Abstract
Pediatric sarcomas represent a diverse group of rare bone and soft tissue malignancies. Though the molecular mechanisms that propel the development of these cancers are not well understood, identification of tumor-specific translocations in many sarcomas has provided significant insight into their tumorigenesis. Each fusion protein resulting from these chromosomal translocations is thought to act as a driving force in the tumor, either as an aberrant transcription factor, constitutively active growth factor, or ligand-independent receptor tyrosine kinase. Identification of transcriptional targets or signaling pathways modulated by these oncogenic fusions has led to the discovery of potential therapeutic targets. Some of these targets have shown considerable promise in pre-clinical models and are currently being tested in clinical trials. This review summarizes the molecular pathology of a subset of pediatric sarcomas with tumor-associated translocations and how increased understanding at the molecular level is being translated to novel therapeutic advances.
INTRODUCTION
Sarcomas are a rare, heterogeneous group of neoplasms presumed to be of mesenchymal origin. Tumors can arise in bone or soft tissues such as muscle and fat and can develop anywhere in the body. They account for only 1% of all malignancies, and although the incidence is higher in adults, sarcomas occur with higher frequency in children. Each year, between 1500 and 1600 children and young adults in the United States develop these malignant bone and soft tissue tumors comprising approximately 13% of cancers afflicting patients below the age of 201,2. The overall five-year survival rate for pediatric sarcomas is approximately 60% and falls closer to 20–30% for recurrent and metastatic cases despite aggressive surgery, multi-agent chemotherapy, and/or radiation.
Sarcomas can be divided into two groups based on the underlying molecular events that initiate tumorigenesis. The first group is characterized by the presence of specific chromosomal translocations (Table 1) or activating mutations while the second is more cytogenically complex. Sarcomas with complex karyotypes predominantly afflict older patients and translocations tend to be observed with higher frequency in pediatric cases. Pediatric sarcomas with tumor-associated translocations include Ewing sarcoma, rhabdomyosarcoma, synovial sarcoma, and dermatofibrosarcoma protuberans. The genetic aberrations in these neoplasms produce defined fusions that are critical for sarcomagenesis. Depending on the genes involved in the fusion, the resulting protein can promote tumor progression through transcriptional modulation, epigenetic modifications, or activation of oncogenic signaling pathways.
Table 1.
Sarcomas with defined chromosomal translocations
| Tumor type | Translocation | Fusion |
|---|---|---|
| ES | t(11;22)(q24;q12) | EWS-FLI1 |
| t(21;22)(q22;q12) | EWS-ERG | |
| t(7;22)(p22;q12) | EWS-ETV1 | |
| t(17;22)(q21;q12) | EWS-ETV4 | |
| t(2;22)(q33;q12) | EWS-FEV | |
| t(16;21)(p11;q22) | FUS-ERG | |
| t(2;16)(q35;p11) | FUS-FEV | |
| (t(1;22)(p36.1;q12) | EWS-ZSG | |
| t(20;22)(q13;q12) | EWS-NFATc2 | |
| ES-like tumors (CD-99 negative) | t(6;22)(p21;q12) | EWS-POU5F1 |
| t(1;22)(q36.1;q12) | EWS-PATZI | |
| t(2;22)(q31;q12) | EWS-SP3 | |
| t(4;19)(q35;q13) | CIC-DUX4 | |
| Clear-cell sarcoma | t(12;22)(p13;q12) | EWS-ATF1 |
| t(2;22)(q33;q12) | EWS-BREB1 | |
| Desmoplastic small round-cell tumor | t(11;22)(p13;q12) | EWS-WT1 |
| t(21;22)(q22;q12) | EWS-ERG | |
| Myxoid liposarcoma | t(12;16)(q13;q11) | FUS-DDIT3 |
| t(12;22)(q13;q12) | EWS-DDIT3 | |
| Extraskeletal myxoid chondrosarcoma | t(9;22)(q22–31;q11–12) | EWS-NR4A3 |
| t(9;17)(q22;q11) | TAF15-NR4A3 | |
| t(9;15)(q22;q21) | TCFI2-NR4A3 | |
| t(9;22)(q22;q15) | TFG-NR4A3 | |
| Low grade fibromyxoid sarcoma | t(7;16)(q33;p11) | FUS-CREB3L2 |
| t(11;16)(p11;p11) | FUS-CREB3L1 | |
| Angiomatoid fibrous histiocytoma | t(12;16)(q13;p11) | FUS-ATF1 |
| t(12;22)(q13;q12) | EWS-ATF1 | |
| t(2;22)(q33;q12) | EWS-CREB1 | |
| Alveolar rhabdomyosarcoma | t(2;13)(q35;q14) | PAX3-FOXO1 |
| t(1;13)(q36;q14) | PAX7-FOXO1 | |
| t(2;2)(p23;q35) | PAX3-NCOA1 | |
| t(2;8)(q35;q13) | PAX3-NCOA2 | |
| t(8;13;9)(p11.2;q14;9q32) | FGFR1-FOXO1 | |
| Alveolar soft part sarcoma | t(X;17)(p11;q25) | ASPSL-TFE3 |
| Congenital fibrosarcoma | t(12;15)(p13;q25) | ETV6-NTRK3 |
| Congenital mesoblastic nephroma | t(12;15)(p13;q25) | ETV6-NTRK3 |
| Inflammatory myofibroblastic tumor | t(1;2)(q25;q23) | TPM3-ALK |
| t(2;19)(q23;q13) | TPM4-ALK | |
| t(2;17)(q23;q23) | CLTC-ALK | |
| t(2;2)(p23;q13) | RANBP2-ALK | |
| Synovial sarcoma | t(X;18)(p11;q11) | SS18-SSX1 |
| t(X;18)(p11;q11) | SS18-SSX2 | |
| t(X;18)(p11;q13) | SS18-SSX4 | |
| t(X;20)(p11;q13) | SS18L1-SSX1 | |
| Endometrial sarcoma | t(7;17)(p15;q21) | JAZF1-SUZ12 |
| t(6;7)(p21;p15) | JAZF1-PHF1 | |
| t(6;10)(p21;p11) | EPC1-PHF1 | |
| Dermatofibrosarcoma protuberans | t(17;22)(q22;q13) | COL1A1-PDGFB |
| Giant cell fibroblastoma | t(17;22)(q22;q13) | COL1A1-PDGFB |
ES, Ewing sarcoma.
Due to the rarity of sarcomas, especially when considering the prevalence of individual subtypes, they are understudied cancers. As the fusion proteins present within translocation-associated sarcomas are inherent to tumor development, they provide an avenue of research for development of improved and targeted treatments. This review focuses on four pediatric sarcomas with tumor-associated translocations and will discuss the molecular genetics of these malignancies, potential therapeutic targets, and the status of agents directed against these targets in clinical trials.
Ewing sarcoma
Ewing sarcoma (ES) is part of a group of bone and soft tissue small round blue cell malignancies that predominantly occur within the second decade of life. ES is the second most common bone malignancy in children and adolescents, with an annual incidence of 2.93 per million in the United States3, and accounts for 3% of pediatric cancers. The 5-year survival rate is nearing 70%, although this number shrinks to approximately 20–30% in patients with metastatic, refractory and/or recurrent disease4. The current treatment of ES is multimodal consisting of intensive multi-agent chemotherapy, surgery and/or high dose radiation therapy. The standard systemic chemotherapeutic regimen includes cycles of vincristine, cyclophosphamide, and doxorubicin with ifosfamide and etoposide.
ES occurs primarily as tumors of the bone, with less than 15% of cases arising in extraosseous locations5. ES primary osseous sites are split between the extremities and the central axis, with an increased tendency for incidence in the shaft of long tubular bones, pelvis, and rib. The histogenesis of ES has long been disputed, as evidence has been provided for both a neural crest and mesenchymal stem cell origin.
Molecular Genetics
The pathognomonic genetic aberration in ES fuses the EWS gene (also known as EWSR1, Ewing sarcoma breakpoint region 1) to one of five ETS (erythroblast transformation-specific) transcription factors (Table 1). FLI1 (Friend leukemia virus integration 1) is the fusion partner in approximately 85% of cases and ERG (v-ets erythroblastosis virus E26 oncogene homolog) in about 10%, while the others each account for less than 1%6. In very rare cases, FUS combines with ERG or FEV (ETS oncogene family) and EWS is juxtaposed to non-ETS genes7 (Table 1). Adding to the complexity of multiple translocations, variation in the location of the chromosomal breakpoint results in numerous types of each fusion. Initial reports suggested EWS-FLI1 type 1 fusions confer a prognostic advantage to patients with localized disease, but more recent studies have demonstrated there is no difference in clinical outcome based on fusion type8.
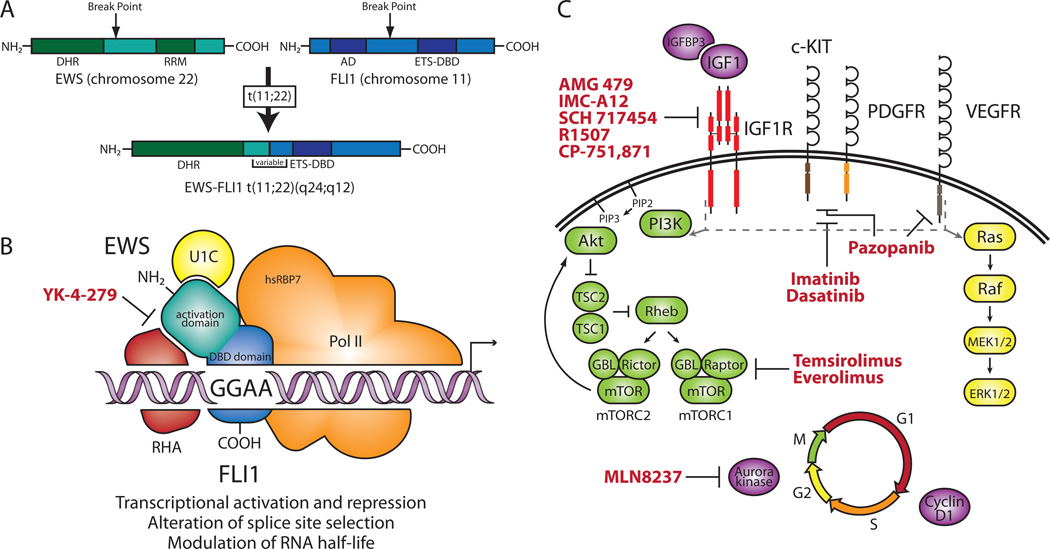
The most common EWS-ETS fusion, EWS-FLI1, is generated from the t(11;22)(q24;q12) reciprocal translocation that combines the N-terminal EWS activation domain and the C-terminal ETS DNA-binding domain of FLI19 (Figure 1A). EWS is a member of the FET (FUS, EWS, and TAF15) family of proteins, which may be involved in transcription and mRNA splicing as they contain both an activation and RNA-binding domain10. The ETS family constitutes a group of 30 transcription factors characterized by the presence of a highly conserved ETS domain that mediates site-specific DNA binding. They also possess either an activation or a repression domain and are involved in various cellular processes such as cell proliferation and differentiation11. Because the more potent EWS activation domain replaces that of FLI1 while the FLI1 DNA-binding domain remains intact, EWS-FLI1 is thought to primarily act as an aberrant transcription factor. As most ETS transcription factors bind to a core consensus (GGAA/T), EWS/FLI1 has also been shown to interact with DNA in a site-specific manner through association with GGAA microsatellites12. Interactions with splicing factors and the ability to alter splice site selection suggest EWS-FLI1 also plays a role in RNA splicing13,14. The capacity of EWS-FLI1 to function post-transcriptionally is further demonstrated by modulation of target gene RNA half-life15. (Figure 1B)
Figure 1. Molecular genetics and targeted therapies in ES.
Molecular genetics and targeted therapies in ES. (A) Schematic of EWS-FLI1 t(11;22)(q24;q12) translocation. The EWS-FLI1 fusion includes the N-terminal activation domain of EWS, which contains multiple degenerate hexapeptide repeats (consensus SYGQQS), and the C-terminal ETS DNA-binding domain (ETS-DBD) of FLI1. The RNA recognition motif (RRM) of EWS and the activation domain (AD) of FLI1 are not retained in the fusion. Variation in the sites of chromosomal break points leads to multiple fusion types (bracketed region). (B) Putative molecular function of the EWS-FLI1 protein and selected protein–protein interactions. As an aberrant transcription factor, EWS-FLI1 regulates genes in part by binding to GGAA microsatellites upstream of target genes. EWS-FLI1 has been shown to interact with the splicing factor U1C (also known as SNRPC, small nuclear ribonucleoprotein polypeptide c), RNA helicase A (RHA), and the hRBP7 subunit of RNA polymerase II (Pol II), which links the protein to splicing and transcription. The small molecule that blocks the EWS-FLI1–RHA interaction is indicated in red. (C) Signaling pathways and targeted therapies in ES. EWS-FLI1 modulation of IGFBP3 and IGF1 and overexpression of IGF1R promote increased IGF1 signaling. ES also expresses PDGFR, c-KIT, and VEGFR. Activation of IGF1R, PDGFR, c-KIT, and VEGFR leads to downstream signaling through the PI3K and MAPK pathways (indicated by gray dashed line and arrows). EWS-FLI1 upregulates Aurora kinase A and cyclin D1, promoting progression through the cell cycle. Targeted therapeutic agents used in recent clinical trials for ES are indicated in red. Genes modulated by EWS-FLI1 are indicated in purple. Receptors overexpressed in ES are indicated in red. ES, Ewing sarcoma; ETS, erythroblast transformation-specific; FLI1, Friend leukemia virus integration 1; IGFBP, insulin-like growth factor binding protein; IGF1, insulin-like growth factor 1; IGF1R, IGF1 receptor; MAPK, mitogen-activated protein kinase; PDGF, platelet-derived growth factor; PDGFR, PDGF receptor; PI3K, phosphoinositide-3-kinase; VEGFR, vascular endothelial growth factor receptor.
Target genes and targeted therapies
Ectopic expression of EWS-FLI1 in heterologous cell types or siRNA-mediated knock down of the fusion in ES cell lines have both been used to discover potential target genes. These studies have identified both up- and down-regulated genes, demonstrating the function of EWS-FLI1 as both a transcriptional activator and repressor. Multiple direct targets have been confirmed through demonstration of EWS-FLI1 binding to their promoters including IGFBP3 (insulin-like growth factor binding protein 3)16 and the Aurora A and B kinases17.
IGFBP3 repression by EWS-FLI1 is one of the multiple connections between the insulin-like growth factor (IGF) pathway and ES pathogenesis. In addition to targeting IGFBP3, EWS-FLI1 has been shown to up-regulate insulin-like growth factor 1 (IGF1) in mesenchymal progenitor cells18. Additionally, ES cell lines ubiquitously express the IGF1 receptor (IGF1R) and display autocrine production of IGF1. Moreover, IGF1R is necessary for EWS/FLI1-mediated cellular transformation and inhibition of IGF1R suppresses tumor growth in vitro and in vivo19. In the clinic, phase I trials of monoclonal antibodies targeting IGF1R (Figure 1C) have shown partial and complete responses in ES patients20. Although these early results were promising, recent phase II studies showed limited response rates of approximately 10% for patients with recurrent or refractory ES21–23. To improve the efficacy of anti-IGF1R therapies, future work is being directed toward identification of predictive biomarkers associated with ES patients that benefit from treatment or combination therapy with other targeted agents (Table 2).
Table 2.
Targeted agents undergoing clinical testing for pediatric sarcomas
| Target | Drug | Phase | NCT Number | Eligible Sarcoma Types | Status |
|---|---|---|---|---|---|
| ALK, c-MET | Crizotinib (PF-02341066) | I/II | NCT01182896 | Sarcoma, OS | Recruiting |
| Aurora kinase A | Alisertib (MLN8237) | II | NCT01154816 | RMS, OS, ES, NRSTS | Recruiting |
| BCL-2 | Oblimersen (G3139) | I | NCT00039481 | ES, OS, SS, DSRCT | Completed |
| c-MET | Tivantinib (ARQ 197) | II | NCT00557609 | CCS, ASPS | Completed |
| Death Receptor-5 | Conatumumab (AMG 655) | I/II | NCT00626704 | Locally advanced, unresectable or metastatic STS | Active |
| EGFR | Cetuximab (IMC-C225) | II | NCT00148109 | EGFR-positive bone and STS | Active |
| HDACs | PCI-24781 | I/II | NCT01027910 | Metastatic or unresectable sarcoma | Recruiting |
| HDACs | SB939 | II | NCT01112384 | Translocation-associated, metastatic sarcomas | Recruiting |
| IGF1R | Cixutumumab (IMC-A12) | II | NCT00668148 | ES, RMS, LMS, LS, SS | Active |
| IGF1R | Cixutumumab (IMC-A12) | II | NCT00831844 | OS, ES, RMS, SS | Recruiting |
| IGF1R | Figitumumab (CP-751,871) | I/II | NCT00560235 | ES | Active |
| IGF1R | Ganitumab (AMG 479) | II | NCT00563680 | ES, DSRCT | Active |
| IGF1R | Teprotumumab (R1507) | II | NCT00642941 | ES, OS, SS, RMS, ASPS, DSRCT, EMC, CCS, MLS | Active |
| IGF1R; mTOR | Cixutumumab; Temsirolimus | I | NCT00880282 | Childhood solid tumor | Recruiting |
| IGF1R; mTOR | Cixutumumab; Temsirolimus | II | NCT01016015 | Mestastatic, locally advanced, or locally recurring bone and STS | Recruiting |
| IGF1R; mTOR | Figitumumab; Everolimus | I | NCT00927966 | Advanced sarcoma | Active |
| mTOR | Everolimus (RAD001) | II | NCT01048723 | Soft tissue extremity and/or retroperitoneal sarcomas | Recruiting |
| mTOR | Everolimus (RAD001) | II | NCT01216839 | RMS and other STS (children and adolescents) | Recruiting |
| PDGFR-α | Olaratumab (IMC-3G3) | I/II | NCT01185964 | STS | Recruiting |
| PDGFR, c-Kit | Imatinib (Gleevec) | II | NCT00031915 | ES, OS, SS, RMS, LS, MPNST, FS, AS | Completed |
| PDGFR, c-Kit | Imatinib (Gleevec) | II | NCT00085475 | DFSP, GCF | Recruiting |
| RAF, VEGFR, PDGFR, c-Kit | Sorafenib | II | NCT00837148 | SS, LMS, MPNST | Recruiting |
| RAF, VEGFR, PDGFR, c-Kit | Sorafenib | II | NCT00330421 | Bone and STS | Completed |
| PDGFR, c-Kit, Src kinases, Eph kinases | Dasatinib | II | NCT00464620 | RMS, MPNST, CS, ES, ASPS, C, EPS, GCTB, HPC, GIST | Active |
| VEGF | Bevacizumab | I/II | NCT01106872 | Locally advanced, unresectable or metastatic STS | Recruiting |
| VEGF | Bevacizumab | II | NCT00643565 | RMS and NRSTS (children and adolescents) | Recruiting |
| VEGF; HDACs | Bevacizumab; Valproic Acid | I/II | NCT01106872 | Locally advanced, unresectable, or metastatic STS | Recruiting |
| VEGFR | Cediranib | II | NCT00942877 | ASPS | Recruiting |
| VEGFR, PDGFR, c-Kit | Sunitinib | II | NCT00400569 | LS, LMS, FS | Active |
| VEGFR, PDGFR, c-Kit; VEGFR | Sunitinib; Cediranib | II | NCT01391962 | ASPS | Recruiting |
| VEGFR, PDGFR, c-Kit | Axitinib | II | NCT01140737 | AS, LMS, SS, RMS, MPNST, fibroblastic, fibrohistiocytic | Recruiting |
| VEGFR, PDGFR, c-Kit | Pazopanib | I | NCT00929903 | STS, DSRCT, extraosseous ES | Recruiting |
| VEGFR, PDGFR, c-Kit | Pazopanib | II | NCT00297258 | STS (LMS, SS, adipocytic tumors) | Active |
| VEGFR, PDGFR, c-Kit | Pazopanib | II | NCT01059656 | DFSP | Not yet recruiting |
| VEGFR, PDGFR, c-Kit | Pazopanib | III | NCT00753688 | Metastatic STS | Active |
AS, angiosarcoma; ALK, anaplastic lymphoma receptor tyrosine kinase; ASPS, alveolar soft part sarcoma; BCL-2, B-cell CLL/lymphoma 2; C, chordoma; CCS, clear-cell sarcoma; CS, chondrosarcoma; c-MET: met proto-oncogene (hepatocyte growth factor receptor); c-KIT, v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog; DFSP, dermatofibrosarcoma protuberans; DSRCT, desmoplastic small-round-cell tumor; EGFR, epidermal growth factor receptor; EMC, extraskeletal myxoid chondrosarcoma; EPS, epitheliod sarcoma; ES, Ewing sarcoma; FS, fibrosarcoma; GCF, giant cell fibrosarcoma; GCTB, giant cell tumor of bone; GIST, gastrointenstinal stromal tumor; HDAC s, histone deacetylases; HPC, hemangiopericytoma; IGF1R, insulin-like growth factor 1 receptor; LMS, leiomyosarcoma; LS, liposarcoma; MLS, myxoid liposarcoma; MPNST, malignant peripheral nerve sheath tumor; mTOR, mammalian target of rapamycin; NRSTS, nonrhabdomyosarcoma soft tissue sarcoma; OS, osteosarcoma; PDGF, platelet-derived growth factor; RAF, v-raf-1 murine leukemia viral oncogene homolog 1; RMS, rhabdomyosarcoma; SS, synovial sarcoma; Src, v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian); STS, soft tissue sarcoma; VEGFR, vascular endothelial growth factor receptor.
Though the IGF pathway has received the most attention, other EWS-FLI1 target genes or interacting proteins provide potential therapeutic targets. EWS-FLI1 up-regulates both Aurora A and Aurora B, cell cycle-regulated serine/threonine kinases that are overexpressed in multiple cancers17. Preclinical testing revealed a maintained complete clinical response for an Aurora kinase A inhibitor (Figure 1C) in an ES xenograft model24. A phase II trial evaluating the effects of this drug in pediatric leukemias and solid tumors, including ES, is currently underway (NCT01154816, Table 2). It has also been shown that RNA helicase A (RHA), a protein involved in the regulation of transcription and splicing, binds to EWS-FLI1 and enhances its transcriptional activity25. Utilization of a small molecule inhibitor to block the RHA-EWS/FLI1 interaction (Figure 1B) induces apoptosis in ES cells and reduces tumor growth in xenografts26. This promising preclinical evidence suggests these agents may be effective in clinical trials.
Alveolar rhabdomyosarcoma
Rhabdomyosarcoma (RMS) is the most common pediatric soft tissue sarcoma, accounting for 5–7% of all malignancies in children and adolescents less than 20 years of age. The overall five-year survival rate is approximately 60%, though is only 30% for those with metastatic disease27. Current therapy for RMS is similar to ES, employing a combination of adjuvant intensive chemotherapy with surgery and/or radiation to the primary and metastatic sites of disease.
RMS is divided into histologic subtypes. Embryonal RMS (ERMS) and alveolar RMS (ARMS) are the two major subtypes, accounting for approximately 60% and 20% of cases, respectively2. RMS tumors can arise anywhere in the body, though frequency of primary sites varies with histological subtype and age of diagnosis. Tumors in unfavorable sites (eg. extremities) are more common in ARMS, which predominantly afflicts adolescents and young adults and confers a worse prognosis than ERMS28. For patients that have refractory, recurrent and/or metastatic ARMS long-term survival is truly the exception rather than the rule. Though the expression of skeletal muscle markers and location of tumors in skeletal muscle suggest a myogenic origin, the exact cell of origin for ARMS remains uncertain. Mouse models and in vitro cell culture systems have provided evidence for both a skeletal muscle and mesenchymal stem cell origin.
Molecular Genetics
In addition to histologic differences, ARMS is distinguished from ERMS by the presence of specific chromosomal translocations present in the majority of ARMS tumors. The predominant translocation, t(2;13)(q35;q14), fuses PAX3 (paired box 3) to FOXO1 (forkhead box O1, also known as FKHR)29. Less commonly, the t(1;13)(p36;q14) translocation fuses another PAX gene, PAX7 (paired box 7), to FOXO130. A recent study also identified rare, noncanonical t(2;2)(p23;35) and t(2:8)(q35;q13) translocations that unite PAX3 with the nuclear receptor transcriptional coactivators NCOA1 or NCOA231 (Table 1).
PAX3 and PAX7 are part of the paired box family of transcription factors, which are involved in embryonic development and myogenesis32. FOXO1 is a member of a subfamily of forkhead transcription factors regulated by the PI3K (phosphoinositide-3-kinase) pathway and believed to play a role in myogenic growth and differentiation33. Both translocation breakpoints consistently occur within the seventh intron of the PAX gene and the first intron of FOXO1, resulting in a chimeric transcription factor that contains the PAX DNA-binding domain and transcriptional activation domain of FOXO1. Fusion type has been found to correlate with clinical outcome, as patients with PAX7-FOXO1-positive tumors had better overall survival rates than those with tumors containing the PAX3-FOXO1 fusion34. More recently, clinical characteristics and prognosis of fusion-negative ARMS were found to be more similar to ERMS than fusion-positive ARMS, implying the presence of the PAX-FOXO1 fusion is more crucial than histology to the underlying biology of the tumor35.
Target genes and targeted therapies
Initial PAX3-FOXO1 target genes were identified by evaluating the expression of known PAX3 target genes in ERMS cells transduced with PAX3-FOXO1. Up-regulation of MET (met proto-oncogene/hepatocyte growth factor receptor) upon PAX3-FOXO1 expression and an observed correlation between MET and PAX3-FOXO1 in tumor samples suggested MET is a downstream target36. The role of MET in ARMS tumorigenesis was further characterized by experiments that demonstrated MET is required for PAX3-FOXO1-mediated transformation of mouse embryonal fibroblasts and shRNA knock down of MET results in decreased tumor growth in vitro and in vivo37. More recently, studies have shown that MET is post-transcriptionally regulated, as low levels of the microRNAs miR-1 and miR-206 result in de-repression and up-regulation of MET in RMS cells. Furthermore, overexpression of these miRNAs promoted myogenic differentiation and inhibited tumor growth in vivo38. Additionally, a preclinical study demonstrated the ability of a c-MET small molecule inhibitor to hinder growth of ARMS cell lines39.
Components of the IGF system are also potential therapeutic targets in ARMS. Heterologous expression of the PAX3-FOXO1 fusion results in an increase in IGF1R levels and overexpression of insulin-like growth factor 2 (IGF2) and IGF1R has been observed in ARMS and ERMS tumors and cell lines40,41. Elevated IGF2 levels in ERMS result from loss of heterozygosity at the 11p15.5 locus while in ARMS it may be transcriptionally up-regulated. Introduction of PAX3-FOXO1 into NIH-3T3 cells identified a myogenic transcriptional signature distinct from PAX3 alone that included genes such as MyoD, myogenin, and IGF242. Multiple studies have demonstrated suppression of tumor growth in vivo and in vitro with small molecular inhibitors and monoclonal antibodies targeting IGF1R. This has led to both phase I and II clinical trials to evaluate anti-IGF1R therapy in RMS. In phase I trials involving multiple tumor types, responses were only observed in Ewing sarcoma patients. However, preliminary phase II data have shown three objective radiological responses for RMS patients41.
The platelet-derived growth factor alpha receptor (PDGFRα) is also transcriptionally up-regulated by PAX3-FOXO143. PDGFRα is overexpressed in human ARMS and ERMS tumors as well as in mouse models of ARMS. SiRNA down-regulation of PDGFRα resulted in decreased cell growth in vitro and PDGFRα inhibition in mouse models using imatinib mesylate or a PDGFRα neutralizing antibody led to disease stabilization and in some cases resistance to therapy44. Resistance has also been observed with anti-IGF1R therapy, which may be due to activation of other growth factor receptors such as HER-2 (human epidermal growth factor receptor 2, also known as ERBB2) and PDGFRα45,46. Because both IGF1R and PDGFRα are potential therapeutic targets that have shown resistance as single agents, combination therapy may enhance patient response. Combination of anti-IGF1R therapy with mTOR (mechanistic target of rapamycin) inhibitors is currently being evaluated in a phase I trial for pediatric solid tumors (NCT00880282) and in combination with cytotoxic chemotherapy in a Children’s Oncology Group Study (ARST08P1) for patients with high-risk rhabdomyosarcoma (Table 2).
Synovial sarcoma
Synovial sarcoma (SS) is the most common non-rhabdomyosarcoma soft tissue sarcoma in adolescents and young adults, accounting for approximately 8% of all soft tissue sarcomas28. The overall five-year survival rate is between 70 and 80%, but as with most sarcomas, this number drops sharply for those patients with metastatic and recurrent disease. Other than complete surgical resection of localized synovial sarcomas, there is no standard of care for treatment and approach to these tumors may vary from center to center. Radiation therapy may be used as an adjuvant therapy to improve local control and chemotherapy is generally reserved for larger tumors or in patients with metastatic disease.
Synovial sarcoma is histologically unique, displaying both biphasic and monophasic tumors. Monophasic tumors are mesenchymal in origin, exhibiting a spindle cell morphology that is difficult to distinguish from fibrosarcoma. Biphasic tumors contain cells of epithelial differentiation that form a glandular component within the mesenchymal spindle cells28. The designation of synovial sarcoma originated from tumor proximity to large joints, as most tumors arise in the extremities, and microscopic resemblance to synovial tissue. However, this is a misnomer as the histogenesis is not of synovial origin. Mouse models utilizing myogenic regulatory factor driven expression of fusion genes suggest SS arises shortly after differentiation from a muscle stem cell47.
Molecular Genetics
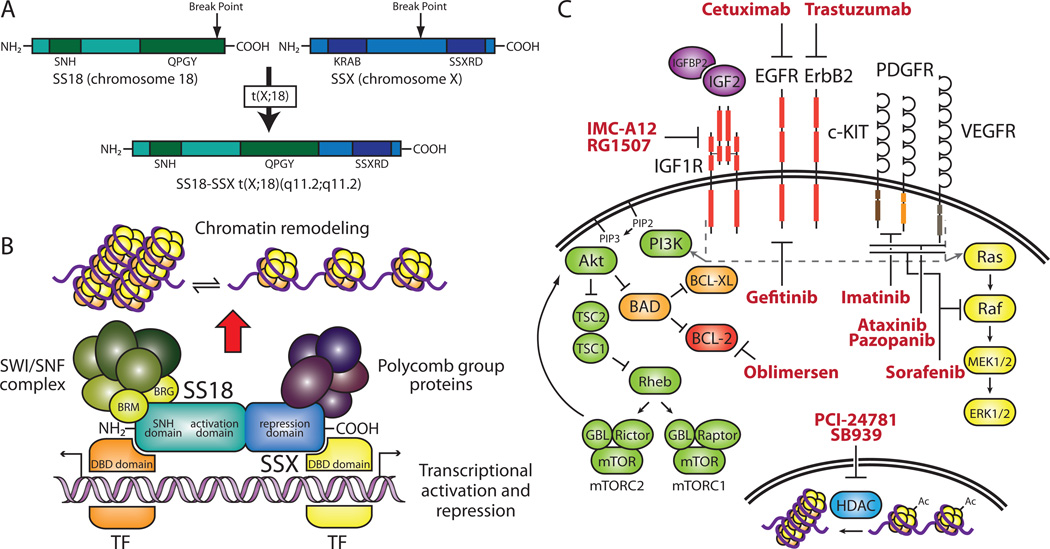
The underlying genetic aberration in SS results from a specific t(X;18)(p11.2;q11.2) translocation that fuses SS18 (synovial sarcoma translocation, chromosome 18, also known as SYT) to either SSX1, SSX2, or SSX4 (synovial sarcoma, X breakpoint 1, 2, or 4)48–51 (Figure 2A). SS18 is a ubiquitously expressed, nuclear protein that contains a novel SNH (SYT N-terminal homology) domain that allows for interaction with chromatin remodeling factors and a C-terminal QPGY domain that resembles the transactivation domain within the FET family of proteins. Despite the absence of a DNA-binding domain, SS18 is thought to function as a transcriptional activator and may play a role in signal transduction via its SH2 (Src homology 2) and SH3 (Src homology 3)-binding motifs. The SSX genes constitute a family of highly homologous proteins located on the X chromosome. They are believed to act as transcription repressors due to the presence of a Kruppel-associated box (KRAB) domain and SSX repression domain (SSXRD). The SSX proteins also lack a DNA-binding domain so must rely on protein-protein interactions to mediate transcriptional repression52.
Figure 2. Molecular genetics and targeted therapies in SS.
(A) Schematic of the SS18-SSX t(X;18)(q11.2;q11.2) translocation. The SS18-SSX fusion contains both the SNH and QPGY activation domains of SS18 as well as the SSXRD. The SSX KRAB domain is not retained in the fusion. (B) Putative molecular function of SS18-SSX and selected protein–protein interactions. The SNH domain facilitates binding to components of the SWI/SNF complex while the SSXRD interacts with polycomb group proteins, which results in chromatin remodeling. Interactions with transcription factors (TFs) may also lead to transcriptional activation and repression. (C) Signaling pathways and targeted therapies in SS. Activation of growth factor receptors leads to downstream signaling through the PI3K and MAPK pathways (indicated by gray dashed line and arrows). Histone deacetylases (HDACs) remove acetyl groups (Ac) from histones, resulting in condensed chromatin. Targeted therapeutic agents used in recent clinical trials for SS are indicated in red. Genes modulated by SS18-SSX are indicated in purple. Receptors overexpressed in SS are indicated in red. EGFR, epidermal growth factor receptor; IGF1, insulin-like growth factor 1; KRAB; Kruppel-associated box; MAPK, mitogen-activated protein kinase; PDGFR, platelet-derived growth factor receptor; PI3K, phosphoinositide-3-kinase; SNH, SYT N-terminal homology; SS, synovial sarcoma; SSXRD, SSX repression domain; SWI/SNF, switch/sucrose nonfermentable; SYT, synovial sarcoma translocation; VEGFR, vascular endothelial growth factor receptor.
The t(X;18) translocation fuses the C-terminus of SSX to all but the last eight amino acids of SS18, generating a chimeric protein that contains both transcriptional activation (QPGY) and repression (SSXRD) domains as well as the SNH domain (Figure 2A). The SSXRD allows SS18-SSX proteins to co-localize with components of the polycomb group (PcG) chromatin remodeling repressor complex while the SNH domain facilitates interaction with members of the SWI/SNF complex53,54 (Figure 2B). This suggests the fusion drives tumor progression though epigenetic chromatin remodeling. The SS18-SSX1 fusion protein is able to transform rat fibroblasts and association the hBRM/hSNF2α (also known as SMARCA2, SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2) chromatin remodeling factor is required for transformation55. Its presence is required for sarcomagenesis as siRNA down-regulation of SS18-SSX expression inhibits tumor growth in vitro and in vivo56.
SS18 is fused to either SSX1 or SSX2 over in over 90% of SS cases and is only rarely observed bound to SSX4. Although initial studies showed patients with SS18-SSX2 fusions had improved survival rates, an expanded study performed more recently concluded there was no correlation between fusion variant and survival57. While fusion type may not determine survival, it is strongly associated with histology as biphasic and monophasic tumors contain the SS18-SSX1 and SS18-SSX2 transcripts, respectively58.
Target genes and targeted therapies
As with RMS, IGF2 plays a role in SS biology. cDNA microarray analysis of SS and closely related spindle cell tumors revealed genes such as insulin-like growth factor binding protein 2 (IGFBP2), HER-2 (ERBB2), and IGF2 are up-regulated in SS59. Evidence for the role of IGF2 in SS pathogenesis is further supported by studies in which SS18-SSX fusions were exogenously expressed in heterologous cell systems. Gene expression profiling determined IGF2 to be the most highly up-regulated gene upon SS18-SSX2 expression in 293T cells60 and SS18-SSX1 expression in human primary lung fibroblasts61.
SS18-SSX2 has been shown to interact with BRG1 (also known as SMARCA4, SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4), a component of the SWI/SNF complex, and BRG1 was found to bind to the IGF2 promoter. This data combined with active chromatin marks observed upon induction of SS18-SSX2 suggests the fusion interacts with BRG1 to epigenetically modulate IGF2 expression60. Additionally, SS18-SSX2 is necessary for maintenance of IGF2 expression and IGF2 is required for SS18-SSX1 mediated tumor formation61. IGF2 expression results in activation of IGF1R and phosphorylation of the downstream proteins Akt (v-akt murine thymoma viral oncogene homolog) and MAPK (mitogen-activated protein kinase). Providing evidence the IGF pathway could be a therapeutic target, treatment of SS cell lines with the IGF1R inhibitor NVP-AEW541 resulted in impaired cell growth and increased apoptosis62.
Immunohistochemical and molecular studies have demonstrated high expression levels of the anti-apoptotic protein BCL-2 (B-cell CLL/lymphoma 2) in the majority of SS tumors63. Due to the absence of genomic amplifications or rearrangements, overexpression of BCL-2 may result from transcriptional activation64. Though BCL-2 has not been shown to be a direct target of SS18-SSX, lower mRNA and protein levels in SS tumors and cell lines lacking the t(X,18) translocation suggests an association does exist65. Furthermore, BCL-2 antisense oligonucleotide treatment of a translocation positive SS cell line resulted in increased sensitivity to doxorubicin treatment, implying BCL-2 may be a promising therapeutic target66. In a phase I trial assessing the effectiveness of BCL-2 antisense therapy (Figure 2C) in combination with chemotherapy in childhood solid tumors, two synovial sarcoma patients displayed prolonged stable disease67.
EGFR (epidermal growth factor receptor) is also overexpressed in SS. Microarray analysis of a set of 41 soft tissue tumors and subsequent clustering analysis identified EGFR as part of cluster that showed SS specific expression68. Immunohistochemical studies and molecular characterization have confirmed the presence of EGFR in SS69,70. This data led to a phase II trial to establish the efficacy of the EGFR inhibitor gefitinib (Figure 2C) in EGFR-positive, chemoresistant synovial sarcoma. In this trial, stable disease was the best observed response, suggesting EGFR is not required for tumorigenesis71. Gene expression profiling and immunohistochemical studies have identified another member of the EGFR family, HER-2 (ERBB2), that is up-regulated in SS that may provide an alternate target for the treatment of this disease59,70 (Figure 2C).
Dermatofibrosarcoma protuberans
Dermatofibrosarcoma protuberans (DFSP) is a relatively rare cutaneous malignancy, accounting for approximately 0.1% of all cancers and 1% of soft tissue sarcomas. DFSP has an annual incidence rate of 4.2 per million and primarily afflicts adults between the ages of 30 and 50 years72. Pediatric cases, both congenital and in young children, do occur, but with much less frequency than adult cases. As suggested by its name, DFSP tumors arise in the dermis, infiltrating the dermal stroma and often breaching the subcutaneous fat. Although studies have provided evidence for a fibroblastic, neural, and histiocytic origin, the histogenesis of DFSP remains uncertain74. Primary site locations can occur throughout the body, though the trunk, proximal extremities, and head and neck are the most common72. The five-year survival rate of patients with DFSP exceeds 95% and metastases occur in less than 5% of patients72,73.
Treatment of DFSP, like other soft tissue sarcomas, centers on achieving a complete surgical resection. Radiation is used when surgical margins are positive and a re-resection is not feasible. Chemotherapy is only used in metastatic cases, but there is increasing evidence that imatinib therapy can be used in an adjuvant setting in patients with recurrent, refractory, and/or metastatic disease.
Molecular Genetics
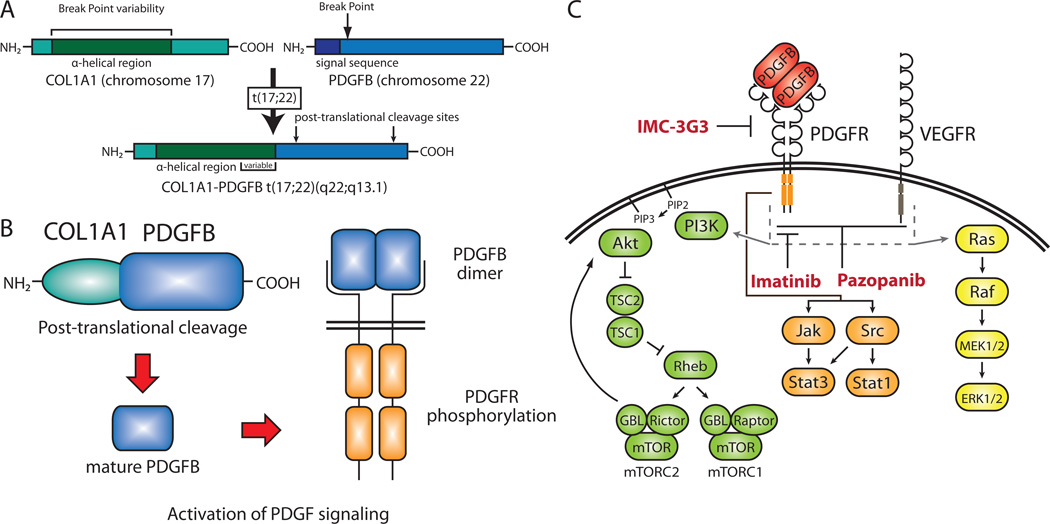
DFSP tumors contain either the t(17;22)(q22;q13.1) reciprocal chromosomal translocation or a supernumerary ring chromosome derived from t(17;22). Both of these karyotypic aberrations result in a fusion of the genes COL1A1 (encoding the pro-alpha1 chain of type I collagen) on 17q21–22 and PDGFB (encoding the platelet-derived growth factor B chain) on 22q13.175 (Figure 3A). Rings are predominantly found in adult cases, though occasionally translocations are identified. In contrast, all pediatric tumors contain translocations. DFSP variants and related malignancies such as giant cell fibroblastoma have also been found to contain the COL1A1-PDGFB fusion.
Figure 3. Molecular genetics and targeted therapies in DFSP.
(A) Molecular genetics and targeted therapies in DFSP. (a) Schematic of the COL1A1-PDGFB t(17;22)(q22;q13.1) translocation. The COL1A1-PDGFB fusion joins the α-helical region of COL1A1 to PDGFB lacking its signal sequence. The COL1A1 N-terminal signal sequence replaces that of PDGFB. Break points occur throughout the α-helical region in COL1A1 but occur only within the first intron of PDGFB. PDGFB post-translational cleavage sites are retained in the fusion. (B) Putative molecular function of the COL1A1-PDGFB fusion. The COL1A1 signal sequence allows for protein export and posttranslational cleavage results in the generation of mature PDGFB. Ligand binding of the PDGFB homodimer dimer results in receptor dimerization, autophosphorylation, and activation. (C) Signaling pathways and targeted therapies in DFSP. Activation of PDGFR by the PDGFB dimer results in downstream signaling through the PI3K, MAPK, and Jak/Stat (Janus kinase/signal transducer and activator of transcription) pathways. VEGFR is also activated in DFSP and signals through the PI3K and MAPK pathways. Targeted therapeutic agents in current clinical trials for DFSP (imatinib and dasatinib) and soft tissue sarcomas expressing PDGFR-α (IMC-3G3) are indicated in red. Growth factors overexpressed in DFSP are indicated in red. DFSP, dermatofibrosarcoma protuberans; MAPK, mitogen-activated protein kinase; PDGFB, platelet-derived growth factor B chain; PDGFR, platelet-derived growth factor receptor; PI3K, phosphoinositide-3-kinase; VEGFR, vascular endothelial growth factor receptor.
The t(17;22) breakpoint occurs upstream of the second exon of PDGFB gene and within the alpha-helical region of COL1A1. This results in the removal of the both the PDGFB inhibitory regulatory elements and signal peptide in the COL1A1-PDGFB fusion and placement of remainder of PDGFB locus, beginning with exon 2, under the control of the COL1A1 promoter. Although the PDGFB breakpoint is invariably located in the first intron, the one in the COL1A1 locus can occur within multiple exons, probably due to the repetitive nature of the alpha-helical region. Most of the COL1A1 coding sequence is postulated to be functionally irrelevant as PDGFB is post-translationally cleaved at sites retained in the fusion to generate the mature growth factor. In contrast, the replacement of the PDGFB repressor elements with the COL1A1 promoter allows for aberrant expression of the protein. The COL1A1 N-terminal signal sequence, also retained in the fusion, permits PDGFB secretion, resulting in constitutive activation of the platelet derived growth factor beta (PDGFβ) pathway76 (Figure 3B). The finding that there is no correlation between COL1A1-PDGFB fusion type and clinical response or histology supports the theory that the COL1A1 portion of the fusion only provides a mechanism for PDGFB overexpression77.
Signaling pathways and targeted therapies
The COL1A1-PDGFB fusion has been shown to transform NIH3T3 cells78. Additionally, stable transfection of the fusion in a Chinese hamster lung fibroblastic line led to growth factor independent growth and tumor formation in nude mice76. Both of these studies demonstrated activation of the PDGFβ receptor, indicating the involvement of PDGF signaling in DFSP tumorigenesis. In order to block this pathway, NIH3T3 cells transformed with COL1A1-PDGFB and DFSP tumor-derived primary cultures were treated with imatinib mesylate (Gleevec or STI-571), which inhibits the PDGF receptor (PDGFR)79 (Figure 3C). Growth inhibitory effects were observed in vitro and in vivo, suggesting imatinib could be an effective therapy in the treatment of DFSP.
Initial case reports demonstrating patient response to imatinib led to a study of its activity in 10 patients with locally advanced or metastatic DFSP. Despite low levels of PDGFR phosphorylation in patient tumors, imatinib was shown to be an effective therapy as four patients with locally advanced disease displayed a complete clinical response80. On a larger scale, the efficacy of imanitib treatment is currently being evaluated in a phase II trial for DFSP and GCF (NCT00085475, Table 2). In addition to imanitib, another tyrosine kinase inhibitor pazopanib, which targets VEGFR (vascular endothelial growth factor receptor) in addition to PDGFR (Figure 3C), has just begun phase II testing for DFSP (NCT01059656, Table 2).
Conclusions
Despite aggressive therapeutic approaches to patients with pediatric sarcomas, we have reached a plateau in the survival rate. Although the molecular genetics of these tumors reveal they are distinct entities, they are often treated as a homogeneous group when it comes to standard therapy. Fusion gene transcriptional targets, downstream signaling pathways, and over-expressed growth factor receptors provide novel therapeutic targets that are currently being investigated in clinical trials. Certain targets have shown promise, but tumor resistance is a common problem, suggesting combination therapy may be required for effective treatment. Although initial trials and in vitro studies have paved the way for advances in targeted therapy in sarcoma, further work is needed to better characterize tumors at the molecular genetic level to tailor therapies to individual tumors.
Acknowledgments
Support: Dr. Federman is supported by a St. Baldrick’s Foundation Career Development Awarda, STOP Cancer Award. Dr. Federman and Dr. Denny are supported by a Hyundai Hope on Wheels Grant. J. Anderson is supported by a Dissertation Year Fellowship from the Graduate Division at UCLA.
References
- 1.Gurney JG, Swensen AR, Bulterys M. Malignant Bone Tumors. In: Ries LAG, Smith MA, Gurney JG, et al., editors. Cancer Incidence and Survival among Children and Adolescents: United States SEER Program 1975–1995. Bethesda, MD: National Cancer Institute, SEER Program; 1999. pp. 99–110. [Google Scholar]
- 2.Gurney JG, Young JL, Jr, Roffers SD, Smith MA, Bunin GR. Soft Tissue Sarcomas. In: Ries LAG, Smith MA, Gurney JG, et al., editors. Cancer Incidence and Survival among Children and Adolescents: United States SEER Program 1975–1995. Bethesda, MD: National Cancer Institute, SEER Program; 1999. pp. 111–124. [Google Scholar]
- 3.Balamuth NJ, Womer RB. Ewing's sarcoma. Lancet Oncol. 2010;11:184–192. doi: 10.1016/S1470-2045(09)70286-4. [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez-Galindo C, Navid F, Khoury J, Krasin M. Ewing sarcoma family of tumors. In: Pappo A, editor. Pediatric Bone and Soft Tissue Sarcomas. Berlin/Heidelberg: Springer-Verlag; 2006. pp. 181–217. [Google Scholar]
- 5.Applebaum MA, Worch J, Matthay KK, et al. Clinical features and outcomes in patients with extraskeletal ewing sarcoma. Cancer. 2011;117:3027–3032. doi: 10.1002/cncr.25840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ludwig JA. Ewing sarcoma: historical perspectives, current state-of-the-art, and opportunities for targeted therapy in the future. Curr Opin Oncol. 2008;20:412–418. doi: 10.1097/CCO.0b013e328303ba1d. [DOI] [PubMed] [Google Scholar]
- 7.Pinto A, Dickman P, Parham D. Pathobiologic markers of the Ewing sarcoma family of tumors: state of the art and prediction of behaviour. Sarcoma. 2011;2011:1–15. doi: 10.1155/2011/856190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barr FG, Meyer WH. Role of fusion subtype in Ewing sarcoma. J Clin Oncol. 2010;28:1973–1974. doi: 10.1200/JCO.2009.27.2161. [DOI] [PubMed] [Google Scholar]
- 9.Delattre O, Zucman J, Plougastel B, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 10.Kovar H. Dr. Jekyll and Mr. Hyde: the two faces of the FUS/EWS/TAF15 protein family. Sarcoma. 2011;2011:837474. doi: 10.1155/2011/837474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol. 2001;2:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- 12.Gangwal K, Sankar S, Hollenhorst P, et al. Microsatellites as EWS/FLI response elements in Ewing's sarcoma. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0801073105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knoop LL, Baker SJ. The splicing factor U1C represses EWS/FLI-mediated transactivation. J Biol Chem. 2000;275:24865–24871. doi: 10.1074/jbc.M001661200. [DOI] [PubMed] [Google Scholar]
- 14.Knoop LL, Baker SJ. EWS/FLI alters 5’-splice site selection. J Biol Chem. 2001;276:22317–22322. doi: 10.1074/jbc.M008950200. [DOI] [PubMed] [Google Scholar]
- 15.France KA, Anderson JL, Park A, Denny CT. Oncogenic fusion protein EWS/FLI1 down-regulates gene expression by both transcriptional and posttranscriptional mechanisms. J Biol Chem. 2011;286:22750–22757. doi: 10.1074/jbc.M111.225433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prieur A, Tirode F, Cohen P, Delattre O. EWS/FLI-1 silencing and gene profiling of Ewing cells reveal downstream oncogenic pathways and a crucial role for repression of insulin-like growth factor binding protein 3. Mol Cell Biol. 2004;24:7275–7283. doi: 10.1128/MCB.24.16.7275-7283.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wakahara K, Ohno T, Kimura M, et al. EWS-Fli1 up-regulates expression of the Aurora A and Aurora B kinases. Mol Cancer Res. 2008;6:1937–1945. doi: 10.1158/1541-7786.MCR-08-0054. [DOI] [PubMed] [Google Scholar]
- 18.Cironi L, Riggi N, Provero P, et al. IGF1 is a common target gene of Ewing's sarcoma fusion proteins in mesenchymal progenitor cells. PLoS ONE. 2008;3:e2634. doi: 10.1371/journal.pone.0002634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scotlandi K, Picci P. Targeting insulin-like growth factor 1 receptor in sarcomas. Curr Opin Oncol. 2008;20:419–427. doi: 10.1097/CCO.0b013e328302edab. [DOI] [PubMed] [Google Scholar]
- 20.Tolcher AW, Sarantopoulos J, Patnaik A, et al. Phase I, pharmacokinetic, and pharmacodynamic study of AMG 479, a fully human monoclonal antibody to insulinlike growth factor receptor 1. J Clin Oncol. 2009;27:5800–5807. doi: 10.1200/JCO.2009.23.6745. [DOI] [PubMed] [Google Scholar]
- 21.Pappo AS, Patel SR, Crowley J, et al. R1507, a monoclonal antibody to the insulinlike growth factor 1 receptor, in patients with recurrent or refractory Ewing sarcoma family of tumors: results of a phase II Sarcoma Alliance for Research through Collaboration study. J Clin Oncol. 2011;29:4541–4547. doi: 10.1200/JCO.2010.34.0000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Juergens H, Daw NC, Geoerger B, et al. Preliminary efficacy of the anti-insulin-like growth factor type 1 receptor antibody figitumumab in patients with refractory Ewing sarcoma. J Clin Oncol. 2011;29:4534–4540. doi: 10.1200/JCO.2010.33.0670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malempati S, Weigel B, Ingle AM, et al. Phase I/II trial and pharmacokinetic study of cixutumumab in pediatric patients with refractory solid tumors and Ewing sarcoma: a report from the Children's Oncology Group. J Clin Oncol. 2012;30:256–262. doi: 10.1200/JCO.2011.37.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maris JM, Morton CL, Gorlick R, et al. Initial testing of the aurora kinase A inhibitor MLN8237 by the Pediatric Preclinical Testing Program (PPTP) Pediatr Blood Cancer. 2010;55:26–34. doi: 10.1002/pbc.22430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toretsky JA, Erkizan V, Levenson A, et al. Oncoprotein EWS-FLI1 activity is enhanced by RNA helicase A. Cancer Res. 2006;66:5574–5581. doi: 10.1158/0008-5472.CAN-05-3293. [DOI] [PubMed] [Google Scholar]
- 26.Erkizan HV, Kong Y, Merchant M, et al. A small molecule blocking oncogenic protein EWS-FLI1 interaction with RNA helicase A inhibits growth of Ewing's sarcoma. Nat Med. 2009;15:750–756. doi: 10.1038/nm.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perez EA, Kassira N, Cheung MC, Koniaris LG, Neville HL, Sola JE. Rhabdomyosarcoma in children: a SEER population based study. J Surg Res. 2011;170:e243–e251. doi: 10.1016/j.jss.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 28.Pizzo PA, Poplack DG. Principles and practice of pediatric oncology. Sixth ed. Vol. 923. Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins; 2011. pp. 980–981. [Google Scholar]
- 29.Barr FG, Galili N, Holick J, Biegel JA, Rovera G, Emanuel BS. Rearrangement of the PAX3 paired box gene in the paediatric solid tumour alveolar rhabdomyosarcoma. Nat Genet. 1993;3:113–117. doi: 10.1038/ng0293-113. [DOI] [PubMed] [Google Scholar]
- 30.Davis RJ, D'Cruz CM, Lovell MA, Biegel JA, Barr FG. Fusion of PAX7 to FKHR by the variant t(1;13)(p36;q14) translocation in alveolar rhabdomyosarcoma. Cancer Res. 1994;54:2869–2872. [PubMed] [Google Scholar]
- 31.Sumegi J, Streblow R, Frayer RW, et al. Recurrent t(2;2) and t(2;8) translocations in rhabdomyosarcoma without the canonical PAX-FOXO1 fuse PAX3 to members of the nuclear receptor transcriptional coactivator family. Genes Chromosomes Cancer. 2010;49:224–236. doi: 10.1002/gcc.20731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Q, Fang W-H, Krupinski J, Kumar S, Slevin M, Kumar P. Pax genes in embryogenesis and oncogenesis. J Cell Mol Med. 2008;12:2281–2294. doi: 10.1111/j.1582-4934.2008.00427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fu Z, Tindall DJ. FOXOs, cancer and regulation of apoptosis. Oncogene. 2008;27:2312–2319. doi: 10.1038/onc.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sorensen PHB, Lynch JC, Qualman SJ, et al. PAX3-FKHR and PAX7-FKHR gene fusions are prognostic indicators in alveolar rhabdomyosarcoma: a report from the children’s oncology group. J Clin Oncol. 2002;20:2672–2679. doi: 10.1200/JCO.2002.03.137. [DOI] [PubMed] [Google Scholar]
- 35.Williamson D, Missiaglia E, de Reyniès A, et al. Fusion gene-negative alveolar rhabdomyosarcoma is clinically and molecularly indistinguishable from embryonal rhabdomyosarcoma. J Clin Oncol. 2010;28:2151–2158. doi: 10.1200/JCO.2009.26.3814. [DOI] [PubMed] [Google Scholar]
- 36.Ginsberg JP, Davis RJ, Bennicelli JL, Nauta LE, Barr FG. Up-regulation of MET but not neural cell adhesion molecule expression by the PAX3-FKHR fusion protein in alveolar rhabdomyosarcoma. Cancer Res. 1998;58:3542–3546. [PubMed] [Google Scholar]
- 37.Taulli R, Scuoppo C, Bersani F, et al. Validation of met as a therapeutic target in alveolar and embryonal rhabdomyosarcoma. Cancer Res. 2006;66:4742–4749. doi: 10.1158/0008-5472.CAN-05-4292. [DOI] [PubMed] [Google Scholar]
- 38.Yan D, Dong XDE, Chen X, et al. MicroRNA-1/206 targets c-Met and inhibits rhabdomyosarcoma development. J Biol Chem. 2009;284:29596–29604. doi: 10.1074/jbc.M109.020511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hou J, Dong J, Sun L, et al. Inhibition of phosphorylated c-Met in rhabdomyosarcoma cell lines by a small molecule inhibitor SU11274. J Transl Med. 2011;9:64. doi: 10.1186/1479-5876-9-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ayalon D, Glaser T, Werner H. Transcriptional regulation of IGF-I receptor gene expression by the PAX3-FKHR oncoprotein. Growth Horm IGF Res. 2001;11:289–297. doi: 10.1054/ghir.2001.0244. [DOI] [PubMed] [Google Scholar]
- 41.Martins AS, Olmos D, Missiaglia E, Shipley J. Targeting the insulin-like growth factor pathway in rhabdomyosarcomas: rationale and future perspectives. Sarcoma. 2011;2011:209736. doi: 10.1155/2011/209736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khan J, Bittner ML, Saal LH, et al. cDNA microarrays detect activation of a myogenic transcription program by the PAX3-FKHR fusion oncogene. Proc Natl Acad Sci U S A. 1999;96:13264–13269. doi: 10.1073/pnas.96.23.13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Epstein JA, Song B, Lakkis M, Wang C. Tumor-specific PAX3-FKHR transcription factor, but not PAX3, activates the platelet-derived growth factor alpha receptor. Mol Cell Biol. 1998;18:4118–4130. doi: 10.1128/mcb.18.7.4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taniguchi E, Nishijo K, McCleish AT, et al. PDGFR-A is a therapeutic target in alveolar rhabdomyosarcoma. Oncogene. 2008;27:6550–6560. doi: 10.1038/onc.2008.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abraham J, Prajapati SI, Nishijo K, et al. Evasion mechanisms to Igf1r inhibition in rhabdomyosarcoma. Mol Cancer Ther. 2011;10:697–707. doi: 10.1158/1535-7163.MCT-10-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mayeenuddin LH, Yu Y, Kang Z, Helman LJ, Cao L. Insulin-like growth factor 1 receptor antibody induces rhabdomyosarcoma cell death via a process involving AKT and Bcl-x(L) Oncogene. 2010;29:6367–6377. doi: 10.1038/onc.2010.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haldar M, Hancock JD, Coffin CM, Lessnick SL, Capecchi MR. A conditional mouse model of synovial sarcoma: insights into a myogenic origin. Cancer Cell. 2007;11:375–388. doi: 10.1016/j.ccr.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 48.Clark J, Rocques PJ, Crew AJ, et al. Identification of novel genes, SYT and SSX, involved in the t(X;18)(p11.2;q11.2) translocation found in human synovial sarcoma. Nat Genet. 1994;7:502–508. doi: 10.1038/ng0894-502. [DOI] [PubMed] [Google Scholar]
- 49.Crew AJ, Clark J, Fisher C, et al. Fusion of SYT to two genes, SSX1 and SSX2, encoding proteins with homology to the Kruppel-associated box in human synovial sarcoma. EMBO J. 1995;14:2333–2340. doi: 10.1002/j.1460-2075.1995.tb07228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Leeuw B, Balemans M, Olde Weghuis D, Geurts van Kessel A. Identification of two alternative fusion genes, SYT-SSX1 and SYT-SSX2, in t(X;18)(p11.2;q11.2)-positive synovial sarcomas. Hum Mol Genet. 1995;4:1097–1099. doi: 10.1093/hmg/4.6.1097. [DOI] [PubMed] [Google Scholar]
- 51.Skytting B, Nilsson G, Brodin B, et al. A novel fusion gene, SYT-SSX4, in synovial sarcoma. J Natl Cancer Inst. 1999;91:974–975. doi: 10.1093/jnci/91.11.974. [DOI] [PubMed] [Google Scholar]
- 52.Haldar M, Randall RL, Capecchi MR. Synovial sarcoma: from genetics to genetic-based animal modeling. Clin Orthop Relat Res. 2008;466:2156–2167. doi: 10.1007/s11999-008-0340-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perani M, Ingram CJE, Cooper CS, Garrett MD, Goodwin GH. Conserved SNH domain of the proto-oncoprotein SYT interacts with components of the human chromatin remodelling complexes, while the QPGY repeat domain forms homo-oligomers. Oncogene. 2003;22:8156–8167. doi: 10.1038/sj.onc.1207031. [DOI] [PubMed] [Google Scholar]
- 54.Soulez M, Saurin AJ, Freemont PS, Knight JC. SSX and the synovial-sarcoma-specific chimaeric protein SYT-SSX co-localize with the human Polycomb group complex. Oncogene. 1999;18:2739–2746. doi: 10.1038/sj.onc.1202613. [DOI] [PubMed] [Google Scholar]
- 55.Nagai M, Tanaka S, Tsuda M, et al. Analysis of transforming activity of human synovial sarcoma-associated chimeric protein SYT-SSX1 bound to chromatin remodeling factor hBRM/hSNF2 alpha. Proc Natl Acad Sci U S A. 2001;98:3843–3848. doi: 10.1073/pnas.061036798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takenaka S, Naka N, Araki N, et al. Downregulation of SS18-SSX1 expression in synovial sarcoma by small interfering RNA enhances the focal adhesion pathway and inhibits anchorage-independent growth in vitro and tumor growth in vivo. Int J Oncol. 2010;36:823–831. doi: 10.3892/ijo_00000559. [DOI] [PubMed] [Google Scholar]
- 57.Guillou L, Benhattar J, Bonichon F, et al. Histologic grade, but not SYT-SSX fusion type, is an important prognostic factor in patients with synovial sarcoma: a multicenter, retrospective analysis. J Clin Oncol. 2004;22:4040–4050. doi: 10.1200/JCO.2004.11.093. [DOI] [PubMed] [Google Scholar]
- 58.Antonescu CR, Kawai A, Leung DH, et al. Strong association of SYT-SSX fusion type and morphologic epithelial differentiation in synovial sarcoma. Diagn Mol Pathol. 2000;9:1–8. doi: 10.1097/00019606-200003000-00001. [DOI] [PubMed] [Google Scholar]
- 59.Allander SV, Illei PB, Chen Y, et al. Expression profiling of synovial sarcoma by cDNA microarrays: association of ERBB2, IGFBP2, and ELF3 with epithelial differentiation. Am J Pathol. 2002;161:1587–1595. doi: 10.1016/S0002-9440(10)64437-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Bruijn DRH, Allander SV, van Dijk AHA, et al. The synovial-sarcoma-associated SS18-SSX2 fusion protein induces epigenetic gene (de)regulation. Cancer Res. 2006;66:9474–9482. doi: 10.1158/0008-5472.CAN-05-3726. [DOI] [PubMed] [Google Scholar]
- 61.Sun Y, Gao D, Liu Y, Huang J, Lessnick S, Tanaka S. IGF2 is critical for tumorigenesis by synovial sarcoma oncoprotein SYT-SSX1. Oncogene. 2006;25:1042–1052. doi: 10.1038/sj.onc.1209143. [DOI] [PubMed] [Google Scholar]
- 62.Friedrichs N, Küchler J, Endl E, et al. Insulin-like growth factor-1 receptor acts as a growth regulator in synovial sarcoma. J Pathol. 2008;216:428–439. doi: 10.1002/path.2438. [DOI] [PubMed] [Google Scholar]
- 63.Krsková L, Kalinová M, Břízová H, Mrhalová M, Sumerauer D, Kodet R. Molecular and immunohistochemical analyses of BCL2, KI-67, and cyclin D1 expression in synovial sarcoma. Cancer Genet. 2009;193:1–8. doi: 10.1016/j.cancergencyto.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 64.Mancuso T, Mezzelani A, Riva C, et al. Analysis of SYT-SSX fusion transcripts and bcl-2 expression and phosphorylation status in synovial sarcoma. Lab Invest. 2000;80:805–813. doi: 10.1038/labinvest.3780085. [DOI] [PubMed] [Google Scholar]
- 65.Albritton KH, Randall RL. Prospects for targeted therapy of synovial sarcoma. J Pediatr Hematol Oncol. 2005;27:219–222. doi: 10.1097/01.mph.0000163713.46762.72. [DOI] [PubMed] [Google Scholar]
- 66.Joyner DE, Albritton KH, Bastar JD, Randall RL. G3139 antisense oligonucleotide directed against antiapoptotic Bcl-2 enhances doxorubicin cytotoxicity in the FU-SY-1 synovial sarcoma cell line. J Orthop Res. 2006;24:474–480. doi: 10.1002/jor.20087. [DOI] [PubMed] [Google Scholar]
- 67.Rheingold SR, Hogarty MD, Blaney SM, et al. Phase I Trial of G3139, a bcl-2 antisense oligonucleotide, combined with doxorubicin and cyclophosphamide in children with relapsed solid tumors: a Children's Oncology Group Study. J Clin Oncol. 2007;25:1512–1518. doi: 10.1200/JCO.2006.09.5125. [DOI] [PubMed] [Google Scholar]
- 68.Nielsen TO, West RB, Linn SC, et al. Molecular characterisation of soft tissue tumours: a gene expression study. Lancet. 2002;359:1301–1307. doi: 10.1016/S0140-6736(02)08270-3. [DOI] [PubMed] [Google Scholar]
- 69.Bozzi F, Ferrari A, Negri T, et al. Molecular characterization of synovial sarcoma in children and adolescents: evidence of akt activation. Transl Oncol. 2008;1:95–101. doi: 10.1593/tlo.08121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thomas DG, Giordano TJ, Sanders D, et al. Expression of receptor tyrosine kinases epidermal growth factor receptor and HER-2/neu in synovial sarcoma. Cancer. 2005;103:830–838. doi: 10.1002/cncr.20847. [DOI] [PubMed] [Google Scholar]
- 71.Ray-Coquard I, Le Cesne A, Whelan JS, et al. A phase II study of gefitinib for patients with advanced HER-1 expressing synovial sarcoma refractory to doxorubicin-containing regimens. Oncologist. 2008;13:467–473. doi: 10.1634/theoncologist.2008-0065. [DOI] [PubMed] [Google Scholar]
- 72.Criscione VD, Weinstock MA. Descriptive epidemiology of dermatofibrosarcoma protuberans in the United States, 1973 to 2002. J Am Acad Dermatol. 2007;56:968–973. doi: 10.1016/j.jaad.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 73.Iqbal CW, St. Peter S, Ishitani MB. Pediatric dermatofibrosarcoma protuberans: multi-institutional outcomes. J Surg Res. 2011;170:69–72. doi: 10.1016/j.jss.2011.01.042. [DOI] [PubMed] [Google Scholar]
- 74.Dominguez-Malagon H, Valdez-Carrillo MdC, Cano-Valdez AM. Dermatofibroma and dermatofibrosarcoma protuberans: a comparative ultrastructural study. Ultrastruct Pathol. 2006;30:283–291. doi: 10.1080/01913120600820468. [DOI] [PubMed] [Google Scholar]
- 75.Simon MP, Pedeutour F, Sirvent N, et al. Deregulation of the platelet-derived growth factor B-chain gene via fusion with collagen gene COL1A1 in dermatofibrosarcoma protuberans and giant-cell fibroblastoma. Nat Genet. 1997;15:95–98. doi: 10.1038/ng0197-95. [DOI] [PubMed] [Google Scholar]
- 76.Simon MP, Navarro M, Roux D, Pouysségur J. Structural and functional analysis of a chimeric protein COL1A1-PDGFB generated by the translocation t(17;22)(q22;q13.1) in Dermatofibrosarcoma protuberans (DP) Oncogene. 2001;20:2965–2975. doi: 10.1038/sj.onc.1204426. [DOI] [PubMed] [Google Scholar]
- 77.Giacchero D, Maire G, Nuin PAS, et al. No correlation between the molecular subtype of COL1A1-PDGFB fusion gene and the clinico-histopathological features of dermatofibrosarcoma protuberans. J Invest Dermatol. 2010;130:904–907. doi: 10.1038/jid.2009.338. [DOI] [PubMed] [Google Scholar]
- 78.Greco A, Fusetti L, Villa R, et al. Transforming activity of the chimeric sequence formed by the fusion of collagen gene COL1A1 and the platelet derived growth factor b-chain gene in dermatofibrosarcoma protuberans. Oncogene. 1998;17:1313–1319. doi: 10.1038/sj.onc.1202051. [DOI] [PubMed] [Google Scholar]
- 79.Greco A, Roccato E, Miranda C, Cleris L, Formelli F, Pierotti MA. Growth-inhibitory effect of STI571 on cells transformed by the COL1A1/PDGFB rearrangement. Int J Cancer. 2001;92:354–360. doi: 10.1002/ijc.1190. [DOI] [PubMed] [Google Scholar]
- 80.McArthur GA, Demetri GD, van Oosterom A, et al. Molecular and clinical analysis of locally advanced dermatofibrosarcoma protuberans treated with imatinib: Imatinib Target Exploration Consortium Study B2225. J Clin Oncol. 2005;23:866–873. doi: 10.1200/JCO.2005.07.088. [DOI] [PubMed] [Google Scholar]