Abstract
Intrachromosomal amplification of chromosome 21 (iAMP21) defines a distinct cytogenetic subgroup of childhood B-cell precursor acute lymphoblastic leukaemia (BCP-ALL). To date, fluorescence in situ hybridisation (FISH), with probes specific for the RUNX1 gene, provides the only reliable detection method (five or more RUNX1 signals per cell). Patients with iAMP21 are older (median age 9 years) with a low white cell count. Previously, we demonstrated a high relapse risk when these patients were treated as standard risk. Recent studies have shown improved outcome on intensive therapy. In view of these treatment implications, accurate identification is essential. Here we have studied the cytogenetics and outcome of 530 iAMP21 patients that highlighted the association of specific secondary chromosomal and genetic changes with iAMP21 to assist in diagnosis, including the gain of chromosome X, loss or deletion of chromosome 7, ETV6 and RB1 deletions. These iAMP21 patients when treated as high risk showed the same improved outcome as those in trial-based studies regardless of the backbone chemotherapy regimen given. This study reinforces the importance of intensified treatment to reduce the risk of relapse in iAMP21 patients. This now well-defined patient subgroup should be recognised by World Health Organisation (WHO) as a distinct entity of BCP-ALL.
Keywords: iAMP21, genetics, outcome, poor prognosis, BCP-ALL, chromosomal abnormalities
INTRODUCTION
Intrachromosomal amplification of chromosome 21 (iAMP21) was originally identified as a distinct cytogenetic subgroup of childhood acute lymphoblastic leukaemia (ALL) in 2003,1,2 following reports of a number of sporadic cases.3–12 In all studies, iAMP21 patients had B-cell precursor ALL (BCP-ALL), were older, with a median age of 9–11 years, and generally had low white cell counts (WCCs). Prospective screening in recent childhood trials has determined the incidence to be 2%.13,14 A significant finding was that patients with iAMP21 had an inferior outcome when treated on standard therapy as compared with other patients treated on the same protocols.15–17 iAMP21 was defined as a primary cytogenetic abnormality with a complex structure of one copy of chromosome 21 comprising multiple regions of gain, amplification, inversion and deletion, identified from cytogenetics, fluorescence in situ hybridisation (FISH) and genomic analysis, which was highly variable between patients.18–20 We identified a common region of highest level amplification spanning 5.1 Mb of chromosome 21 from 32.8 to 37.9 Mb, within which the RUNX1 gene is located.18 We proposed that the abnormal chromosome 21 arose through a breakage-fusion-bridge mechanism,19 supported by the observation of anaphase bridges involving chromosome 21 in some iAMP21 patients.21 Further studies pointed to clustered breakpoints within the PDE9A gene in a number of patients involved in complex events around microhomology-mediated end joining as preceding or initiating the breakage-fusion-bridge cycles.22 As the search for the initiating event continues, FISH, by using probes directed to RUNX1 to determine the number of copies of the most highly amplified region, provides the most reliable detection method.23 Three or more extra copies of RUNX1 on a single abnormal chromosome 21 (a total of five or more RUNX1 signals per cell) defines iAMP21; a definition that has now been adopted internationally.24
The Ponte di Legno International Childhood ALL Group has published a series of manuscripts on relatively rare, high-risk patient subsets, most recently Philadelphia chromosome-positive ALL treated without tyrosine kinase inhibitors and children with induction failure.25,26 In this study, the group has collected cytogenetic and associated data from 530 pediatric ALL patients with iAMP21, which has further characterised this subgroup in relation to its cytogenetic profile. We showed the same improved response as trial-based studies when iAMP21 patients are treated as high risk in multiple treatment centres, regardless of the backbone chemotherapy regimen given. The findings from this study have improved the definition that, in view of the poor outcome of this subgroup, will assist in ensuring that all iAMP21 patients are correctly identified.
PATIENTS AND METHODS
Patient information
Patients included in this study originated from 18 international study groups (Supplementary Table 1) and were diagnosed between February 1987 and December 2011. They were all diagnosed with BCP-ALL using standard morphological and immunophenotypic criteria. Individual patient data on age, sex, WCC, immunophenotype and outcome as well as karyotype were collected from each clinical study group. We classified patients into National Cancer Institute (NCI) standard-risk (NCI-SR) and high-risk groups (NCI-HR) according to age and WCC; NCI-SR: age ≤ 10 years and WCC ≤ 50 × 109/l, NCI-HR: age ≥ 10 years or WCC ≥ 50 × 109/l. Because of the range of treatment protocols, information collected from each study group was restricted to whether the patient was treated as nonhigh risk or high risk according to their individual protocols.
Cytogenetics, FISH, Multiplex Ligation-dependent Probe Amplification and SNP 6.0 array analysis
Patients in this study were classified as iAMP21using the established criteria of three or more additional RUNX1 signals (five or more signals per cell in total) with a FISH probe targeting the RUNX1 gene. The usual probe is one designed for the detection of the ETV6-RUNX1 fusion: a number of them are available commercially.24 Where metaphases were available, the extra signals were located on a single abnormal chromosome 21. All patients except five were diagnosed using this standard FISH approach: two cases with a positive cytogenetic result were included because either the abnormal chromosome 21 was confirmed by chromosome painting (wcp21; patient 450) or the region surrounding RUNX1 (21q21.3–q22.3 (32.0–43.70) was shown to be amplified by 1 Mb BAC arrays (array comparative genomic hybridization; no. 455 and no. 3698 in Strefford et al.20), and three other cases were included based on having a single-nucleotide polymorphism (SNP) profile consistent with iAMP21 (nos. 365, 366, and 367; and nos. 1765, 1896 and 2012 in Bungaro et al.27), as previously defined.18,22 The cytogenetic data were written according to the International System for Human Cytogenetic Nomenclature (ISCN).28 A total of 10 or usually 20 normal cells were analysed to classify a normal karyotype when no chromosomal abnormalities were identified. FISH signal patterns and other relevant FISH or molecular data were collected to indicate the presence of the common established chromosomal abnormalities.24,29 Multiplex ligation-dependent probe amplification (MLPA) data were generated for a subset of patients from two study groups using the SALSA MLPA kit P335 IKZF1 (MRC Holland, Amsterdam, The Netherlands) as previously described.30 This kit evaluates the copy number of IKZF1, CDKN2A/B, PAX5, EBF1, ETV6, BTG1, RB1 and genes within the pseudoautosomal region (PAR1): CRLF2, CSF2RA, IL3RA. RB1 deletions were verified using the P047 RB1 SALSA MLPA kit with a higher probe density covering RB1. SNP arrays for patients 365–368 were prepared and analysed as previously described.27
Statistics and end points
Survival analysis considered three end points: event-free survival (EFS) defined as time to relapse or death, censoring at last contact; relapse rate defined as time from complete remission to relapse, censoring at death in remission or last contact; and overall survival (OS) defined as time to death, censoring at last contact. Kaplan–Meier methods and curves were used to assess and compare survival differences between groups. Other comparisons were performed using the χ2, Fisher’s exact or Wilcoxon rank-sum tests, as appropriate. All analyses were performed using Intercooled Stata 11.0 (Stata Corporation, College Station, TX, USA).
RESULTS
iAMP21 has a distinct cytogenetic/genetic profile
This study included 530 patients with a confirmed diagnosis of iAMP21. The total number of RUNX1 signals observed for each patient by FISH ranged from 5 to 20 per cell, although in many cases a range was provided or the number noted as ‘many’ because of difficulties in scoring closely clustered signals in interphase, as previously noted.19 Details of the demographic and cytogenetic data (SNP data for patients 365–368) are provided in Supplementary Table 2. A total of 119 patients with a failed cytogenetic result or a normal karyotype, without cytogenetic evidence of the acquired iAMP21 chromosome, were included in the study based on their abnormal FISH, array comparative genomic hybridization and/or SNP result that defined iAMP21 (Table 1). Among the 411 patients with an abnormal cytogenetic result, the majority of karyotypes were near diploid (44–48 chromosomes; Supplementary Table 2). There were only four patients each with 49 and 50–56 chromosomes. The cytogenetic nomenclature used to describe iAMP21 was highly varied, for example, add(21), dup(21) and der(21), with loss of chromosome 21 and associated gain of a marker chromosome being the most common, reflecting the highly variable morphology of this abnormal chromosome. Where FISH testing had been carried out to exclude the presence of other established cytogenetic changes, iAMP21 was mutually exclusive of MLL rearrangements (412 tested) and TCF3 rearrangements (to rule out TCF3-PBX1 and TCF3-HLF, 156 tested). There was evidence of classical high hyperdiploidy in two patients based on their karyotype (nos. 139 and 359, with triple trisomy: gains of chromosomes 4, 10 and 17, as well as abnormalities of 1q).31 The one patient with 52 chromosomes showed atypical gains with an inconclusive karyotype (no. 247). The presence of a hidden high hyperdiploid clone was excluded by FISH among 156 patients tested for evidence of triple trisomy32 that included patients with failed cytogenetic results (n = 6) and a normal karyotype (n = 14). This test confirmed gain of chromosomes 10 and 17 in two (nos. 167 and 170) and one (no. 171) patients, respectively, and gain of chromosome 10 in two patients (nos. 134 and 160 with a normal karyotype), where it was not seen by cytogenetic analysis. Three patients with iAMP21 were positive for ETV6-RUNX1 fusion among 528 tested (no. 525 reported in Haltrich et al.,33 and nos. 528 and 530). In these cases it was possible to ascertain that the fusion and amplification of the RUNX1 signals were present in the same cells, as they are detected using the same probes, but as they were always seen to occur together (apart from a single cell with the ETV6-RUNX1 fusion only in one patient, no. 530), it was not possible to determine whether iAMP21 arose as a primary event or a secondary event following ETV6-RUNX1 fusion. During this study period, among 2465 and 1200 BCP-ALL patients tested for ETV6-RUNX1 fusion in the Children’s Oncology Group (COG) and UK childhood ALL treatment trials, respectively, no ETV6-RUNX1-positive cases were found with iAMP21 (although a single case has recently been found by COG from outside the reporting period, in which iAMP21 was clearly a secondary change to ETV6-RUNX1 fusion; NA Heerema and AJ Carroll, personal communication). These observations emphasise the rarity of this association.
Table 1.
Summary of cytogenetic data
| No. | %a | |
|---|---|---|
| Total patients | 530 | 100 |
| Fail | 80 | 15 |
| Normal | 39 | 7 |
| Abnormal | 411 | 78 |
| Abnormality | No. | %b |
|---|---|---|
| iAMP21 alone | 84 | 20 |
| Gain of 21 | 10 | 3 |
| Gain of iAMP21 | 4 | 1 |
| Gain of X | 86c | 21 |
| Monosomy 7 | 19 | 5 |
| Trisomy 10 | 18 | 4 |
| Trisomy 14 | 17 | 4 |
| Monosomy 15 | 3 | |
| abn1q | 43 | 11 |
| abn6q | 18 | 4 |
| abn7q | 46 | 11 |
| abn7q/-7 | 65d | 16 |
| abn9p | 42e | 10 |
| abn11q | 50f | 12 |
| abn12p | 46g | 11 |
| abn13q | 22 | 6 |
| abn16q | 26 | 6 |
Abbreviations: abn1q, abnormalities involving 1q; abn6q, abnormalities involving 6q; abn7q/-7, monosomy 7 plus abnormalities involving 7q; abn9p, abnormalities involving 9p; abn11q, abnormalities involving 11q; abn12p, abnormalities involving 12p; abn13q, abnormalities involving 13q; abn16q, abnormalities involving 16q; iAMP21, intrachromosomal amplification of chromosome 21.
Percentage of total cases.
Percentage of cases with an abnormal karyotype.
A total of 49 males and 27 females.
Combined numbers for abn7q and monosomy 7.
Fluorescence in situ hybridisation (FISH) confirmed deletion of 9p in two cases with an abnormal karyotype.
FISH confirmed deletion of 11q in four cases with an abnormal karyotype.
FISH confirmed deletion of 12p in 14 cases with an abnormal karyotype.
Four patients (<1%) among 415 tested were positive for the BCR-ABL1 fusion, and this correlated with a translocation, t(9;22)(q34;q11.2), within the karyotype in 3 of them (nos. 151, 296 and 307). In these three patients, the percentages of cells with additional RUNX1 signals and BCR-ABL1 fusion were identical, implying that both iAMP21 and BCR-ABL1 were present together in all abnormal cells. In the fourth patient (no. 103), the der(22)t(9;22) only was visible in the karyotype and the BCR-ABL1 fusion was present in only a subpopulation (30%) of the iAMP21-positive cells, indicating that it likely arose as a secondary event in this patient. Among 287 and 124 BCR-ABL1-positive cases, which had also been screened by FISH for evidence of ETV6-RUNX1 by COG and United Kingdom, respectively, only the 3 cases reported here were found with BCR-ABL1 and iAMP21, reinforcing this association also as a rare event.
iAMP21 was seen as the sole cytogenetic change in 20% patients with an abnormal karyotype (Table 1). Only a small number of the 327 patients with additional chromosomal abnormalities had two copies of the normal chromosome 21 in addition to iAMP21 (n = 10), whereas the iAMP21 chromosome was duplicated in four patients. Other whole chromosomes commonly gained or lost among the 407 patients with abnormal karyotypes were: +X (21%) (57% were male and 43% were females), −7 (5%), +10 (4%), +14 (4%) and −15 (3%) (Table 1). Abnormalities of certain chromosome arms were also frequent: 1q (11%), 6q (4%), 7q (11%), 9p (10%), 11q (12%), 12p (11%), 13q (6%) and 16q (6%), that most frequently involved loss of chromosomal material. Monosomy 7 was mutually exclusive of abnormalities of 7q. Therefore, by adding these together, 7q was shown to be involved in a total of 16% of patients. From FISH studies used to identify MLL rearrangements and the ETV6-RUNX1 fusion, deletions of the MLL and ETV6 genes were found in 10% (42 positive/406 tested) and 10% (44/458 where the number of ETV6 signals was recorded) patients, respectively. As these deletions detected by FISH correlated well with the respective cytogenetically visible deletions, these FISH tests may be used as indicators of 11q and 12p deletions in patients with a failed or normal cytogenetic result.
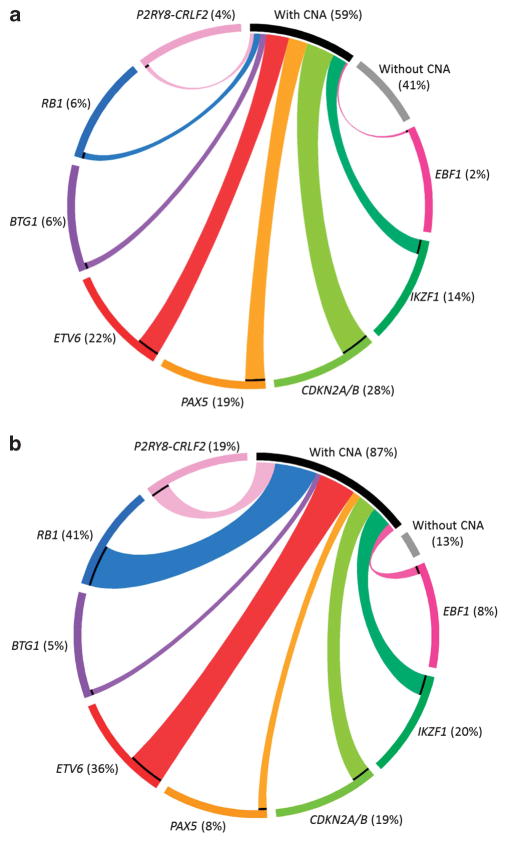
Table 2 and Figure 1 indicate the relative proportions of copy number abnormalities in those genes tested by MLPA among iAMP21 patients compared with a group of 1427 childhood BCP-ALL reported in Schwab et al.34 Those genes with a significantly higher incidence of deletions among the iAMP21 patients were ETV6 if compared with ETV6-RUNX1-negative patients (Table 2), P2RY8-CRLF2 and RB1. The incidence of ETV6 deletions at 37% detected by MLPA is much higher than the incidence detected by FISH, because of the deletion of individual exons either not covered by the FISH probe or below the resolution of FISH, as we have previously noted.30 For example, two of the four cases with SNP data showed deletions of exons 2 and 3 and 2–4 only, which would not have been visible by FISH.27 We have previously reported the higher incidence of CRLF2 rearrangements and RB1 deletions in iAMP21 patients.18,34 Only 7/27 of the MLPA tested cases with a RB1 deletion showed a visible 13q abnormality in the karyotype.
Table 2.
Summary of MLPA data
| Genetic abnormality | iAMP21 cohort
|
ALL97/ALL2003a
|
P-value | ||
|---|---|---|---|---|---|
| Yes | No | Yes | No | ||
| No. (%) | No. (%) | No. (%) | No. (%) | ||
| BTG1 deletion | 4 (5) | 70 (95) | 89 (6) | 1338 (94) | 0.794 |
| CDKN2A/B deletion | 16 (21) | 61 (79) | 395 (28) | 1032 (72) | 0.161 |
| EBF1 deletion | 6 (8) | 68 (92) | 30 (2) | 1397 (98) | 0.004 |
| ETV6 deletion | 30 (37)b | 51 (63)b | 312 (22)c | 1115 (78) | 0.013 |
| IKZF1 deletion | 15 (20) | 59 (80) | 196 (14) | 1231 (86) | 0.118 |
| CRLF2-R | 18 (18)d | 84 (82)d | 63 (4) | 1364 (96) | <0.001 |
| PAX5 aberration | 6 (8) | 68 (92) | 272 (19) | 1155 (81) | 0.037 |
| RB1 deletion | 31 (41) | 44 (59) | 92 (6) | 1335 (94) | <0.001 |
Abbreviations: CRLF2-R, CRLF2 rearrangement; iAMP21, intrachromosomal amplification of chromosome 21; MLPA, Multiplex Ligation-dependent Probe Amplification.
Data from Schwab et al.34
Includes two cases tested by single-nucleotide polymorphism (SNP) arrays.
Of the 312 cases, 203 were found within the ETV6-RUNX1-positive group, accounting for 54% of the ETV6-RUNX1-positive cases. If the ETV6-RUNX1-positive cases are removed then the percentage among ETV6-RUNX1-negative patients is 10% (109/1048).
Includes data from 38 cases tested by fluorescence in situ hybridisation (FISH).
Figure 1.
Circos plots showing the relative distribution of the significant copy number changes in a comparator cohort of childhood BCP-ALL (a) and iAMP21 patients (b). In both (a) and (b) the black bar indicates the proportion of patients in which a copy number abnormality (CNA) was detected compared to the proportion without (grey bar). The proportions of CNA affecting the genes tested are colour coded as indicated. PAR1 deletions represent P2RY8-CRLF2.
iAMP21 and constitutional abnormalities involving chromosome 21 Interestingly, there were several patients with constitutional abnormalities involving chromosome 21 in the iAMP21 cohort: Down syndrome (n = 1) (no. 20), ring chromosome 21, r(21)c (n = 3; nos. 177, 195 and 217) and Robertsonian translocation between chromosomes 15 and 21, rob(15;21)(q10;q10)c (n = 5; nos. 371, 416, 438 and 486).
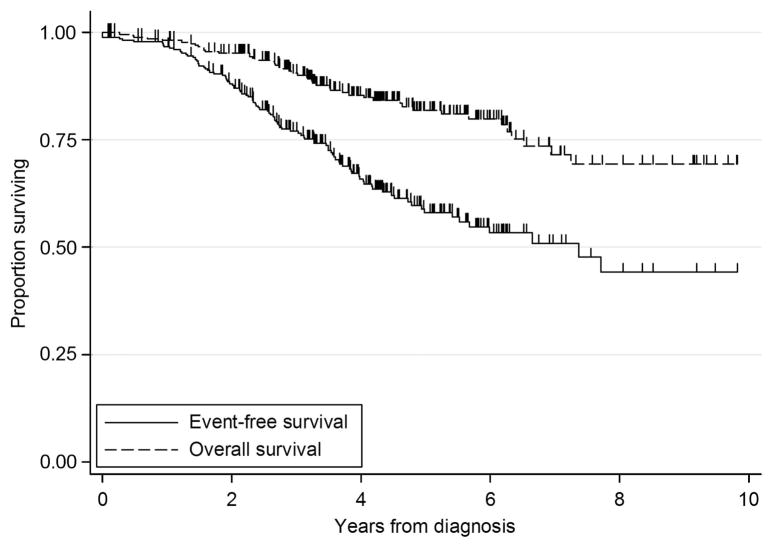
iAMP21 patients have a distinct demographic and clinical profile Patients with iAMP21 had a distinctive demographic and clinical profile: almost equal numbers of females and males, older age (median 9 years) and low WCC (median 5 × 109/l; Table 3). Half of the patients (52%) were classified as NCI-HR (Table 3). To ensure adequate patient follow-up, we limited EFS/OS analyses to the cohort of 283 patients diagnosed before 01 January 2009 (Table 3). Complete remission was achieved in 99%. Among the 280 remitters, 90 (32%) suffered a relapse. Site of relapse was isolated bone marrow (62%), isolated central nervous system (22%), combined (11%) and other/unknown (4%). Approximately half of the relapses (56%) occurred within 3 years of diagnosis but a substantial proportion occurred more than 4 years after treatment had started (20%) (Table 3). This continued drop in EFS beyond 4 years is an uncommon finding in pediatric ALL. A total of 47 (17%) patients died, after relapse (n = 40), in first remission (n = 5) plus nonremitters (n = 2). One nonremitter was alive at last follow-up. The 5-year EFS was 58% (95% confidence interval 51× 65%) and 5-year OS was 82% (95% confidence interval 76–87%; Table 4 and Figure 2).
Table 3.
Demographic and clinical details of 530 patients with iAMP21-positive ALL
| No. | Percent | |
|---|---|---|
| Total number | 530 | 100% |
| Sex | ||
| Female | 271 | 51% |
| Male | 255 | 48% |
| Unknown | 4 | 1% |
| Age (years)a | ||
| Mean | 10 | |
| Median | 9 | |
| Range | 2–30 | |
| <10 years, n (%) | 245 | 50% |
| >10 years, n (%) | 242 | 50% |
| White cell count (× 109/l)b | ||
| Mean | 17.1 | |
| Median | 5 | |
| Range | 0.3–900 | |
| <20 × 109/l | 384 | 83% |
| 20–50 × 109/l | 43 | 9% |
| >50 × 109/l | 31 | 7% |
| NCI risk statusc | ||
| Standard | 211 | 48% |
| High | 232 | 52% |
| Complete remissiond | ||
| Yes | 280 | 99% |
| <35 days | 214 | 76% |
| ≥35 days | 66 | 24% |
| Relapsed | ||
| Total patients | 90 | 32% |
| Site of relapse | ||
| Bone marrow | 56 | 62% |
| Isolated CNS | 20 | 22% |
| Combined | 10 | 11% |
| Other | 4 | 4% |
| Time to relapse | ||
| <18 months | 13 | 14% |
| 18 month to 3 years | 38 | 42% |
| 3–4 years | 21 | 23% |
| 4–5 years | 11 | 12% |
| 5 + years | 7 | 8% |
| Deathd | ||
| Total patients | 47 | 17% |
| Post relapse | 40 | 85% |
| In first CR | 5 | 11% |
| Nonremitters | 2 | 4% |
| Treatment schedulee | ||
| High risk | 121 | 43% |
| Standard risk | 158 | 57% |
Abbreviations: ALL, acute lymphoblastic leukaemia; CNS, central nervous system; CR, complete remission; iAMP21, intrachromosomal amplification of chromosome 21; NCI, National Cancer Institute.
Age available for 493 patients.
White cell count (WCC) available for 458 patients.
NCI risk status available for 443 patients.
Outcome data were assessed for 283 patients.
Treatment schedule was known for 279 patients.
Table 4.
Outcome data
| 3 Years
|
5 Years
|
HR (95% CI), P-value | |||
|---|---|---|---|---|---|
| Estimate % | 95% Confidence Interval | Estimate % | 95% Confidence Interval | ||
| RFS | |||||
| Total | 79 | 74–84 | 60 | 52–67 | |
| Treated HR | 87 | 79–92 | 73 | 62–81 | 0.68 (0.54, 0.86), P = 0.001 |
| Not treated HR | 74 | 66–80 | 51 | 40–60 | |
| NCI-SR | 76 | 67–83 | 48 | 35–59 | 0.65 (0.43, 1.00), P = 0.048 |
| NCI-HR | 83 | 76–89 | 69 | 59–77 | |
| EFS | |||||
| Total | 77 | 71–82 | 58 | 51–65 | |
| Treated HR | 84 | 75–89 | 70 | 59–78 | 0.73 (0.59, 0.91), P = 0.005 |
| Not treated HR | 72 | 64–79 | 50 | 39–59 | |
| NCI-SR | 74 | 65–81 | 47 | 35–58 | 0.71 (0.47, 1.05), P = 0.089 |
| NCI-HR | 80 | 72–86 | 66 | 57–74 | |
| OS | |||||
| Total | 91 | 86–94 | 82 | 76–87 | |
| Treated HR | 89 | 81–93 | 83 | 74–90 | 0.97 (0.72, 1.30), P = 0.829 |
| Not treated HR | 92 | 86–95 | 81 | 72–87 | |
| NCI-SR | 92 | 85–96 | 81 | 71–88 | 1.12 (0.63, 2.00), P = 0.688 |
| NCI-HR | 89 | 83–94 | 82 | 74–88 | |
Abbreviations: CI, confidence interval; EFS, event-free survival; HR, hazard ratio; NCI, National Cancer Institute; NCI-HR, NCI high risk; OS, overall survival; RFS, relapse-free survival; SR, standard risk. RFS (median follow-up time 3.5 years). EFS (median follow-up time 3.4 years). OS (median follow-up time 4.0 years).
Figure 2.
Kaplan–Meier survival graph showing the EFS and OS of 283 patients with iAMP21.
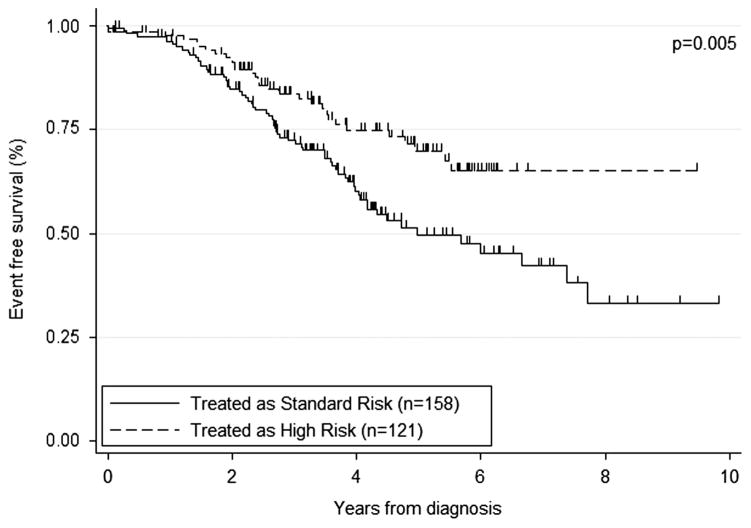
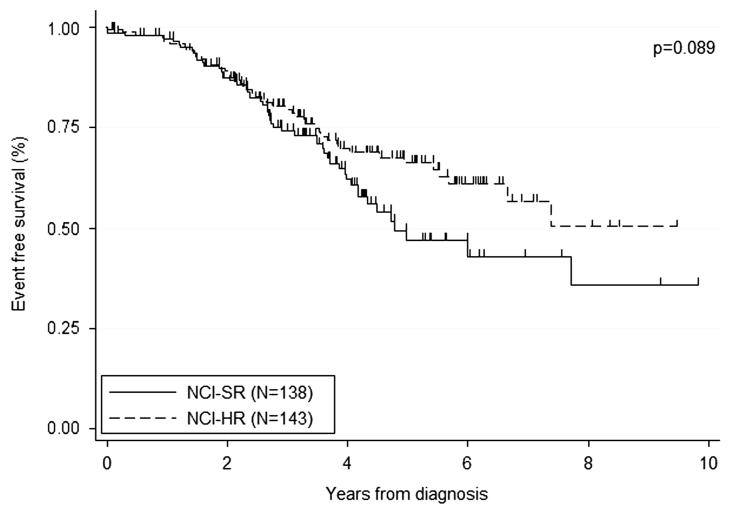
Whether the patients were treated as high risk or not within their protocols was known for 99% patients: 121 (43%) were treated as high whereas the remainder (158, 57%) were treated as nonhigh risk. Most NCI-HR patients were treated as high risk (97/141, 69%), whereas only a small number of NCI-SR were treated as high risk (23/137, 17%; Table 4). There was a significant improvement in outcome for patients treated as high-risk compared with nonhigh-risk protocols with 5-year EFS of 70% and 50%, respectively (P = 0.005; Figure 3 and Table 4). However, there was no difference in 5-year OS rates for those treated on high and nonhigh risk protocols, 83 vs 81% (P = 0.829), suggesting that many of the relapses following nonhigh-risk treatment were salvaged with retrieval therapy. There was a nonsignificant trend toward improved outcome for NCI-HR patients, most of whom received high-risk therapy, as compared with NCI-SR patients, many of whom received nonhigh-risk therapy (5-year EFS 66% vs 47%, P = 0.089; Figure 4).
Figure 3.
Kaplan–Meier survival graph showing the proportion of iAMP21 surviving event free according to whether or not they were treated as high risk on their respective protocols.
Figure 4.
Kaplan–Meier survival graph showing the proportion of iAMP21 surviving event free according to whether or not they were classified as NCI-SR (<10 years old and WCC <50 × 109/l) or NCI-HR (≥ 10years old or WCC >50 × 109/l).
DISCUSSION
This large international study of iAMP21 patients with BCP-ALL has confirmed the demographic and clinical findings from other series involving smaller patient cohorts.2,16,17 The abnormality is found predominantly in older children with a low WCC. The oldest patient identified in this study was 30 years old. This upper age limit remained consistent even when adult ALL series were scrutinised. The male-to-female ratio was 1:1, indicating a slightly higher incidence of females compared with other BCP-ALL subtypes in which the male-to-female ratio was 1.4:1. Significantly, this study showed overall that patients treated as high risk had a significantly reduced risk of relapse compared with those treated as nonhigh risk. As this study involved patients treated on a number of different protocols, in-depth analyses were not possible. However, these observations reflect the findings of two studies involving iAMP21 patients treated on defined protocols. First, a retrospective study from the United Kingdom had previously shown greatly reduced EFS for iAMP21 patients treated as standard risk on a single treatment trial.16 As a result of these observations, in the subsequent UK trial, iAMP21 patients were prospectively identified, stratified as high risk and treated on the most intensive treatment arm. The consequence was a significant improvement in EFS and reduction in relapse rate.13 Similar findings were seen in a pair of contemporary COG trials.14 When children with NCI-SR ALL were treated less intensively by COG, those with iAMP21 had a significantly inferior outcome compared with those patients without iAMP21. In contrast, children with NCI-HR ALL were treated more intensively by COG and there was no significant difference in the outcome of those with/without iAMP21. Many of the patients investigated in these two publications are included in this study. However, by excluding those patients treated by COG and NCRI, we showed the same trend in EFS for patients treated on the remaining protocols, indicating that the survival data presented here were not solely because of the predominating COG and UK cases. These combined findings suggest that patients with iAMP21 should not receive less intensive therapy regimens and would likely benefit from intensification of treatment regardless of the backbone chemotherapy regimen to which they are assigned. Indeed, the level of minimal residual disease (MRD) could be taken into consideration for risk stratification. We showed that in the UK trial, UKALL2003, only 50% of iAMP21 patients were MRD positive after induction, and there was no clear link to outcome. However, because most of the MRD-positive patients (16 of 23 patients in this trial) received transplantation, whereas most MRD-negative patients did not receive transplantation (20 of 22 patients), it was not possible to assess the prognostic significance of MRD in iAMP21 patients. Other studies have produced conflicting results. The BFM group found that iAMP21 patients who were MRD positive had a poorer outcome compared with MRD-negative patients,17 whereas the results from the recent COG study suggested that MRD may not be prognostically relevant in this subgroup.14 However, the COG and UK studies concluded that future iAMP21 patients should be treated intensively irrespective of MRD. In the United Kingdom, it is recommended that patients with iAMP21 are enrolled on our current pediatric trial, UKALL2011. In accordance with the protocol, iAMP21 are transferred to regimen C where they receive eight additional doses of pegylated asparaginase and two courses of Capizzi interim maintenance, which includes escalating doses of intravenous methotrexate.13
As a result of these treatment implications, it is imperative that this chromosomal abnormality is accurately identified. The current recommendation is to use FISH with probes targeting the RUNX1 gene that is located within the region on chromosome 21 with the highest level of amplification.24 This approach is reliable when metaphases are available to indicate that the additional signals are located on a single abnormal chromosome 21. If only interphase cells are available for analysis, additional FISH tests are required to identify iAMP21 from high hyperdiploidy with multiple copies of intact chromosomes 21.24 This study has provided the opportunity for in-depth karyotype analysis to determine the complete cytogenetic profile of iAMP21 patients. Although largely exclusive of known established chromosomal abnormalities, a small number of patients showed an association between iAMP21 and ETV6-RUNX1, BCR-ABL1 or high hyperdiploidy. However, these cases are extremely rare. It is known that gain of RUNX1 is often associated with ETV6-RUNX1, either in the form of gain of a normal or derived chromosome 21. There have also been reports of duplication of RUNX1 on the same chromosome 21 in association with the fusion.35 To our knowledge, there is one case report of iAMP21 associated with the fusion.33 This case is included in this study (no. 525) along with two newly identified patients (nos. 528 and 530). A second case in the literature described the ETV6-RUNX1 fusion as a secondary event to amplification of RUNX1; case 4 in Ma et al.36 Although this report was published before the first definition of iAMP21,1 this patient showed the characteristic iAMP21 array comparative genomic hybridization profile.
This study has shown that patients with iAMP21 display a unique spectrum of secondary genetic abnormalities. These include gain of chromosomes X, 10 or 14 in the absence of high hyperdiploidy, or monosomy 7/deletion of 7q, deletions of 11q, including the MLL gene, P2RY8-CRLF2, deletions of ETV6 and RB1. The characteristic additional cytogenetic abnormalities in association with the characteristic profile may now be used for improved diagnosis. Although Down syndrome was rare, with only one Down syndrome patient identified, other constitutional abnormalities of chromosome 21, including rings and Robertsonian translocations, specifically rob(15;21)(q10;q10)c, were related to iAMP21 in these patients.
This study has confirmed that treatment of iAMP21 patients as high risk provides a significant improvement in outcome. In view of the treatment implications, accurate detection of iAMP21 is important, indicating a need for World Health Organisation (WHO) to recognise this subgroup of BCP-ALL as a distinct entity. Particular chromosomal abnormalities are associated with iAMP21 that assist in improving identification of iAMP21 patients. In the absence of cytogenetics, FISH with a probe for ETV6-RUNX1 will identify the additional RUNX1 signals and at the same time give an indication of ETV6 status. MLL FISH will identify associated 11q deletions, whereas MLPA will highlight the P2RY8-CRLF2 and RB1, as well as ETV6 deletions commonly associated with iAMP21.
Supplementary Material
Acknowledgments
We thank the member laboratories of the UK Cancer Cytogenetics Group, the National Cancer Research Institute and Childhood Leukaemia Subgroup, UK, those cytogenetics laboratories contributing to the COG trails in the United States and all other participating cytogenetics laboratories and treating physicians worldwide. We are grateful for financial support from Leukaemia and Lymphoma Research, Grants CA13539 and CA98543 from the National Institutes of Health to the COG, Swedish Childhood Cancer Foundation, FWO-Vlaanderen, Vlaamse liga tegen Kanker and NKP 29-020, Jubilämsfonds Österreichische Nationalbank (ÖNB No. 14133).
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
CJH, OAH and AVM designed the study. CJH and AVM organised data collection. CJH and CS analysed the cytogenetic, FISH and MLPA data. CS, AJC and NAH reviewed the cytogenetic data. CS, SS and KN carried out and interpreted the MLPA data. CS, AJC, EAR, MD, SS, KN, JH, AT-S, MZ, ND, AB, JS, M-FA, AA, GM, BS, GC, LC, PV, EF, IH, SCR, MP, J-PB, JT, CH, AV, SPH, NAH and OAH contributed cytogenetic, FISH and/or SNP data and/or clinical and follow-up data. CJH, AVM, SPH and OAH wrote the manuscript that was critically reviewed by all authors.
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
References
- 1.Harewood L, Robinson H, Harris R, Al Obaidi MJ, Jalali GR, Martineau M, et al. Amplification of AML1 on a duplicated chromosome 21 in acute lymphoblastic leukemia: a study of 20 cases. Leukemia. 2003;17:547–553. doi: 10.1038/sj.leu.2402849. [DOI] [PubMed] [Google Scholar]
- 2.Soulier J, Trakhtenbrot L, Najfeld V, Lipton JM, Mathew S, Avet-Loiseau H, et al. Amplification of band q22 of chromosome 21, including AML1, in older children with acute lymphoblastic leukemia: an emerging molecular cytogenetic subgroup. Leukemia. 2003;17:1679–1682. doi: 10.1038/sj.leu.2403000. [DOI] [PubMed] [Google Scholar]
- 3.Alvarez Y, Coll MD, Bastida P, Ortega JJ, Caballin MR. AML1 amplification in a child with acute lymphoblastic leukemia. Cancer Genet Cytogenet. 2003;140:58–61. doi: 10.1016/s0165-4608(02)00639-8. [DOI] [PubMed] [Google Scholar]
- 4.Dube ID, el Solh H. An apparent tandem quadruplication of chromosome 21 in a case of childhood acute lymphoblastic leukemia. Cancer Genet Cytogenet. 1986;23:253–256. doi: 10.1016/0165-4608(86)90185-8. [DOI] [PubMed] [Google Scholar]
- 5.Le Coniat M, Romana SP, Berger R. Partial chromosome 21 amplification in a child with acute lymphoblastic leukemia. Genes Chromosomes Cancer. 1995;14:204–209. doi: 10.1002/gcc.2870140308. [DOI] [PubMed] [Google Scholar]
- 6.Busson-Le Coniat M, Nguyen KF, Daniel MT, Bernard OA, Berger R. Chromosome 21 abnormalities with AML1 amplification in acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2001;32:244–249. doi: 10.1002/gcc.1188. [DOI] [PubMed] [Google Scholar]
- 7.Baialardo EM, Felice MS, Rossi J, Barreiro C, Gallego MS. Tandem triplication and quadruplication of chromosome 21 in childhood acute lymphoblastic leukemia. Cancer Genet Cytogenet. 1996;92:43–45. doi: 10.1016/s0165-4608(96)00151-3. [DOI] [PubMed] [Google Scholar]
- 8.Niini T, Kanerva J, Vettenranta K, Saarinen-Pihkala UM, Knuutila S. AML1 gene amplification: a novel finding in childhood acute lymphoblastic leukemia. Haematologica. 2000;85:362–366. [PubMed] [Google Scholar]
- 9.Dal Cin P, Atkins L, Ford C, Ariyanayagam S, Armstrong SA, George R, et al. Amplification of AML1 in childhood acute lymphoblastic leukemias. Genes Chromosomes Cancer. 2001;30:407–409. doi: 10.1002/1098-2264(2001)9999:9999<::aid-gcc1107>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 10.Mathew S, Rao PH, Dalton J, Downing JR, Raimondi SC. Multicolor spectral karyotyping identifies novel translocations in childhood acute lymphoblastic leukemia. Leukemia. 2001;15:468–472. doi: 10.1038/sj.leu.2401989. [DOI] [PubMed] [Google Scholar]
- 11.Morel F, Herry A, Le Bris MJ, Douet-Guilbert N, Le Calvez G, Marion V, et al. AML1 amplification in a case of childhood acute lymphoblastic leukemia. Cancer Genet Cytogenet. 2002;137:142–145. doi: 10.1016/s0165-4608(02)00566-6. [DOI] [PubMed] [Google Scholar]
- 12.Penther D, Preudhomme C, Talmant P, Roumier C, Godon A, Mechinaud F, et al. Amplification of AML1 gene is present in childhood acute lymphoblastic leukemia but not in adult, and is not associated with AML1 gene mutation. Leukemia. 2002;16:1131–1134. doi: 10.1038/sj.leu.2402479. [DOI] [PubMed] [Google Scholar]
- 13.Moorman AV, Robinson H, Schwab C, Richards SM, Hancock J, Mitchell CD, et al. Risk-directed treatment intensification significantly reduces the risk of relapse among children and adolescents with acute lymphoblastic leukemia and intrachromosomal amplification of chromosome 21: a comparison of the MRC ALL97/99 and UKALL2003 trials. J Clin Oncol. 2013;31:3389–3396. doi: 10.1200/JCO.2013.48.9377. [DOI] [PubMed] [Google Scholar]
- 14.Heerema NA, Carroll AJ, Devidas M, Loh ML, Borowitz MJ, Gastier-Foster JM, et al. Intrachromosomal amplification of chromosome 21 is associated with inferior outcomes in children with acute lymphoblastic leukemia treated in contemporary standard-risk Children’s Oncology Group studies: a report from the Children’s Oncology Group. J Clin Oncol. 2013;31:3397–3402. doi: 10.1200/JCO.2013.49.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinson HM, Broadfield ZJ, Cheung KL, Harewood L, Harris RL, Jalali GR, et al. Amplification of AML1 in acute lymphoblastic leukemia is associated with a poor outcome. Leukemia. 2003;17:2249–2250. doi: 10.1038/sj.leu.2403140. [DOI] [PubMed] [Google Scholar]
- 16.Moorman AV, Richards SM, Robinson HM, Strefford JC, Gibson BE, Kinsey SE, et al. Prognosis of children with acute lymphoblastic leukemia (ALL) and intrachromosomal amplification of chromosome 21 (iAMP21) Blood. 2007;109:2327–2330. doi: 10.1182/blood-2006-08-040436. [DOI] [PubMed] [Google Scholar]
- 17.Attarbaschi A, Mann G, Panzer-Grumayer R, Rottgers S, Steiner M, Konig M, et al. Minimal residual disease values discriminate between low and high relapse risk in children with B-cell precursor acute lymphoblastic leukemia and an intrachromosomal amplification of chromosome 21: the Austrian and German acute lymphoblastic leukemia Berlin-Frankfurt-Munster (ALL-BFM) trials. J Clin Oncol. 2008;26:3046–3050. doi: 10.1200/JCO.2008.16.1117. [DOI] [PubMed] [Google Scholar]
- 18.Rand V, Parker H, Russell LJ, Schwab C, Ensor H, Irving J, et al. Genomic characterization implicates iAMP21 as a likely primary genetic event in childhood B-cell precursor acute lymphoblastic leukemia. Blood. 2011;117:6848–6855. doi: 10.1182/blood-2011-01-329961. [DOI] [PubMed] [Google Scholar]
- 19.Robinson HM, Harrison CJ, Moorman AV, Chudoba I, Strefford JC. Intrachromosomal amplification of chromosome 21 (iAMP21) may arise from a breakage-fusion-bridge cycle. Genes Chromosomes Cancer. 2007;46:318–326. doi: 10.1002/gcc.20412. [DOI] [PubMed] [Google Scholar]
- 20.Strefford JC, Van Delft FW, Robinson HM, Worley H, Yiannikouris O, Selzer R, et al. Complex genomic alterations and gene expression in acute lymphoblastic leukemia with intrachromosomal amplification of chromosome 21. Proc Natl Acad Sci USA. 2006;103:8167–8172. doi: 10.1073/pnas.0602360103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuchinskaya E, Nordgren A, Heyman M, Schoumans J, Corcoran M, Staaf J, et al. Tiling-resolution array-CGH reveals the pattern of DNA copy number alterations in acute lymphoblastic leukemia with 21q amplification: the result of telomere dysfunction and breakage/fusion/breakage cycles? Leukemia. 2007;21:1327–1330. doi: 10.1038/sj.leu.2404628. [DOI] [PubMed] [Google Scholar]
- 22.Sinclair PB, Parker H, An Q, Rand V, Ensor H, Harrison CJ, et al. Analysis of a breakpoint cluster reveals insight into the mechanism of intrachromosomal amplification in a lymphoid malignancy. Hum Mol Genet. 2011;20:2591–2602. doi: 10.1093/hmg/ddr159. [DOI] [PubMed] [Google Scholar]
- 23.Harrison CJ. Cytogenetics of paediatric and adolescent acute lymphoblastic leukaemia. Br J Haematol. 2009;144:147–156. doi: 10.1111/j.1365-2141.2008.07417.x. [DOI] [PubMed] [Google Scholar]
- 24.Harrison CJ, Haas O, Harbott J, Biondi A, Stanulla M, Trka J, et al. Detection of prognostically relevant genetic abnormalities in childhood B-cell precursor acute lymphoblastic leukaemia: recommendations from the Biology and Diagnosis Committee of the International Berlin-Frankfurt-Munster study group. Br J Haematol. 2010;151:132–142. doi: 10.1111/j.1365-2141.2010.08314.x. [DOI] [PubMed] [Google Scholar]
- 25.Arico M, Schrappe M, Hunger SP, Carroll WL, Conter V, Galimberti S, et al. Clinical outcome of children with newly diagnosed Philadelphia chromosome-positive acute lymphoblastic leukemia treated between 1995 and 2005. J Clin Oncol. 2010;28:4755–4761. doi: 10.1200/JCO.2010.30.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schrappe M, Hunger SP, Pui CH, Saha V, Gaynon PS, Baruchel A, et al. Outcomes after induction failure in childhood acute lymphoblastic leukemia. N Engl J Med. 2012;366:1371–1381. doi: 10.1056/NEJMoa1110169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bungaro S, Dell’Orto MC, Zangrando A, Basso D, Gorletta T, Lo Nigro L, et al. Integration of genomic and gene expression data of childhood ALL without known aberrations identifies subgroups with specific genetic hallmarks. Genes Chromosomes Cancer. 2009;48:22–38. doi: 10.1002/gcc.20616. [DOI] [PubMed] [Google Scholar]
- 28.Shaffer LG, Slovak ML, Campbell LJ, editors. ISCN. An International System for Human Cytogenetic Nomenclature. S. Karger; Basel: 2009. [Google Scholar]
- 29.Harrison CJ, Moorman AV, Barber KE, Broadfield ZJ, Cheung KL, Harris RL, et al. Interphase molecular cytogenetic screening for chromosomal abnormalities of prognostic significance in childhood acute lymphoblastic leukaemia: a UK Cancer Cytogenetics Group Study. Br J Haematol. 2005;129:520–530. doi: 10.1111/j.1365-2141.2005.05497.x. [DOI] [PubMed] [Google Scholar]
- 30.Schwab CJ, Jones LR, Morrison H, Ryan SL, Yigittop H, Schouten JP, et al. Evaluation of multiplex ligation-dependent probe amplification as a method for the detection of copy number abnormalities in B-cell precursor acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2010;49:1104–1113. doi: 10.1002/gcc.20818. [DOI] [PubMed] [Google Scholar]
- 31.Moorman AV, Richards SM, Martineau M, Cheung KL, Robinson HM, Jalali GR, et al. Outcome heterogeneity in childhood high-hyperdiploid acute lymphoblastic leukemia. Blood. 2003;102:2756–2762. doi: 10.1182/blood-2003-04-1128. [DOI] [PubMed] [Google Scholar]
- 32.Sutcliffe MJ, Shuster JJ, Sather HN, Camitta BM, Pullen J, Schultz KR, et al. High concordance from independent studies by the Children’s Cancer Group (CCG) and Pediatric Oncology Group (POG) associating favorable prognosis with combined trisomies 4, 10, and 17 in children with NCI standard-risk B-precursor acute lymphoblastic leukemia: a Children’s Oncology Group (COG) initiative. Leukemia. 2005;19:734–740. doi: 10.1038/sj.leu.2403673. [DOI] [PubMed] [Google Scholar]
- 33.Haltrich I, Csoka M, Kovacs G, Torok D, Alpar D, Ottoffy G, et al. Six cases of rare gene amplifications and multiple copy of fusion gene in childhood acute lymphoblastic leukemia. Pathol Oncol Res. 2013;19:123–128. doi: 10.1007/s12253-012-9533-9. [DOI] [PubMed] [Google Scholar]
- 34.Schwab CJ, Chilton L, Morrison H, Jones L, Al-Shehhi H, Erhorn A, et al. Genes commonly deleted in childhood B-cell precursor acute lymphoblastic leukemia: association with cytogenetics and clinical features. Haematologica. 2013;98:1081–1088. doi: 10.3324/haematol.2013.085175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Broadfield ZJ, Hain RD, Harrison CJ, Reza JG, McKinley M, Michalova K, et al. Complex chromosomal abnormalities in utero, 5 years before leukaemia. Br J Haematol. 2004;126:307–312. doi: 10.1111/j.1365-2141.2004.05036.x. [DOI] [PubMed] [Google Scholar]
- 36.Ma SK, Wan TS, Cheuk AT, Fung LF, Chan GC, Chan SY, et al. Characterization of additional genetic events in childhood acute lymphoblastic leukemia with TEL/AML1 gene fusion: a molecular cytogenetics study. Leukemia. 2001;15:1442–1447. doi: 10.1038/sj.leu.2402202. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.