tra1 FAT domain and a region of Taf12 that is N-terminal to the histone fold domain. While the absence of other crosslinks does not rule out additional Tra1-SAGA interactions, this limited region of crosslinking suggests a peripheral position for the majority of Tra1, perhaps on the opposite side of the TFIID-like core from the other modules. Interestingly, the FAT domain also crosslinks to a small segment of Sgf73, which in turn interacts with the HAT module subunits Ada2, Ada3, and Gcn5. It was previously shown that a small deletion within the Tra1 FAT domain caused inhibition of HAT activity (Knutson & Hahn, 2011). From our crosslinking results, this inhibition is likely an indirect effect. One explanation for this inhibitory effect is that the FAT domain mutation alters its interaction with Sgf73, causing an aberrant interaction between Sgf73 and the HAT module. It is also interesting that many small deletions and other mutations throughout Tra1 cause dissociation of Tra1 from other SAGA components (Knutson & Hahn, 2011). Given the domain topology derived from the crosslinking results, most of these mutations likely affect the FAT domain-SAGA interaction. Intramolecular Tra1 crosslinks show interactions between the FAT and HEAT repeat domains as well as interactions between the Tra1 PI3K, FRB, and FAT domains (Supplementary Fig S2). We propose that disrupting these intramolecular FAT domain interactions alters the conformation of the FAT domain and disrupts Tra1-SAGA contacts.
TBP-SAGA interactions
The two TBP-binding subunits, Spt3 and Spt8, play complex roles in gene regulation and, depending on the regulated gene, can have either positive or negative effects on transcription (Bhaumik, 2011). For example, deletion of Spt8 causes an increase in basal transcription at some genes (Belotserkovskaya et al, 2000) and Spt8 can compete with TFIIA for binding to TBP in vitro (Warfield et al, 2004). Our crosslinking and biochemical results provide an explanation for this competition, as both the unstructured region of the TFIIA large subunit and Spt8 bind a positively charged groove on TBP. Since DNA was found to compete with Spt8 for TBP binding (Sermwittayawong & Tan, 2006), it is likely that the isolated Spt8 subunit additionally interacts with TBP near its DNA-binding surface. Although Spt3-TBP binding is genetically the most important SAGA-TBP interaction for gene activation, Spt3 alone cannot bind TBP. The crosslinking data show that Spt20 is positioned either side of the Spt3 histone fold domain involved in TBP binding. This finding suggests that Spt20 may enhance Spt3-TBP binding by promoting a conformation of Spt3 that is active for TBP binding. Our combined results show a complex network of interactions between TBP and Spt3, 7, 8, and 20.
SAGA DUB-HAT interactions
The human DUB module subunit Ataxin7 (orthologous to yeast Sgf73) contains a region that can undergo poly-glutamine expansion, causing the disease spinocerebellar ataxia type 7. Human Ataxin7 can partially complement a SGF73 deletion, but the Poly Q form inhibits HAT activity in vivo and in vitro (McMahon et al, 2005; Palhan et al, 2005; Burke et al, 2013). Here, we have shown that mutations in the DUB module subunit Sgf73 have modest effects on SAGA nucleosomal HAT activity and stability of the HAT module within SAGA. This relationship is consistent with our observed protein crosslinking as well as inhibition of HAT activity by the mutant form of Ataxin7.
Taken together, our results have led to a model for SAGA architecture and revealed new and unexpected interactions between the SAGA subunits and modules. Our model reveals insights into how the disparate functions of SAGA are coordinated to activate a wide range of genes with different coactivator requirements and suggest new directions for understanding the molecular interactions of SAGA with transcription regulators and components of the transcription machinery.
Materials and Methods
See Supplementary Methods for strains and plasmids used, methods of protein purification, HAT and IP assays and structure modeling methods.
BS3 crosslinking
Wild-type SAGA complex was mixed with 20% molar excess of recombinant TBP. 50 μg of this complex were incubated with 2 mM or 5 mM BS3 (Thermo Scientific) in 200 μl total volume at room temperature for 2 h. Reactions were quenched with 10 μl 1 M Tris pH 7.5. Small-scale titrations were performed with ˜3 μg of SAGA complex in 20 μl total volume with various amount of BS3 as shown in Supplementary Fig S1B. Reactions were analyzed by SDS-PAGE and silver stain (Invitrogen).
MS sample preparation and data analysis
Protein samples were denatured in 50% trifluoroethanol (TFE) at 60°C for 30 min and reduced by addition of 5 mM TCEP at 37°C for 30 min. The samples were then alkylated in the dark at room temperature with 10 mM iodoacetamide for 30 min. The samples were diluted tenfold with 100 mM ammonium bicarbonate and digested with trypsin (3:50 w/w) overnight at 37°C. Tryptic peptides were then purified by C18 chromatography (Waters) and dried in a speedvac. For crosslinked samples, dried peptides were resuspended in 30 μl buffer A (25 mM ammonium formate, 20% acetonitrile [ACN], pH 2.8) and then fractionated by microcapillary partisphere strong cation exchange (SCX, 5 μm, 200 Å, GE Healthcare). For separation of crosslinked peptides by HPLC, peptides were loaded onto the capillary column equilibrated in 5% ACN/0.1% trifluoroacetic acid (TFA) and washed with 20% ACN/0.1% formic acid (FA). Bound peptides were eluted with 50 μl of buffer A with 10, 30, 50, 70 and 90% buffer B (800 mM ammonium formate, 20% ACN, pH 2.8), followed by 50 μl 100% buffer B and 50 μl buffer C (1 M ammonium acetate, 10% ACN, 0.1% FA, pH 8). All fractions were then dried in a speedvac. Most crosslinked peptides were eluted in fractions with 50–90% buffer B.
All dried peptides were analyzed on a Thermo Scientific Orbitrap Elite at the Proteomics facility at the Fred Hutchinson Cancer Research Center (FHCRC, Seattle, Washington, USA) with HCD fragmentation and serial MS events that included one FTMS1 event at 30,000 resolution followed by 10 FTMS2 events at 15,000 resolution. Other instrument settings included: Charge state rejection: +1, +2, +3 and unassigned charges; Monoisotopic precursor selection enabled; Dynamic exclusion enabled: repeat count 1, exclusion list size 500, exclusion duration 30 s; HCD normalized collision energy 35%, isolation width 3 Da, minimum signal count 5,000; FTMS MSn AGC target 50,000, FTMS MSn Max ion time 250 ms. HPLC uses a 90 min gradient from 5% ACN to 40% ACN for crosslinked fractions and a 60 min gradient from 5% ACN to 35% ACN for normal non-crosslinked peptides.
Crosslinking data were analyzed using pLink (Yang et al, 2012) with default settings (precursor monoisotopic mass tolerance: ± 10 ppm; fragment mass tolerance: ± 20 ppm; up to 4 isotopic peaks; max evalue 1; static modification on cysteines; 57.0215 Da; differential oxidation modification on methionines; 15.9949 Da) against a database containing 22 proteins (21 SAGA subunits and TBP), and the search engine outputs results with 5% FPR. Each spectrum was manually evaluated for the quality of the match to each peptide using the COMET/Lorikeet Spectrum Viewer (Trans-Proteomic Pipeline, TPP) (Supplementary Fig S6). The crosslinked peptides were considered confidently identified if at least 3 consecutive b or y ions for each peptide were observed with minimum peptide length ≥ 4 and the majority of the observed ions are accounted for. Fourteen percent of spectra were removed after manual inspection. For iBAQ analysis, MS raw files were loaded to MaxQuant (Cox & Mann, 2008) (version 1.3.0.5) and searched against the yeast whole genome database with deletions in Sgf73 or Ada3 under default settings with the following exceptions: group-specific parameters tab, multiplicity = 1, enzyme = trypsin; identification & quantification tab, Min. ration count = 1; Misc. tab, check re-quantify and keep low-scoring versions of identified peptides only within parameter groups, check match between runs, check label-free quantification, LFQ min. ratio count = 1, uncheck Fast LFQ, check iBAQ and log fit, check calculate peak properties. Alternatively, MS data were first searched against the yeast whole genome database using X!Tandem on the computer cluster at the Institute for Systems Biology (ISB, Seattle, Washington, USA). The search results were then analyzed using PeptideProphet and ProteinProphet in TPP on the computer cluster at ISB. Adjusted NSAF (adjNSAF) values were calculated using Abacus (Fermin et al, 2011) with default parameters.
Acknowledgments
We thank Steve Henikoff (FHCRC) for mono-nucleosomes, Toshio Tsukiyama (FHCRC) for H3K14, K23 acetylation antibodies, Patrick Schultz (IGBMC, Strasbourg, France) for sharing the coordinates of the human TFIID core model and Phil Gafken and the FHCRC proteomics resource for sample analysis. We also thank the members of the Hahn Lab (FHCRC) and the Ranish Lab (ISB) for help throughout the course of this project and B. Knutson and S. Grünberg for comments on the manuscript. This work was supported by NIH grants 2RO1GM057114 to SH and 2P50GM076547 and R21CA175849 to JR.
Author contributions
All authors designed the research, analyzed the data, and wrote the paper. YH and JL performed the research.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information
Supplementary information for this article is available online: http://emboj.embopress.org
References
- Anandapadamanaban M, Andresen C, Helander S, Ohyama Y, Siponen MI, Lundström P, Kokubo T, Ikura M, Moche M, Sunnerhagen M. High-resolution structure of TBP with TAF1 reveals anchoring patterns in transcriptional regulation. Nat Struct Mol Biol. 2013;20:1008–1014. doi: 10.1038/nsmb.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanassov BS, Evrard YA, Multani AS, Zhang Z, Tora L, Devys D, Chang S, Dent SYR. Gcn5 and SAGA regulate shelterin protein turnover and telomere maintenance. Mol Cell. 2009;35:352–364. doi: 10.1016/j.molcel.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auble DT, Hahn S. An ATP-dependent inhibitor of TBP binding to DNA. Genes Dev. 1993;7:844–856. doi: 10.1101/gad.7.5.844. [DOI] [PubMed] [Google Scholar]
- Bagby S, Mal TK, Liu D, Raddatz E, Nakatani Y, Ikura M. TFIIA-TAF regulatory interplay: NMR evidence for overlapping binding sites on TBP. FEBS Lett. 2000;468:149–154. doi: 10.1016/s0014-5793(00)01213-8. [DOI] [PubMed] [Google Scholar]
- Balasubramanian R, Pray-Grant MG, Selleck W, Grant PA, Tan S. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J Biol Chem. 2002;277:7989–7995. doi: 10.1074/jbc.M110849200. [DOI] [PubMed] [Google Scholar]
- Basehoar AD, Zanton SJ, Pugh BF. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. doi: 10.1016/s0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- Belotserkovskaya R, Sterner DE, Deng M, Sayre MH, Lieberman PM, Berger SL. Inhibition of TATA-binding protein function by SAGA subunits Spt3 and Spt8 at Gcn4-activated promoters. Mol Cell Biol. 2000;20:634–647. doi: 10.1128/mcb.20.2.634-647.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik SR, Green MR. Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol Cell Biol. 2002;22:7365–7371. doi: 10.1128/MCB.22.21.7365-7371.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik SR. Distinct regulatory mechanisms of eukaryotic transcriptional activation by SAGA and TFIID. Biochim Biophys Acta. 2011;1809:97–108. doi: 10.1016/j.bbagrm.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian C, Xu C, Ruan J, Lee KK, Burke TL, Tempel W, Barsyte D, Li J, Wu M, Zhou BO, Fleharty BE, Paulson A, Allali-Hassani A, Zhou J-Q, Mer G, Grant PA, Workman JL, Zang J, Min J. Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation. EMBO J. 2011;30:2829–2842. doi: 10.1038/emboj.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniossek C, Papai G, Schaffitzel C, Garzoni F, Chaillet M, Scheer E, Papadopoulos P, Tora L, Schultz P, Berger I. The architecture of human general transcription factor TFIID core complex. Nature. 2013;493:699–702. doi: 10.1038/nature11791. [DOI] [PubMed] [Google Scholar]
- Birck C, Poch O, Romier C, Ruff M, Mengus G, Lavigne AC, Davidson I, Moras D. Human TAF(II)28 and TAF(II)18 interact through a histone fold encoded by atypical evolutionary conserved motifs also found in the SPT3 family. Cell. 1998;94:239–249. doi: 10.1016/s0092-8674(00)81423-3. [DOI] [PubMed] [Google Scholar]
- Buratowski S, Zhou H. Transcription factor IID mutants defective for interaction with transcription factor IIA. Science. 1992;255:1130–1132. doi: 10.1126/science.1546314. [DOI] [PubMed] [Google Scholar]
- Burke TL, Miller JL, Grant PA. Direct inhibition of Gcn5 protein catalytic activity by polyglutamine-expanded ataxin-7. J Biol Chem. 2013;288:34266–34275. doi: 10.1074/jbc.M113.487538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZA, Jawhari A, Fischer L, Buchen C, Tahir S, Kamenski T, Rasmussen M, Lariviere L, Bukowski-Wills J-C, Nilges M, Cramer P, Rappsilber J. Architecture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry. EMBO J. 2010;29:717–726. doi: 10.1038/emboj.2009.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X-F, Lehmann L, Lin JJ, Vashisht A, Schmidt R, Ferrari R, Huang C, McKee R, Mosley A, Plath K, Kurdistani SK, Wohlschlegel J, Carey M. Mediator and SAGA have distinct roles in Pol II preinitiation complex assembly and function. CELREP. 2012;2:1061–1067. doi: 10.1016/j.celrep.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Cui S, Viswanathan R, Berninghausen O, Wells MN, Moldt M, Witte G, Butryn A, Wendler P, Beckmann R, Auble DT, Hopfner K-P. Structure and mechanism of the Swi2/Snf2 remodeller Mot1 in complex with its substrate TBP. Nature. 2011;475:403–407. doi: 10.1038/nature10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durso RJ, Fisher AK, Albright-Frey TJ, Reese JC. Analysis of TAF90 mutants displaying allele-specific and broad defects in transcription. Mol Cell Biol. 2001;21:7331–7344. doi: 10.1128/MCB.21.21.7331-7344.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann DM, Arndt KM, Ricupero SL, Rooney JW, Winston F. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev. 1992;6:1319–1331. doi: 10.1101/gad.6.7.1319. [DOI] [PubMed] [Google Scholar]
- Fermin D, Basrur V, Yocum AK, Nesvizhskii AI. Abacus: a computational tool for extracting and pre-processing spectral count data for label-free quantitative proteomic analysis. Proteomics. 2011;11:1340–1345. doi: 10.1002/pmic.201000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant PA, Schieltz D, Pray-Grant MG, Steger DJ, Reese JC, Yates JR, Workman JL. A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell. 1998;94:45–53. doi: 10.1016/s0092-8674(00)81220-9. [DOI] [PubMed] [Google Scholar]
- Grant PA, Eberharter A, John S, Cook RG, Turner BM, Workman JL. Expanded lysine acetylation specificity of Gcn5 in native complexes. J Biol Chem. 1999;274:5895–5900. doi: 10.1074/jbc.274.9.5895. [DOI] [PubMed] [Google Scholar]
- Gangloff YG, Werten S, Romier C, Carre L, Poch O, Moras D, Davidson I. The human TFIID components TAF(II)135 and TAF(II)20 and the yeast SAGA components ADA1 and TAF(II)68 heterodimerize to form histone-like pairs. Mol Cell Biol. 2000;20:340–351. doi: 10.1128/mcb.20.1.340-351.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff YG, Sanders SL, Romier C, Kirschner D, Weil PA, Tora L, Davidson I. Histone folds mediate selective heterodimerization of yeast TAF(II)25 with TFIID components yTAF(II)47 and yTAF(II)65 and with SAGA component ySPT7. Mol Cell Biol. 2001;21:1841–1853. doi: 10.1128/MCB.21.5.1841-1853.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmlinger D. New insights into the SAGA complex from studies of the Tra1 subunit in budding and fission yeast. Transcription. 2012;3:13–18. doi: 10.4161/trns.3.1.19271. [DOI] [PubMed] [Google Scholar]
- Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- Kalkhof S, Sinz A. Chances and pitfalls of chemical cross-linking with amine-reactive N-hydroxysuccinimide esters. Anal Bioanal Chem. 2008;392:305–312. doi: 10.1007/s00216-008-2231-5. [DOI] [PubMed] [Google Scholar]
- Kim TK, Roeder RG. Involvement of the basic repeat domain of TATA-binding protein (TBP) in transcription by RNA polymerases I, II, and III. J Biol Chem. 1994;269:4891–4894. [PubMed] [Google Scholar]
- Kirschner DB, Baur vom E, Thibault C, Sanders SL, Gangloff Y-G, Davidson I, Weil PA, Tora L. Distinct mutations in yeast TAF(II)25 differentially affect the composition of TFIID and SAGA complexes as well as global gene expression patterns. Mol Cell Biol. 2002;22:3178–3193. doi: 10.1128/MCB.22.9.3178-3193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson BA, Hahn S. Domains of Tra1 important for activator recruitment and transcription coactivator functions of SAGA and NuA4 complexes. Mol Cell Biol. 2011;31:818–831. doi: 10.1128/MCB.00687-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler A, Schneider M, Cabal GG, Nehrbass U, Hurt E. Yeast Ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nat Cell Biol. 2008;10:707–715. doi: 10.1038/ncb1733. [DOI] [PubMed] [Google Scholar]
- Köhler A, Zimmerman E, Schneider M, Hurt E, Zheng N. Structural basis for assembly and activation of the heterotetrameric SAGA histone H2B deubiquitinase module. Cell. 2010;141:606–617. doi: 10.1016/j.cell.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokubo T, Swanson MJ, Nishikawa JI, Hinnebusch AG, Nakatani Y. The yeast TAF145 inhibitory domain and TFIIA competitively bind to TATA-binding protein. Mol Cell Biol. 1998;18:1003–1012. doi: 10.1128/mcb.18.2.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G, Bonnet J, Umlauf D, Karmodiya K, Koffler J, Stierle M, Devys D, Tora L. The Tightly Controlled Deubiquitination Activity of the Human SAGA Complex Differentially Modifies Distinct Gene Regulatory Elements. Mol Cell Biol. 2011;31:3734–3744. doi: 10.1128/MCB.05231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laprade L, Rose D, Winston F. Characterization of new Spt3 and TATA-binding protein mutants of Saccharomyces cerevisiae: Spt3 TBP allele-specific interactions and bypass of Spt8. Genetics. 2007;177:2007–2017. doi: 10.1534/genetics.107.081976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larschan E, Winston F. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 2001;15:1946–1956. doi: 10.1101/gad.911501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Causton HC, Holstege FC, Shen WC, Hannett N, Jennings EG, Winston F, Green MR, Young RA. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature. 2000;405:701–704. doi: 10.1038/35015104. [DOI] [PubMed] [Google Scholar]
- Lee KK, Swanson SK, Florens L, Washburn MP, Workman JL. Yeast Sgf73/Ataxin-7 serves to anchor the deubiquitination module into both SAGA and Slik(SALSA) HAT complexes. Epigenetics & Chromatin. 2009;2:2. doi: 10.1186/1756-8935-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, Sardiu ME, Swanson SK, Gilmore JM, Torok M, Grant PA, Florens L, Workman JL, Washburn MP. Combinatorial depletion analysis to assemble the network architecture of the SAGA and ADA chromatin remodeling complexes. Mol Syst Biol. 2011;7:503. doi: 10.1038/msb.2011.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mädler S, Bich C, Touboul D, Zenobi R. Chemical cross-linking with NHS esters: a systematic study on amino acid reactivities. J Mass Spectrom. 2009;44:694–706. doi: 10.1002/jms.1544. [DOI] [PubMed] [Google Scholar]
- McMahon SJ, Pray-Grant MG, Schieltz D, Yates JR, Grant PA. Polyglutamine-expanded spinocerebellar ataxia-7 protein disrupts normal SAGA and SLIK histone acetyltransferase activity. Proc Natl Acad Sci USA. 2005;102:8478–8482. doi: 10.1073/pnas.0503493102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkley ED, Rysavy S, Kahraman A, Hafen RP, Daggett V, Adkins JN. Distance restraints from crosslinking mass spectrometry: mining a molecular dynamics simulation database to evaluate lysine–lysine distances. Protein Sci. 2014;23:747–759. doi: 10.1002/pro.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohibullah N, Hahn S. Site-specific cross-linking of TBP in vivo and in vitro reveals a direct functional interaction with the SAGA subunit Spt3. Genes Dev. 2008;22:2994–3006. doi: 10.1101/gad.1724408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller MQ, Sinz A. Chemical cross-linking and high-resolution mass spectrometry to study protein-drug interactions. Methods Mol Biol. 2012;803:205–218. doi: 10.1007/978-1-61779-364-6_14. [DOI] [PubMed] [Google Scholar]
- Palhan VB, Chen S, Peng G-H, Tjernberg A, Gamper AM, Fan Y, Chait BT, La Spada AR, Roeder RG. Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration. Proc Natl Acad Sci USA. 2005;102:8472–8477. doi: 10.1073/pnas.0503505102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pray-Grant MG, Daniel JA, Schieltz D, Yates JR, Grant PA. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- Reddy P, Hahn S. Dominant negative mutations in yeast TFIID define a bipartite DNA-binding region. Cell. 1991;65:349–357. doi: 10.1016/0092-8674(91)90168-x. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Navarro S. Insights into SAGA function during gene expression. EMBO Rep. 2009;10:843–850. doi: 10.1038/embor.2009.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samara NL, Datta AB, Berndsen CE, Zhang X, Yao T, Cohen RE, Wolberger C. Structural insights into the assembly and function of the SAGA deubiquitinating module. Science. 2010;328:1025–1029. doi: 10.1126/science.1190049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samara NL, Wolberger C. A new chapter in the transcription SAGA. Curr Opin Struct Biol. 2011;21:767–774. doi: 10.1016/j.sbi.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Sermwittayawong D, Tan S. SAGA binds TBP via its Spt8 subunit in competition with DNA: implications for TBP recruitment. EMBO J. 2006;25:3791–3800. doi: 10.1038/sj.emboj.7601265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Grant PA, Roberts SM, Duggan LJ, Belotserkovskaya R, Pacella LA, Winston F, Workman JL, Berger SL. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol Cell Biol. 1999;19:86–98. doi: 10.1128/mcb.19.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warfield L, Ranish JA, Hahn S. Positive and negative functions of the SAGA complex mediated through interaction of Spt8 with TBP and the N-terminal domain of TFIIA. Genes Dev. 2004;18:1022–1034. doi: 10.1101/gad.1192204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake VM, Workman JL. Inducible gene expression: diverse regulatory mechanisms. Nat Rev Genet. 2010;11:426–437. doi: 10.1038/nrg2781. [DOI] [PubMed] [Google Scholar]
- Weake VM, Workman JL. SAGA function in tissue-specific gene expression. Trends Cell Biol. 2012;22:177–184. doi: 10.1016/j.tcb.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P-YJ, Winston F. Analysis of Spt7 function in the Saccharomyces cerevisiae SAGA coactivator complex. Mol Cell Biol. 2002;22:5367–5379. doi: 10.1128/MCB.22.15.5367-5379.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P-YJ, Ruhlmann C, Winston F, Schultz P. Molecular architecture of the S. cerevisiae SAGA complex. Mol Cell. 2004;15:199–208. doi: 10.1016/j.molcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Wyce A, Xiao T, Whelan KA, Kosman C, Walter W, Eick D, Hughes TR, Krogan NJ, Strahl BD, Berger SL. H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex. Mol Cell. 2007;27:275–288. doi: 10.1016/j.molcel.2007.01.035. [DOI] [PubMed] [Google Scholar]
- Yang B, Wu Y-J, Zhu M, Fan S-B, Lin J, Zhang K, Li S, Chi H, Li Y-X, Chen H-F, Luo S-K, Ding Y-H, Wang L-H, Hao Z, Xiu L-Y, Chen S, Ye K, He S-M, Dong M-Q. Identification of cross-linked peptides from complex samples. Nat Meth. 2012;9:904–906. doi: 10.1038/nmeth.2099. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.