Abstract
“Time-lapse markers,” which are defined by time-lapse imaging and correlated with clinical outcomes, may provide embryologists with new opportunities for improving embryo selection. This article provides an overview of noninvasive biomarkers defined by time-lapse imaging studies. In addition to comprehensively reviewing the discovery of each time-lapse marker, it focuses on the criteria necessary for their successful integration into clinical practice, including [1] statistical and biological significance, [2] validation through prospective clinical studies, and [3] development of reliable technology to measure and quantify the time-lapse marker. Because manual analysis of time-lapse images is labor intensive and limits the practical use of the image data in the clinic, automated image analysis software platforms may contribute substantially to improvements in embryo selection accuracy. Ultimately, time-lapse markers that are based on a foundation of basic research, validated through prospective clinical studies, and enabled by a reliable quantification technology may improve IVF success rates, encourage broader adoption of single-embryo transfer, and reduce the risks associated with multiple gestation pregnancies.
Keywords: Biomarker, embryo selection, noninvasive, prediction, time-lapse imaging
Single-embryo transfer (SET) is the preferred practice in in vitro fertilization (IVF) treatment today to reduce the risk for adverse outcomes associated with multiple gestation pregnancy (1). However, to improve the pregnancy rate for SET, embryologists need reliable biomarkers to aid their selection of embryos with the highest developmental potential. Biomarkers identified by time-lapse imaging have been under investigation for use in clinical embryo selection and have unique potential advantages. Derived from continuous monitoring of human embryos, time-lapse markers are inherently non-invasive. Further, several reports have shown promising correlations between time-lapse markers and embryo development, embryo quality, and implantation potential (2–14).
Although establishing correlations between biomarkers and clinical outcomes is an important first step in the discovery of new biomarkers, successful application to clinical embryo selection requires additional criteria. First, the correlation between a biomarker and outcome should be statistically significant, reproducible, and preferably based on sound science. Second, the biomarker and its detection should be validated for safety, efficacy, and practical utility through well-designed clinical trials, preferably prospective, randomized, and controlled trials. Third, the biomarker should be quantified using technology that is reliable and compatible with generic clinical settings and workflow.
Here, we review the current status of candidate time-lapse markers identified for human embryos. We also discuss their practical application to clinical embryo selection using the criteria we have listed. Biomarkers that are able to meet all the criteria may improve embryo selection, increase pregnancy rates, and ultimately enable broader practice of SET.
BIOMARKERS IDENTIFIED WITH TIME-LAPSE IMAGING
Time-Lapse Imaging of Human Embryos
For more than a decade, time-lapse imaging technologies have allowed researchers to capture embryo images at defined intervals over time. Payne et al. (2) used a laboratory-made time-lapse system with a videocassette recorder to document the exact sequence and timing of events occurring every 1 minute for 17 to 20 hours after intracytoplasmic sperm injection (ICSI), including the extrusion of the second polar body and the appearance of pronuclei (PN), for 50 human oocytes. Cytoplasmic flares and periodic waves of granulation within the ooplasm were also recorded. The investigators noted that good quality embryos appeared to arise from oocytes with more uniform timing from ICSI to PN abuttal, and moreover tended to exhibit cytoplasmic waves at slightly longer periodicity. Subsequently, Mio et al. (15) recorded the development of 286 human embryos every 2 minutes for 2 to 5 days, capturing the sequence and timing of events during the fertilization process, the development of a 2-cell stage embryo into a hatched blastocyst, and the splitting of the inner cell mass. Forty-six of the imaged embryos were transferred to patients and resulted in four healthy live births. In another study by Pribenszky et al. (4), five embryos from a single patient were imaged for 5 days at 10-minute intervals, and a single live birth after blastocyst transfer was reported.
Time-Lapse Markers Correlated to Embryo Development
Beyond human embryo time-lapse observations, researchers have statistically correlated embryo development outcomes with diverse dynamic embryo phenomena. Lemmen et al. (3) evaluated 102 2PN embryos at 5-minute intervals for 20 to 24 hours after fertilization and found that embryos with a higher number of blastomeres on day 2 tended to have similar morphological characteristics—including early disappearance of PN, early first cleavage, and early appearance of nuclei after the first cleavage. It is interesting that the embryos that implanted successfully appeared to have more synchronous nuclei appearances in both blastomeres after first cleavage. These observations were for a small sample size (19 transferred embryos resulting in six pregnancies), and measurable biomarkers that could quantifiably predict embryo development were not described.
In a later study, Wong et al. (5) combined imaging and high-throughput gene expression technologies to extract the first time-lapse markers with defined predictive ability. In their study, a total of 242 frozen human embryos were thawed and cultured for 5 days while images were taken at 5-minute intervals, and subsets of embryos for single-cell or whole embryo gene expression analysis were collected every 24 hours. Among several parameters evaluated, three time-lapse markers with distinct time windows were demonstrated to predict blastocyst formation by the 4-cell stage with high sensitivity and specificity. The time-lapse markers and their timings for blastocyst formation were: P1 = duration of the first cytokinesis 14.3 ± 6.0 minutes; P2 = time between the first and second mitosis (or 2- to 3-cell stage) 11.1 ± 2.2 hours; and P3 = time between or synchrony of the second and third mitosis (or 3- to 4-cell stage) 1.0 ± 1.6 hours.
Several groups have extended the search for predictive time-lapse markers by evaluating further stages of developmental success or adding new potential parameters. In 2011, Meseguer et al. (6) reported on a set of 522 embryos transferred on day 3, imaged for 64 hours at 15-minute intervals, including 247 embryos with known implantation. The results of the study showed that implantation success was strongly correlated with the reported timings for two time-lapse markers described earlier, namely, cc2 (or P2 = time between the first and second mitosis, or the 2- to 3-cell stage) ≤ 11.9 hours, and s2 (or P3 = time between or synchrony of the second and third mitosis, or the 3- to 4-cell stage) ≤ 0.76 hours. In a second study from the same group, Cruz et al. (9) imaged 834 embryos at 20-minute intervals for 120 hours and confirmed that cc2 (P2) and s2 (P3) were statistically significant indicators of blastocyst development. The P1 marker was not evaluated in these two studies, possibly because the imaging at 15- to 20-minute intervals was an insufficient frequency to capture the cytokinesis event, which on average lasts approximately 15 minutes (5). The researchers also reported strong outcome correlations for an additional time-lapse marker, the time between ICSI and the 5-cell embryo stage. Together with P2 and/or P3 as reported by Wong and colleagues, the parameter was correlated with development to good-quality blastocysts and implantation (6, 9).
Several reports from separate clinics have further confirmed that statistically significant differences between time-lapse markers could be observed for embryos that develop to blastocysts of varying quality. Cruz et al. (9) and Hashimoto et al. (7) both reported that better quality blastocysts develop with significantly shorter times for synchrony of the second-generation cell divisions (P3, or 3- to 4 cell stage). In addition to P3, Cruz et al. (9) additionally described the time from ICSI to the 5-cell stage and the time from ICSI to the morula stage as indicative of good blastocyst quality; Hashimoto et al. (7) highlighted the third generation of cell division (5- to-8 cell stage). Dal Canto et al. (11) also found that embryos that developed into expanded blastocysts have significantly shorter P2, P3, 4– to 8-cell–stage duration, and 5- to 8-cell–stage duration than those that developed into nonexpanded blastocysts, after performing systematic measurements for cell-cycle timings during embryo-cleavage divisions. Finally, in another recent, retrospective analysis of 180 embryo time-lapse movies, Hlinka et al. (10) used cell-cycle timings to grade embryos and predict pregnancy. Again, the durations of interphase 2 (or P2, 2- to-3-cell stage), cleavage 2 (or P3, 3- to-4-cell stage), and interphase 3 (4- to 5-cell stage), along with the durations of cleavage 3 (5- to 8-cell stage), interphase 4 (8- to 9-cell stage), and cleavage 4 (9-to 16 cell stage), were determined to be useful selection criteria for embryo outcomes.
Besides studies focusing on defining the parameter range for selecting the best embryos, two publications used time-lapse markers to exclude embryos with undesired outcomes. Azzarello et al. (13) examined the timing of PN breakdown relative to ICSI in 159 transferred embryos, and reported that no embryo with PN breakdown earlier than 20.75 hours resulted in a live birth. Elsewhere, Rubio et al. (12) found that embryos with direct cleavage of 2- to 3-cells (termed “DC2–3”, but identical to P2 <5 hours) have very low implantation rate (1.2%). Based on their findings, the authors suggested PN breakdown or DC2–3 (P2) as novel exclusion criteria for embryo selection.
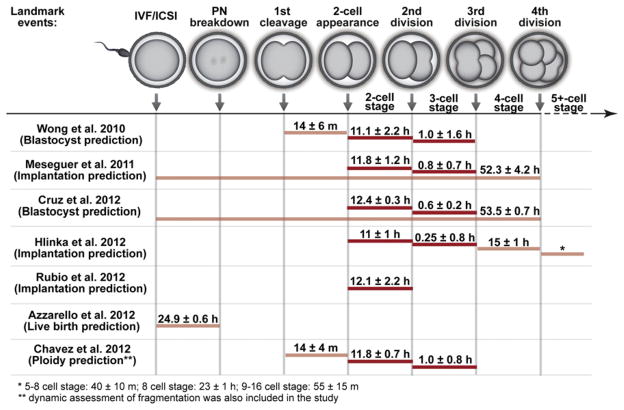
Overall, the time-lapse markers that have exhibited correlations with human embryo developmental outcomes are summarized in Table 1. The results of these studies highlight remarkable reproducibility for only a few specific time-lapse markers, for different embryo outcomes and across independent clinics and labs. For example, P2 and/or P3 are included in 8 out of 11 of the studies listed. A comparative analysis of the findings for these and other reproducible time-lapse marker studies is provided in Figure 1, where a subset of time-lapse studies that had precisely defined windows for embryo development prediction are plotted along a map of pre-implantation embryo development. Currently most of these biomarkers cluster at early stages of cleavage divisions (before the 5-cell stage). Timing parameters extracted from later stages—such as the morula, blastocyst, and blastocyst expansion stages—are still under investigation and could potentially provide additional or complementary time-lapse marker candidates. Upon future clinical validation, parameters with continued statistical robustness may prove to be viable time-lapse markers that can significantly improve embryo selection.
TABLE 1.
Human embryo time-lapse observations and markers, and the status of their practical application in the clinical IVF laboratory.
| Study | Time-lapse observation or marker | Outcomes | Biological study in humans | Technology to quantify time-lapse marker | Validation through prospective clinical trial |
|---|---|---|---|---|---|
| Payne et al. 1997 (2) | Time from ICSI to extrusion of PB2 | Day-3 embryo quality (n = 50) | No | Manual review | No |
| Synchrony of formation of gametic PN | |||||
| Time from ICSI to PN abuttal | |||||
| Periodicity of the cytoplasmic wave | |||||
| Lemmen et al. 2008 (3) | Time from fertilization to PN disappearancea | No | Manual review | No | |
| Time from fertilization to first cleavage (2-cell)b | Day-2 cell number (n = 102)a,b Ongoing pregnancy (n = 29)b |
||||
| Time and synchrony of nuclei appearance after first cleavageb | |||||
| Pribenszky et al. 2010 (4) | Time of cell cleavages and lack of fragmentation | Live birth (n = 5) | No | Manual review | No |
| Wong et al. 2010 (5) | Duration of first cytokinesis | Blastocyst formation (n = 100) | Gene expression of whole embryos and single blastomeres (n = 142) | Automated cell tracking measurements and outcome prediction | Nod |
| Time between first and second mitosis (2- to 3-cell stage) | |||||
| Synchrony of second and third mitosis (3- to 4-cell stage) | |||||
| Meseguer et al. 2011 (6) | Time between first and second mitosis (2- to 3-cell stage) | Implantation for Day-3 transfer (n = 247) | No | Manual review | Nod |
| Synchrony of second and third mitosis (3- to 4-cell stage) | |||||
| Time from ICSI to 5-cell stage | |||||
| Hashimoto et al. 2012 (7) | Synchrony of second and third mitosis (3- to 4-cell stage) | Blastocyst formation and quality (n = 80) | No | Manual review | No |
| Time of third cell division (5- to 8-cell stage) | |||||
| Swann et al. 2012 (8) | Cytoplasmic waves | Ca2+ oscillations (n = 10) | No | Automated velocity measurements | No |
| Cruz et al. 2012 (9) | Time between first and second mitosis (2- to 3-cell stage)a | No | Manual review | No | |
| Synchrony of second and third mitosis (3- to 4-cell stage)a,b | Blastocyst formation (n = 834)a | ||||
| Time from ICSI to 5-cell stageb | Blastocyst quality (n = 293)b | ||||
| Time to morula formationb | |||||
| Hlinka et al. 2012 (10) | Time between first and second mitosis (2- to 3-cell stage) | No | Manual review | No | |
| Synchrony of second and third mitosis (3- to 4-cell stage) | |||||
| Time between third and forth mitosis (4- to 5-cell stage) | Blastocyst formation (n = 180) | ||||
| Synchrony of fourth to seventh mitosis (5- to 8-cell stage) | Implantation for Day-5 transfer (n = 114) | ||||
| Time between seventh and eighth mitosis (8- to 9-cell stage) | |||||
| Synchrony of 8th to 15th mitosis (9- to 16-cell stage) | |||||
| Dal Canto et al. 2012 (11) | Time from insemination to 7-cell stagea,b | No | Manual review | No | |
| Time from insemination to 8-cell stagea,b,c | |||||
| Time from 4- to 8-cell stagea,b | |||||
| Time from 5- to 8-cell stagea,b | Blastocyst formation (n = 459)a | ||||
| Time from insemination to 3-cell stageb | Blastocyst expansion (n = 151)b | ||||
| Time from insemination to 4-cell stageb | Implantation for Day-3 or Day-5 transfer (n = 134)c | ||||
| Time from insemination to 5-cell stageb | |||||
| Time from insemination to 6-cell stageb | |||||
| Time from 2- to 3-cell stage | |||||
| Time from 3- to 4-cell stage | |||||
| Time from 2- to 4-cell stage | |||||
| Rubio et al. 2012 (12) | Time between first and second mitosis (2- to 3-cell stage) | Implantation for Day-3 transfer (n = 1,659) | No | Manual review | No |
| Azzarello et al. 2012 (13) | Time from ICSI to PN breakdown | Live birth for 4-cell transfer (n = 159) | No | Manual review | Yes |
| Chavez et al. 2012 (14) | Duration of first cytokinesis | Chromosomes were found in fragments | Manual review and automation method developed | No | |
| Time between first and second mitosis (2- to 3-cell stage) | Ploidy of blastomeres at 4-cell stage (n = 45) | ||||
| Synchrony of second and third mitosis (3- to 4-cell stage) | |||||
| Dynamic assessment of fragmentation |
Note: Matching superscript letters “a,b,c” indicate correlation between a time-lapse observation or marker and an outcome. Implantation was confirmed by ultrasound scan at 6 to 7 weeks for presence of gestational sac [Dal Canto et al. 2012 (11) and Rubio et al. 2012 (12)] or fetal heartbeat [Meseguer et al. 2011 (6) and Hlinka et al. 2012 (10)]. ICSI = intracytoplasmic sperm injection; PB = polar body; PN = pronuclei.
Matching superscript letters indicate correlation between a time-lapse observation or marker and an outcome.
FIGURE 1.
Time-lapse markers used for clinical outcome predictions in published studies. Landmark events captured by time-lapse imaging are mapped to the progression of preimplantation embryo development. Time-lapse markers that have been used for prediction in at least three publications are colored dark red while others are colored light red. Average values for embryo outcomes within the prediction windows are labeled above colored bars.
Novel Time-Lapse Markers and Underlying Biology
Understanding the scientific underpinnings of a novel biomarker provides confidence in achieving clinical significance and generates hypotheses for further clinical validation (16–18). However, due to ethical and resource constraints, it is challenging to perform the basic research required to elucidate the biological mechanisms of new prognostic factors. Mechanistic studies have been performed in mouse and other mammalian embryos for a few potential time-lapse markers of embryo fertilization and development (19, 20); however, only a limited number of research studies have succeeded in correlating time-lapse markers with both embryo outcome and molecular data for the human embryo (5, 14). Wong et al. (5) collected single embryos for gene expression analysis and revealed that embryos with time-lapse markers P1, P2, and P3 outside of the optimal ranges exhibited abnormal RNA patterns for embryo cytokinesis, micro RNA (miRNA) biogenesis, and maternal mRNA reserve. Their molecular findings suggest that embryo fate may be predetermined and inherited very early in development (by the 4-cell stage).
Chavez et al. (14) subsequently observed that euploid embryos clustered tightly in the P1, P2, or P3 time-lapse marker window that was predictive of blastocyst formation. Performing further molecular analysis, Chavez et al. (14) discovered that fragmentation dynamics detected by time-lapse imaging, together with P1, P2, and P3, could potentially distinguish euploid from aneuploid embryos at the 4-cell stage, as the fragments contained nuclear DNA, kinetochore proteins, and whole chromosomes detected by fluorescence in situ hybridization (FISH). Although these basic research findings are not prerequisites for clinical use of new time-lapse markers, and biologic validation of markers in IVF clinics is rare, the addition of biologic validation supports their use in the clinic and further extends our understanding of human embryology.
CLINICAL VALIDATION AND APPLICATION
Clinical validation and practical application of time-lapse markers for IVF requires demonstration of safety and efficacy in properly designed clinical trials. In this section, we review the current efforts to address the safety, efficacy, and ultimately practical use of time-lapse markers for human IVF embryos.
Safety of Time-Lapse Markers
Time-lapse imaging is considered noninvasive to the embryo, as no biopsies or other physical/chemical manipulations are involved. Although time-lapse imaging requires periodic exposure to light during image acquisitions, the calculated total dose of light exposure is less than what is commonly used in traditional morphology assessment and micromanipulation, both of which have been routinely used in IVF clinics for decades (5, 21, 22).
Several studies, including a handful of randomized and controlled clinical trials, have examined the safety of using time-lapse imaging. Pribenszky et al. (4) reported the first successful delivery of a baby after selection of a blastocyst following time-lapse imaging. Several other reports systematically examined fertilization rate (2, 22), blastocyst formation rate (5, 23, 24), pregnancy rate (15, 23, 24), quality (3, 22), and gene expression (5), and have shown comparable results between embryos exposed to time-lapse imaging and control embryos that were not exposed. In these studies, both bright-field and dark-field imaging modalities have been tested; both use long light wavelengths, low light intensity, and short durations of image acquisition (5, 6). Overall, current findings are promising and suggest time-lapse imaging is safe for clinical use. Future studies examining live-birth rates and more subtle light-inducible alterations in embryo development (e.g., epigenomic alterations) need to be performed to further confirm the safety of time-lapse imaging in clinical use.
Efficacy of Time-Lapse Markers
For the clinical application of time-lapse markers to embryo selection, robust validation of their efficacy is currently under close examination. After Wong et al.’s initial discovery of time-lapse markers that predict clinically relevant outcomes, Meseguer et al. (25) performed a retrospective clinical study and found that selecting embryos with time-lapse markers and morphologic exclusion of poor quality embryos could improve pregnancy rates an average of ~20% across 10 clinics. The improvement is likely to be dominated by the use of key, predictive time-lapse markers, as previous studies from the same group found no statistically significant difference for pregnancy rates of embryos cultured in enclosed chambers compared with standard incubators (24). Therefore, the published retrospective studies suggest promising efficacy of time-lapse markers in embryo selection.
A more critical standard for proving efficacy of time-lapse markers is a multicenter, prospective clinical trial, and a few are underway or completed (Conaghan et al., submitted) (26–28). We recently completed a prospective, multicenter clinical study that evaluated the effectiveness and utility of the Eeva Test, a noninvasive, computer-automated test of blastocyst formation based on time-lapse markers discovered by Wong et al. (ClinicalTrials.gov NCT01369446) (Conaghan et al., submitted) (5, 26, 27). The ability to predict blastocyst formation has clinical value as it could potentially avoid the risks associated with extended culture for blastocyst transfer (29–34).
We first evaluated Eeva against blastocyst formation as the outcome to examine whether the time-lapse markers predictive of blastocyst formation discovered in Wong et al. are applicable to fresh human embryos cultured in IVF clinical settings. We assessed the effectiveness and utility by evaluating [1] Eeva’s ability to predict blastocyst formation on independent data sets, and [2] whether Eeva predictions could aid embryologists in embryo selection on day 3. Our study results showed that blastocyst formation in clinical IVF settings could be predicted at the 4-cell stage using similar time-lapse markers to those previously discovered using cryopreserved human embryos donated to research (5).
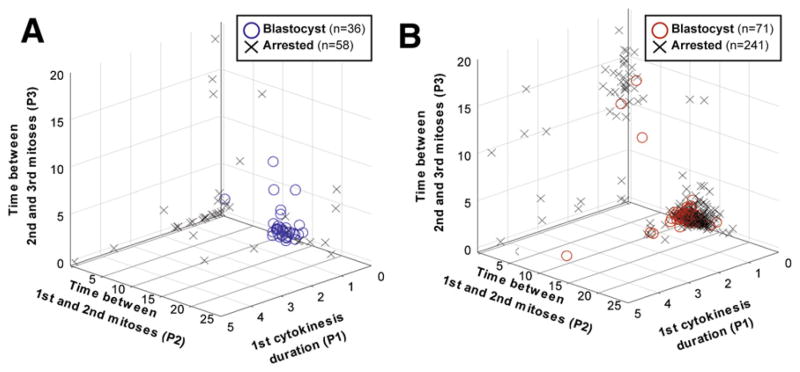
In this review, we present a side-by-side comparison of research and clinical human embryos plotted along early time-lapse markers P1, P2, and P3. Our results demonstrate that the blastocyst prediction window discovered in the original Wong et al. study (Fig. 2A) largely extends to fresh human embryos cultured according to the standard practices of five IVF clinics (see Fig. 2B) (Conaghan et al., submitted) (26, 27). An increased overlap between blastocysts and arrested embryos in the window for the clinical study data may represent differences between frozen versus fresh embryos. However, despite the slight biological variation, the time-lapse markers discovered by Wong et al. were consistently dominant for blastocysts among independently tested data sets.
FIGURE 2.
Time-lapse markers used for blastocyst prediction by the 4-cell stage in basic and molecular research (A) and a multicenter clinical validation study (B). Plots show that blastocysts mostly cluster tightly in a specific region for first cytokinesis duration (P1), the time between first and second mitosis (P2), and the time between the second and third mitosis (P3), while arrested embryos are mostly scattered. The time-lapse markers are plotted in units of hours.
When using the time-lapse markers P2 and P3 (9.33 ≤ P2 ≤ 11.45 hours and 0 ≤ P3 ≤ 1.73 hours), Eeva could distinguish blastocysts from arrested embryos with statistically significantly improved diagnostic specificity (85%) compared with traditional morphology (57%, P<.0001) (Conaghan et al., submitted) (27). Specificity measures the ability to correctly predict which embryos will arrest, and is particularly important because traditional morphology is most limited in selecting among “good morphology” embryos. Indeed, when Eeva was used in combination with day 3 morphology, the embryologists’ likelihood of selecting embryos that would develop to blastocysts was particularly improved among those embryos with good morphologic profiles (Conaghan et al., submitted) (26, 27).
We performed a secondary analysis to examine whether the time-lapse markers used by Eeva correlate with implantation and pregnancy outcomes. Importantly, as this study was a blastocyst prediction validation study, embryos were transferred at the blastocyst stage using the standard procedures of the participating clinics, and Eeva predictions were not made available at the time of transfer. We observed that, of 141 embryos transferred at the blastocyst stage, those with both P2 and P3 markers within range (Eeva High) had a statistically higher chance of implantation than embryos with P2 or P3 out of range (Eeva Low) (49% vs. 21%, P<.001) (Table 2). Similarly, for these 77 patients, those with at least one Eeva High embryo transferred were more likely to achieve clinical pregnancy (60% vs. 40%) and ongoing pregnancy (56% vs. 37%) than those with only Eeva Low embryos transferred. Although the limited patient sample size in this retrospective analysis for pregnancy is not enough for us to draw statistically significant conclusions, recent retrospective reports have suggested that the key time-lapse markers used by Eeva correlate both with implantation (6) and with pregnancy outcomes (25). Future studies will expand this data set for prospectively selected embryos, evaluate whether additional time-lapse markers could further improve implantation and pregnancy outcomes, and also address the hypothesis that day-3 transfer outcomes will be significantly improved using Eeva as an adjunct to morphologic grading.
TABLE 2.
Analysis of implantation and pregnancy rates for two populations of patients with blastocyst transfer suggest a correlation between time-lapse markers used by Eeva and implantation and pregnancy outcomes.
| Patient population | No. of patients | No. of embryos | Age (y) | Implantation rate | Clinical pregnancy rate | Ongoing pregnancy rate |
|---|---|---|---|---|---|---|
| At least 1 Eeva High transferred | 47 | 89 | 32.1 ± 5.2 | 49% (44/89) | 60% (28/47) | 55% (26/47) |
| Only Eeva Low transferred | 30 | 52 | 32.2 ± 5.1 | 21% (11/52) | 40% (12/30) | 37% (11/30) |
| P value | .9 | <.001 | .09 | .11 |
Note: Eeva High versus Low denominations are based on whether time-lapse markers P2 and P3 are within defined time windows (9.33 ≤ P2 ≤ 11.45 hours and 0 ≤ P3 ≤ 1.73 hours).
TECHNOLOGIES TO MEASURE AND QUANTIFY TIME-LAPSE MARKERS IN THE CLINIC
Time-Lapse Hardware
Basic advances and increased availability of time-lapse imaging hardware make it relatively simple to collect image data from a human embryo in a safe and noninvasive manner. Time-lapse imaging hardware now encompasses both homemade technologies and commercial devices. These technologies maintain an optimal embryo culture environment by either enclosing the incubation system around the image platform (2, 6, 15, 35) or by integrating miniaturized imaging systems inside conventional incubators (5, 14, 36). Both hardware strategies typically provide automated optical alignment, focusing, image capture, and image storage capabilities. Increasingly, specialized slides or dishes are used to facilitate the identification of embryos over the course of the study. For some systems, embryos are cultured in a slide with individualized wells and environments, and the slide is constantly moved into the field of optical view to visualize embryos (6). For other systems, embryos are cultured in a multiwell dish where media is shared, and the arrayed embryos are tracked under a single field of view (5, 36, 37).
Automated Image Analysis Software
There is growing interest in analyzing the abundant image data that have been gathered from time-lapse imaging systems. Accompanying this interest is a need to improve image data processing so that studies can be performed efficiently and their results translated into clinical practice. Currently, manual analysis of biomarkers captured with time-lapse imaging is hindering its routine use in clinics for several reasons. First, manual analysis is laborious and requires extensive training and practice for each time-lapse user. Second, the time needed for even highly trained users to perform manual analysis of large stacks of images in the limited time available before embryo transfer is prohibitive in the workflow common to IVF clinics. Finally, potential interobserver and intraobserver variability may impact time-lapse marker interpretation, similar to what has been found with manual embryo morphology grading (38).
Automated analysis of time-lapse image data through new “computer vision” software is emerging with the potential to enable reproducible, real-time, and quantitative assessment of time-lapse embryo image sequences (39). To date, several areas of reproductive medicine have benefited from computer vision-based image analysis software, such as computer-assisted semen analysis (CASA) (40, 41) and computer-assisted cervical cytology (42, 43). For the analysis of embryo time-lapse image data, only two software systems have been described (see Table 1). Recently, cytoplasmic waves have been quantified by a software system using particle image velocimetry methods to measure cytoplasmic waves (8, 19). However, despite elegant studies that probed Ca2+ oscillation activity, cytoskeleton integrity, and mouse embryo development, the detection of cytoplasmic waves in human embryos has not been correlated with clinical outcomes. Further, the image frame rate required (every 10 seconds for 2 hours immediately after fertilization) may not be clinically practical with existing hardware for many embryos at a time.
Another software system uses computer vision image analysis techniques to quantify cell-division dynamics. This software is based on cell tracking, and it leverages probabilistic model estimation techniques to infer the number of cells as well as cell size and shape as a function of time (5, 44). In addition to quantifiably tracking cell divisions, the software can be programmed with specific, predictive time-lapse markers, allowing a computer to both measure and identify embryos with the highest likelihood of developmental success. Currently, this software (termed Eeva) is the only prediction software that has been tested on human embryos in a prospective clinical study (Conaghan et al., submitted) (44). Recent work has suggested that this software could be further extended to include an automated fragmentation detector, which Chavez et al. (14) suggested may aid in the selection of euploid embryos together with P1, P2, and P3. Given the strong and reproducible predictive value of time-lapse markers that are based on cell-cycle timings and have been reported in many independent studies, software that can track individual cells and their division patterns is likely to be useful in clinical settings.
CONCLUSIONS
Time-lapse markers hold promise for aiding clinicians in determining which embryos are most suitable for transfer. However, the adoption of any new embryo selection biomarker requires a sound scientific basis, prospective clinical validation, and reliable quantification technology. Many time-lapse imaging studies have found parameters that have statistical significance between embryos with different clinical outcomes. However, few of these studies have succeeded in extracting predictive time-lapse markers with defined ranges for embryo selection, and fewer still have investigated the biology underlying these biomarkers. Further, the manual extraction of new or promising time-lapse markers adds prohibitive constraints to their use in actual clinical settings. Therefore, before time-lapse markers are to be implemented in the clinic, additional clinical validation of their safety and efficacy and their measurement/quantification technologies is urgently needed. Randomized controlled trials should also be performed to definitively confirm their clinical value. Altogether, should time-lapse markers successfully meet the criteria described here, it is likely that their implementation will improve embryo selection, reduce the number of embryos required for transfer, and ultimately increase the success of IVF treatment.
Acknowledgments
The authors thank the physicians who participated in the Eeva Clinical Validation study, including Drs. Susan P. Willman, Philip E. Chenette, Robert Boostanfar, Valerie L. Baker, and Mary E. Abusief, and also thank the clinical study embryologists Kristen Ivani, Joe Conaghan, Anh Le, Mariluz Suarez, and Marina Gvakharia for their input; and the Auxogyn scientists Kelly Wirka, Farshid Moussavi, and Mahnaz Maddah for their technical contributions.
Footnotes
A.A.C. is an employee and shareholder of Auxogyn. L.T. is an employee and shareholder of Auxogyn. V.S. is an employee and shareholder of Auxogyn. R.R.P. is a cofounder, employee, and shareholder of Auxogyn and receives patent royalties from Stanford for an invention related to this publication (Auxogyn is the licensee of this patent). S.S. is an employee and shareholder of Auxogyn.
References
- 1.van Montfoort AP, Dumoulin JC, Land JA, Coonen E, Derhaag JG, Evers JL. Elective single embryo transfer (eSET) policy in the first three IVF/ICSI treatment cycles. Hum Reprod. 2005;20:433–6. doi: 10.1093/humrep/deh619. [DOI] [PubMed] [Google Scholar]
- 2.Payne D, Flaherty SP, Barry MF, Matthews CD. Preliminary observations on polar body extrusion and pronuclear formation in human oocytes using time-lapse video cinematography. Hum Reprod. 1997;12:532–41. doi: 10.1093/humrep/12.3.532. [DOI] [PubMed] [Google Scholar]
- 3.Lemmen JG, Agerholm I, Ziebe S. Kinetic markers of human embryo quality using time-lapse recordings of IVF/ICSI-fertilized oocytes. Reprod Biomed Online. 2008;17:385–91. doi: 10.1016/s1472-6483(10)60222-2. [DOI] [PubMed] [Google Scholar]
- 4.Pribenszky C, Matyas S, Kovacs P, Losonczi E, Zadori J, Vajta G. Pregnancy achieved by transfer of a single blastocyst selected by time-lapse monitoring. Reprod Biomed Online. 2010;21:533–6. doi: 10.1016/j.rbmo.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 5.Wong C, Loewke K, Bossert N, Behr B, De Jonge C, Baer T, et al. Non-invasive imaging of human embryos before embryonic genome activation predicts development to the blastocyst stage. Nat Biotechnol. 2010;28:1115–21. doi: 10.1038/nbt.1686. [DOI] [PubMed] [Google Scholar]
- 6.Meseguer M, Herrero J, Tejera A, Hilligsoe KM, Ramsing NB, Remohi J. The use of morphokinetics as a predictor of embryo implantation. Hum Reprod. 2011;26:2658–71. doi: 10.1093/humrep/der256. [DOI] [PubMed] [Google Scholar]
- 7.Hashimoto S, Kato N, Saeki K, Morimoto Y. Selection of high-potential embryos by culture in poly(dimethylsiloxane) microwells and time-lapse imaging. Fertil Steril. 2012;97:332–7. doi: 10.1016/j.fertnstert.2011.11.042. [DOI] [PubMed] [Google Scholar]
- 8.Swann K, Windsor S, Campbell K, Elgmati K, Nomikos M, Zernicka-Goetz M, et al. Phospholipase C-zeta-induced Ca2+ oscillations cause coincident cytoplasmic movements in human oocytes that failed to fertilize after intracytoplasmic sperm injection. Fertil Steril. 2012;97:742–7. doi: 10.1016/j.fertnstert.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cruz M, Garrido N, Herrero J, Perez-Cano I, Munoz M, Meseguer M. Timing of cell division in human cleavage-stage embryos is linked with blastocyst formation and quality. Reprod Biomed Online. 2012;25:371–81. doi: 10.1016/j.rbmo.2012.06.017. [DOI] [PubMed] [Google Scholar]
- 10.Hlinka D, Kalatova B, Uhrinova I, Dolinska S, Rutarova J, Rezacova J, et al. Time-lapse cleavage rating predicts human embryo viability. Physiol Res. 2012;61:513–25. doi: 10.33549/physiolres.932287. [DOI] [PubMed] [Google Scholar]
- 11.Dal Canto M, Coticchio G, Mignini Renzini M, De Ponti E, Novara PV, Brambillasca F, et al. Cleavage kinetics analysis of human embryos predicts development to blastocyst and implantation. Reprod Biomed Online. 2012;25:474–80. doi: 10.1016/j.rbmo.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 12.Rubio I, Kuhlmann R, Agerholm I, Kirk J, Herrero J, Escribá M-J, et al. Limited implantation success of direct-cleaved human zygotes: a time-lapse study. Fertil Steril. 2012;98:1458–63. doi: 10.1016/j.fertnstert.2012.07.1135. [DOI] [PubMed] [Google Scholar]
- 13.Azzarello A, Hoest T, Mikkelsen AL. The impact of pronuclei morphology and dynamicity on live birth outcome after time-lapse culture. Hum Reprod. 2012;27:2649–57. doi: 10.1093/humrep/des210. [DOI] [PubMed] [Google Scholar]
- 14.Chavez SL, Loewke KE, Han J, Moussavi F, Colls P, Munne S, et al. Dynamic blastomere behaviour reflects human embryo ploidy by the four-cell stage. Nat Commun. 2012;3:1251. doi: 10.1038/ncomms2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mio Y, Maeda K. Time-lapse cinematography of dynamic changes occurring during in vitro development of human embryos. Am J Obstet Gynecol. 2008;199:660, e1–5. doi: 10.1016/j.ajog.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 16.Brown R, Harper J. The clinical benefit and safety of current and future assisted reproductive technology. Reprod Biomed Online. 2012;25:108–17. doi: 10.1016/j.rbmo.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 17.Harper J, Cristina Magli M, Lundin K, Barratt CLR, Brison D. When and how should new technology be introduced into the IVF laboratory? Hum Reprod. 2012;27:303–13. doi: 10.1093/humrep/der414. [DOI] [PubMed] [Google Scholar]
- 18.Palmer SS, Barnhart KT. Biomarkers in reproductive medicine: the promise, and can it be fulfilled? Fertil Steril. 2012;13:02431–4. doi: 10.1016/j.fertnstert.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ajduk A, Ilozue T, Windsor S, Yu Y, Seres KB, Bomphrey RJ, et al. Rhythmic actomyosin-driven contractions induced by sperm entry predict mammalian embryo viability. Nat Commun. 2011;9:417. doi: 10.1038/ncomms1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piotrowska K, Wianny F, Pedersen RA, Zernicka-Goetz M. Blastomeres arising from the first cleavage division have distinguishable fates in normal mouse development. Development. 2001;128:3739–48. doi: 10.1242/dev.128.19.3739. [DOI] [PubMed] [Google Scholar]
- 21.Ottosen LM, Hindkjær J, Ingerslev J. Light exposure of the ovum and preimplantation embryo during ART procedures. J Assist Reprod Genet. 2007;24:99–103. doi: 10.1007/s10815-006-9081-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakahara T, Iwase A, Goto M, Harata T, Suzuki M, Ienaga M, et al. Evaluation of the safety of time-lapse observations for human embryos. J Assist Reprod Genet. 2010;27:93–6. doi: 10.1007/s10815-010-9385-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirkegaard K, Hindkjaer JJ, Grondahl ML, Kesmodel US, Ingerslev HJ. A randomized clinical trial comparing embryo culture in a conventional incubator with a time-lapse incubator. J Assist Reprod Genet. 2012;29:565–72. doi: 10.1007/s10815-012-9750-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cruz M, Gadea B, Garrido N, Pedersen KS, Martinez M, Perez-Cano I, et al. Embryo quality, blastocyst and ongoing pregnancy rates in oocyte donation patients whose embryos were monitored by time-lapse imaging. J Assist Reprod Genet. 2011;28:569–73. doi: 10.1007/s10815-011-9549-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meseguer M, Rubio I, Cruz M, Basile N, Marcos J, Requena A. Embryo incubation and selection in a time-lapse monitoring system improves pregnancy outcome compared with a standard incubator: a retrospective cohort study. Fertil Steril. 2012;98:1481–9. e10. doi: 10.1016/j.fertnstert.2012.08.016. [DOI] [PubMed] [Google Scholar]
- 26.Chen AA, Ivani K, Conaghan J, Gvakharia M, Le A, Shen S. Improved embryo selection accuracy using cell division characteristics defined by time-lapse and automated image analysis. Fertil Steril. 2012;98(Suppl):S17. [Google Scholar]
- 27.Shen S, Chen AA, Willman SP, Chenette PE, Boostanfar R, Baker VL, et al. Early cell cycle durations detected by time-lapse imaging predict embryo developmental potential. Hum Reprod. 2012;27:ii22–4. [Google Scholar]
- 28.Meseguer M, Herrero J, Tejera A, Perez S, Hilligsøe KM, De los Santos MJ. Preliminary evaluation of an embryo quality classification system based in a multivariate analysis of morphokinetic data from time-lapse. Fertil Steril. 2011;96:S244. [Google Scholar]
- 29.Papanikolaou EG, D’Haeseleer E, Verheyen G, Van de Velde H, Camus M, Van Steirteghem A, et al. Live birth rate is significantly higher after blastocyst transfer than after cleavage-stage embryo transfer when at least four embryos are available on day 3 of embryo culture. A randomized prospective study. Hum Reprod. 2005;20:3198–203. doi: 10.1093/humrep/dei217. [DOI] [PubMed] [Google Scholar]
- 30.Papanikolaou EG, Camus M, Kolibianakis EM, Van Landuyt L, Van Steirteghem A, Devroey P. In vitro fertilization with single blastocyst-stage versus single cleavage-stage embryos. N Engl J Med. 2006;354:1139–46. doi: 10.1056/NEJMoa053524. [DOI] [PubMed] [Google Scholar]
- 31.Blake DA, Farquhar CM, Johnson N, Proctor M. Cleavage stage versus blastocyst stage embryo transfer in assisted conception. Cochrane Database Syst Rev. 2007;4:CD002118. doi: 10.1002/14651858.CD002118.pub3. [DOI] [PubMed] [Google Scholar]
- 32.Kallen B, Finnstrom O, Lindam A, Nilsson E, Nygren KG, Olausson PO. Blastocyst versus cleavage stage transfer in in vitro fertilization: differences in neonatal outcome? Fertil Steril. 2010;94:1680–3. doi: 10.1016/j.fertnstert.2009.12.027. [DOI] [PubMed] [Google Scholar]
- 33.Kalra SK, Ratcliffe SJ, Barnhart KT, Coutifaris C. Extended Embryo Culture and an Increased Risk of Preterm Delivery. Obstetrics & Gynecology. 2012;120:69–75. doi: 10.1097/AOG.0b013e31825b88fc. http://dx.doi.org/10.1097/AOG.0b013e31825b88fc. [DOI] [PubMed] [Google Scholar]
- 34.Diamond MP, Willman S, Chenette P, Cedars MI. The clinical need for a method of identification of embryos destined to become a blastocyst in assisted reproductive technology cycles. J Assist Reprod Genet. 2012;29:391–6. doi: 10.1007/s10815-012-9732-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hardarson T, Lofman C, Coull G, Sjogren A, Hamberger L, Edwards RG. Internalization of cellular fragments in a human embryo: time-lapse recordings. Reprod Biomed Online. 2002;5:36–8. doi: 10.1016/s1472-6483(10)61594-5. [DOI] [PubMed] [Google Scholar]
- 36.Vajta G, Peura TT, Holm P, Paldi A, Greve T, Trounson AO, et al. New method for culture of zona-included or zona-free embryos: the Well of the Well (WOW) system. Mol Reprod Dev. 2000;55:256–64. doi: 10.1002/(SICI)1098-2795(200003)55:3<256::AID-MRD3>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 37.Dai S-J, Xu C-L, Wang J, Sun Y-P, Chian R-C. Effect of culture medium volume and embryo density on early mouse embryonic development: Tracking the development of the individual embryo. J Assist Reprod Genet. 2012;29:617–23. doi: 10.1007/s10815-012-9744-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baxter Bendus AE, Mayer JF, Shipley SK, Catherino WH. Interobserver and intraobserver variation in day 3 embryo grading. Fertil Steril. 2006;86:1608–15. doi: 10.1016/j.fertnstert.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 39.Danuser G. Computer vision in cell biology. Cell. 2011;147:973–8. doi: 10.1016/j.cell.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 40.Abaigar T, Cano M, Pickard AR, Holt WV. Use of computer-assisted sperm motility assessment and multivariate pattern analysis to characterize ejaculate quality in Mohor gazelles (Gazella dama mhorr): effects of body weight, electroejaculation technique and short-term semen storage. Reproduction. 2001;122:265–73. doi: 10.1530/rep.0.1220265. [DOI] [PubMed] [Google Scholar]
- 41.Krause W, Viethen G. Quality assessment of computer-assisted semen analysis (CASA) in the andrology laboratory. Andrologia. 1999;31:125–9. [PubMed] [Google Scholar]
- 42.Dziura B, Quinn S, Richard K. Performance of an imaging system vs. manual screening in the detection of squamous intraepithelial lesions of the uterine cervix. Acta Cytol. 2006;50:309–11. doi: 10.1159/000325959. [DOI] [PubMed] [Google Scholar]
- 43.Lozano R. Comparison of computer-assisted and manual screening of cervical cytology. Gynecol Oncol. 2007;104:134–8. doi: 10.1016/j.ygyno.2006.07.025. [DOI] [PubMed] [Google Scholar]
- 44.Loewke K, Moussavi F, Maddah M, Ivani K, Behr B, Suraj V. Development and validation of an automated computer vision algorithm for real-time embryo viability prediction. Fertil Steril. 2012;98(Suppl):S288–9. [Google Scholar]