Abstract
Cysteine cathepsins are a group of enzymes normally found in the endolysosomes where they are primarily involved in intracellular protein turnover but also have a critical role in MHC II-mediated antigen processing and presentation. However, in a number of pathologies cysteine cathepsins were found to be heavily upregulated and secreted into extracellular milieu, where they were found to degrade a number of extracellular proteins. A major role in modulating cathepsin activities play glycosaminoglycans, which were found not only to facilitate their autocatalytic activation including at neutral pH, but also to critically modulate their activities such as in the case of the collagenolytic activity of cathepsin K. The interaction between cathepsins and glycosaminoglycans will be discussed in more detail.
1. Introduction
Cysteine cathepsins are members of the papain-like cysteine peptidase family [1]. Despite the fact that the eleven cysteine cathepsins found in man [2] represent only a small fraction of the human proteolytic repertoire, these enzymes have been attracting a lot of attention for their diverse roles in physiological and pathological processes that range from nonspecific protein turnover within the endolysosomal pathway to highly specialized functions in tissue homeostasis. A number of excellent reviews have been published recently, summarizing the structural and functional characteristics of cysteine cathepsins in health and disease [3–5].
All the cathepsins share the same structural scaffold, also called the papain-like fold. The structure consists of two subdomains which have been termed the L- and R-domains referring to their position when the molecule is shown in the standard orientation (Figure 1). The active site cleft is at the top of the molecule between the L- and R-domains and contains the conserved catalytic dyad Cys-His (marked by yellow and blue spheres in Figure 1, resp.). In general, papain-like peptidases can act as endo- or exopeptidases. In a typical endopeptidase the primary specificity determinant is the S2 site [6] and well-determined sites on the enzyme interact with residues P3 through P2′ of the substrate [7]. Five of the eleven human members of the family (cathepsins F, K, L, S, and V) are exclusively endopeptidases, cathepsin B is also a peptidyl dipeptidase, cathepsin X is a carboxypeptidase, cathepsin H is an aminopeptidase, and cathepsin C is a dipeptidyl peptidase. The proteolytic activity of the remaining two members, cathepsins O and W, remains to be determined [4]. Most cysteine cathepsins are ubiquitously expressed in the human body, while some (cathepsins K, S, V, and W) are expressed in more restricted patterns [3]. Cathepsin K is abundantly expressed in osteoclasts and synovial fibroblasts [8, 9] but is also found in other cells of the hematopoietic, epithelial, and fibroblast lineages [10]. Highest expression levels of cathepsin S are found in antigen-presenting cells [11], cathepsin V is expressed predominantly in thymus and testis [12], and the expression of cathepsin W is restricted to CD8+ lymphocytes and natural killer cells [13].
Figure 1.
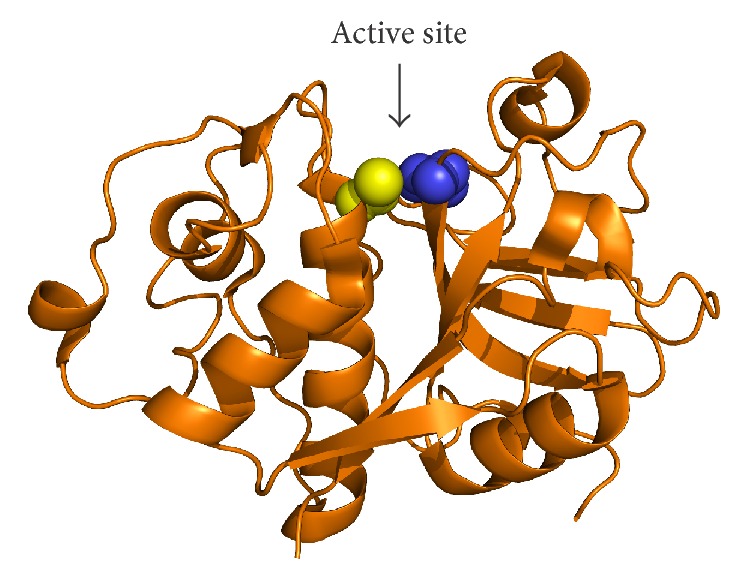
The papain-like peptidase fold illustrated on the crystal structure of papain. The protein is shown in cartoon representation and the position of the active site cleft is marked by an arrow. Catalytic residues Cys and His are shown as yellow and blue spheres, respectively. Coordinates were obtained from the Protein Data Bank under accession code 1PPN. The image was created with PyMOL (Schrödinger, LLC, Portland, OR, USA).
2. Regulation of Cysteine Cathepsins Activity
Zymogen activation is one of the major means of regulations of cathepsin activity. All the cathepsins are namely synthesized as inactive zymogens and activated in the acidic milieu of the endolysosomal vesicles. The molecular mechanism of their activation was puzzling for a long time. The critical information came from the combination of structural studies of procathepsins B, K, and L, which showed that the propeptide runs through the active site of cathepsins in the opposite direction of the substrate, thus excluding the cleavage of the propeptide in the molecule without enormous and energetically unfavorable structural movements of the propeptide [15–19], thereby eliminating the unimolecular mechanism initially suggested, and detailed kinetic studies, which clearly demonstrated that the activation of cathepsin B is a bimolecular process [20]. The current model, which is mostly based on the cathepsin B studies, suggests that the propeptide in the cathepsin zymogen switches between two conformations, the so-called “closed” and “open.” In the “closed” conformation, favored at neutral to slightly acidic pH, the propeptide blocks the active site and prevents substrate hydrolysis, whereas, in the “open” form, favored at acidic pH below pH 5.0, the propeptide is removed from the active side cleft, resulting in a low catalytic activity of the zymogen. This activity is sufficient to activate another cathepsin zymogen in one or several steps, thereby starting the chain reaction, where such fully active mature cathepsin B processes the majority of zymogen molecules [21].
The other major regulators of cysteine cathepsins are macromolecular inhibitors that bind into the active site and thereby prevent association of the peptidase with its substrate. They belong to several distinct families including the cystatins, the thyropins, and the serpins that can, in addition to serine proteases, also inhibit several cathepsins [3, 22–25].
3. Glycosaminoglycans as Major Regulators of Cysteine Cathepsin Activity
Glycosaminoglycans (GAGs) are heteropolysaccharides composed of repeating disaccharide units with a high negative charge. This is a result of the presence of multiple carboxyl groups and sulfate substitutions. Most of GAGs are sulfated, including chondroitin sulfates (CS), keratan sulfate (KS), dermatan sulfate (DS), heparan sulfate (HS), and heparin, whereas hyaluronan (HA) is the only nonsulfated GAG. In recent years GAGs have been emerging as important regulators of cysteine cathepsins with diverse effects on their targets. Traditionally, cysteine cathepsins had been viewed as lysosomal proteases and, like other lysosomal enzymes, had been known to be inhibited by intralysosomal GAGs in the resting lysosome [26]. Today, however, cysteine cathepsins are established as major players in extracellular proteolysis [27]. Their action in the glycosaminoglycan-rich extracellular environment raised questions about the interplay between cysteine cathepsins and GAGs outside of the lysosome. The two groups of endogenous human cysteine cathepsins most commonly associated with extracellular proteolysis are cathepsin L-like proteases (cathepsins K, L, S, and V in humans) and cathepsin B [27]. Data accumulated over the past two decades shows that the interplay between these peptidases and GAGs goes both ways; cysteine cathepsins are capable of cleaving proteoglycan core proteins and thereby release GAGs from their support, whereas GAGs in turn affect both the activity and stability of cysteine cathepsins in the extracellular space.
The regulation of papain-like cysteine peptidases by GAGs was first described for cathepsin L [28, 29]. In these early works it was found that GAGs and various negatively charged surfaces substantially accelerate activation of the cathepsin L zymogen into the mature form, including at pH close to the neutral, such as also found in the extracellular milieu in various disease conditions. This has been confirmed for several other cathepsins, the most important being cathepsins B and S [30, 31], and even for a T. congolense parasite homologue congopain [32], suggesting that GAGs and other negatively charged surfaces may play a major role in extracellular cathepsin activation in disease. However, recent findings with cathepsin S at high GAG concentration suggest that this enzyme may behave somewhat differently under such conditions with chondroitin-4-sulfate (C4S) even exhibiting a decelerating effect [33]. Nevertheless, facilitating autocatalytic activation of cathepsins by the negatively charged polysaccharide dextran sulfate also became a routine method in preparation of recombinant cathepsins [20, 34–36].
The majority of information about the molecular mechanism of GAG-assisted cathepsin activation came from a study by Caglič et al. using human cathepsin B as a model [31]. As shown, GAGs seem to contribute to the processing in two ways. First, upon binding they seem to convert the cathepsin zymogen into a better substrate. Second, binding of GAGs apparently favors the open conformation of the zymogen, thereby promoting activation not only at acidic pH but also at pH values closer to neutral. This seems to be the case for most GAGs and does not depend critically on the charge density of GAGs, as also HA was able to accelerate the activation, although to a lower extent, which is unusual for a protein-GAG interaction. Moreover, already a tetrasaccharide was sufficient for a prominent acceleration of cathepsin B autoactivation, which is substantially smaller than found for a number of other GAG-mediated reactions [37, 38]. The interaction is mediated by ionic interactions; however, there seems to be no conserved GAG-binding surface on the zymogens as completely unrelated residues in procathepsins L and B, which were largely located on the prodomains, were found to govern the interaction [31, 39].
The other important role of GAGs in the regulation of cathepsin activity came from the studies on papain, the archetypal representative of the family [40]. This mode of regulation quickly received more attention with the discovery that chondroitin-sulfate from cartilage prominently increased the collagenolytic activity of cathepsin K [41]. This particular peptidase had been discovered a few years prior as the sole protease responsible for collagen degradation in bone remodeling and immediately recognized as a potential drug candidate for the treatment of metabolic bone diseases, such as osteoporosis [42]. The interaction of cathepsin K with chondroitin-sulfate and other glycosaminoglycans was later examined in detail from both the structural and functional perspectives and glycosaminoglycans have been recognized as the first known allosteric regulators of a cysteine cathepsin peptidase [43, 44], as described in detail in the following sections. In parallel, functionally relevant interactions with glycosaminoglycans have also been documented for other members of the cysteine cathepsin family. Taken together, glycosaminoglycans have been found to affect both the activity and the stability of cysteine cathepsins. Kinetic profiles are usually consistent with hyperbolic mechanisms, indicating interactions with enzymes outside of the active site, possibly via allosteric mechanisms. The stabilizing effect is important especially because of the relative instability of cysteine cathepsins at neutral pH found in the extracellular matrix.
4. Regulation of Cathepsin K Activity and Stability
Of all papain-like peptidases, cathepsin K has been established as the cathepsin most tightly linked to glycosaminoglycans. It was originally identified as a protease expressed predominantly in osteoclasts [8] and its impaired activity was shown to result in severe bone disorders [42, 45]. Cathepsin K is a collagenase with unique activity among mammalian peptidases [46–48], which is specifically modulated by glycosaminoglycans via allosteric mechanisms [41, 44]. Due to its central role in bone turnover, cathepsin K is currently considered one of the most promising targets for the treatment of osteoporosis [49]. Apart from bone remodeling, cathepsin K is involved in diverse physiological and pathological processes (for a recent review see [10]). It can cleave a number of extracellular substrates, including proteoglycans, to release active glycosaminoglycans [50], which in turn modulate its activity. In cartilage, cathepsin K degrades both type I and type II collagens and thereby contributes to the development of various inflammatory joint diseases [9, 51, 52]. Moreover, cathepsin K has been associated with cardiovascular diseases, obesity, schizophrenia, and cancer [10].
The interaction of cathepsin K with different glycosaminoglycans has been the subject of in-depth investigation. Though several aspects of these interactions remain elusive, accumulated data suggest that the interactions are heterogeneous and diverse and probably involve multiple binding sites on the enzyme. Chondroitin-4-sulfate (C4S) was initially identified as the GAG with the most dramatic effect on cathepsin K [41] while the effects of chondroitin-6-sulfate (C6S), dermatan sulfate (DS), and hyaluronan (HA) were weaker. All tested GAGs increased the stability of cathepsin K over a broad pH range. C4S had the most prominent effect on the degradation of types I and II collagens by cathepsin K [41], whereas its effect on the hydrolysis of a synthetic substrate was virtually identical to that of C6S and DS and resulted in a twofold increase in the values of the specificity constant (k cat/K m). In a later study, keratan sulfate (KS) and C6S were found to have a stimulatory effect on cathepsin K similar to that of C4S, whereas heparin and HS had a limited effect on the collagenolytic activity of cathepsin K [53]. Early experiments have also suggested that complex formation with CS is necessary for the collagenolytic activity of cathepsin K [54]; however, recent findings have shown that type I collagen can also be efficiently degraded in the absence of glycosaminoglycans [55]. Nonetheless, bone-resident GAGs have been shown to potentiate the collagenolytic activity of cathepsin K and endogenous GAG concentrations in bone were sufficient for a maximal effect on cathepsin K activity [55].
The kinetic mechanism of the effect of CS, DS, and heparin (HP) on cathepsin K was also investigated in detail at physiological plasma pH of 7.4. Under these conditions, CS and DS were characterized as nonessential activators with a predominant effect on the affinity for the substrate K m [44]. DS was more effective than CS, which was attributed to its greater flexibility due to fewer intramolecular hydrogen bonds [56]. Intrinsic fluorescence indicated that binding of GAGs affects the conformation of cathepsin K. In contrast to experiments performed at pH 5.5, CS and DS acted as inhibitors of collagen degradation at physiological plasma pH. In contrast, the kinetic mechanism of heparin was biphasic, indicating interaction with two distinct sites on the enzyme. Altogether, heparin was a strong activator of cathepsin K at physiological plasma pH, increasing both its collagenolytic and elastinolytic activities. Moreover, heparin had a strong stabilizing effect on cathepsin K under these conditions, resulting in a more than 5-fold increase in the half-life of the enzyme [44].
5. Structural Basis for the Interaction between Cathepsin K and GAGs
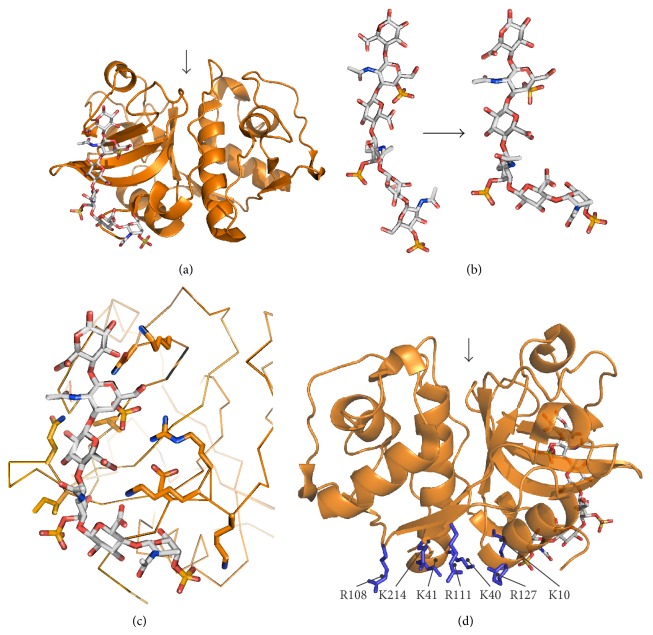
The crystal structure of cathepsin K and C4S revealed the structural basis for the interaction [43]. The binding site is located on the back of cathepsin K and interacts with three disaccharide units of CS in the crystal structure (Figure 2(a)). As usual, the glycosaminoglycan/protein interaction is mediated mostly by electrostatic interactions between the negatively charged GAG chain and positively charged residues on the enzyme. Binding of chondroitin-sulfate does not cause significant conformational changes in cathepsin K compared to the CS-free cathepsin K. Conversely, the CS chain is bent upon binding to cathepsin K (Figure 2(b)). The bulk of the conformational change can be attributed to the interaction with a short helical region Arg8-Lys9-Lys10 which interacts with four negatively charged groups on CS (Figure 2(c)). Further close contacts include Asp6, Ile171, Gln172, Asn190, Lys191, and Leu195 and a few additional water-mediated contacts [43].
Figure 2.
Interactions between human cathepsin K and GAGs. (a) Crystal structure of the cathepsin K/chondroitin-4-sulfate (C4S) complex. The protein is shown in cartoon representation and C4S is shown as sticks. (b) Conformational change in C4S upon binding to cathepsin K. (c) Detailed representation of the interaction in panel (a). C4S is shown as sticks. The backbone of cathepsin K is shown as ribbons and residues that interact with C4S are shown as sticks. (d) Location of the predicted second heparin-binding site in cathepsin K. Positively charged residues proposed to interact with heparin are shown as blue sticks. For orientation, C4S bound at the first binding site is shown as sticks. The position of the active site cleft is marked by an arrow. Coordinates of the cathepsin K/C4S complex were retrieved from the Protein Data Bank under accession code 3C9E. The solution structure of C4S was modeled using data from [14]. All images were created with PyMOL.
The kinetic behavior of DS was analogous to that of CS: it was hence proposed that it interacts with cathepsin K in the same manner as CS [44]. Heparin, on the other hand, was proposed to bind to two sites on cathepsin K according to its kinetic profile. While the first binding site was proposed to be identical with that for CS/DS, the second binding site was predicted on the bottom of the molecule by chemical cross-linking experiments and computational modeling [44]. From a structural perspective, the predicted binding site is a continuation of the CD/DS-binding site and consists of several basic residues (Lys10, Lys40, Lys41, Arg108, Arg111, Arg127, and Lys214) organized in a ring-shaped structure (Figure 2(d)). Kinetic experiments have confirmed that heparin can be bound to both sites at the same time [44]. However, it remains to be determined whether this requires one HP chain that interacts with both sites at the same time or two separate HP chains. This is neither clear for the interaction of other GAGs with the second HP-binding site.
6. Interactions of GAGs with Cathepsins S and B
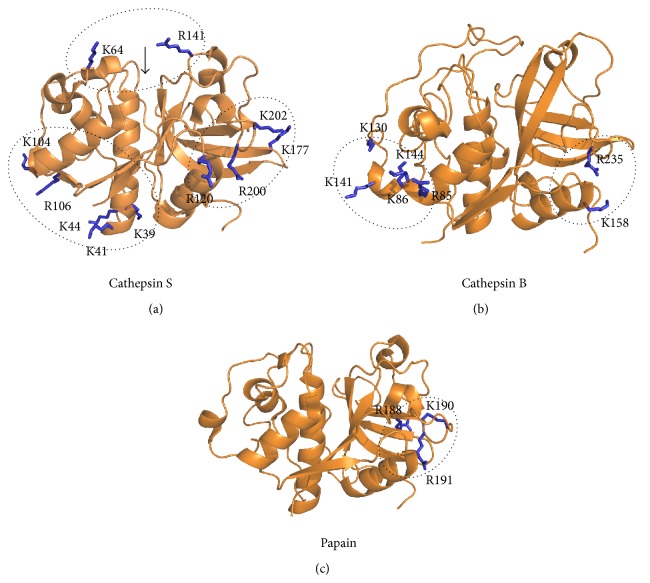
Apart from cathepsin K, two other human papain-like peptidases, cathepsins S and B, have been shown to be regulated by glycosaminoglycans in their mature forms [33, 57]. Cathepsin S, the closest relative of cathepsin K, is unusual among cysteine cathepsins for being stable at neutral pH [58]. Cathepsin S has major physiological roles in antigen-presenting cells as the most important protease in antigen processing [59–61] and was recently found to be regulated by C4S [33]. In contrast to the activation effect observed with cathepsin K, C4S acted as an inhibitor of type IV collagen degradation by cathepsin S. Inhibition was also observed with HS, whereas HP, DS, C6S, and HA slightly increased the proteolytic activity of cathepsin S using type IV collagen as the substrate. C4S, C6S, and HS also inhibited the hydrolysis of Z-Phe-Arg-AMC via a partial, mixed mechanism. Similar to cathepsin K, subtle conformational changes in cathepsin S were observed upon C4S binding by intrinsic fluorescence spectroscopy. Three binding sites for C4S were predicted on cathepsin S by molecular docking (Figure 3(a)). One of the proposed sites is the active site, which, however, is not in agreement with the observed mixed inhibition profile of C4S; the second is located on the bottom right side of the molecule and corresponds to the recently identified allosteric site in cathepsin K [62], while the third is located at the bottom of the molecule and roughly corresponds to the secondary heparin-binding site identified in cathepsin K [44].
Figure 3.
Predicted GAG-binding sites in papain-like peptidases. (a) Three predicted CS-binding sites in cathepsin S. (b) Two predicted HS/HP-binding sites in cathepsin B. (c) The conserved GAG-binding motif in papain. Predicted sites are shown in circles and positively charged residues at each site are shown as blue sticks and labeled. The position of the active site cleft is marked by an arrow. All coordinates were obtained from the Protein Data Bank (accession codes: 1NQC for cathepsin S, 3AI8 for cathepsin B, and 1PPN for papain, resp.). All images were created with PyMOL.
Cathepsin B is unique among cysteine cathepsins for being both an endopeptidase and a peptidyl dipeptidase, depending on the conformation of the occluding loop, a cathepsin B-specific structure that provides a pH-specific switch between both activities [63]. In the lysosome, the low pH restricts the protease to a closed, exopeptidase conformation, whereas the near-neutral pH of the extracellular environment promotes endopeptidase activity of cathepsin B [63]. Extracellular cathepsin B is most commonly associated with cancer and various types of arthritis [64, 65]. The protease localized to the cell surface in several studies and was found to be involved in cell migration under both physiological and pathological conditions [66, 67]. At the molecular level, it was shown to cleave a number of extracellular substrates, including laminin, type IV collagen, and fibronectin [68, 69]. Recently, cathepsin B has also been suggested to be a β-secretase that produces amyloid β peptides in secretory vesicles of neuronal chromaffin cells [70]. However, it has also been shown to degrade amyloid deposits in an animal model [71] and the overall outcome has been suggested to be determined by the balance between cathepsin B and its endogenous inhibitor cystatin C [72].
Binding of HP or HS has been shown to increase the stability of the otherwise unstable enzyme at alkaline pH (8.0), while slightly reducing the activity of the enzyme along the entire pH profile of the enzyme [57]. Computational simulations have predicted that heparin stabilizes the conformation of the molecule under these conditions and have predicted two putative GAG-binding sites (Figure 3(b)), one on each side of the enzyme [73]. The putative binding site in the L domain consists of five basic residues (Arg85, Lys86, Lys130, Lys141, and Lys144), while the one in the R domain contains only two (Lys158 and Arg235). The authors have suggested that the binding site in the R domain has higher affinity for shorter GAG fragments, such as the heparin disaccharide used in their docking simulations, whereas the one in the L domain is likely more relevant for the binding of longer GAG fragments [73].
7. Interactions of GAGs with Other Papain-Like Peptidases
Interestingly, papain has also been shown to interact with GAGs. Despite bearing no physiological relevance, these interactions point to evolutionarily conserved mechanisms of regulation within the family. HP inhibited papain by a hyperbolic mixed mechanism and affected its conformation [40]. A classical heparin-binding consensus sequence was identified in papain, in the form of the sequence 187-Ile-Arg-Ile-Lys-Arg-Gly-192. Structurally, this sequence is located on the right side of the molecule (Figure 3(c)) in a region that lies between both allosteric sites known in cathepsin K.
In addition, a few examples of proteases from protozoan parasites have been described that interact with GAGs, suggesting the possibility of their influence on host-parasite interactions. The cathepsin L homolog brucipain, a crucial virulence factor of the protozoan parasite Trypanosoma brucei, has been found to be allosterically modulated by HS. The effect of HS in this study was subtle and it had the ability to reverse substrate inhibition by a small dipeptide substrate (Z-Phe-Arg-AMC) [74]. A stronger effect of HS was observed for cruzipain from the related parasite Trypanosoma cruzi. In this case, HS was an activator of the peptidase that caused a significant (up to 6-fold) increase in the activity of the peptidase measured with a synthetic substrate. Moreover, HS increased the release of kinin from high molecular weight kininogen by cruzipain in vitro as well as by living trypomastigotes and reduced the inhibitory properties of kininogen towards cruzipain [75]. Similarly, HP was recently shown to modulate the activity of the cathepsin L-like peptidase rCPB2.8 from Leishmania mexicana [76]. In this case, HP and HS, but not CS or DS, inhibited the hydrolysis of Z-Phe-Arg-AMC by a hyperbolic mixed mechanism and affected the conformation of the protein [76]. Altogether these examples show that interactions with GAGs are not restricted to endogenous cysteine cathepsins but can also play diverse roles in host-pathogen interactions and can act either as part of the body's defense against invading pathogens or as factors contributing to the invasive mechanisms of the pathogen.
8. Pharmacological Targeting
Cathepsin K currently represents the most attractive drug target among the cathepsins, although cathepsin S is also a relevant target in diseases associated with elevated immune response, such as bronchial asthma and psoriasis [5, 27]. Several cathepsin K inhibitors are currently under development, which target the active site of the enzyme (collected in [10, 27]). At the moment, the most promising inhibitor is odanacatib (Merck & Co., Inc., Whitehouse Station, NJ, USA), a nitrile warhead-containing inhibitor, highly selective for cathepsin K [77]. Phase III clinical trials for odanacatib have been successfully concluded and applications for approval are expected to be filed soon. If approved, the drug will position itself in the market against other new generation drugs, such as the anti-RANK-ligand antibody denosumab (Amgen, Inc., Thousand Oaks, CA, USA) and the teriparatide, a recombinant form of parathyroid hormone (Eli Lilly and Company, Indianapolis, IN, USA), as well as the well-established bisphosphonates [78]. Targeting the cathepsin K/chondroitin-sulfate interaction would represent an alternative to these treatments. Endogenous chondroitin-sulfate is sufficient to exhibit a maximum activation effect on cathepsin K and its digestion reduces the activity of cathepsin K by 40% [55]. A reduction in bone turnover of this magnitude would likely suffice for the treatment of patients with less severe bone density reduction. An added benefit would be that cathepsin K activity per se as well as the viability and cell count of osteoclasts and osteoblasts would remain undisturbed.
Acknowledgment
The work has been supported by Grants from the Slovene Research Agency (P1-0140 and J1-3602) to Boris Turk.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Rawlings N. D., Barrett A. J., Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Research. 2012;40(1):D343–D350. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rossi A., Deveraux Q., Turk B., Sali A. Comprehensive search for cysteine cathepsins in the human genome. Biological Chemistry. 2004;385(5):363–372. doi: 10.1515/BC.2004.040. [DOI] [PubMed] [Google Scholar]
- 3.Turk V., Stoka V., Vasiljeva O., Renko M., Sun T., Turk B., Turk D. Cysteine cathepsins: from structure, function and regulation to new frontiers. Biochimica et Biophysica Acta—Proteins and Proteomics. 2012;1824(1):68–88. doi: 10.1016/j.bbapap.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Novinec M., Lenarčič B. Papain-like peptidases: Structure, function, and evolution. Biomolecular Concepts. 2013;4(3):287–308. doi: 10.1515/bmc-2012-0054. [DOI] [PubMed] [Google Scholar]
- 5.Vasiljeva O., Reinheckel T., Peters C., Turk D., Turk V., Turk B. Emerging roles of cysteine cathepsins in disease and their potential as drug targets. Current Pharmaceutical Design. 2007;13(4):387–403. doi: 10.2174/138161207780162962. [DOI] [PubMed] [Google Scholar]
- 6.Schechter I., Berger A. On the size of the active site in proteases. I. Papain. Biochemical and Biophysical Research Communications. 1967;27(2):157–162. doi: 10.1016/S0006-291X(67)80055-X. [DOI] [PubMed] [Google Scholar]
- 7.Turk D., Gunčar G., Podobnik M., Turk B. Revised definition of substrate binding sites of papain-like cysteine proteases. Biological Chemistry. 1998;379(2):137–147. doi: 10.1515/bchm.1998.379.2.137. [DOI] [PubMed] [Google Scholar]
- 8.Inaoka T., Bilbe G., Ishibashi O., Tezuka K.-I., Kumegawa M., Kokubo T. Molecular cloning of human cDNA for cathepsin K: novel cysteine proteinase predominantly expressed in bone. Biochemical and Biophysical Research Communications. 1995;206(1):89–96. doi: 10.1006/bbrc.1995.1013. [DOI] [PubMed] [Google Scholar]
- 9.Hou W.-S., Li Z., Gordon R. E., Chan K., Klein M. J., Levy R., Keysser M., Keyszer G., Brömme D. Cathepsin K is a critical protease in synovial fibroblast-mediated collagen degradation. The American Journal of Pathology. 2001;159(6):2167–2177. doi: 10.1016/S0002-9440(10)63068-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Novinec M., Lenarčic B. Cathepsin K: a unique collagenolytic cysteine peptidase. Biological Chemistry. 2013;394(9):1163–1179. doi: 10.1515/hsz-2013-0134. [DOI] [PubMed] [Google Scholar]
- 11.Zavašnik-Bergant T., Turk B. Cysteine cathepsins in the immune response. Tissue Antigens. 2006;67(5):349–355. doi: 10.1111/j.1399-0039.2006.00585.x. [DOI] [PubMed] [Google Scholar]
- 12.Brömme D., Li Z., Barnes M., Mehler E. Human cathepsin V functional expression, tissue distribution, electrostatic surface potential, enzymatic characterization, and chromosomal localization. Biochemistry. 1999;38(8):2377–2385. doi: 10.1021/bi982175f. [DOI] [PubMed] [Google Scholar]
- 13.Stoeckle C., Gouttefangeas C., Hammer M., Weber E., Melms A., Tolosa E. Cathepsin W expressed exclusively in CD8+ T cells and NK cells, is secreted during target cell killing but is not essential for cytotoxicity in human CTLs. Experimental Hematology. 2009;37(2):266–275. doi: 10.1016/j.exphem.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 14.Almond A., Sheehan J. K. Glycosaminoglycan conformation: do aqueous molecular dynamics simulations agree with x-ray fiber diffraction? Glycobiology. 2000;10(3):329–338. doi: 10.1093/glycob/10.3.329. [DOI] [PubMed] [Google Scholar]
- 15.Cygler M., Sivaraman J., Grochulski P., Coulombe R., Storer A. C., Mort J. S. Structure of rat procathepsin B: model for inhibition of cysteine protease activity by the proregion. Structure. 1996;4(4):405–416. doi: 10.1016/S0969-2126(96)00046-9. [DOI] [PubMed] [Google Scholar]
- 16.Coulombe R., Grochulski P., Sivaraman J., Ménard R., Mort J. S., Cygler M. Structure of human procathepsin L reveals the molecular basis of inhibition by the prosegment. EMBO Journal. 1996;15(20):5492–5503. [PMC free article] [PubMed] [Google Scholar]
- 17.Sivaraman J., Lalumière M., Ménard R., Cygler M. Crystal structure of wild-type human procathepsin K. Protein Science. 1999;8(2):283–290. doi: 10.1110/ps.8.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turk D., Podobnik M., Kuhelj R., Dolinar M., Turk V. Crystal structures of human procathepsin B at 3.2 and 3.3 Å resolution reveal an interaction motif between a papain-like cysteine protease and its propeptide. FEBS Letters. 1996;384(3):211–214. doi: 10.1016/0014-5793(96)00309-2. [DOI] [PubMed] [Google Scholar]
- 19.Podobnik M., Kuhelj R., Turk V., Turk D. Crystal structure of the wild-type human procathepsin B at 2.5 Å resolution reveals the native active site of a papain-like cysteine protease zymogen. Journal of Molecular Biology. 1997;271(5):774–788. doi: 10.1006/jmbi.1997.1218. [DOI] [PubMed] [Google Scholar]
- 20.Rozman J., Stojan J., Kuhelj R., Turk V., Turk B. Autocatalytic processing of recombinant human procathepsin B is a bimolecular process. FEBS Letters. 1999;459(3):358–362. doi: 10.1016/S0014-5793(99)01302-2. [DOI] [PubMed] [Google Scholar]
- 21. Rozman Pungerčar J., Caglič D., Sajid M., Dolinar M., Vasiljeva O., Požgan U., Turk D., Bogyo M., Turk V., Turk B. Autocatalytic processing of procathepsin B is triggered by proenzyme activity. FEBS Journal. 2009;276(3):660–668. doi: 10.1111/j.1742-4658.2008.06815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dubin G. Proteinaceous cysteine protease inhibitors. Cellular and Molecular Life Sciences. 2005;62(6):653–669. doi: 10.1007/s00018-004-4445-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turk B., Turk D., Salvesen G. S. Regulating cysteine protease activity: essential role of protease inhibitors as guardians and regulators. Current Pharmaceutical Design. 2002;8(18):1623–1637. doi: 10.2174/1381612023394124. [DOI] [PubMed] [Google Scholar]
- 24.Turk B., Turk V., Turk D. Structural and functional aspects of papain-like cysteine proteinases and their protein inhibitors. Biological Chemistry. 1997;378(3-4):141–150. [PubMed] [Google Scholar]
- 25.Lenarčič B., Bevec T. Thyropins—new structurally related proteinase inhibitors. The Biological Chemistry. 1998;379(2):105–111. [PubMed] [Google Scholar]
- 26.Avila J. L., Convit J. Inhibition of leucocytic lysosomal enzymes by glycosaminoglycans in vitro. Biochemical Journal. 1975;152(1):57–64. doi: 10.1042/bj1520057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fonović M., Turk B. Cysteine cathepsins and extracellular matrix degradation. Biochimica et Biophysica Acta—General Subjects. 2014;1840(8):2560–2570. doi: 10.1016/j.bbagen.2014.03.017. [DOI] [PubMed] [Google Scholar]
- 28.Ishidoh K., Kominami E. Procathepsin L degrades extracellular matrix proteins in the presence of glycosaminoglycans in vitro. Biochemical and Biophysical Research Communications. 1995;217(2):624–631. doi: 10.1006/bbrc.1995.2820. [DOI] [PubMed] [Google Scholar]
- 29.Mason R. W., Massey S. D. Surface activation of pro-cathepsin L. Biochemical and Biophysical Research Communications. 1992;189(3):1659–1666. doi: 10.1016/0006-291X(92)90268-P. [DOI] [PubMed] [Google Scholar]
- 30.Vasiljeva O., Dolinar M., Rozman Pungerčar J., Turk V., Turk B. Recombinant human procathepsin S is capable of autocatalytic processing at neutral pH in the presence of glycosaminoglycans. The FEBS Letters. 2005;579(5):1285–1290. doi: 10.1016/j.febslet.2004.12.093. [DOI] [PubMed] [Google Scholar]
- 31.Caglič D., Rozman Pungerčar J., Pejler G., Turk V., Turk B. Glycosaminoglycans facilitate procathepsin B activation through disruption of propeptide-mature enzyme interactions. The Journal of Biological Chemistry. 2007;282(45):33076–33085. doi: 10.1074/jbc.M705761200. [DOI] [PubMed] [Google Scholar]
- 32.Serveau C., Boulangé A., Lecaille F., Gauthier F., Authié E., Lalmanach G. Procongopain from Trypanosoma congolense is processed at basic pH: an unusual feature among cathepsin L-like cysteine proteases. Biological Chemistry. 2003;384(6):921–927. doi: 10.1515/BC.2003.103. [DOI] [PubMed] [Google Scholar]
- 33.Sage J., Mallèvre F., Barbarin-Costes F., Samsonov S. A., Gehrcke J.-P., Pisabarro M. T., Perrier E., Schnebert S., Roget A., Livache T., Nizard C., Lalmanach G., Lecaille F. Binding of chondroitin 4-sulfate to cathepsin S regulates its enzymatic activity. Biochemistry. 2013;52(37):6487–6498. doi: 10.1021/bi400925g. [DOI] [PubMed] [Google Scholar]
- 34.Brömme D., Nallaseth F. S., Turk B. Production and activation of recombinant papain-like cysteine proteases. Methods. 2004;32(2):199–206. doi: 10.1016/S1046-2023(03)00212-3. [DOI] [PubMed] [Google Scholar]
- 35.Kuhelj R., Dolinar M., Rozman Pungerčar J., Turk V. The preparation of catalytically active human cathepsin B from its precursor expressed in Escherichia coli in the form of inclusion bodies. European Journal of Biochemistry. 1995;229(2):533–539. doi: 10.1111/j.1432-1033.1995.0533k.x. [DOI] [PubMed] [Google Scholar]
- 36.Kopitar G., Dolinar M., Štrukelj B., Pungerčar J., Turk V. Folding and activation of human procathepsin S from inclusion bodies produced in Escherichia coli. European Journal of Biochemistry. 1996;236(2):558–562. doi: 10.1111/j.1432-1033.1996.00558.x. [DOI] [PubMed] [Google Scholar]
- 37.Hallgren J., Spillmann D., Pejler G. Structural requirements and mechanism for heparin-induced activation of a recombinant mouse mast cell tryptase, mouse mast cell protease-6: formation of active tryptase monomers in the presence of low molecular weight heparin. Journal of Biological Chemistry. 2001;276(46):42774–42781. doi: 10.1074/jbc.M105531200. [DOI] [PubMed] [Google Scholar]
- 38.Vanwildemeersch M., Olsson A.-K., Gottfridsson E., Claesson-Welsh L., Lindahl U., Spillmann D. The anti-angiogenic His/Pro-rich fragment of histidine-rich glycoprotein binds to endothelial cell heparan sulfate in a Zn2+-dependent manner. The Journal of Biological Chemistry. 2006;281(15):10298–10304. doi: 10.1074/jbc.M508483200. [DOI] [PubMed] [Google Scholar]
- 39.Fairhead M., Kelly S. M., van der Walle C. F. A heparin binding motif on the pro-domain of human procathepsin L mediates zymogen destabilization and activation. Biochemical and Biophysical Research Communications. 2008;366(3):862–867. doi: 10.1016/j.bbrc.2007.12.062. [DOI] [PubMed] [Google Scholar]
- 40.Almeida P. C., Nantes I. L., Rizzi C. C. A., Júdice W. A. S., Chagas J. R., Juliano L., Nader H. B., Tersariol I. L. S. Cysteine proteinase activity regulation: a possible role of heparin and heparin-like glycosaminoglycans. The Journal of Biological Chemistry. 1999;274(43):30433–30438. doi: 10.1074/jbc.274.43.30433. [DOI] [PubMed] [Google Scholar]
- 41.Li Z., Hou W. S., Brömme D. Collagenolytic activity of cathepsin K is specifically modulated by cartilage-resident chondroitin sulfates. Biochemistry. 2000;39(3):529–536. doi: 10.1021/bi992251u. [DOI] [PubMed] [Google Scholar]
- 42.Gelb B. D., Shi G.-P., Chapman H. A., Desnick R. J. Pycnodysostosis, a lysosomal disease caused by cathepsin K deficiency. Science. 1996;273(5279):1236–1239. doi: 10.1126/science.273.5279.1236. [DOI] [PubMed] [Google Scholar]
- 43.Li Z., Kienetz M., Cherney M. M., James M. N. G., Brömme D. The crystal and molecular structures of a cathepsin K:chondroitin sulfate complex. Journal of Molecular Biology. 2008;383(1):78–91. doi: 10.1016/j.jmb.2008.07.038. [DOI] [PubMed] [Google Scholar]
- 44.Novinec M., Kovačič L., Lenarĉič B., Baici A. Conformational flexibility and allosteric regulation of cathepsin K. Biochemical Journal. 2010;429(2):379–389. doi: 10.1042/BJ20100337. [DOI] [PubMed] [Google Scholar]
- 45.Saftig P., Hunziker E., Wehmeyer O., Jones S., Boyde A., Rommerskirch W., Moritz J. D., Schu P., Von Figura K. Impaired osteoclastic bone resorption leads to osteopetrosis in cathepsin-K-deficient mice. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(23):13453–13458. doi: 10.1073/pnas.95.23.13453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kafienah W., Brömme D., Buttle D. J., Croucher L. J., Hollander A. P. Human cathepsin K cleaves native type I and II collagens at the N-terminal end of the triple helix. Biochemical Journal. 1998;331(part 3):727–732. doi: 10.1042/bj3310727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garnero P., Borel O., Byrjalsen I., Ferreras M., Drake F. H., McQueney M. S., Foged N. T., Delmas P. D., Delaissé J.-M. The collagenolytic activity of cathepsin K is unique among mammalian proteinases. The Journal of Biological Chemistry. 1998;273(48):32347–32352. doi: 10.1074/jbc.273.48.32347. [DOI] [PubMed] [Google Scholar]
- 48.Brömme D., Okamoto K., Wang B. B., Biroc S. Human cathepsin O2, a matrix protein-degrading cysteine protease expressed in osteoclasts: functional expression of human cathepsin O2 in Spodoptera frugiperda and characterization of the enzyme. Journal of Biological Chemistry. 1996;271(4):2126–2132. doi: 10.1074/jbc.271.4.2126. [DOI] [PubMed] [Google Scholar]
- 49.Boonen S., Rosenberg E., Claessens F., Vanderschueren D., Papapoulos S. Inhibition of cathepsin K for treatment of osteoporosis. Current Osteoporosis Reports. 2012;10(1):73–79. doi: 10.1007/s11914-011-0085-9. [DOI] [PubMed] [Google Scholar]
- 50.Hou W. S., Li Z., Büttner F. H., Bartnik E., Brömme D. Cleavage site specificity of cathepsin K toward cartilage proteoglycans and protease complex formation. Biological Chemistry. 2003;384(6):891–897. doi: 10.1515/BC.2003.100. [DOI] [PubMed] [Google Scholar]
- 51.Dejica V. M., Mort J. S., Laverty S., Antoniou J., Zukor D. J., Tanzer M., Poole A. R. Increased type II collagen cleavage by cathepsin K and collagenase activities with aging and osteoarthritis in human articular cartilage. Arthritis Research and Therapy. 2012;14(3, article R113) doi: 10.1186/ar3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dejica V. M., Mort J. S., Laverty S., Percival M. D., Antoniou J., Zukor D. J., Poole A. R. Cleavage of type II collagen by cathepsin K in human osteoarthritic cartilage. The American Journal of Pathology. 2008;173(1):161–169. doi: 10.2353/ajpath.2008.070494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Z., Yasuda Y., Li W., Bogyo M., Katz N., Gordon R. E., Fields G. B., Brömme D. egulation of collagenase activities of human cathepsins by glycosaminoglycans. The Journal of Biological Chemistry. 2004;279(7):5470–5479. doi: 10.1074/jbc.M310349200. [DOI] [PubMed] [Google Scholar]
- 54.Li Z., Hou W.-S., Escalante-Torres C. R., Gelb B. D., Brömme D. Collagenase activity of cathepsin K depends on complex formation with chondroitin sulfate. The Journal of Biological Chemistry. 2002;277(32):28669–28676. doi: 10.1074/jbc.M204004200. [DOI] [PubMed] [Google Scholar]
- 55.Borel O., Gineyts E., Bertholon C., Garnero P. Cathepsin K preferentially solubilizes matured bone matrix. Calcified Tissue International. 2012;91(1):32–39. doi: 10.1007/s00223-012-9604-7. [DOI] [PubMed] [Google Scholar]
- 56.Casu B., Petitou M., Provasoli M., Sinaÿ P. Conformational flexibility: a new concept for explaining binding and biological properties of iduronic acid-containing glycosaminoglycans. Trends in Biochemical Sciences. 1988;13(6):221–225. doi: 10.1016/0968-0004(88)90088-6. [DOI] [PubMed] [Google Scholar]
- 57.Almeida P. C., Nantes I. L., Chagas J. R., Rizzi C. C. A., Faljoni-Alario A., Carmona E., Julianoi L., Nader H. B., Tersariol I. L. S. Cathepsin B activity regulation. Heparin-like glycosaminoglycans protect human cathepsin B from alkaline pH-induced inactivation. Journal of Biological Chemistry. 2001;276(2):944–951. doi: 10.1074/jbc.M003820200. [DOI] [PubMed] [Google Scholar]
- 58.Kirschke H., Wiederanders B., Brömme D., Rinne A. Cathepsin S from bovine spleen. Purification, distribution, intracellular localization and action on proteins. Biochemical Journal. 1989;264(2):467–473. doi: 10.1042/bj2640467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nakagawa T. Y., Brissette W. H., Lira P. D., Griffiths R. J., Petrushova N., Stock J., McNeish J. D., Eastman S. E., Howard E. D., Clarke S. R. M., Rosloniec E. F., Elliott E. A., Rudensky A. Y. Impaired invariant chain degradation and antigen presentation and diminished collagen-induced arthritis in cathepsin S null mice. Immunity. 1999;10(2):207–217. doi: 10.1016/S1074-7613(00)80021-7. [DOI] [PubMed] [Google Scholar]
- 60.Guo-Ping S., Villadangos J. A., Dranoff G., Small C., Lijuan G., Haley K. J., Riese R., Ploegh H. L., Chapman H. A. Cathepsin S required for normal MHC class II peptide loading and germinal center development. Immunity. 1999;10(2):197–206. doi: 10.1016/S1074-7613(00)80020-5. [DOI] [PubMed] [Google Scholar]
- 61.Driessen C., Bryant R. A. R., Lennon-Duménil A. M., Villadangos J. A., Bryant P. W., Shi G.-P., Chapman H. A., Ploegh H. L. Cathepsin S controls the trafficking and maturation of MHC class II molecules in dendritic cells. Journal of Cell Biology. 1999;147(4):775–790. doi: 10.1083/jcb.147.4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Novinec M., Korenč M., Caflisch A., Ranganathan R., Lenarčič B., Baici A. A novel allosteric mechanism in the cysteine peptidase cathepsin K discovered by computational methods. Nature Communications. 2014;5 doi: 10.1038/ncomms4287.3287 [DOI] [PubMed] [Google Scholar]
- 63.Musil D., Zučič D., Turk D., Engh R. A., Mayr I., Huber R., Popovič T., Turk Towatari V. T., Katunuma N., Bode W. The refined 2.15 Å X-ray crystal structure of human liver cathepsin B: the structural basis for its specificity. EMBO Journal. 1991;10(9):2321–2330. doi: 10.1002/j.1460-2075.1991.tb07771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mohamed M. M., Sloane B. F. Cysteine cathepsins: multifunctional enzymes in cancer. Nature Reviews Cancer. 2006;6(10):764–775. doi: 10.1038/nrc1949. [DOI] [PubMed] [Google Scholar]
- 65.Baici A., Lang A., Zwicky R., Müntener K. Cathepsin B in osteoarthritis: uncontrolled proteolysis in the wrong place. Seminars in Arthritis and Rheumatism. 2004;34(2):24–28. doi: 10.1016/j.semarthrit.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 66.Cavallo-Medved D., Mai J., Dosescu J., Sameni M., Sloane B. F. Caveolin-1 mediates the expression and localization of cathepsin B, pro-urokinase plasminogen activator and their cell-surface receptors in human colorectal carcinoma cells. Journal of Cell Science. 2005;118(7):1493–1503. doi: 10.1242/jcs.02278. [DOI] [PubMed] [Google Scholar]
- 67.Cavallo-Medved D., Rudy D., Blum G., Bogyo M., Caglic D., Sloane B. F. Live-cell imaging demonstrates extracellular matrix degradation in association with active cathepsin B in caveolae of endothelial cells during tube formation. Experimental Cell Research. 2009;315(7):1234–1246. doi: 10.1016/j.yexcr.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lah T. T., Buck M. R., Honn K. V., Crissman J. D., Rao N. C., Liotta L. A., Sloane B. F. Degradation of laminin by human tumor cathepsin B. Clinical & Experimental Metastasis. 1989;7(4):461–468. doi: 10.1007/BF01753666. [DOI] [PubMed] [Google Scholar]
- 69.Buck M. R., Karustis D. G., Day N. A., Honn K. V., Sloane B. F. Degradation of extracellular-matrix proteins by human cathepsin B from normal and tumour tissues. Biochemical Journal. 1992;282(1):273–278. doi: 10.1042/bj2820273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hook V., Toneff T., Bogyo M., Greenbaum D., Medzihradszky K. F., Neveu J., Lane W., Hook G., Reisine T. Inhibition of cathepsin B reduces beta-amyloid production in regulated secretory vesicles of neuronal chromaffin cells: evidence for cathepsin B as a candidate beta-secretase of Alzheimer's disease. Biological Chemistry. 2005;386(12):931–940. doi: 10.1515/BC.2005.151. [DOI] [PubMed] [Google Scholar]
- 71.Wang C., Sun B., Zhou Y., Grubb A., Gan L. Cathepsin B degrades amyloid-β in mice expressing wild-type human amyloid precursor protein. The Journal of Biological Chemistry. 2012;287(47):39834–39841. doi: 10.1074/jbc.M112.371641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun B., Zhou Y., Halabisky B., Lo I., Cho S.-H., Mueller-Steiner S., Devidze N., Wang X., Grubb A., Gan L. Cystatin C-cathepsin B axis regulates amyloid beta levels and associated neuronal deficits in an animal model of Alzheimer's disease. Neuron. 2008;60(2):247–257. doi: 10.1016/j.neuron.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Costa M. G. S., Batista P. R., Shida C. S., Robert C. H., Bisch P. M., Pascutti P. G. How does heparin prevent the pH inactivation of cathepsin B? Allosteric mechanism elucidated by docking and molecular dynamics. BMC Genomics. 2010;11(supplement 5, article S5) doi: 10.1186/1471-2164-11-S5-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Costa T. F. R., Reis F. C. G. D., Lima A. P. C. A. Substrate inhibition and allosteric regulation by heparan sulfate of Trypanosoma brucei cathepsin L. Biochimica et Biophysica Acta. 2012;1824(3):493–501. doi: 10.1016/j.bbapap.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 75.Lima A. P., Almeida P. C., Tersariol I. L., Schmitz V., Schmaier A. H., Juliano L., Hirata I. Y., Müller-Esterl W., Chagas J. R., Scharfstein J. Heparan sulfate modulates kinin release by Trypanosoma cruzi through the activity of cruzipain. The Journal of Biological Chemistry. 2002;277(8):5875–5881. doi: 10.1074/jbc.M108518200. [DOI] [PubMed] [Google Scholar]
- 76.Judice W. A. S., Manfredi M. A., Souza G. P., Sansevero T. M., Almeida P. C., Shida C. S., Gesteira T. F., Juliano L., Westrop G. D., Sanderson S. J., Coombs G. H., Tersariol I. L. S. Heparin modulates the endopeptidase activity of Leishmania mexicana cysteine protease cathepsin L-like rCPB2.8. PLoS ONE. 2013;8(11) doi: 10.1371/journal.pone.0080153.e80153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gauthier J. Y., Chauret N., Cromlish W., Desmarais S., Duong L. T., Falgueyret J.-P., Kimmel D. B., Lamontagne S., Léger S., LeRiche T., Li C. S., Massé F., McKay D. J., Nicoll-Griffith D. A., Oballa R. M., Palmer J. T., Percival M. D., Riendeau D., Robichaud J., Rodan G. A., Rodan S. B., Seto C., Thérien M., Truong V.-L., Venuti M. C., Wesolowski G., Young R. N., Zamboni R., Black W. C. The discovery of odanacatib (MK-0822), a selective inhibitor of cathepsin K. Bioorganic and Medicinal Chemistry Letters. 2008;18(3):923–928. doi: 10.1016/j.bmcl.2007.12.047. [DOI] [PubMed] [Google Scholar]
- 78.Lippuner K. The future of osteoporosis treatment—a research update. Swiss Medical Weekly. 2012;142 doi: 10.4414/smw.2012.13624.w13624 [DOI] [PubMed] [Google Scholar]