Abstract
G-protein coupled receptors (GPCRs) mediate numerous physiological processes and represent the targets for a vast array of therapeutics for diseases ranging from depression to hypertension to reflux. Despite the recognition that GPCRs can act as oncogenes and tumor suppressors by regulating oncogenic signaling networks, few drugs targeting GPCRs are utilized in cancer therapy. Recent large-scale genome-wide analyses of multiple human tumors have uncovered novel GPCRs altered in cancer. However, the work of determining which GPCRs from these lists are drivers of tumorigenesis, and hence valid therapeutic targets, remains a formidable challenge. In this review I will highlight recent studies providing evidence that GPCRs are relevant targets for cancer therapy through their effects on known cancer signaling pathways, tumor progression, invasion and metastasis, and the microenvironment. Furthermore, I will explore how genomic analysis is beginning to shine a light on GPCRs as therapeutic targets in the age of personalized medicine.
Keywords: GPCR, cancer, bioinformatics, signal transduction, microenvironment, metastasis, genomics
Introduction
G-protein coupled receptors (GPCRs) transduce extracellular signals from a variety of ligands through the plasma membrane, resulting in the modulation of intracellular signaling pathways (1). This is accomplished in large measure by the activation of heterotrimeric G-proteins and downstream second messengers. Composed of over 900 members in humans, GPCRs are seven-transmembrane proteins that regulate many physiological processes including vision, olfaction, taste, behavior and autonomic nervous system transmission. This wide array of functions has resulted in the extensive utilization of GPCR targeted therapeutics, accounting for 30–50% of all currently used drugs, both prescription and over-the-counter. The wide use of GPCR drugs can also be attributed to GPCR localization on the cell surface, abrogating the requirement for a drug to be cell permeable, as well as the ability of GPCRs to bind a variety of ligands, including antibodies, peptides and small molecules. Furthermore, GPCR signaling can be tightly regulated through the utilization of agonists, antagonists and inverse agonists. However, drugs targeting GPCRs are rarely utilized in cancer treatment, despite evidence that GPCRs mediate many aspects of tumorigenesis (2). This evidence has been accumulating since the discovery in 1986 of the transforming activity of the GPCR mas (3). More recently, genomic analyses have uncovered GPCR mutations, copy number alterations and gene expression and methylation changes in a wide variety of cancers (4–6). Determining the biological implication of these genomic alterations will allow utilization of GPCR targeted therapeutics in those patients with GPCR-driven tumors. In the first section of this review I will highlight recent genomic analyses of large-scale tumor sets and discuss how these studies are providing insight into GPCR-driven tumorigenesis. In the next several sections I will discuss recent advances implicating GPCR signaling in various stages of tumorigenesis, including regulation of known oncogenic signaling networks. Finally, I will discuss the implications of this work on the future of personalized cancer medicine. As several comprehensive reviews have detailed the varied roles for GPCRs in mitogenic signaling and cancer, I will focus on recent advances in the field (2, 7).
Identification of GPCRs in large-scale human tumor analyses
Advances in sequencing technologies and bioinformatics tools have resulted in the recent explosion in genomic information from human tumor samples (8, 9). These data have been coupled with information on copy number alterations, gene expression and DNA methylation to provide a glimpse into the complex world of human cancer. However, merely providing lists of genomic alterations in a specific disease is only the first step in the process of translating this knowledge into cancer therapeutics. Due to the inherent complexity of the data obtained from such analyses it is critical to utilize computational methods to identify statistically significant alterations. One must then perform biological experiments in the proper model system to determine the oncogenic (i.e., cancer promoting) or tumor suppressive (i.e., cancer suppressing) activity of any candidate.
As whole genome analyses can provide an overwhelming deluge of information, several methods for decreasing this complexity have been utilized. These include focused analysis of a specific gene family, cancer from a specific tissue of origin, or specific genomic alteration, or combining data from multiple analyses to find commonalities. The efficacy of the gene family approach was displayed in a study that sequenced 734 GPCRs in 11 melanoma samples (10). Mutations in GRM3 (glutamate receptor, metabotropic) were found to be frequent in melanoma and subsequent functional analysis provided a mechanism of action (increased MEK phosphorylation) and possible therapeutic intervention (MEK inhibition). In addition to the obvious implications for patients with GRM3 mutations, this study highlights an important facet of GPCR biology with respect to cancer therapeutics: as mediators of diverse cell signaling pathways, the effect of mutations in GPCRs can often be overcome through inhibition of downstream signaling effectors. Therefore, although GPCRs are themselves good drug targets, knowledge of GPCR alterations may be quickly translated to the clinic even in the absence of specific agonists or antagonists. As GPCRs are known to be highly druggable targets, they are often included in screens designed to detect the “druggable genome.” One such analysis was performed on 441 mixed primary tumors, looking for mutations in known cancer genes, protein kinases, E3 ligases, deubiquitinases, GPCRs and other enzymes (4). Of the 157 GPCRs analyzed, mutations were detected in 13, including GRM1, GRM8 and BAI3 (brain-specific angiogenesis inhibitor) in lung cancer. This information was then coupled with copy number analysis to identify further putative cancer genes. The cancer relevance of GRM8 was reinforced in a study coupling whole-exome sequencing and high-throughput screening for detection of candidate driver genes in endometrial cancer (11). Candidate genes from the sequencing effort were targeted for knockdown in endometrial cancer cell lines, providing functional validation. A novel method for the unbiased detection of tumor suppressor genes, in which loss of heterozygosity and allele-specific detection were uncovered via exome and transcriptome sequencing, revealed 21 potential tumor suppressive GPCRs in two cancer cell lines (12). Although no validation was performed, this type of analysis can provide a starting point for selection of candidates for biological analysis. In addition to genetic mutations, differential DNA methylation can drive tumorigenesis. A screen designed to identify genes whose DNA methylation-mediated silencing is required for cancer cell survival uncovered several GPCRs, including the purinergic receptor P2RY14 (6). Reexpression of P2RY14 in colon cancer cell lines decreased cell viability, suggesting that this method can detect novel tumor suppressors.
Large-scale whole genome analyses, such as those undertaken by The Cancer Genome Atlas (TCGA) seek to provide a comprehensive map of genomic and epigenomic alterations in a number of cancers (5). Mutations in GPCRs were found to be a frequent event in lung squamous cell carcinoma. Genotyping of single nucleotide polymorphisms (SNPs) among thousands of breast cancer samples has recently uncovered novel loci associated with increased risk (13, 14). Among these, PTH1R (parathyroid hormone 1 receptor) was identified as a plausible candidate gene from a newly identified rick locus (13), and LGR6 was found to associate specifically with ER-negative breast cancer (14). These datasets can be mined for specific GPCRs and again provide a starting point to pick genes for functional validation in the appropriate model systems. A recent analysis has utilized this information to attempt to fully characterize the mutational landscape of GPCRs in human tumors (9). As the use of targeted therapeutics and sequencing of tumors expands, cancer treatment may well focus not on the organ or origin, but on the specific genetic alterations present in the tumor. Therefore, uncovering the presence, and biological relevance, of these mutations in distinct tumor types is of great value. To this end, it was noted that TSHR (thyroid stimulating hormone receptor), known to be frequently mutated in thyroid cancer, is also mutated in several other cancers, including those of the large intestine, lung and ovary (9). This analysis also uncovered the unexpected finding that adhesion family GPCRs are frequently mutated in a variety of cancers. These receptors are important for interactions with neighboring cells and the extracellular matrix. Importantly, as GPCRs are known to regulate processes such as cell migration and immune cell function, assays focusing solely on cell viability will fail to capture the full extent of GPCR involvement in cancer pathogenesis.
Novel bioinformatics tools are being developed to objectively prioritize gene lists for biological analysis. One such tool utilizes multiple parameters, including structural information, drug target homology and chemoinformatics to identify the most chemically tractable targets (15). As a proof-of-concept, an analysis was performed on the Cancer Gene Census list, containing 488 genes implicated in human tumorigenesis, including several GPCRs. TSHR was identified as a putative druggable target in thyroid cancer and Smoothened (a Hedgehog pathway component) as a target in glioblastoma multiforme. Extension of this analysis to the entire GPCR family may elucidate novel drug targets, as well as expand the clinical utility of FDA-approved therapeutics into new patient cohorts. These recent papers have clearly demonstrated the power of large-scale genomic analysis on the identification of putative cancer-associated GPCRs. Many of them, however, lack the functional validation necessary for further clinical evaluation. The following sections will highlight studies focused on biological target validation, a critical step in translating genomic information to the clinic.
GPCR regulation of cancer signaling pathways
Once a cancer-associated GPCR has been identified, it is critical to determine the underlying biology of how alterations in these GPCRs contribute to tumorigenesis. GPCRs have been known for many years to regulate rapid signaling events through the generation of short-lived second messengers. However, the mechanisms by which GPCRs can sustain long-term biological responses are still being elucidated. One such mechanism involves the transactivation of other signaling molecules, including the epidermal growth factor receptor (EGFR). Several distinct mechanisms for GPCR-induced EGFR transactivation have been uncovered. One pathway involves second messenger-stimulated signaling, including accumulation of intracellular Ca2+, protein kinase C (PKC) activation and reactive oxygen species generation. For example, angiotensin II requires Gαq-mediated intracellular Ca2+ release for EGFR phosphorylation and subsequent MAP kinase activation (16). GPCRs can also transactivate EGFR through the action of nonreceptor tyrosine kinases, including Src and Pyk2 (17). This is accomplished both through direct phosphorylation of EGFR, as well as phosphorylation of adaptor proteins (including Shc) important for downstream pathway activation (18). A third mechanism involves the GPCR-induced metalloproteinase-mediated cleavage of EGFR ligands, including proHB-EGF (19). GPCRs can also transactivate EGFR through TACE/ADAM17-dependent TGF-α (transforming growth factor-α) shedding, a process that requires the mitochondrial production of reactive oxygen species (20). Direct association between GPCRs and EGFR has also been reported. In prostate cancer cells, the proinflammatory chemokine receptor CXCR7 associates with EGFR, and CXCR7 overexpression promotes EGFR phosphorylation (21). Similarly, the Kisspeptin (Kp) receptor GPR54 interacts directly with EGFR in a ligand-dependent manner (22). Stimulation of breast cancer cells with Kp transactivates EGFR and promotes invasive behavior. Therefore, GPCRs can transactivate growth factor signaling through a variety of mechanisms, creating opportunities for therapeutic intervention.
A recurring theme in the utilization of molecularly targeting therapeutics is the development of acquired resistance. Therefore, an intense focus is being placed on the identification of genes capable of overcoming these resistance mechanisms. To determine the contribution of GPCRs to gefitinib (EGFR inhibitor) resistance in non-small-cell lung cancer (NCSLC), a GPCR-specific microarray was used to measure GPCR expression in a resistant cell line as compared with normal human lung fibroblasts (23). Next, agonists and antagonists of differentially expressed GPCRs were tested for their ability to alter cancer cell growth. This screening technique identified inhibition of adenosine A2a receptors as a mechanism for therapeutic intervention of EGFR mutant NSCLC. These results suggest that various GPCRs can regulate EGFR signaling and may provide novel avenues for treatment of EGFR-driven tumors.
In addition to EGFR, cooperation between GPCRs and oncogenes or tumor suppressors has also been described in several human tumors. For example, cyclooxygenase-2 (COX-2)-induced prostaglandin E2 promotes colorectal tumorigenesis in the absence of functional APC (adenomatous polyposis coli) through nuclear accumulation of β-catenin (24). Similarly, Ras upregulates prostaglandin E2 through COX-2 and inhibition of the prostaglandin receptor blocks Ras-mediated transformation (25). Therefore, a greater understanding of the interactions between GPCR and oncogene/tumor suppressor-mediated signaling may uncover novel drug targets.
GPCRs can regulate other cancer-associated signaling pathways, including signal transducer and activator of transcription-3 (STAT3) (26, 27), Hippo (28, 29), phosphoinositide 3-kinase (PI3K) (30), and RhoA/Rac1 (31). The IL-6/JAK/STAT3 signaling axis is a major mediator of cancer initiation and progression (32). Under normal physiological conditions, this pathway is tightly regulated through negative feedback loops (33). However, the mechanism by which STAT3 is persistently activated in cancer remained elusive. An analysis of tumor-derived myeloid cells uncovered the elevated expression of the sphingosine-1-phosphate receptor S1PR1 (a GPCR) as compared with normal splenic myeloid cells (26). STAT3-induced S1PR1 expression promotes persistent STAT3 activation through JAK2, and S1PR1 knockdown inhibits STAT3-mediated tumorigenesis. Persistent STAT3 activation can also be accomplished through depletion of GPRC5a, a tumor suppressor in NSCLC, via the destabilization of the STAT3 inhibitor Socs3 (27). The Hippo pathway, in which the Lats1/2 (large tumor suppressor) kinases phosphorylate and inactivate the transcriptional coactivator YAP (Yes-associated protein), controls organ size and has been implicated in human tumorigenesis (34). Recent evidence suggests that GPCRs can both activate and inactivate Lats1/2, depending on the coupled G-protein, in a process mediated by Rho GTPases and the actin cytoskeleton (28, 29). Activation of Rho and Rac has also been implicated in sustained proliferative signaling downstream of Gαq-linked receptors (31). This novel pathway results in the activation of c-Jun-mediated transcription in a PLC (phospholipase C)-independent manner.
GPCRs have also been implicated in the activation of PI3K, through direct binding of the catalytic p110β to the Gβγ subunits (30). Disruption of this interaction can impair proliferation and invasion in PTEN (phosphatase and tensin homolog)-null prostate cancer cells, suggesting a possible avenue for therapeutic intervention. In uterine fibroids, GPR10 is frequently overexpressed through loss of the tumor suppressor REST/NRSF (RE1 suppressing transcription factor/neuron-restrictive silencing factor) (35). GPR10 activates PI3K/mTOR (mammalian target of rapamycin) signaling, and mice overexpressing GPR10 in the myometrium develop a uterine fibroid phenotype. This novel mouse model will allow the development of therapeutics for fibroids, including GPR10 antagonists. Finally, CXCR4/CXCL12 has been shown to promote growth of malignant peripheral nerve sheath tumors (MPNST) through activation of PI3K and β-catenin signaling (36). Inhibition of CXCR4 activity attenuates tumorigenesis in mouse models of MPNST, providing a novel target for human disease intervention.
GPCRs regulate the Wnt and Hedgehog pathways through the receptors Frizzled and Smoothened, respectively. The role of these pathways in human tumorigenesis has been covered in recent reviews (37, 38). The regulation of multiple cancer-associated signaling pathways by GPCRs presents many opportunities for therapeutic intervention. In tumors with GPCR alterations, chemical inhibitors of downstream pathways may provide clinical benefit, without the need for specific GPCR antagonists. Conversely, GPCR regulation of oncogenic receptor tyrosine kinase signaling can provide a distinct mechanism for pathway inhibition. Therefore, understanding the molecular mechanisms by which GPCRs influence cancer progression is of critical importance to their utility as drug targets.
GPCR regulation of hormone-sensitive tumors
A wide variety of hormones bind and activate GPCRs. Therefore, it is unsurprising that GPCRs, and their coupled G-proteins, have been shown to play critical roles in the pathogenesis of hormone responsive tumors. Among the first examples of this phenomenon was the identification of somatic mutations in the thyrotropin receptor, resulting in constitutive activation of adenylyl cyclase and hyperfunctioning thyroid adenomas (39). Similarly, activating mutations have also been found in the luteinizing hormone receptor in Leydig-cell testicular tumors (40). Recently, much attention has focused on the role of the novel estrogen receptor GPER-1 (formerly GPR30) in cancers of the reproductive system. It has been well established that estrogen exerts transcriptional activation through the action of the estrogen receptors ERα and ERβ (41). However, the molecular mechanisms by which estrogen induced nongenomic signaling, including Ca2+ and NO generation and tyrosine kinase activation, remained largely unknown (42). These questions were answered with the discovery that the orphan GPCR GPER-1 binds to estrogen, resulting in intracellular Ca2+ accumulation and PI3K activation (43). Recent data has begun to elucidate the role of GPER-1 in estrogen-responsive cancers. GPER-1 activation increases cancer cell proliferation from several reproductive organs, including that of the breast, endometrium, ovary and testis, and expression correlates with disease progression (44). GPER-1 may also regulate tumor cell invasion through increasing the expression and proteolytic activity of MMP-9 in ovarian cancer cells (45) and enhance fatty acid metabolism through upregulation of fatty acid synthase and EGFR transactivation (46). Furthermore, a role for GPER-1 in regulation of the tumor microenvironment has recently been uncovered. GPER-1 is localized to the nucleus in cancer-associated fibroblasts, activates transcription, and regulates estrogen-induced migration (47). Therefore, GPCRs can function both at the cell surface and the nucleus to mediate cellular responses. A greater understanding of the contribution of GPER-1 to cancer pathogenesis may create opportunities for therapeutic intervention in both ER positive and negative tumors.
GPCR regulation of invasion, metastasis and the tumor microenvironment
The control of metastatic dissemination and growth at the secondary site are critical nodes in the development of novel cancer drugs. Several recent studies have highlighted the role of GPCRs in controlling cancer cell invasion and metastasis, providing targets for therapeutic intervention. In a mouse model of melanoma, expression of endothelin receptor B (EDNRB) enhanced spontaneous central nervous system metastases (48). Importantly, inhibition of EDNRB increased life span in mice with visceral metastases and shrunk intracranial melanoma tumors when combined with cyclosporin A to increase drug levels in the brain parenchyma. The thrombin receptor (PAR-1) is also associated with metastatic melanoma and PAR-1 silencing can decrease tumor growth and lung metastases (49). PAR-1 exerts these effects through transcriptional repression of the tumor suppressor Mapsin (50). Similarly, knockdown of EMR2 (EGF-module containing mucin-like receptor 2) in glioblastoma cell lines reduced migration (51). This correlates with the observation that EMR2 is associated with poor prognosis in glioblastoma patients. In triple negative breast cancer (TNBC) cells, a lactoferrin/endothelin-1 (ET-1) pathway promotes cell invasion, and inhibition of the ET-1 receptor blocks motility (52). This pathway may be clinically relevant as patients with TNBC display elevated levels of ET-1. In kidney cancer, the von Hippel-Lindau (VHL) tumor suppressor gene regulates expression of the chemokine receptor CXCR4, a regulator of metastatic progression, through hypoxia-inducible factor 2 (HIF2-α) (53). While loss of VHL can initiate tumorigenesis, further epigenetic events are required for CXCR4 expression and metastatic colonization (54). This suggests that chromatin-modifying drugs, or those targeting CXCR4, may inhibit kidney cancer progression. GPCRs can also modulate cell invasive behavior by regulating expression of matrix metalloproteinases (MMPs) (55), and activation of Rho GTPases (56–58).
Preparation and maintenance of the secondary niche are critical steps during metastatic progression and both processes are regulated by GPCRs. The S1PR1-STAT3 axis supports myeloid cell colonization at the premetastatic niche, providing factors necessary for tumor cell growth (59). Furthermore, STAT3 activity is elevated in tumor-free lymph nodes from cancer patients, suggesting that pathway inhibition may disrupt the ability of tumor cells to grow at secondary sites. In metastatic breast cancer, production of tenascin C (TNC) promotes lung metastasis through the expression of the Wnt target gene LGR5, and knockdown of LGR5 decreased the ability of breast cancer cells to colonize the lung (60). The chemokines CXCL1 and CXCL2 are amplified in invasive breast cancer and recruit myeloid cells to the primary tumor, which in turn provide factors required for cell survival and invasion (61). This axis is stimulated by chemotherapy, providing a mechanism for chemoresistance. However, combining chemotherapy with CXCR2 inhibition in mice with lung metastases reduced metastatic burden, providing preclinical evidence for the utility of CXCR2 inhibition in the clinic. GPCRs can also regulate the metastatic switch through disruption of cell adhesion. S1P1 (sphingosine 1-phosphate) expression underlies the progression of T-lymphoblastic lymphoma (T-LBL) to acute T-lymphoblastic leukemia (T-ALL) (62). Increased S1P1 signaling promotes cell adhesion and inhibits intravasation and dissemination in T-LBL, while a selective S1P1 antagonist promotes intravasation in vivo. GPCRs can regulate lymphatic vessel dilation and metastatic progression via crosstalk between the lymphangiogenic growth factor VEGF-D and prostaglandins. Finally, several GPCRs regulate tumor-associated angiogenesis, including PAR-2 (63), vGPCR (64), BAI1 (65), and GPR56 (66).
As tumors are highly proinflammatory, regulation of the tumor microenvironment has become an important target for therapeutic intervention. GPCRs, most specifically the inflammatory chemokine receptors, control recruitment and activity of leukocytes within the tumor microenvironment and provide a means for regulating tumor-stroma interactions. Excitingly, several chemokine receptor antagonists are in clinical development for inflammatory diseases and may be co-opted for use in cancer treatment. However, as immune cells within the tumor stroma can play both positive and negative roles in tumor progression, novel inhibitors must be first tested in the appropriate cancer models. Two such studies have provided evidence that inhibition of CXCR2 may be of clinical utility in both inflammation-driven and spontaneous skin, intestinal and pancreatic tumorigenesis (67, 68). In oral squamous cell carcinoma, CXCL13/CXCR5 signaling can activate the osteoclastogenic factor RANKL (receptor activator of NF-κB ligand) in stromal cells through c-Myc, resulting in bone invasion (69). GPCRs have also been shown to regulate resistance to chemotherapy through effects on immune cell infiltration (70). Upon doxorubicin treatment, myeloid cells infiltrate the tumor in a CCR2-dependent manner, and CCR2 null mice showed enhanced response to doxorubicin or cisplatin (70). Therefore, these signaling axes may represent promising therapeutic targets.
GPCR signaling also provides a link between obesity and inflammation in breast cancer, as the proinflammatory prostaglandin E2 regulates aromatase expression, the enzyme necessary for estrogen synthesis, through the cAMP (cyclic adenosine monophosphate)/PKA (protein kinase A) pathway (71). As elevated aromatase activity is known to increase the risk for post-menopausal hormone receptor positive breast cancer, strategies aimed at reducing inflammation may reduce breast cancer risk. Collectively, these reports highlight the importance of studying GPCR function in many contexts beyond regulation of cell proliferation. While more difficult to study in a high-throughput manner, effects on cell migration and invasion, as well as immune function and secondary niche formation must be taken into account when probing the cancer-associated activity of a GPCR. Furthermore, these results suggest that drugs targeting GPCRs may have utility against metastatic progression, the deadliest aspect of human cancer.
Implications and future directions
The advent of targeted therapeutics in cancer has begun to lessen our reliance on non-specific chemotherapy. Large-scale genomic analyses of tumors have provided potential novel targets for targeted therapeutic interventions; however, separating the tumor-initiators from bystanders remains a critical challenge for both bioinformaticans and those at the bench. It is this biological validation that will truly expand the pool of targeted therapies in the clinic. It is apparent that targeted therapies will be beset by multiple mechanisms of resistance. Therefore, finding novel druggable targets is of critical importance. The GPCRs represent a class of highly druggable, yet understudied molecules in cancer. As transmembrane proteins, GPCRs can be targeted by antibodies, small molecules and peptides, obviating nicely the drug development issues associated with crossing the cell membrane. Furthermore, GPCR signaling can be modulated by agonists, antagonists and inverse agonists, allowing precise control over signaling pathways. Importantly, drug companies have extensive experience developing GPCR-based therapeutics for a wide range of diseases, providing a knowledge base for novel target development.
An analysis of GPCR signaling in cancer reveals several emerging concepts. Advances in sequencing technologies have recently uncovered widespread GPCR alterations in human cancer (9). However, the functional outcome of these changes remains largely unexplored. GPCRs are known to regulate multiple aspects of tumorigenesis, both tumor cell-intrinsic (proliferation, migration) and extrinsic (regulation of the tumor microenvironment, metastatic niche). Therefore, it is crucial that functional assays take into account these diverse biological processes. Furthermore, it has become evident that GPCRs can regulate cancer-associated signaling networks, resulting in pathway activation and acquired resistance. A deeper understanding of this molecular crosstalk will expand the potential for GPCR-targeted therapeutics. Advances in drug design have introduced the concept of biased ligands, which selectively regulate signaling pathways downstream of GPCRs. In order for these novel compounds to reach their potential we must elucidate the multitude of possible signaling events triggered upon GPCR activation, as well as develop robust high-throughout assays to monitor signaling output. As the sequencing of the human genome provided the raw material for functional genomic annotation, major tumor sequencing efforts have begun to provide a comprehensive view of human cancer. We must now use this information to intelligently select and validate targets for translation of these data into real clinical advances in cancer treatment.
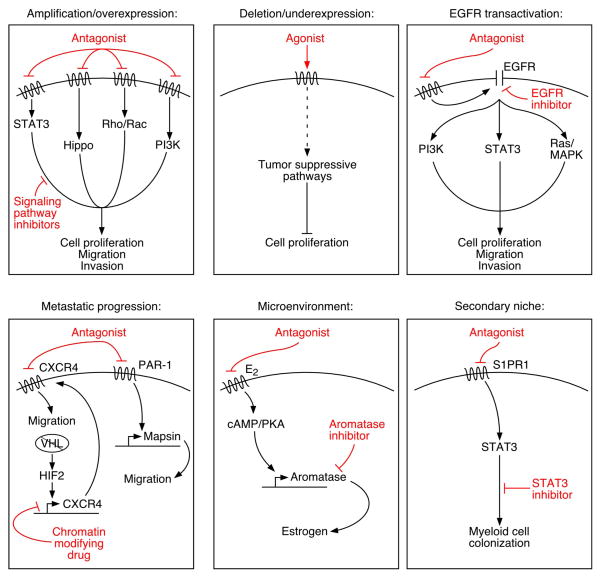
Figure.
GPCRs regulate multiple steps in cancer progression, including initiation, metastatic dissemination and secondary niche maintenance, through a wide array of signaling effectors. This diversity may provide novel avenues for GPCR-targeted therapeutics in the treatment of cancer. Abbreviations: STAT3 (signal transducer and activator of transcription 3), PI3K (phosphoinositide 3-kinase), MAPK (mitogen activated protein kinase), EGFR (epidermal growth factor receptor), PAR-1 (protease activated receptor 1), VHL (von Hippel-Lindau), HIF2 (hypoxia-inducible factor 2), cAMP (cyclic adenosine monophosphate), PKA (protein kinase A), S1PR1 (sphingosine-1-phosphate receptor 1).
Acknowledgments
I would like to thank Dr. Craig Malbon, Dr. Senthil Muthuswamy, and past and current members of the Muthuswamy laboratory for helpful discussions, and Jim Duffy for figure preparation. This work was supported by an American Cancer Society postdoctoral fellowship to MF (PF-11-026-01-CSM) and NCI grant CA098830 to Dr. Muthuswamy.
Abbreviations
- GPCR
G-protein coupled receptor
- GRM
glutamate receptor, metabotropic
- BAI
brain-specific angiogenesis inhibitor
- P2RY
purinergic receptor
- TCGA
The Cancer Genome Atlas
- SNP
single nucleotide polymorphism
- PTH1R
parathyroid hormone 1 receptor
- TSHR
thyroid stimulating hormone receptor
- EGFR
epidermal growth factor receptor
- PKC
protein kinase C
- TGF-α
transforming growth factor-α
- Kp
kisspeptin
- NSCLC
non-small-cell lung cancer
- COX-20
cyclooxygenase-2
- APC
adenomatous polyposis coli
- STAT3
signal transducer and activator of transcription-3
- PI3K
phosphoinositide 3-kinase
- S1PR1
sphingosine-1-phosphate receptor
- Lats
large tumor suppressor kinase
- Yes
Yes-associated protein
- PLC
phospholipase C
- PTEN
phosphatase and tensin homolog
- REST/NRSF
RE1 suppressing transcription factor/neuron-restrictive silencing factor
- mTOR
mammalian target of rapamycin
- MPNST
malignant peripheral nerve tumor
- GPER
G-protein coupled estrogen receptor
- ER
estrogen receptor
- MMP
matrix metalloproteinase
- EDNRB
endothelin receptor B
- PAR-1
protease activated receptor
- EMR2
EGF-module containing mucin-like receptor 2
- TNBC
triple negative breast cancer
- ET-1
lactoferrin/endothelin-1
- VHL
von Hippel-Lindau
- HIF-2
hypoxia-inducible factor 2
- T-LBL
T-lymphoblastic lymphoma
- T-ALL
Tlymphoblastic leukemia
- RANKL
receptor activator of NF-κB ligand
- cAMP
cyclic adenosine monophosphate
- PKA
protein kinase A
- MAPK
mitogen activated protein kinase
References
- 1.Morris AJ, Malbon CC. Physiological regulation of G protein-linked signaling. Physiol Rev. 1999;79:1373–1430. doi: 10.1152/physrev.1999.79.4.1373. [DOI] [PubMed] [Google Scholar]
- 2.Lappano R, Maggiolini M. G protein-coupled receptors: novel targets for drug discovery in cancer. Nat Rev Drug Discov. 2011;10:47–60. doi: 10.1038/nrd3320. [DOI] [PubMed] [Google Scholar]
- 3.Young D, Waitches G, Birchmeier C, Fasano O, Wigler M. Isolation and characterization of a new cellular oncogene encoding a protein with multiple potential transmembrane domains. Cell. 1986;45:711–719. doi: 10.1016/0092-8674(86)90785-3. [DOI] [PubMed] [Google Scholar]
- 4.Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, Stern HM, Yue P, Haverty PM, Bourgon R, Zheng J, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 5.Hammerman PS, Hayes DN, Wilkerson MD, Schultz N, Bose R, Chu A, Collisson EA, Cope L, Creighton CJ, Getz G, et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Carvalho DD, Sharma S, You JS, Su SF, Taberlay PC, Kelly TK, Yang X, Liang G, Jones PA. DNA methylation screening identifies driver epigenetic events of cancer cell survival. Cancer Cell. 2012;21:655–667. doi: 10.1016/j.ccr.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dorsam RT, Gutkind JS. G-protein-coupled receptors and cancer. Nat Rev Cancer. 2007;7:79–94. doi: 10.1038/nrc2069. [DOI] [PubMed] [Google Scholar]
- 8.Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153:17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 9.O’Hayre M, Vazquez-Prado J, Kufareva I, Stawiski EW, Handel TM, Seshagiri S, Gutkind JS. The emerging mutational landscape of G-proteins and G-protein-coupled receptors in cancer. Nat Rev Cancer. 2013;13:412–424. doi: 10.1038/nrc3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prickett TD, Wei X, Cardenas-Navia I, Teer JK, Lin JC, Walia V, Gartner J, Jiang J, Cherukuri PF, Molinolo A, et al. Exon capture analysis of G protein-coupled receptors identifies activating mutations in GRM3 in melanoma. Nat Genet. 2011;43:1119–1126. doi: 10.1038/ng.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang H, Cheung LW, Li J, Ju Z, Yu S, Stemke-Hale K, Dogruluk T, Lu Y, Liu X, Gu C, et al. Whole-exome sequencing combined with functional genomics reveals novel candidate driver cancer genes in endometrial cancer. Genome Res. 2012;22:2120–2129. doi: 10.1101/gr.137596.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao Q, Kirkness EF, Caballero OL, Galante PA, Parmigiani RB, Edsall L, Kuan S, Ye Z, Levy S, Vasconcelos AT, et al. Systematic detection of putative tumor suppressor genes through the combined use of exome and transcriptome sequencing. Genome Biol. 2010;11:R114. doi: 10.1186/gb-2010-11-11-r114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, Schmidt MK, Chang-Claude J, Bojesen SE, Bolla MK, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45:353–361. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia-Closas M, Couch FJ, Lindstrom S, Michailidou K, Schmidt MK, Brook MN, Orr N, Rhie SK, Riboli E, Feigelson HS, et al. Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nat Genet. 2013;45:392–398. doi: 10.1038/ng.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patel MN, Halling-Brown MD, Tym JE, Workman P, Al-Lazikani B. Objective assessment of cancer genes for drug discovery. Nat Rev Drug Discov. 2012;12:35–50. doi: 10.1038/nrd3913. [DOI] [PubMed] [Google Scholar]
- 16.Eguchi S, Numaguchi K, Iwasaki H, Matsumoto T, Yamakawa T, Utsunomiya H, Motley ED, Kawakatsu H, Owada KM, Hirata Y, et al. Calcium-dependent epidermal growth factor receptor transactivation mediates the angiotensin II-induced mitogen-activated protein kinase activation in vascular smooth muscle cells. J Biol Chem. 1998;273:8890–8896. doi: 10.1074/jbc.273.15.8890. [DOI] [PubMed] [Google Scholar]
- 17.Andreev J, Galisteo ML, Kranenburg O, Logan SK, Chiu ES, Okigaki M, Cary LA, Moolenaar WH, Schlessinger J. Src and Pyk2 mediate G-protein-coupled receptor activation of epidermal growth factor receptor (EGFR) but are not required for coupling to the mitogen-activated protein (MAP) kinase signaling cascade. J Biol Chem. 2001;276:20130–20135. doi: 10.1074/jbc.M102307200. [DOI] [PubMed] [Google Scholar]
- 18.Luttrell LM, Della Rocca GJ, van Biesen T, Luttrell DK, Lefkowitz RJ. Gbetagamma subunits mediate Src-dependent phosphorylation of the epidermal growth factor receptor. A scaffold for G protein-coupled receptor-mediated Ras activation. J Biol Chem. 1997;272:4637–4644. doi: 10.1074/jbc.272.7.4637. [DOI] [PubMed] [Google Scholar]
- 19.Prenzel N, Zwick E, Daub H, Leserer M, Abraham R, Wallasch C, Ullrich A. EGF receptor transactivation by G-protein-coupled receptors requires metalloproteinase cleavage of proHB-EGF. Nature. 1999;402:884–888. doi: 10.1038/47260. [DOI] [PubMed] [Google Scholar]
- 20.Myers TJ, Brennaman LH, Stevenson M, Higashiyama S, Russell WE, Lee DC, Sunnarborg SW. Mitochondrial reactive oxygen species mediate GPCR-induced TACE/ADAM17-dependent transforming growth factor-alpha shedding. Mol Biol Cell. 2009;20:5236–5249. doi: 10.1091/mbc.E08-12-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh RK, Lokeshwar BL. The IL-8-regulated chemokine receptor CXCR7 stimulates EGFR signaling to promote prostate cancer growth. Cancer Res. 2011;71:3268–3277. doi: 10.1158/0008-5472.CAN-10-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zajac M, Law J, Cvetkovic DD, Pampillo M, McColl L, Pape C, Di Guglielmo GM, Postovit LM, Babwah AV, Bhattacharya M. GPR54 (KISS1R) transactivates EGFR to promote breast cancer cell invasiveness. PLoS One. 2011;6:e21599. doi: 10.1371/journal.pone.0021599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuzumaki N, Suzuki A, Narita M, Hosoya T, Nagasawa A, Imai S, Yamamizu K, Morita H, Suzuki T, Okada Y, et al. Multiple Analyses of G-Protein Coupled Receptor (GPCR) Expression in the Development of Gefitinib-Resistance in Transforming Non-Small-Cell Lung Cancer. PLoS One. 2012;7:e44368. doi: 10.1371/journal.pone.0044368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X, Lazenby AJ, Siegal GP. Signal transduction cross-talk during colorectal tumorigenesis. Adv Anat Pathol. 2006;13:270–274. doi: 10.1097/01.pap.0000213046.61941.5c. [DOI] [PubMed] [Google Scholar]
- 25.Repasky GA, Zhou Y, Morita S, Der CJ. Ras-mediated intestinal epithelial cell transformation requires cyclooxygenase-2-induced prostaglandin E2 signaling. Mol Carcinog. 2007;46:958–970. doi: 10.1002/mc.20333. [DOI] [PubMed] [Google Scholar]
- 26.Lee H, Deng J, Kujawski M, Yang C, Liu Y, Herrmann A, Kortylewski M, Horne D, Somlo G, Forman S, et al. STAT3-induced S1PR1 expression is crucial for persistent STAT3 activation in tumors. Nat Med. 2010;16:1421–1428. doi: 10.1038/nm.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y, Deng J, Fujimoto J, Kadara H, Men T, Lotan D, Lotan R. Gprc5a deletion enhances the transformed phenotype in normal and malignant lung epithelial cells by eliciting persistent Stat3 signaling induced by autocrine leukemia inhibitory factor. Cancer Res. 2010;70:8917–8926. doi: 10.1158/0008-5472.CAN-10-0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu FX, Zhao B, Panupinthu N, Jewell JL, Lian I, Wang LH, Zhao J, Yuan H, Tumaneng K, Li H, et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150:780–791. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mo JS, Yu FX, Gong R, Brown JH, Guan KL. Regulation of the Hippo-YAP pathway by protease-activated receptors (PARs) Genes Dev. 2012;26:2138–2143. doi: 10.1101/gad.197582.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dbouk HA, Vadas O, Shymanets A, Burke JE, Salamon RS, Khalil BD, Barrett MO, Waldo GL, Surve C, Hsueh C, et al. G Protein-Coupled Receptor-Mediated Activation of p110beta by Gbetagamma Is Required for Cellular Transformation and Invasiveness. Sci Signal. 2012;5:ra89. doi: 10.1126/scisignal.2003264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaque JP, Dorsam RT, Feng X, Iglesias-Bartolome R, Forsthoefel DJ, Chen Q, Debant A, Seeger MA, Ksander BR, Teramoto H, et al. A Genome-wide RNAi Screen Reveals a Trio-Regulated Rho GTPase Circuitry Transducing Mitogenic Signals Initiated by G Protein-Coupled Receptors. Mol Cell. 2012 doi: 10.1016/j.molcel.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu H, Jove R. The STATs of cancer--new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 33.Yoshimura A, Naka T, Kubo M. SOCS proteins, cytokine signalling and immune regulation. Nat Rev Immunol. 2007;7:454–465. doi: 10.1038/nri2093. [DOI] [PubMed] [Google Scholar]
- 34.Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Varghese BV, Koohestani F, McWilliams M, Colvin A, Gunewardena S, Kinsey WH, Nowak RA, Nothnick WB, Chennathukuzhi VM. Loss of the repressor REST in uterine fibroids promotes aberrant G protein-coupled receptor 10 expression and activates mammalian target of rapamycin pathway. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1215759110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mo W, Chen J, Patel A, Zhang L, Chau V, Li Y, Cho W, Lim K, Xu J, Lazar AJ, et al. CXCR4/CXCL12 Mediate Autocrine Cell-Cycle Progression in NF1-Associated Malignant Peripheral Nerve Sheath Tumors. Cell. 2013;152:1077–1090. doi: 10.1016/j.cell.2013.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2012;13:11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- 38.Kar S, Deb M, Sengupta D, Shilpi A, Bhutia SK, Patra SK. Intricacies of hedgehog signaling pathways: a perspective in tumorigenesis. Exp Cell Res. 2012;318:1959–1972. doi: 10.1016/j.yexcr.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 39.Parma J, Duprez L, Van Sande J, Cochaux P, Gervy C, Mockel J, Dumont J, Vassart G. Somatic mutations in the thyrotropin receptor gene cause hyperfunctioning thyroid adenomas. Nature. 1993;365:649–651. doi: 10.1038/365649a0. [DOI] [PubMed] [Google Scholar]
- 40.Liu G, Duranteau L, Carel JC, Monroe J, Doyle DA, Shenker A. Leydig-cell tumors caused by an activating mutation of the gene encoding the luteinizing hormone receptor. N Engl J Med. 1999;341:1731–1736. doi: 10.1056/NEJM199912023412304. [DOI] [PubMed] [Google Scholar]
- 41.Hall JM, Couse JF, Korach KS. The multifaceted mechanisms of estradiol and estrogen receptor signaling. J Biol Chem. 2001;276:36869–36872. doi: 10.1074/jbc.R100029200. [DOI] [PubMed] [Google Scholar]
- 42.Kelly MJ, Levin ER. Rapid actions of plasma membrane estrogen receptors. Trends Endocrinol Metab. 2001;12:152–156. doi: 10.1016/s1043-2760(01)00377-0. [DOI] [PubMed] [Google Scholar]
- 43.Revankar CM, Cimino DF, Sklar LA, Arterburn JB, Prossnitz ER. A transmembrane intracellular estrogen receptor mediates rapid cell signaling. Science. 2005;307:1625–1630. doi: 10.1126/science.1106943. [DOI] [PubMed] [Google Scholar]
- 44.Filardo EJ, Thomas P. Minireview: G protein-coupled estrogen receptor-1, GPER-1: its mechanism of action and role in female reproductive cancer, renal and vascular physiology. Endocrinology. 2012;153:2953–2962. doi: 10.1210/en.2012-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan Y, Liu H, Wen H, Jiang X, Cao X, Zhang G, Liu G. The novel estrogen receptor GPER regulates the migration and invasion of ovarian cancer cells. Mol Cell Biochem. 2013;378:1–7. doi: 10.1007/s11010-013-1579-9. [DOI] [PubMed] [Google Scholar]
- 46.Santolla MF, Lappano R, De Marco P, Pupo M, Vivacqua A, Sisci D, Abonante S, Iacopetta D, Cappello AR, Dolce V, et al. G protein-coupled estrogen receptor mediates the up-regulation of fatty acid synthase induced by 17beta-estradiol in cancer cells and cancer-associated fibroblasts. J Biol Chem. 2012;287:43234–43245. doi: 10.1074/jbc.M112.417303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pupo M, Vivacqua A, Perrotta I, Pisano A, Aquila S, Abonante S, Gasperi-Campani A, Pezzi V, Maggiolini M. The nuclear localization signal is required for nuclear GPER translocation and function in breast Cancer-Associated Fibroblasts (CAFs) Mol Cell Endocrinol. 2013;376:23–32. doi: 10.1016/j.mce.2013.05.023. [DOI] [PubMed] [Google Scholar]
- 48.Cruz-Munoz W, Jaramillo ML, Man S, Xu P, Banville M, Collins C, Nantel A, Francia G, Morgan SS, Cranmer LD, et al. Roles for endothelin receptor B and BCL2A1 in spontaneous CNS metastasis of melanoma. Cancer Res. 2012;72:4909–4919. doi: 10.1158/0008-5472.CAN-12-2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Villares GJ, Zigler M, Wang H, Melnikova VO, Wu H, Friedman R, Leslie MC, Vivas-Mejia PE, Lopez-Berestein G, Sood AK, et al. Targeting melanoma growth and metastasis with systemic delivery of liposome-incorporated protease-activated receptor-1 small interfering RNA. Cancer Res. 2008;68:9078–9086. doi: 10.1158/0008-5472.CAN-08-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Villares GJ, Zigler M, Dobroff AS, Wang H, Song R, Melnikova VO, Huang L, Braeuer RR, Bar-Eli M. Protease activated receptor-1 inhibits the Maspin tumor-suppressor gene to determine the melanoma metastatic phenotype. Proc Natl Acad Sci U S A. 2011;108:626–631. doi: 10.1073/pnas.1006886108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rutkowski MJ, Sughrue ME, Kane AJ, Kim JM, Bloch O, Parsa AT. Epidermal growth factor module-containing mucin-like receptor 2 is a newly identified adhesion G protein-coupled receptor associated with poor overall survival and an invasive phenotype in glioblastoma. J Neurooncol. 2011;105:165–171. doi: 10.1007/s11060-011-0576-7. [DOI] [PubMed] [Google Scholar]
- 52.Ha NH, Nair VS, Reddy DN, Mudvari P, Ohshiro K, Ghanta KS, Pakala SB, Li DQ, Costa L, Lipton A, et al. Lactoferrin-endothelin-1 axis contributes to the development and invasiveness of triple-negative breast cancer phenotypes. Cancer Res. 2011;71:7259–7269. doi: 10.1158/0008-5472.CAN-11-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Staller P, Sulitkova J, Lisztwan J, Moch H, Oakeley EJ, Krek W. Chemokine receptor CXCR4 downregulated by von Hippel-Lindau tumour suppressor pVHL. Nature. 2003;425:307–311. doi: 10.1038/nature01874. [DOI] [PubMed] [Google Scholar]
- 54.Vanharanta S, Shu W, Brenet F, Hakimi AA, Heguy A, Viale A, Reuter VE, Hsieh JJ, Scandura JM, Massague J. Epigenetic expansion of VHL-HIF signal output drives multiorgan metastasis in renal cancer. Nat Med. 2012 doi: 10.1038/nm.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park SY, Jeong KJ, Panupinthu N, Yu S, Lee J, Han JW, Kim JM, Lee JS, Kang J, Park CG, et al. Lysophosphatidic acid augments human hepatocellular carcinoma cell invasion through LPA1 receptor and MMP-9 expression. Oncogene. 2011;30:1351–1359. doi: 10.1038/onc.2010.517. [DOI] [PubMed] [Google Scholar]
- 56.Castellone RD, Leffler NR, Dong L, Yang LV. Inhibition of tumor cell migration and metastasis by the proton-sensing GPR4 receptor. Cancer Lett. 2011;312:197–208. doi: 10.1016/j.canlet.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 57.Nithipatikom K, Gomez-Granados AD, Tang AT, Pfeiffer AW, Williams CL, Campbell WB. Cannabinoid receptor type 1 (CB1) activation inhibits small GTPase RhoA activity and regulates motility of prostate carcinoma cells. Endocrinology. 2012;153:29–41. doi: 10.1210/en.2011-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ward Y, Lake R, Yin JJ, Heger CD, Raffeld M, Goldsmith PK, Merino M, Kelly K. LPA receptor heterodimerizes with CD97 to amplify LPA-initiated RHO-dependent signaling and invasion in prostate cancer cells. Cancer Res. 2011;71:7301–7311. doi: 10.1158/0008-5472.CAN-11-2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deng J, Liu Y, Lee H, Herrmann A, Zhang W, Zhang C, Shen S, Priceman SJ, Kujawski M, Pal SK, et al. S1PR1-STAT3 signaling is crucial for myeloid cell colonization at future metastatic sites. Cancer Cell. 2012;21:642–654. doi: 10.1016/j.ccr.2012.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oskarsson T, Acharyya S, Zhang XH, Vanharanta S, Tavazoie SF, Morris PG, Downey RJ, Manova-Todorova K, Brogi E, Massague J. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat Med. 2011;17:867–874. doi: 10.1038/nm.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Acharyya S, Oskarsson T, Vanharanta S, Malladi S, Kim J, Morris PG, Manova-Todorova K, Leversha M, Hogg N, Seshan VE, et al. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell. 2012;150:165–178. doi: 10.1016/j.cell.2012.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Feng H, Stachura DL, White RM, Gutierrez A, Zhang L, Sanda T, Jette CA, Testa JR, Neuberg DS, Langenau DM, et al. T-lymphoblastic lymphoma cells express high levels of BCL2, S1P1, and ICAM1, leading to a blockade of tumor cell intravasation. Cancer Cell. 2010;18:353–366. doi: 10.1016/j.ccr.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang L, Chen G, Mohanty S, Scott G, Fazal F, Rahman A, Begum S, Hynes RO, Xu L. GPR56 Regulates VEGF production and angiogenesis during melanoma progression. Cancer Res. 2011;71:5558–5568. doi: 10.1158/0008-5472.CAN-10-4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Svensson KJ, Kucharzewska P, Christianson HC, Skold S, Lofstedt T, Johansson MC, Morgelin M, Bengzon J, Ruf W, Belting M. Hypoxia triggers a proangiogenic pathway involving cancer cell microvesicles and PAR-2-mediated heparin-binding EGF signaling in endothelial cells. Proc Natl Acad Sci U S A. 2011;108:13147–13152. doi: 10.1073/pnas.1104261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ma T, Jham BC, Hu J, Friedman ER, Basile JR, Molinolo A, Sodhi A, Montaner S. Viral G protein-coupled receptor up-regulates Angiopoietin-like 4 promoting angiogenesis and vascular permeability in Kaposi’s sarcoma. Proc Natl Acad Sci U S A. 2010;107:14363–14368. doi: 10.1073/pnas.1001065107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu D, Hunter SB, Vertino PM, Van Meir EG. Overexpression of MBD2 in glioblastoma maintains epigenetic silencing and inhibits the antiangiogenic function of the tumor suppressor gene BAI1. Cancer Res. 2011;71:5859–5870. doi: 10.1158/0008-5472.CAN-11-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ijichi H, Chytil A, Gorska AE, Aakre ME, Bierie B, Tada M, Mohri D, Miyabayashi K, Asaoka Y, Maeda S, et al. Inhibiting Cxcr2 disrupts tumorstromal interactions and improves survival in a mouse model of pancreatic ductal adenocarcinoma. J Clin Invest. 2011;121:4106–4117. doi: 10.1172/JCI42754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jamieson T, Clarke M, Steele CW, Samuel MS, Neumann J, Jung A, Huels D, Olson MF, Das S, Nibbs RJ, et al. Inhibition of CXCR2 profoundly suppresses inflammation-driven and spontaneous tumorigenesis. J Clin Invest. 2012;122:3127–3144. doi: 10.1172/JCI61067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sambandam Y, Sundaram K, Liu A, Kirkwood KL, Ries WL, Reddy SV. CXCL13 activation of c-Myc induces RANK ligand expression in stromal/preosteoblast cells in the oral squamous cell carcinoma tumor-bone microenvironment. Oncogene. 2012 doi: 10.1038/onc.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nakasone ES, Askautrud HA, Kees T, Park JH, Plaks V, Ewald AJ, Fein M, Rasch MG, Tan YX, Qiu J, et al. Imaging tumor-stroma interactions during chemotherapy reveals contributions of the microenvironment to resistance. Cancer Cell. 2012;21:488–503. doi: 10.1016/j.ccr.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Subbaramaiah K, Morris PG, Zhou XK, Morrow M, Du B, Giri D, Kopelovich L, Hudis CA, Dannenberg AJ. Increased levels of COX-2 and prostaglandin E2 contribute to elevated aromatase expression in inflamed breast tissue of obese women. Cancer Discov. 2012;2:356–365. doi: 10.1158/2159-8290.CD-11-0241. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]