Abstract
Epstein–Barr virus-encoded latent membrane protein-1 (LMP1) plays a fundamental role in the malignant transformation of nasopharyngeal carcinoma (NPC), although the mechanism is not well understood. Here, we showed that Limb-bud and Heart (LBH) is considerably downregulated in patient NPC tissues. The expression of LBH in biopsies of 40 consecutive NPC patients devoid of initial distant metastasis and treated according to consistent guidelines was also analyzed, and we found the LBH expression level was correlated with some of clinicopathological features, disease-specific survival (DSS), distant metastasis-free survival (DMFS). We further determined that LBH normally induces NPC cell cycle arrest at the G1/S transition, and LBH can suppress the growth of transplanted NPC tumors in vivo by downregulating LMP1-mediated NF-κB transcriptional activity. Transforming growth factor-beta 1 (TGF-β1) normally protects against tumor development by suppressing cell proliferation, but NPC cells acquire resistance to TGF-β1–mediated inhibition. We found that TGF-β1 inhibits NF-κB transcriptional activity and nasopharyngeal epithelial cell proliferation through upregulating LBH expression. These data reveal a previously unknown NPC transformation mechanism and provide a new concept and treatment strategy for LMP1-driven oncogenesis in NPC.
Nasopharyngeal carcinoma (NPC) is prevalent in Southeast Asian populations. Genetic predisposition, Epstein-Barr virus (EBV) infection, and environmental conditions are major contributing factors to malignant transformation1, but the precise mechanisms are unclear. NPC is not associated with any point mutations in known tumor suppressor genes (TSGs)2,3, although P16 and Rb2 downregulation may play a role4,5. Polymorphisms of Toll-like receptor 9 are associated with nasopharyngeal carcinoma susceptibility6. The polymorphism interleukin-8 -251A/T7 and Cyclin D1 G870A8 are associated with a significantly increased risk of NPC.
Several factors influence NPC tumorigenesis. EBV infection plays a crucial role through EBV-encoded latent genes, such as LMP1, that act as oncogenes by affecting normal cellular functions like cell cycle progression1,9. Transforming growth factor-beta (TGF-β) is also implicated10,11, as NPC cells acquire resistance to normal TGF-β1–mediated inhibition of cell growth10,12, similar to other tumors13. Interestingly, EBV-encoded LMP1 confers resistance to TGF-β–mediated inhibition by upregulating Cyclin D1 and downregulating P15, which normally function to maintain cell cycle arrest at the G1 phase. However, the molecular mechanism underlying how these cell cycle regulators lead to NPC transformation is unclear.
Previously, we identified that expression of a new expressed sequence tag (EST) NP9 (Genbank No. BF718797) was considerably downregulated in both NPC cell lines and biopsy tissues compared to normal nasopharyngeal epithelial tissue14,15,16, implicating it as a candidate TSG for NPC. Sequence blast analysis showed complete homology between NP9 and the Limb-bud and heart (Lbh) gene (Genbank No. NM_030915). LBH is a highly conserved, tissue-specific transcription cofactor in vertebrates normally implicated in embryonic development17,18, although its role in adult tissues is unclear. Structurally, LBH exhibits a high degree of disorder, suggesting that conformational plasticity may influence LBH-dependent transcriptional activity. Aberrant LBH gain-of-function during heart development can lead to congenital heart disease, similar to that observed in partial trisomy 2p patients, and various other growth defects19. LBH is also overexpressed in some basal-like breast cancers, where Wnt signaling can directly regulate LBH during both development and cancer progression20,21. Despite this association with breast cancer tumorigenesis, how the observed LBH downregulation in NPC tissues may relate to NPC tumorigenesis is unknown.
To address the potential functional relationship between LBH and NPC, we tested the effect of LBH overexpression on proliferation and cell cycle progression in the CNE1 NPC cell line. Furthermore, we tested the effect of small-interfering RNA (siRNA) knockdown of LBH in the normal nasopharyngeal epithelial cell line, NP69. Since EBV and TGF-β play crucial roles in NPC transformation, we also investigated the relationships among EBV infection, TGF-β, and LBH expression in normal NP69 or CNE1 NPC cells. Collectively, our findings provide further insight into the function of LBH in NPC transformation.
Results
LBH is downregulated in NPC tissue, and LBH overexpression inhibits growth of NPC xenografted tumors in mice and in vitro
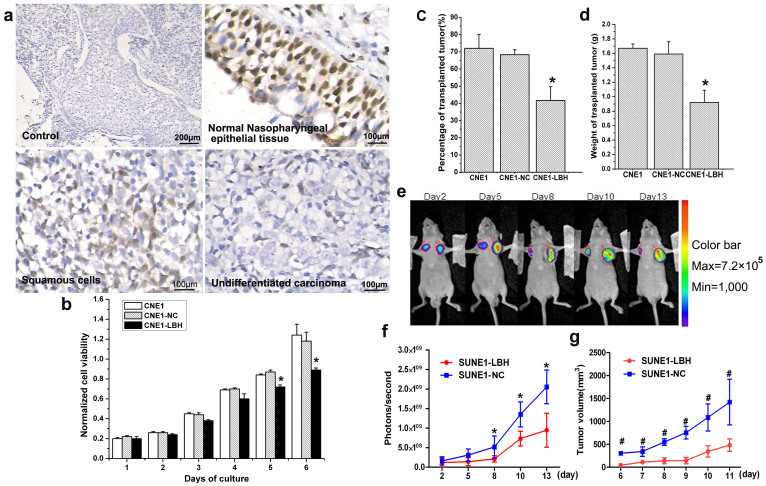
To confirm downregulation of LBH protein expression in NPC tissues, immunohistochemical (IHC) staining showed that while 100% of normal epidermal cells in nasopharyngeal tissue strongly expressed LBH, much lower LBH expression was observed in almost all NPC tissues, especially in undifferentiated nasopharyngeal carcinoma (Fig. 1a). These results indicated that LBH was significantly downregulated in NPC tissues. Determining the functional consequences of LBH expression in vitro, we found that LBH overexpression inhibited CNE1-LBH proliferation by 24% compared to the non-transfected CNE1 or control-transfected CNE1-NC cells on day 6 by MTT assay (*p<0.05, Fig. 1b). Similar results were obtained in vivo in BALB/c nude mice, as the tumorigenicity rate and average tumor weight in mice transplanted with CNE1-LBH cells were significantly lower than in mice transplanted with either of the control cells (*p<0.05, Fig. 1c,d). In vivo bioluminescent imaging also monitored tumor growth in nude mice over time. While the luciferase-containing SUNE1-LBH transplanted tumor cells (left flank) showed decreasing signal density over time, signal density gradually increased in control SUNE1-NC transplanted tumors (right flank) beginning 2 days post-injection (p.i.) (Fig. 1e). Confirming this result, the photon signals (photons/s) from SUNE1-LBH tumors decreased 53.9% from that observed in control tumors on day 13 p.i. (*p<0.05, Fig. 1f), and SUNE1-LBH tumor size also decreased (*p<0.05, Fig. 1g). The above in vitro and in vivo results suggest that re-expressing LBH in NPC cells can inhibit NPC growth.
Figure 1. LBH is expressed in NPC tissues and inhibits the proliferation and growth of xenografted NPC tumors in BALB/c nude mice.
(a) Negative control (no primary antibody) of nasopharyngeal epidermal tissue showed no positive staining in IHC. Almost all normal epidermal cells in normal nasopharyngeal epidermal tissue exhibited strong positive LBH staining. Partially cancerous cells of the squamous cell NPC tissue type exhibited moderately positive LBH staining. Almost all cancer cells of the nasopharyngeal undifferentiated NPC tissue type were negative for LBH staining. (b) MTT assay showed that LBH expression inhibited CNE1 cell proliferation (*P < 0.05 vs. CNE1 or CNE1-NC group). (c) Tumor-formation rate after CNE1, CNE1-NC, or CNE1-LBH cell (5 × 106) injection into the axilla of BALB/c nude mice was 72%, 68% and 42%, respectively (*P < 0.05 vs. CNE1 or CNE1-NC group). (d) The average weight of transplanted tumor in nude mice injected with CNE1, CNE1-NC, and CNE1-LBH cells was 1.67 g, 1.59 g, and 0.92 g, respectively. (*P < 0.05 vs. CNE1 or CNE1-NC group) (e) Representative image of the inhibition effect on SUNE1 tumor xenograft growth by LBH expression detected by bioluminescent imaging. SUNE1-NC and SUNE1-LBH cells were injected subcutaneously into the left and right armpits, respectively, in nude mice. (f) Photons/sec analysis showed that the growth of the SUNE1-LBH xenografted tumors was slower than SUNE1-NC control tumors (*P < 0.05). (g) The volume of transplanted tumors in nude mice also showed that the SUNE1-LBH grew slower than the control SUNE1-NC tumors (#P < 0.01).
We employed the standard method of constructing a recombinant expression vector to create the pEGFP-C1-LBH plasmid encoding a GFP-LBH fusion protein. Forty-eight hours after transfecting pEGFP-C1-LBH or the pEGFP-C1 control plasmid (GFP alone) into CNE1 cells, we observed by confocal microscopy that GFP-LBH fusion protein predominantly localized inside the nucleus of CNE1 cells (Fig. SI 1a), while signals from GFP alone were diffused throughout the entire cell (Fig. SI1b).
LBH inhibits cell cycle progression and expression of certain cell cycle-related genes
We confirmed LBH upregulation in CNE1-LBH cells or LBH downregulation in siRNA-treated NP69 cells by qRT-PCR. The relative mRNA level of LBH in CNE1-LBH cells was approximately 8-fold higher 24 and 48 hours after cell passage than that in control CNE1-NC cells (Fig. SI2a). Also, LBH mRNA expression in NP69 cells transfected with LBH-siRNA decreased to approximately 30% and 10% of NC-siRNA control-transfected NP69 cells 24 and 48 hours after transfection, respectively (Fig. SI2b).
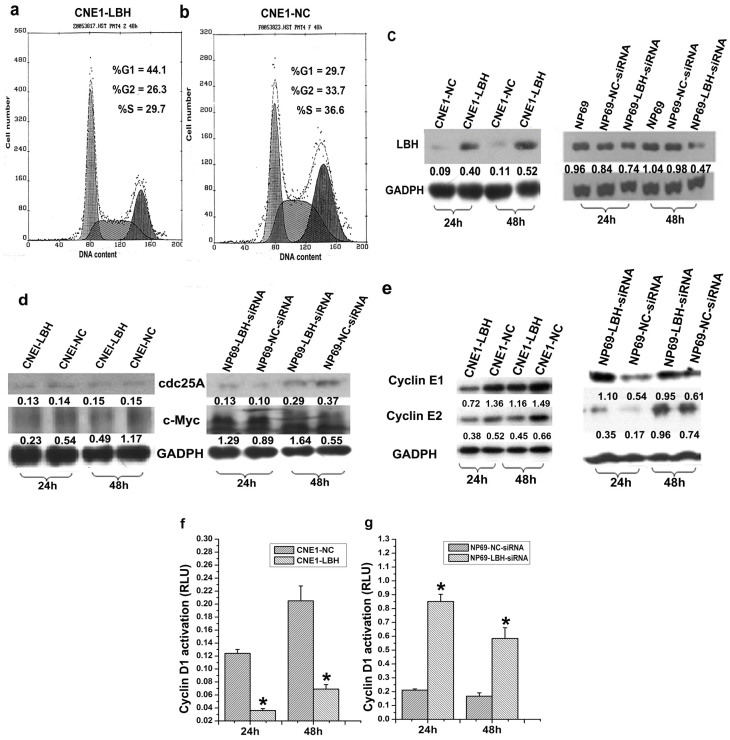
Determining the effect of LBH on cell cycle progression by flow cytometry, CNE1-LBH cells had a lower ratio S-phase cells but a higher ratio of G1-phase cells than CNE1-NC control cells (S: 29.7% vs. 36.6%; G1: 44.1% vs. 29.7%, respectively) (Fig. 2a,b), correlating with significantly increased LBH protein expression 24 and 48 h after cell passage (Fig. 2c). CNE1-LBH cells exhibited lower protein expression of the cell cycle-related proteins c-Myc as well as Cyclin E1 and E2, but not cdc25A, than control cells (Fig. 2d,e). Accordingly, expression of c-Myc as well Cyclin E1 and E2, but not cdc25, increased after siRNA-mediated LBH knockdown in NP69 cells (Fig. 2d,e), correlating with a ~50% decrease in LBH protein expression 48 h after LBH-siRNA transfection (Fig. 2c). Similar results were obtained for Cyclin D1 expression (*p<0.05, Fig. 2f,g). These results suggest that LBH functions to inhibit cell cycle progression in NPC cells.
Figure 2. Effect of LBH on cell cycle progression and expression of cell cycle-related genes.
Cell cycle assay by flow cytometry showed that (a) CNE1-LBH cells, but not (b) control CNE1-NC cells transfected with pRC/CMV2, were arrested at the G1/S transition. (c) Western blot showed significant increase in LBH protein expression in CNE1-LBH after subculturing. In NP69 cells transfected with LBH-siRNA (NP69-LBH-siRNA), LBH protein expression was approximately 50% of that in NP69 cells and NP69 cells transfected with NC-siRNA (NP69-NC- siRNA) 48 h after transfection. (d, e) c-Myc and Cyclin E1/E2 expression were downregulated after LBH was re-expressed in CNE1 cells. c-Myc and Cyclin E1/E2 expression were both increased in NP69 cells after LBH knockdown. In contrast, cdc-25A levels remained the same regardless of LBH expression. (f) Cyclin D1 transcriptional activity decreased in CNE1-LBH cells (*p < 0.05 vs. CNE1-NC group). (g) Cyclin D1 transcriptional activity increased after LBH knockdown in NP69 cells (*p < 0.05 vs. NP69-NC-siRNA group).
LBH inhibits the transcriptional activity of NF-κB
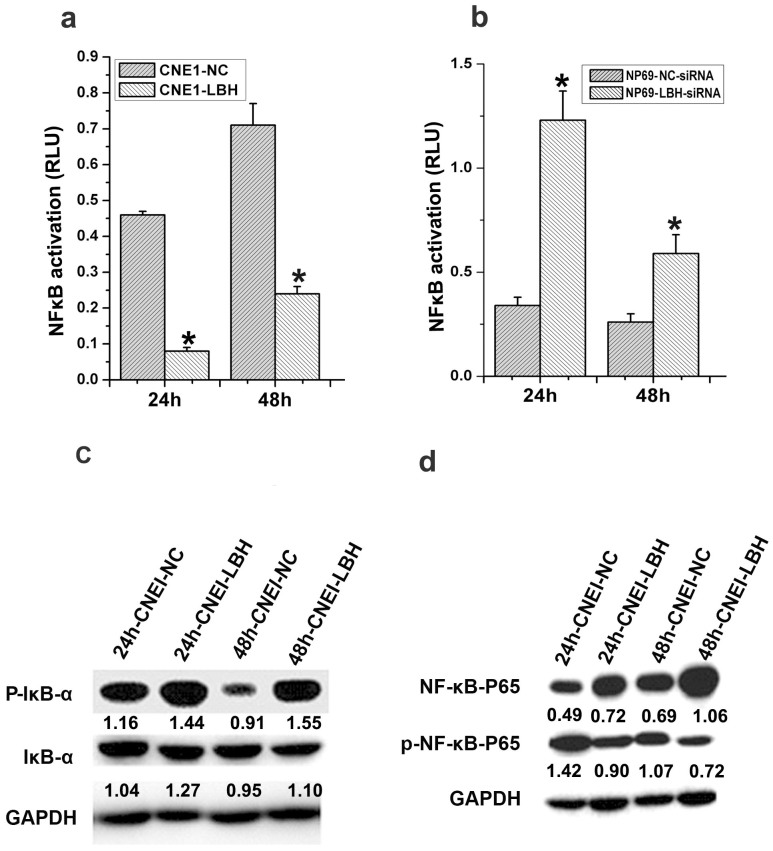
We next evaluated the effect of LBH on the cell cycle regulators NF-κB and Cyclin D1. The NF-κB relative luciferase unit (RLU) in NF-κB-Luc–transfected CNE1-LBH cells significantly decreased compared to CNE1-NC cells at 24 and 48 h post-transfection (*p<0.05, Fig. 3a). Consistent with this result, the NF-κB RLU in LBH-siRNA–treated NP69 cells significantly increased compared to control cells 24 and 48 h after knockdown (*p<0.05, Fig. 3b). Additionally, LBH expression increased phosphorylation of the NF-κB inhibitor IκBα and decreased phosphorylation of the NF-κB P65 subunit (Fig. 3c,d). Together, these results suggest that LBH expression can inhibit NF-κB and Cyclin D1 activation and further implicate LBH in regulating the G1/S transition.
Figure 3. LBH inhibits NF-κB transcriptional activity.
(a) NF-κB-Luc reporter plasmid was co-transfected with the pRL-SV40 plasmid into CNE1-LBH and CNE1-NC control cells. The relative luciferase unit (RLU) at indicated time points post-transfection showed that the NF-κB transcriptional activity decreased in CNE1-LBH cells (*P < 0.05 vs. CNE1-NC group). (b) NF-κB transcriptional activity significantly increased after LBH knockdown in NP69 cells (*P < 0.05 vs. NP69-NC-siRNA group). (c) IκB phosphorylation was increased in CNE1-LBH compared to CNE1-NC, and (d) P65 phosphorylation was correspondingly downregulated.
EBV infection downregulates LBH expression in nasopharyngeal epithelial cells
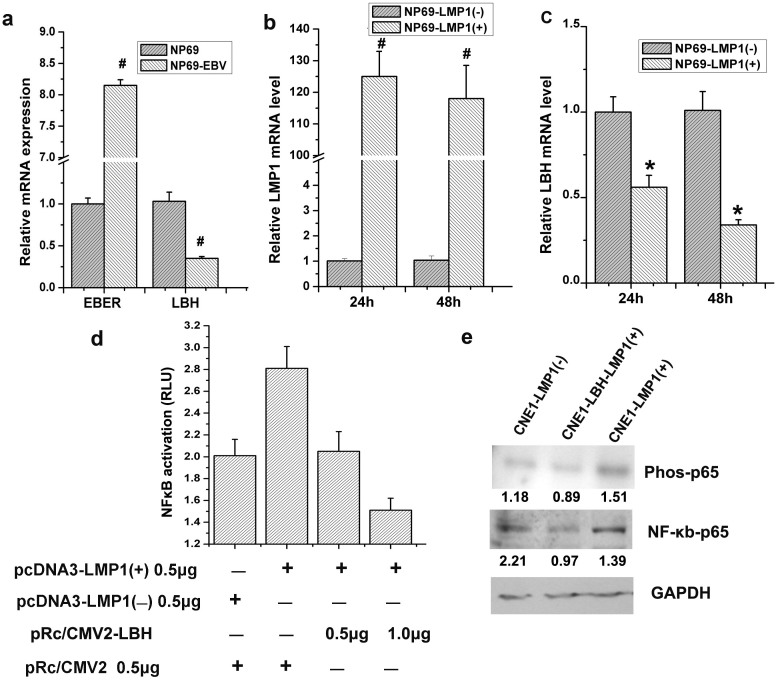
Testing whether EBV infection regulates LBH expression, EBV-infected normal nasopharyngeal NP69 cells had 8-fold higher EBER mRNA than non-infected control cells, confirming EBV infection, but had a > 60% decrease in LBH mRNA (#p<0.01, Fig. 4a). Specifically testing whether EBV-encoded LMP1 regulated LBH, pCDNA3-LMP1 transfection remarkably inhibited LBH mRNA >60% over control-plasmid transfection 48 h post-transfection (*p<0.05, #p<0.01, Fig. 4b,c). A dual-reporter system showed that while NF-κB transcriptional activity increased by 40% in LMP1-overexpressed CNE1 cells, simultaneous LBH overexpression significantly inhibited this LMP1-induced NF-κB (Fig. 4d), likely by decreasing P65 phosphorylation (Fig. 4e). Thus, EBV-encoded LMP1 can downregulate LBH expression in NPC.
Figure 4. EBV infection induces downregulation of LBH in NP69 cells, and LMP1 plays a fundamental role.
(a) EBER mRNA levels in NP69 cells increased approximately 8-fold after EBV infection, and LBH expression correspondingly decreased; LBH mRNA levels were approximately 1/3 of that in NP69 cells (#P < 0.01 vs. NP69 group). (b) LMP1 mRNA levels significantly increased after pcDNA3-LMP1 was transfected into NP69 cells. (#P < 0.01 vs. NP69-LMP1(-) group). (c) LBH mRNA levels decreased approximately 66% 48 h after LMP1 expression (*P < 0.05 vs. NP69-LMP1(-) group). (d) LBH inhibited NF-κB transcriptional activity induced by LMP1 in CNE1 cells. (e) LBH inhibited the LMP1-mediated phosphorylation NF-κB-P65 in CNE1 cells.
TGF-β1 inhibits the growth of NP69 cells through regulating LBH expression
As a gene regulated by NF-κB, C-Myc was selected for gene expression detection with qRT-PCR. After treatment of TGF-β1, the level of C-Myc mRNA and protein decreased significantly (Fig. SI3).
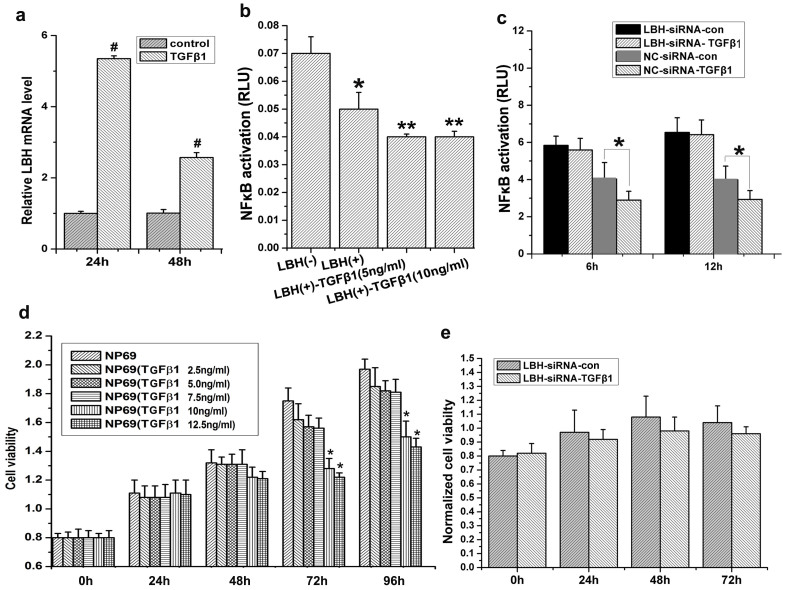
Testing the relationship between normal TGF-β1–mediated suppression of cell growth and LBH, TGF-β1 treatment significantly increased LBH mRNA levels (>4-fold at 24 h; >2-fold at 48 h) in cultured NP69 cells (#p<0.01, Fig. 5a). Although TGF-β1 could inhibit NF-κB transcriptional activity by itself or in synergy with LBH overexpression (*P<0.05, LBH(+) group vs. LBH(−) group; **P<0.05, LBH(+)-TGFβ1 group vs. LBH(+) group, Fig. 5b), TGF-β1 could not significantly downregulate NF-κB transcriptional activity in LBH-siRNA–treated NP69 cells (*p<0.05, Fig. 5c), suggesting that TGF-β1 inhibition of NF-κB was mediated through LBH. Finally, we analyzed the effect of TGF-β1 on NP69 proliferation in the presence or absence of LBH expression. TGF-β1 lost the ability to inhibit NP69 proliferation after LBH knockdown (*p<0.05, Fig. 5d,e), indicating that normal LBH expression is an important factor in TGF-β1–mediated suppression of NP69 cell growth.
Figure 5. TGF-β1 inhibits NF-κB transcriptional activity by upregulating LBH expression in NP69 cells.
(a) qRT-PCR analysis showed that TGF-β1 induced LBH mRNA expression in NP69 cells (#P < 0.01 vs. control group). (b) A dual-reporter assay showed that LBH expression or TGF-β1 treatment downregulated NF-κB transcriptional activity in NP69 cells and that LBH expression and TGF-β1 demonstrated some synergistic effects on inhibiting NF-κB transcriptional activity (*P < 0.05, LBH(+) group vs. LBH(−) group; **P < 0.05, LBH(+)-TGFβ1 group vs. LBH(+) group). (c) NF-κB transcriptional activity was inhibited in NP69 cells after TGF-β1 treatment, but this inhibition effect was no longer significant after LBH knockdown (*P < 0.05, NC-siRNA-TGF-β1 vs. NC- siRNA-con group). (d) TGF-β1 at the 10 ng/mL and 12.5 ng/mL doses inhibited NP69 cell proliferation (*P < 0.05 vs. NP69 group). (e) TGF-β1 had no effect on NP69 cell proliferation after LBH knockdown.
LBH expression restores the ability of TGF-β1 to inhibit NPC cell growth
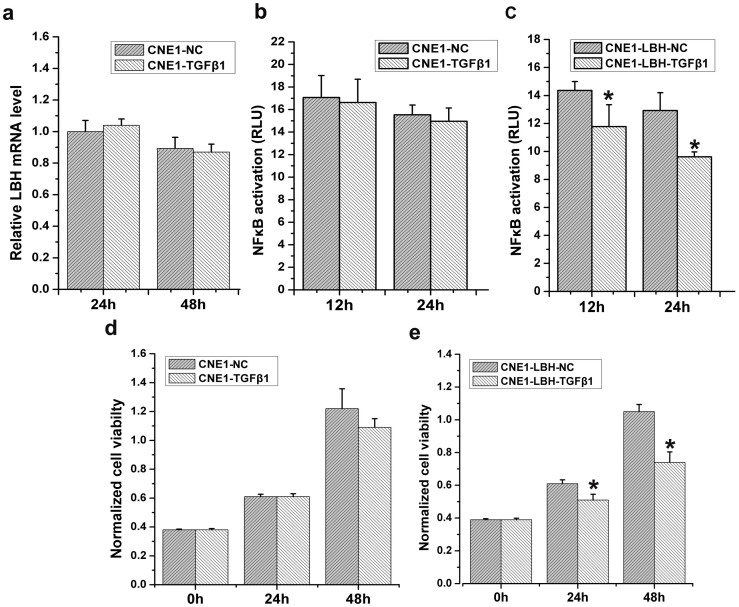
In contrast to NP69 cells, TGF-β1 had no significant effect on LBH mRNA and NF-κB transcriptional activity in CNE1 NPC cells (Fig. 6a,b). After upregulating LBH expression, however, TGF-β1 remarkably regained the ability to significantly inhibit NF-κB transcriptional activity at both 24 and 48 h after TGF-β1 treatment (*p<0.05, Fig. 6c). Accordingly, similar results were observed with regard to the effect of TGF-β1 on CNE1 proliferation (*p<0.05, Fig. 6d,e). These results suggest not only that resistance of CNE1 cells to TGF-β1 correlates with LBH downregulation and dysfunction but also that TGF-β1 regains the ability to inhibit NPC cell growth upon restoring LBH expression.
Figure 6. Re-induction of LBH in CNE1 cells restores TGF-β1 inhibition of cell growth.
(a) TGF-β1 had no effect on LBH expression in CNE1 cells. (b) TGF-β1 had no effect on NF-κB transcriptional activity in CNE1 cells. (c) TGF-β1 significantly inhibited NF-κB transcriptional activity after re-induction of LBH in the CNE1-LBH-TGF-β1 group (*P < 0.05 vs. CNE1-LBH-NC group). (d) TGF-β1 had no effect on CNE1 cell proliferation. (e) TGF-β1 inhibited CNE1 cell proliferation by re-induction of LBH in the CNE1-LBH-TGF-β1 group (*P < 0.05 vs. CNE1-LBH-NC group).
LBH expression and its association with clinicopathological factors in NPC
As shown in Table 1, LBH immunoexpression was successfully assessed in all 40 cases of NPC, consisting of one keratinizing squamous cell carcinomas, 17 nonkeratinizing differentiated carcinomas, and 22 nonkeratinizing undifferentiated carcinomas. There were 30 men and 10 women with a mean age of 50.3 years (range, 22–84). Three cases were classified as stage I, 4 as stage II, 17 as stage III, and 16 as stage IV. LBH low-expression was present in 26 (65%) of the cases and was significantly associated with NPC cases featuring pT status 3, 4 (p = 0.0096), N status 2, 3 (p = 0.0313), and AJCC stage 3, 4 (p = 0.0261). However, we found no association between LBH expression and other clinicopathological factors.
Table 1. Associations between LBH expression and other important clinicopathologic variables.
| LBH expression | ||||
|---|---|---|---|---|
| Parameters | Category | Low | High | P value |
| Gender | Male | 20 | 10 | 0.7019 |
| Female | 6 | 4 | ||
| Age (years) | <60 | 18 | 14 | 0.7491 |
| ≥60 | 5 | 3 | ||
| Primary tumor (T) | T1–T2 | 4 | 7 | 0.0096 a |
| T3–T4 | 23 | 6 | ||
| Nodal status (N) | N0–N1 | 6 | 11 | 0.0313 a |
| N2–N3 | 16 | 7 | ||
| Stage | I–II | 2 | 5 | 0.0261 a |
| III–IV | 24 | 9 | ||
| Histological grade | Keratinizing | 4 | 5 | 0.4577 |
| Nonkeratinizing | 8 | 7 | ||
| Undifferentiated | 11 | 5 | ||
aStatistically significant.
Prognostic significance of LBH expression in NPCs
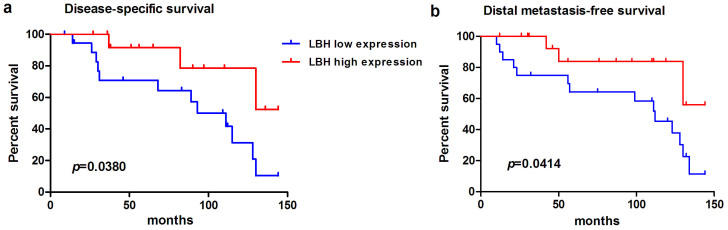
LBH low expression correlated with a more aggressive clinical course and a significantly shorter DSS in patients with NPC (p = 0.0380, Fig. 7a). In addition, LBH low expression was strongly predictive of distal metastasis (p = 0.0414 for DMeFS) (Fig. 7b).
Figure 7. By using log-rank test, the LBH overexpression is univariately predictive of inferior disease-specific survival (a) and distant metastasis-free survival (b).
Discussion
Although malignant transformation normally involves oncogenes and loss of TSG function, very few of these genes are associated with NPC2,4. We previously identified that the LBH gene was downregulated in NPC tissues. In the current study, we showed that LBH protein levels were significantly downregulated in NPC biopsies compared to normal nasopharyngeal epithelial cells; moreover, almost no LBH was detected in undifferentiated NPC tissues, suggesting that LBH might inhibit normal cell transformation into NPC. We further confirmed in vivo and in vitro that LBH re-expression in both CNE1 and SUNE1 NPC cell lines significantly inhibited proliferation, tumorigenesis, and tumor growth.
Cell cycle analysis revealed that LBH expression induced arrest at the G1/S transition. The core cell-cycle regulation mechanism relies on the temporal activation of cyclin-dependent kinase (CDK) promotion of cell cycle progression22. Cyclin D1, Rb phosphorylation, and the Cyclin E subtypes (E1 and E2) are involved in the G1/S phase progression. Cyclin D1 overexpression induces cell growth dysregulation and tumorigenesis23,24. Cyclin E1 and E2 are contributing factors in G1-phase arrest during terminal differentiation, and their overexpression correlates with tumorigenesis of several cancers25,26; while Cyclin E1 is expressed during hyperplasia of most normal cells and some tumor cells, Cyclin E2 is highly expressed in tumor cells, but not non-transformed cells27. Here, LBH expression in NPC cells decreased transcriptional activation of the Cyclin D1 promoter and downregulated Cyclin E1 and E2 expression. Supported by results from the LBH knockdown in NP69 cells, our data collectively suggest that downregulating LBH promotes Cyclin D1, E1, and E2 expression in nasopharyngeal epithelial cells, which leads to excessive Rb phosphorylation, finally resulting in release from cell cycle arrest at the G1 phase. The precise molecular mechanism downstream of how LBH regulates CDKs to lead to cell cycle arrest, however, needs further investigation.
Since Cyclin D1 is crucial for progression through G1 and NF-κB is a direct transcriptional activator of Cyclin D1, NF-κB-mediated Cyclin D1 activation is necessary to induce progression from G1 into S phase28. Thus, cell cycle arrest observed in CNE1-LBH cells was likely due to decreased Cyclin D1 transcriptional activity downstream of downregulated NF-κB transcriptional activity; moreover, it implicated LBH as an important NF-κB regulator. Consistent with NF-κB negative regulation of cell differentiation29, transplanted CNE1-LBH tumors were more differentiated than control CNE1 tumors30. Further studies are underway to better understand the relationship between LBH and CNE1-NC cell differentiation.
LBH likely plays a functional role beyond development in adult tissues, as some diseases are associated with gain-of-function mutations in LBH, although its precise role is unclear. Our data now reveal that LBH limits cell growth and proliferation in adult tissues by inhibiting cell cycle regulators at the G1/S transition. Interestingly, LBH is already linked to aggressive basal-subtype breast cancers; unlike in NPC, however, LBH is overexpressed. Despite this seeming contradiction, LBH regulation of cell cycle may also contribute to breast carcinogenesis. Reiger et al. suggested that LBH suppressed mammary cell differentiation, allowing for Wnt-induced carcinogenesis in these relatively undifferentiated cells, although this remains to be formally tested21. Also in this previous study, Wnt signaling directly regulates LBH. Interestingly, since EBV-positive NPC tissues have elevated Wnt signaling31, our finding that LBH is downregulated in EBV-positive NPC tissues may contradict the finding that Wnt regulates LBH, although it remains possible that LBH may be differentially regulated in different cells. Further study into upstream LBH mediators may help to resolve these seemingly opposing results. Based on our study, it would be interesting to determine whether the recently described LBH-deficient mice, which only show a delay in postnatal mammary gland development, are prone to developing tumors.
EBV infection and NPC pathogenesis are closely related, and EBV-encoded LMP1 functions like an oncogene32 by activating several signaling pathways to induce carcinogenesis, including NF-κB, cell cycle, proliferation, and apoptosis33,34. Our data now reveal LBH involvement in this mechanism downstream of LMP1 and strongly suggest that ubiquitous EBV infection in NPC explains their low LBH expression. To our knowledge, our study is the first to link LBH to LMP1-mediated activation of the NF-κB pathway, an essential developmental and oncogenic signaling network.
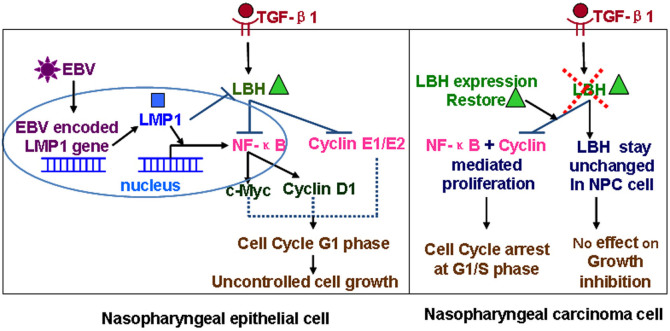
TGF-β inhibits the growth of most epithelial cells, and developing resistance to this inhibition is regarded as an important step toward malignant transformation35,36. EBV-encoded LMP1 somewhat enables CNE1 NPC cells to acquire this resistance, and NPC-derived LMP1 can induce complete resistance in CNE1 cells12,37. Here, we identified LBH as new target in the TGF-β1 signaling pathway. After confirming that TGF-β1 had no effect on NPC cell proliferation or cell cycle progression, we showed that re-expression of LBH in CNE1 cells led to synergistic inhibition by TGF-β1 and LBH on NF-κB transcriptional activity, proliferation, and cell cycle progression. These results suggest the intriguing possibility that nasopharyngeal epithelia may initiate malignant transformation upon losing the protection afforded by TGF-β1–mediated inhibition of cell growth through a mechanism involving EBV-mediated LBH downregulation. Overall (Fig. 8), this study not only helps to further understand normal LBH function in nasopharyngeal cells and the mechanism underlying how it may be regulated upon malignant transformation but also may provide a novel strategy for treating NPC by activating TGF-β1–mediated protection through restoring LBH function.
Figure 8. The mechanism of LBH in the pathogenesis of nasopharyngeal carcinoma.
Methods
Patients and tumor specimens
Available paraffin-embedded tissue blocks were retrieved from 40 consecutive NPC patients who underwent biopsies between January 2004 and December 2010. All these patients were free of distant metastasis at initial diagnosis. The histological subtypes were reappraised by two pathologists, according to the current WHO classification. Tumor staging was reevaluated with the 7th American Joint Committee on Cancer (AJCC) system. All of the individuals participating in this project gave informed consent, and the study was approved by the Sun Yat-sen University Cancer Center Medical Ethics Board.
Treatment and follow-up
All 40 cases accepted a complete course of radiotherapy (RT), total dose ≧7,000 cGy, and cisplatin-based chemotherapy was performed in those with stage II–IV disease. The method of RT was generally uniform over this period. After RT, all 40 patients were regularly monitored until death or their last appointment, with the mean follow-up duration being 67.9 months (range 5 to 121).
Cell lines and plasmids
NP69, an immortalized human NPC cell line (ATCC), was cultured in keratinocyte-SFM medium (Invitrogen). Both the CNE1 (previously established at the Chinese University of Hong Kong) and SUNE1 (a poorly differentiated nasopharyngeal squamous carcinoma established at Sun Yat-sen University Cancer Center) cell lines were cultured in RPMI-1640 containing 10% fetal bovine serum (FBS).
The human Cyclin D1 promoter plasmid (-1745-CD1-Luc), provided by RG Pestell (Albert Einstein College of Medicine, USA), contained a luciferase reporter under control of the human Cyclin D1 promoter. The NF-κB-Luc plasmid contained three tandem NF-κB binding sites. Control pRL-sv40 plasmid was purchased from Promega.
Immunohistochemistry
NPC tissue from patients at the Sun Yat-sen University Cancer Center were biopsied under fiberoptic nasopharyngoscopy, snap-frozen in LN2, and embedded in paraffin. Normal nasopharyngeal tissues from 20 control subjects and tumors from 40 NPC patients (diagnosed by pathology) were used. Tissue sections were incubated overnight at 4°C with an anti-LBH mAb (Sigma). Nonspecifically stained control sections were incubated in Tris-buffered saline (TBS) without primary antibody. Biotinylated anti-rabbit secondary antibody was added (40 min, room temperature), followed by the avidin-biotinylated peroxidase complex (40 min). Sections were washed with distilled water (10 min), treated with DAB to visualize positive staining, counterstained with Mayer's hemalum, and mounted.
Construction of recombinant plasmids and establishment of stable cell lines
Total RNA was reverse-transcribed with superscript III RT (Invitrogen). The LBH CDS PCR product (forward: 5′-CCCGTGTCATCCTCACTCG-3′; reverse: 5′-CAGATGCTGGCTGGTATGACC-3′) was ligated separately into the pEGFP-C1 and pRC/CMV2 vectors. Recombinant pRC/CMV2-LBH plasmid was transfected into CNE1 cells by Lipofectamine2000 (Invitrogen). Transfected CNE1 cells were screened with G418 (400 μg/mL) (Invitrogen) for ~2 weeks to obtain a single-cell clone. RT-PCR identified stable CNE1-LBH cell lines. Control CNE1-NC cells were transfected with a pRC/CMV2 vector, screened with G418, and identified by PCR (forward: 5′-TGACGCAAATGGGCGGTAG-3′; reverse: 5′-GCACCTTCCAGGGTCAAG-3′).
MTT assay
CNE1-LBH and CNE1-NC (5 × 103 cells/well) were cultured in 100 μL RPMI-1640 containing 10% FBS in 96-well plates. After 24 h, 10 μL/well of MTT (5 mg/mL, Sigma) was added (37°C, 4 h). Supernatant was removed, and formazan was dissolved in DMSO (100 μL/well, 37°C, 10 min). Absorbance (570 nm) was read by a microplate reader (Thermo Scientific) and monitored every other day for 6 days.
Tumorigenicity in BALB/c nude mice
CNE1-LBH, CNE1-NC, and CNE1 cells (5 × 106) in 100 μL of D-Hank's buffer were injected into the bilateral axilla of 4-week-old BALB/c nude mice. After tumors emerged, tumor size was measured every 6 days. Thirty days after inoculation, mice were sacrificed, tumors were removed and weighed, and tumorigenicity rates were calculated.
In vivo imaging system
Equal numbers of male and female 4-week-old BALB/c nu/nu mice (weight, 16 ± 2 g) were used. SUNE1-LBH-Luc NPC cells (constructed by stable integration of LBH and luciferase in a lentiviral system) in logarithmic phase and control SUNE1-NC-Luc cells were injected subcutaneously (100 μL; 2 × 107 cells/mL) into the forelimb of mice near the armpit (n = 6). Live image detection was performed on days 2, 5, 8, and 13. After anesthetizing mice, 150 mg/mL Luciferin per 150 mg/kg weight was intraperitoneally injected. Signal intensity was recorded 10 min later (In Vivo Imaging system, Berthold Corp.). After tumors emerged, tumor volume was measured every day using a vernier caliper. Mice were sacrificed 2 weeks later, and tumors were analyzed for pathology.
Cell cycle analysis
CNE1-LBH and CNE1-NC cells (5 × 105) were seeded into a 25 cm2 flasks, harvested by trypsinization 48 h after propagation, washed twice with ice-cold PBS, fixed with cold 70% ethanol, and stained with 50 μg/mL propidium iodide (Sigma) containing 5 mg/mL RNaseA. DNA synthesis and cell cycle status were analyzed (Coulter Elite flow cytometer, Beckman-Coulter).
siRNA transfection and qRT-PCR analysis
LBH-siRNA and negative control siRNA duplexes were designed (http://www.invitrogen.com/rnai) and synthesized (Shanghai GenePharma). NP69 cells (5 × 105) grown in triplicate 10 cm2 dishes were transfected with siRNA (30 nM) the following day with Lipofectamine RNAiMAX (Invitrogen). Total RNA was isolated 1–2 days later with TRIzol (Invitrogen). Reverse transcription was performed using PrimeScript RT (Takara). PCR primers were designed by DNA Club software (Table S1). mRNA levels were analyzed by the 2−ΔΔCt method, normalized by GAPDH.
Western blot
Cells (1 × 106) were seeded in 25 cm2 dishes in complete medium for 24–48 h. Lysates were centrifuged (10,000 g, 10 min, 4°C), run on SDS-PAGE, transferred onto nitrocellulose, blocked with 5% blotting-grade NFDM (60 min), and stained with the following antibodies: rabbit anti-Cyclin D1 and E2 mAbs (1:500; Cell Signaling Technology), rabbit anti-GAPDH mAb (1:1000; Cell Signaling Technology), anti-rabbit IgG HRP-conjugated secondary antibodies (1:2000). All the gels have been run under the same experimental conditions.
Statistical analysis
Data are shown as the mean ± SEM. Significance (P < 0.05) was determined by Student's t-test or analysis of variance (ANOVA) followed by assessment of differences (SPSS 12.0). Survival analysis was performed using the Kaplan-Meier method. Univariate survival analyses were performed using KaplanMeier plots and compared with the log-rank test.
Author Contributions
L.Q., G.X., L.J., L.X. and W.Y. performed the experiments in vitro and in vivo, L.Q. and G.X. wrote the manuscript, L.L. supervised the project.
Supplementary Material
Supplemental Material
Acknowledgments
We thank Professor Musheng Zeng for assistance with Epstein–Barr virus infection of nasopharyngeal epithelial cells. This work was funded by the National Nature Science Foundation of China (No.81271622; 81471711), the Guangdong Province of Higher School “Thousand Hundred Ten Talents Project” (No.84000-52010004) and the Science and Technology Planning Project of Guangdong Province, China (No.2013B021800161).
References
- Hildesheim A. & Levine P. H. Etiology of nasopharyngeal carcinoma: a review. Epidemiol Rev 15, 466–485 (1993). [DOI] [PubMed] [Google Scholar]
- Yung W. C., Ng M. H., Sham J. S. & Choy D. T. p53 codon 72 polymorphism in nasopharyngeal carcinoma. Cancer Genet Cytogenet 93, 181–182 (1997). [DOI] [PubMed] [Google Scholar]
- Sun Y. et al. The von Hippel-Lindau (VHL) disease tumor-suppressor gene is not mutated in nasopharyngeal carcinomas. Int J Cancer 61, 437–438 (1995). [DOI] [PubMed] [Google Scholar]
- Sun Y. et al. No point mutation but a codon 31ser-->arg polymorphism of the WAF-1/CIP-1/p21 tumor suppressor gene in nasopharyngeal carcinoma (NPC): the polymorphism distinguishes Caucasians from Chinese. Cancer Epidemiol Biomarkers Prev 4, 261–267 (1995). [PubMed] [Google Scholar]
- Lo K. W., Huang D. P. & Lau K. M. p16 gene alterations in nasopharyngeal carcinoma. Cancer Res 55, 2039–2043 (1995). [PubMed] [Google Scholar]
- Dai Q., Li X. P., Chai L., Long H. A. & Yang Z. H. Polymorphisms of Toll-like receptor 9 are associated with nasopharyngeal carcinoma susceptibility. Tumour Biol 35, 3247–3253 (2014). [DOI] [PubMed] [Google Scholar]
- Wang Z. et al. The polymorphism interleukin-8 -251A/T is associated with a significantly increased risk of cancers from a meta-analysis. Tumour Biol 35, 7115–7123 (2014). [DOI] [PubMed] [Google Scholar]
- Li M., Dai W. & Zhou H. Cyclin D1 G870A polymorphism and risk of nasopharyngeal carcinoma: a meta-analysis. ScientificWorldJournal 2013, 689048 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., Li L. L., Hu D. S., Deng X. Y. & Cao Y. Role of Epstein-Barr virus encoded latent membrane protein 1 in the carcinogenesis of nasopharyngeal carcinoma. Cell Mol Immunol 4, 185–196 (2007). [PubMed] [Google Scholar]
- Xiao J. et al. The effect of transforming growth factor-beta1 on nasopharyngeal carcinoma cells: insensitive to cell growth but functional to TGF-beta/Smad pathway. J Exp Clin Cancer Res 29, 35 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Menezes J., Prasad U. & Ahmad A. Elevated serum levels of transforming growth factor beta1 in Epstein-Barr virus-associated nasopharyngeal carcinoma patients. Int J Cancer 84, 396–399 (1999). [DOI] [PubMed] [Google Scholar]
- Huang H. & Huang P. C. [Effects of two LMP1 variants on resistance of CNE1 cell strain to TGF-beta1]. Ai Zheng 22, 1254–1259 (2003). [PubMed] [Google Scholar]
- Teicher S., Crawford K., Williams B., Nelson B. & Andrews C. Emerging role of the pediatric nurse practitioner in acute care. Pediatr Nurs 27, 387–390 (2001). [PubMed] [Google Scholar]
- Ma X., Ling K. W. & Dzierzak E. Cloning of the Ly-6A (Sca-1) gene locus and identification of a 3' distal fragment responsible for high-level gamma-interferon-induced expression in vitro. Br J Haematol 114, 724–730 (2001). [DOI] [PubMed] [Google Scholar]
- Lindley L. E. & Briegel K. J. Generation of mice with a conditional Lbh null allele. Genesis 51, 491–497 (2013). [DOI] [PubMed] [Google Scholar]
- Liu Q. C., Fang Y., Li X. Y., Liang W. J. & Zeng Y. X. [Cloning of NP9 gene and its influence on cyclin D1 transcription activity]. Ai Zheng 22, 725–728 (2003). [PubMed] [Google Scholar]
- Briegel K. J. & Joyner A. L. Identification and characterization of Lbh, a novel conserved nuclear protein expressed during early limb and heart development. Dev Biol 233, 291–304 (2001). [DOI] [PubMed] [Google Scholar]
- Conen K. L., Nishimori S., Provot S. & Kronenberg H. M. The transcriptional cofactor Lbh regulates angiogenesis and endochondral bone formation during fetal bone development. Dev Biol 333, 348–358 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briegel K. J., Baldwin H. S., Epstein J. A. & Joyner A. L. Congenital heart disease reminiscent of partial trisomy 2p syndrome in mice transgenic for the transcription factor Lbh. Development 132, 3305–3316 (2005). [DOI] [PubMed] [Google Scholar]
- Al-Ali H. et al. Biophysical characterization reveals structural disorder in the developmental transcriptional regulator LBH. Biochem Biophys Res Commun 391, 1104–1109 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieger M. E., Sims A. H., Coats E. R., Clarke R. B. & Briegel K. J. The embryonic transcription cofactor LBH is a direct target of the Wnt signaling pathway in epithelial development and in aggressive basal subtype breast cancers. Mol Cell Biol 30, 4267–4279 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavletich N. P. Mechanisms of cyclin-dependent kinase regulation: structures of Cdks, their cyclin activators, and Cip and INK4 inhibitors. J Mol Biol 287, 821–828 (1999). [DOI] [PubMed] [Google Scholar]
- Velasco-Velazquez M. A. et al. Examining the role of cyclin D1 in breast cancer. Future Oncol 7, 753–765 (2011). [DOI] [PubMed] [Google Scholar]
- Ortega S., Malumbres M. & Barbacid M. Cyclin D-dependent kinases, INK4 inhibitors and cancer. Biochim Biophys Acta 1602, 73–87 (2002). [DOI] [PubMed] [Google Scholar]
- Moroy T. & Geisen C. Cyclin E. Int J Biochem Cell Biol 36, 1424–1439 (2004). [DOI] [PubMed] [Google Scholar]
- Payton M., Scully S., Chung G. & Coats S. Deregulation of cyclin E2 expression and associated kinase activity in primary breast tumors. Oncogene 21, 8529–8534 (2002). [DOI] [PubMed] [Google Scholar]
- Muller-Tidow C. et al. Cyclin E is the only cyclin-dependent kinase 2-associated cyclin that predicts metastasis and survival in early stage non-small cell lung cancer. Cancer Res 61, 647–653 (2001). [PubMed] [Google Scholar]
- Torricelli C. et al. Rottlerin inhibits the nuclear factor kappaB/cyclin-D1 cascade in MCF-7 breast cancer cells. Life Sci 82, 638–643 (2008). [DOI] [PubMed] [Google Scholar]
- Hinz M. et al. NF-kappaB function in growth control: regulation of cyclin D1 expression and G0/G1-to-S-phase transition. Mol Cell Biol 19, 2690–2698 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q. C., Li X. Y., Wei B. X. & Zeng Y. X. [NP9 gene inhibits tumorigenicity of nasopharyngeal carcinoma]. Zhonghua Zhong Liu Za Zhi 27, 641–644 (2005). [PubMed] [Google Scholar]
- Zeng Z. Y. et al. Gene expression profiling of nasopharyngeal carcinoma reveals the abnormally regulated Wnt signaling pathway. Hum Pathol 38, 120–133 (2007). [DOI] [PubMed] [Google Scholar]
- Wang D., Liebowitz D. & Kieff E. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell 43, 831–840 (1985). [DOI] [PubMed] [Google Scholar]
- Ardila-Osorio H. et al. TRAF interactions with raft-like buoyant complexes, better than TRAF rates of degradation, differentiate signaling by CD40 and EBV latent membrane protein 1. Int J Cancer 113, 267–275 (2005). [DOI] [PubMed] [Google Scholar]
- Song X. et al. Epstein-Barr virus-encoded latent membrane protein 1 modulates cyclin D1 by c-Jun/Jun B heterodimers. Sci China C Life Sci 48, 385–393 (2005). [DOI] [PubMed] [Google Scholar]
- Kretschmer A. et al. Differential regulation of TGF-beta signaling through Smad2, Smad3 and Smad4. Oncogene 22, 6748–6763 (2003). [DOI] [PubMed] [Google Scholar]
- Mourskaia A. A. et al. Transforming growth factor-beta1 is the predominant isoform required for breast cancer cell outgrowth in bone. Oncogene 28, 1005–1015 (2009). [DOI] [PubMed] [Google Scholar]
- Lo A. K., Dawson C. W., Lo K. W., Yu Y. & Young L. S. Upregulation of Id1 by Epstein-Barr virus-encoded LMP1 confers resistance to TGFbeta-mediated growth inhibition. Mol Cancer 9, 155 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material