Abstract
A simple method to select disomic (N + 1) strains that should be applicable for almost any chromosome in Saccharomyces cerevisiae is presented. A diploid heterozygous for a KanMX knock-out mutation in an essential gene is sporulated and viable geneticin (G418)-resistant colonies selected. Disomic products of a missegregation or non-disjunction event containing a copy of both the wild-type essential gene and its complementary KanMX knock-out allele make up most of the viable colonies. This method has been used to isolate disomic haploids for a variety of chromosomes. It is appropriately named MARV (for missegregation-associated restoration of viability) and is easily adaptable to virtually any strain.
Keywords: Yeast, aneuploid
Introduction
Disomic strains, haploids containing an extra copy of one chromosome (N + 1), have proved useful in genetic mapping, investigating chromosome stability, isolating recombination mutants and studying aneuploid physiology (Ajimura et al., 1993; Campbell et al., 1981; Mortimer and Hawthorne, 1973; Torres et al., 2007). However, disomic strain isolation can be laborious, involving multiple crosses, sporulation, dissection of many asci and selection of candidate strains, followed by time-consuming screenings that might also entail additional ascus dissections. To facilitate the isolation of disomic strains for virtually any chromosome in any strain background, a method was developed that involves only two polymerase chain reaction (PCR)-based DNA transformations, mating, sporulation and a single selection for disomic candidates.
The method involves the construction and sporulation of a starter diploid strain containing a heterozygous essential gene (ESG) inactivated with KanMX (esg : KanMX/ESG), which confers resistance to geneticin (G418) (Wach et al., 1994). When this strain is sporulated and chromosomes have undergone proper disjunction, all spores are non-viable on G418-containing medium. Missegregation or non-disjunction of the KanMX-containing chromosome or its homologue produces disomic spores that contain both esg::KanMX and the essential gene (ESG) and are viable on G418-containing medium (Figure1A). In addition, a heterozygous can1::STE2pr-HIS3 gene is employed to facilitate selection of haploid canavanine-resistant (CanR) meiotic products using random spores (Tong and Boone, 2007). As di- some production using this method involves chromosome non-disjunction or missegregation, the method has been aptly termed missegregation-associated restoration of viability (MARV).
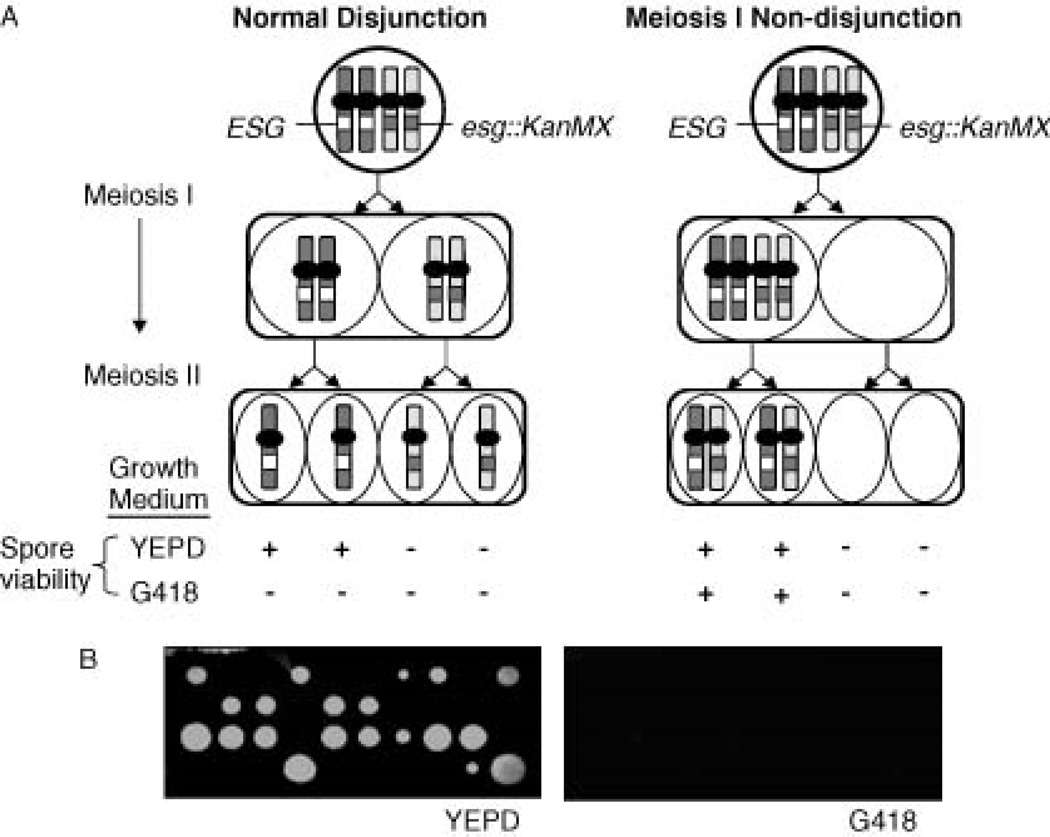
Figure 1.
(A) Meiotic disjunction and non-disjunction of a heterozygous KanMX insertion mutation in an essential gene. The dominant selectable markerKanMX (G418 resistance) inactivates an essential gene (ESG) in a diploid (esg::KanMX). Normal meiosis produces 2 : 2 (esg::KanMX : ESG) segregation of spore viability on YEPD media and 0 : 4 segregation of spore viability on G418-containing media. Meiosis I non-disjunction results in co-segregation of ESG and esg::KanMX producing two disomic spores that are viable on G418 medium. (B) Asci from starter diploid strain DZ11 that were dissected onto YEPD medium show normal chromosome I disjunction: 2 : 2 segregation of viability and 0 : 4 segregation of viability when replica-plated onto G418-containing media
Materials and Methods
Yeast cell growth, sporulation and ascus dissection
All strains were maintained, grown and sporulated on standard media and asci were dissected and analysed as previously described (Sherman et al., 1986).
Yeast transformation and DNA analysis
DNA molecules were introduced into yeast by one-step gene replacement (Rothstein, 1983a). Intact chromosomes were separated by pulsed-field gel electrophoresis, as previously described (Guerra and Kaback, 1999). Integrative transformation was confirmed by DNA blot hybridization (Southern, 1975) and colony polymerase chain reaction (PCR; Liang and Richardson, 1992). Gels were stained with ethidium bromide and fluorescent gel bands were quantitated using Image Quant 5.2 (Molecular Dynamics, University of Virginia, USA).
Selection of disomic strains
Appropriately marked diploids were sporulated on 2% KAc plates for 5 days at 30 °C. The cells were harvested using a sterile spatula and suspended in 1 ml 0.05 mg/ml Zymolyase 20-T (MP Biomedical, Solon, OH, USA) dissolved in sterile distilled water and incubated for 3 h at 30°C on a shaker. The treated spores were then sonicated for 60 s using a Microson ultrasonic cell disruptor (Misonic Inc., Farmingdale, NY, USA) set at 100 W, and plated on His−Arg− synthetic dropout medium containing 0.8 mg/ml G418 (MP Biomedical) and 60 µg/ml canavanine (Sigma-Aldrich, St. Louis, MO, USA) and incubated for 2 days at 30 °C.
Results
To construct the strains used in all studies presented here, PCR-amplified can1::STE2pr-HIS3 was introduced into a MATa haploid from which a bona fide CanRHis+ transformant was selected and crossed to a MATα strain to produce starter diploid DZ10 (genotype MATa/MATα, can1::STE2pr- HIS3/CAN1, his3-11,15/his3-11, 15, ade1/ADE1). Next, a centromere-linked essential gene from the desired chromosome was chosen and esg::KanMX copies of it PCR-amplified from the appropriate yeast gene knock-out strain (Winzeler et al., 1999). The PCR product was introduced into strain DZ10 and a bona fide G418-resistance (G418R) transformant selected (Figure 2). To isolate disomic strains, the starter diploid was sporulated and the resultant asci treated with zymolyase, which kills most unsporulated cells (Herman and Rine, 1997). Spores were then dissociated by sonication and plated on synthetic His−Arg− medium containing G418 and canavanine and colonies arising after 2 days analysed.
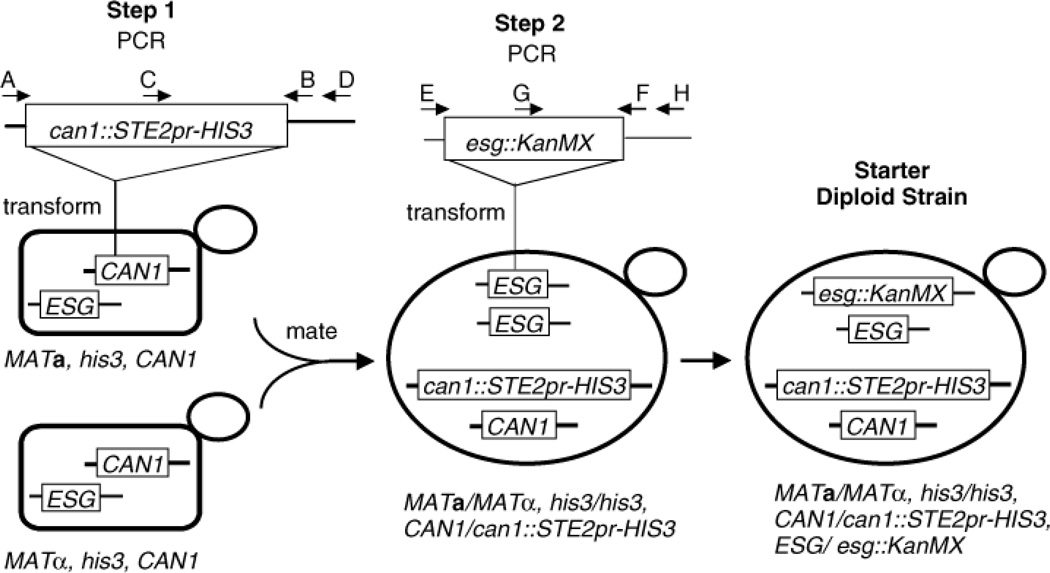
Figure 2.
Construction of strains used to generate disomes. In Step 1, PCR-amplified can1::STE2pr-HIS3 templated from S. cerevisiae strain Y7029 (courtesy of Charles Boone), using primers A (5′-ATCAGGGAATCCCTTTTTGC) and B (5′- ACTCCGTCTGCTTTCTTTTCG) was integrated in place of CAN1 in strain DZ3-4A by one-step gene replacement. Proper integration in His+CanR transformants was verified by colony PCR, using primers C (5′-GAACTTGTTTTGCAAAGGGA) and D (5′- AAAGAGTTCTGCCCTTGGCTT) and a transformant was mated as shown. In Step 2, PCR-amplified esg::KanMX for the noted chromosome (efb1::KanMX, chromosome I; sqt1::KanMX, chromosome IX; mpp10::KanMX, chromosome X; and rlp7::KanMX, chromosome XIV) templated from Yeast Gene Knock-out Strains 20404, 22348, 26799 and 25330 (Open Biosystems, Huntsville, AL, USA), using primers E and F (see below), was integrated in place of one copy of the wild-type ESG by one-step gene replacement and G418R transformants selected. Integration was verified by colony PCR, using primers G and H (sequence listed below). PCR primers used: E, (EFB1, 5′-TTCCGCCATTTTCAACCA; SQT1, 5′- TGCTGGGTTTAAAACCCCAT; MPP10, 5′-TTCCACATCTCGTCATGAAT; RLP7, 5′- TTCATCACGCAGTACGGATT). F, (EFB1, 5′-ACACCGCGTATTTTCGACAA; SQT1, 5′- ATCCCCAAAGAAC TTACCGGA; MPP10, 5′-TTCCACATCTCGTCATGAAT; RLP7, 5′- AAGCGAGGAAAAAAAGAGGC). G, (5′-TGATTTT GATGACGAGCGTAAT). H, (EFB1, 5′- TGTCCTTTTTCACGCAGGTA; SQT1, 5′-CCAACGGAACAGCCTTCAAA; MPP10, 5′— CAGATCTTGCTGAAAGTGACG; RLP7, 5′-TTTGCTGATATATGGCCTTGA)
Strains disomic for chromosome I, which were expected to be obtained easily (Tauro et al., 1968), were the first isolated using diploid strain DZ11, which contains the heterozygous chromosome I essential gene mutation efb1::KanMX/EFB1 (Figure 2). Colony PCR (not shown) and blot hybridization of pulsed-field gel electrophoresis (PFGE)-separated chromosomes confirmed proper integration of efb1::KanMXon chromosome I (Figure 3A). In addition, ascus dissection demonstrated 2 : 2 segregation of spore viability on YEPD medium, where all viable spores were G418-sensitive (Figure 1B). DZ11 is also heterozygous for ADE1, which is tightly linked to EFB1, and all viable spores were Ade+, confirming this linkage in the transformant shown. Microscopic observation of YEPD dissection plates showed that non-viable spores did not produce microcolonies.
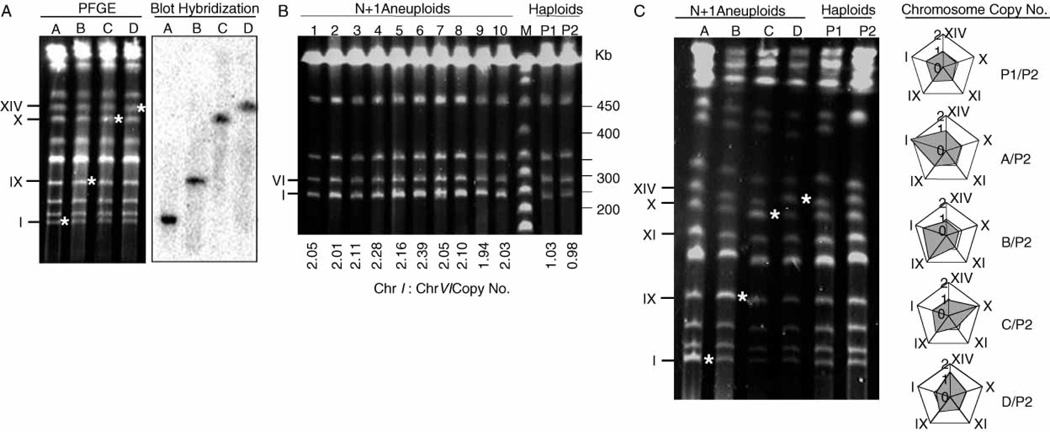
Figure 3.
PFGE karyotypes of marked diploids, haploid parents and disomic strains selected by MARV. (A) Proper localization of the KanMX marker in heterozygous esg::KanMX/ESG starter diploids for chromosomes I, IX, X or XIV. Intact chromosomes were separated using 60 s pulses and stained with ethidium bromide (left). The gel was blotted and hybridized to 32P multi-prime™(GE Healthcare, Piscataway, NJ, USA) -labelled KanMX DNA probe (right). Probe DNA was amplified from plasmid PJT, using forward 5′-TCAGGTGCGACAATCTATCGAT and reverse 5′- CTCACCGAGGCAGTTCCATAG primer sequences. Lane A, strain DZ11 (Chr. I); lane B, strain DZ12 (Chr. IX); lane C, strain DZ13 (Chr. X); lane D, strain DZ14 (Chr. XIV). Asterisks denote esg::KanMXtargeted chromosomes. (B) PFGE analysis of Chr. I disomic strains. Intact chromosomes from G418RCanRHis+ spore colonies were separated using 25 s pulses and the fluorescent band intensities quantitated. Chr. I copy number shown below each lane was determined by comparing the relative band intensities of Chr. I and VI within the same strain and correcting for the size difference between the two chromosomes. Lanes 1–10, MARV-selected Chr. I disomes; lane M, Lambda Ladder PFG Marker (New England BioLabs, Ipswich, MA, USA); lanes P1 and P2, haploid parents of starter diploid DZ10, showing euploid karyotypes. (C) PFGE analysis of Chr. I, IX, X and XIV disomic strains. Intact chromosomes from G418RCanRHis+ spore colonies were separated using 60 s pulses and the fluorescent band intensities quantitated. Lane A, Chr. I; lane B, Chr. IX; lane C, Chr. X; lane D, Chr. XIV. P1 and P2 are the same as in lane B. Determination of chromosome copy number: for each aneuploid (lanes A–D), band intensities of five chromomes (I, IX, XI, X, XIV) were quantitated and the results normalized against the corresponding band intensities in the haploid parent control (lane P2). Band intensities of the two haploid parents (P1 and P2) were similarly compared to ensure euploidy P1/P2. The adjacent Radar chart constructed using Excel (Microsoft, Redmond, WA, USA) shows the ratio of each quantitated chromosome for aneuploids (A–D) and haploid (P1) with respect to the haploid control (P2).
To isolate chromosome I disomes, ca. 106 spores were plated, producing ca. 200 G418RCanRHis+ colonies. 10 colonies were randomly selected and all were MATa and none sporulated. All contained a disomic copy of chromosome I, as demonstrated by PFGE, which showed an increased intensity of the chromosome I band (Figure 3B). PFGE using 60 s pulses indicated that all other resolved chromosomes were present at ca. 1 copy/cell (Figure 3C). Disomic haploids for chromosome I were also isolated with similar frequency, using three other essential gene knock-out mutations, cdc24::KanMX, pta1::KanMX and tfc3::KanMX.
To determine whether this method could be used to isolate strains disomic for other chromosomes, diploid strains DZ12, DZ13 and DZ14, each heterozygous for an essential gene KanMX knock-out mutation on chromosomes IX, X and XIV, respectively, were constructed. Colony PCR (not shown) and blot hybridization of PFGE-separated chromosomes confirmed proper integration of esg::KanMX constructs (Figure 3A). Dissection of ca. 20 asci from each showed 2 : 2 segregation for spore viability on YEPD, where all viable spores were G418-sensitive (not shown). To isolate disomes, diploids were sporulated and treated as described above. All G418RCanRHis+ colonies tested were MATa and did not sporulate. Singly disomic strains were found for each chromosome, as demonstrated by PFGE (Figure 3C). 90% (9/10) of DZ12 spores were singly disomic for chromosome IX, 50% (4/8) of DZ13 spores were singly disomic for chromosome X and 22% (2/9) of DZ14 spores were singly disomic for chromosome XIV. Remaining colonies were either disomic for several chromosomes or homozygous MATa diploids. Based on these studies, every essential gene tried produced disomic products, making it likely that most essential genes in the yeast genome knock-out collection can be used. However, it should be noted that this method cannot be employed as described above for chromosomes III and V, which contain MATand CAN1, respectively.
Discussion
A new method called MARV was developed for isolating haploids that are disomic for almost any chromosome. Since this method involves only two transformations, sporulation and selection, it is quick and easily adaptable to virtually any strain. While the method described excludes chromosomes III and V, transplanting MATa to a different chromosome (Wu et al., 1997) should enable selection of chromosome III disomes, while substituting lyp1::MFα1pr-LEU2 (Tong and Boone, 2006) for can1::STE2pr-HIS3 and selecting for thialysine rather than canavanine resistance should enable selection of chromosome V disomes.
Disomic haploids can arise due to non- disjunction during mitosis, meiosis I or meiosis II. It appears that most of the disomes isolated here were from meiosis I non-disjunction. Selection of disomes that arose during meiosis II requires a prior crossover between the centromere and the heterozygous esg::KanMX gene. Most of the heterozygous esg::KanMX genes used here were tightly linked to centromeres, limiting the frequency of disomes that arose during meiosis II. When either cdc24::KanMX or pta1::KanMX which are not tightly linked to the chromosome I centromere was used, the frequency of disome isolation increased by ~30%, a fraction consistent with the contribution of additional meiosis II non-disjunction events (data not shown). In addition, mitotic non-disjunction is too rare in yeast to account for the frequency of disomic colonies isolated (Chan and Botstein, 1993). Accordingly, meiosis I non-disjunction is likely to be responsible for most of the disomes selected using this method.
Strains isolated for the two smaller chromosomes tested were almost always singly disomic. In contrast, single disomes for the two larger chromosomes were not found 100% of the time and required screening of multiple colonies. Furthermore, it appears that the overall frequency of disome isolation decreased as chromosome size increased. This observation is in agreement with prior results suggesting that small chromosomes segregate less efficiently during meiosis (Goldway et al., 1993).
In summary, MARV has been shown to be useful for isolating haploids that are singly disomic for a desired chromosome. It appears to work using many different essential genes and indeed might be useful in genomic screens using all essential genes. Once stable disomes are isolated, they can be used as is or the esg::KanMX mutation can be removed, using crosses or DNA transformation. This method should be applicable to a wide variety of studies where disomic strains are required.
Acknowledgements
The authors are grateful to Arnold Barton and the laboratory of Carol Newlon for all their help and suggestions. This work was supported by the National Institutes of Health under award number: T32HL007572. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health; and the National Science Foundation.
References
- Ajimura M, Leem S, Ogawa H. Identification of new genes required for meiotic recombination in Saccharomyces cerevisiae. Genetics. 1993;133(1):51–66. doi: 10.1093/genetics/133.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell D, Doctor J, Feuersanger J, Doolittle M. Differential mitotic stability of yeast disomes derived from triploid meiosis. Genetics. 1981;98(2):239–255. doi: 10.1093/genetics/98.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S, Botstein D. Isolation and characterization of chromosome-gain and increase-in-ploidy mutants in yeast. Genetics. 1993;135(3):667–691. doi: 10.1093/genetics/135.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldway M, Arbel T, Simchen G. Meiotic non-disjunction and recombination of chromosome III and homologous fragments in Saccharomyces cerevisiae. Genetics. 1993;133(2):149–158. doi: 10.1093/genetics/133.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra CE, Kaback DB. The role of Centromere Alignment in Meiosis I Segregation of Homologous Chromosomes in Saccharomyces cerevisae. Genetics. 1999;153:1547–1560. doi: 10.1093/genetics/153.4.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman P, Rine J. Yeast spore germination: a requirement for Ras protein activity during re-entry into the cell cycle. EMBO J. 1997;16(20):6171–6181. doi: 10.1093/emboj/16.20.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q, Richardson T. A simple and rapid method for screening transformant yeast colonies using PCR. BioTechniques. 1992;13:730–732. 735. [PubMed] [Google Scholar]
- Mortimer R, Hawthorne D. Genetic mapping in Saccharomyces IV. Mapping of temperature-sensitive genes and use of disomic strains in localizing genes. Genetics. 1973;74(1):33–54. doi: 10.1093/genetics/74.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sherman F, Fink G, Hicks J. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- Southern E. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;3:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Tauro P, Rodarte-Y-Ramon U, McKey T, Mortimer R. Donner Laboratory Semiannual Report. Berkeley, CA: Lawrence Radioation Laboratory; 1968. Isolation of diomics in Saccharomyces cerevisiae; pp. 19–24. [Google Scholar]
- Tong A, Boone C. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol Biol. 2006;313:171–192. doi: 10.1385/1-59259-958-3:171. [DOI] [PubMed] [Google Scholar]
- Tong A, Boone C. High-throughput strain construction and systematic synthetic lethal screening in Saccharomyces cerevisiae. Methods Microbiol. 2007;36 (in press). [Google Scholar]
- Torres E, Sokolsky T, Tucker C, et al. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007a;317(5840):916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Poehlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Winzeler E, Shoemaker D, Astromoff A, et al. Functional characterization of the S.cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285(5429):901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Wu X, Wu C, Haber J. Rules of donor preference in Saccharomycesmating-type gene switching revealed by a competition assay involving two types of recombination. Genetics. 1997;147(2):399–407. doi: 10.1093/genetics/147.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]