The spindle checkpoint ensures proper chromosome segregation during cell division. Unraveling checkpoint signaling has been a longstanding challenge due to the complexity of the structures and forces that regulate chromosome segregation. New reports have now made significant inroads to understanding the checkpoint signaling mechanisms at the kinetochore, the nexus point for microtubules and chromatin. In contrast to the traditional view of a switch-like checkpoint response, new findings indicate it is graded. This revised perspective aids in understanding how failures in the checkpoint can lead to aneuploidy and informs strategies to exploit these errors for cancer treatments.
Accurate chromosome segregation is essential for genome inheritance and cellular fitness. Chromosome missegregation results in lethality or aneuploidy, the state where cells have an aberrant number of chromosomes. Aneuploidy leads to abnormal gene dosage and exposes detrimental recessive mutations, potentially causing birth defects and promoting cancer cell proliferation (for reviews, see1,2). Accurate segregation is achieved by linking sister chromatids following replication and then segregating them to opposite spindle poles prior to cytokinesis. Segregation is mediated by spindle microtubules that attach to chromosomes through kinetochores, large protein complexes that assemble on centromeric DNA. Microtubule disassembly provides the force to segregate chromosomes at anaphase3.
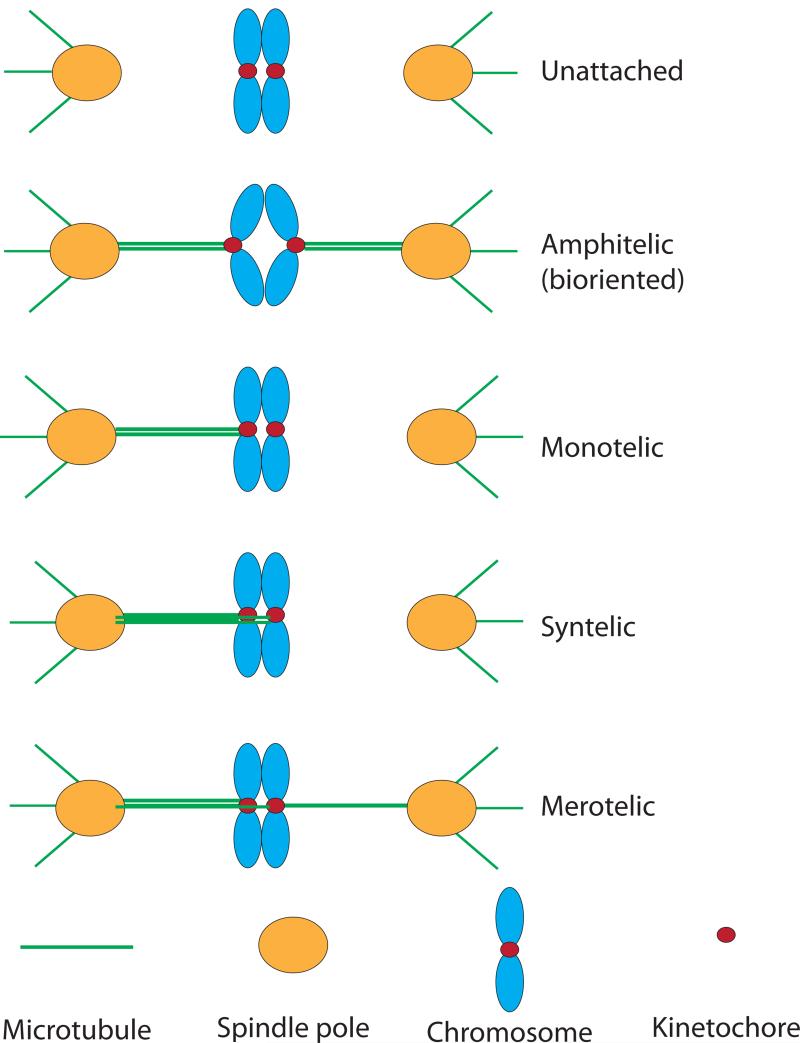
Several different attachment states are possible within the mitotic spindle because sister kinetochores are equivalently capable of binding to microtubules from either pole (Fig. 1). Sister kinetochores may biorient by making attachments to microtubules from opposite poles (amphitelic), or they may make mono-oriented attachments. These occur when microtubules from the same pole attach to both sister kinetochores (syntelic), or when only one of the two sister kinetochores attaches (monotelic). Individual kinetochores typically bind multiple microtubules (from ~3 in fission yeast to ~30 in mammalian cells), while the unusual budding yeast kinetochore binds to a single microtubule. Most organisms are therefore also capable of attaching some microtubules from the same spindle pole to both sister kinetochores (merotelic). However, only bioriented attachments will reliably lead to correct segregation, and cell must therefore attain biorientation before anaphase onset. To monitor biorientation, cells need to sense forces at the kinetochore. Prior to anaphase, sister chromatids are linked by the cohesin complex, which resists microtubule pulling forces (Fig. 1). Evidence suggests that the tension generated on sister kinetochores by the pulling forces of microtubules signals proper biorientation.
Figure 1. Kinetochore-microtubule attachment states on the mitotic spindle.
Kinetochores can bind to microtubules attached to either spindle pole, making several different configurations possible. See text for description of each. Spindle poles nucleate spindle microtubules, shown attaching to kinetochores, as well as astral microtubules. Sister chromosomes are linked by the protein complex cohesin until anaphase onset. Microtubules tend to pull chromosomes towards spindle poles, resulting in tension across bioriented sister chromosomes. Syntelic attachments lead to spindle checkpoint activation whereas merotelic attachments do not necessarily do so because some tension is applied across sister kinetochores134.
Cells utilize at least two central mechanisms to ensure bioriented attachments. First, error correction mechanisms detect and correct mono-oriented attachments. These mechanisms destabilize incorrect microtubule attachments, thus allowing cells another chance to achieve biorientation. The second major mechanism is the spindle checkpoint signaling cascade (also called the spindle assembly checkpoint (SAC) or mitotic checkpoint) that senses the attachment state of kinetochores. Kinetochores that lack tension or attachment induce a spindle checkpoint arrest prior to anaphase, giving cells time to resolve incorrect attachments. Because tension defects generate unattached kinetochores through error correction mechanisms, it has been unclear whether tension and attachment utilize different upstream pathways to trigger the checkpoint. In addition, a precise tension signal has not been conclusively identified4.
The spindle checkpoint was discovered in budding yeast in two landmark studies, which screened for mutants that failed to arrest in response to microtubuledestabilization5,6. These screens identified the Budding Uninhibited by Benzimizadole (BUB1-3)5 and Mitotic Arrest-Deficient (MAD1-3)6 genes. The majority of the corresponding proteins exhibit a hierarchical recruitment to kinetochores (Fig. 2A), indicating that the kinetochore serves as the hub for checkpoint activation. However, because kinetochores have a complex composition, it has been difficult to elucidate the molecular details underlying the recruitment of these proteins. Seminal observations established that phosphorylation is essential for the checkpoint7,8 (see for review9), and at least one kinetochore phospho-epitope persists on both unattached and low-tension kinetochores but is extinguished upon biorientation10-13. Although numerous kinases have direct or indirect roles in the checkpoint (Box 1), the conserved kinetochore kinase Mps1 has emerged as a dominant effector of the checkpoint. Aurora B plays a central role in both error correction and the checkpoint (Box 1), and is reviewed elsewhere9,14,15. New studies have now succeeded in reconstituting kinetochore signaling events in vitro and in vivo, showing that a specific set of kinetochore proteins termed the KMN network (named for the subcomplexes Knl1/Blinkin/Spc105/Spc7, Mis12, and Ndc8016 (Fig. 2A)) are the scaffold for checkpoint signaling. KMN is also a major microtubule-binding interface of the kinetochore and is therefore primed to link microtubule attachment states to checkpoint signaling14,16,17.
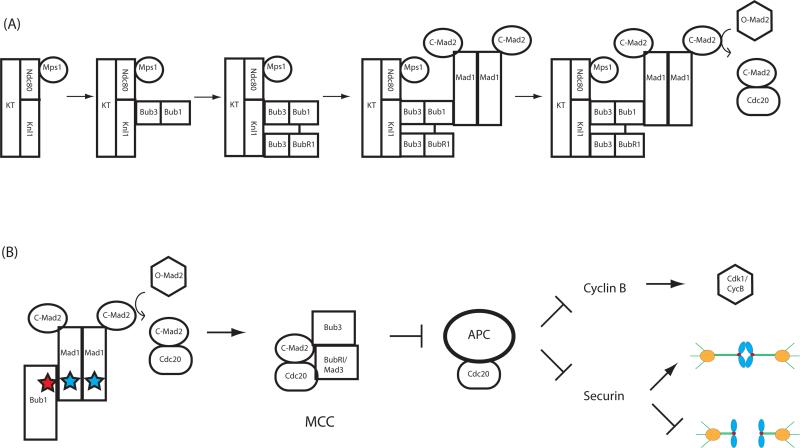
Figure 2. Kinetochore activation of the checkpoint through hierarchical checkpoint protein recruitment.
(A) Assembly of a functional checkpoint-signaling complex on kinetochores is stepwise. Bub3 forms separate complexes with Bub1 and BubR1/Mad3. Mps1 activity recruits Bub1/3, and Bub1 permits localization of BubR1/Mad3. Next, a Mad1/2 heterodimer binds to the kinetochore through a Mad1-Bub1 interaction and possibly through an additional receptor. Kinetochore-bound Mad1/2 facilitates checkpoint activation through catalytic conversion of soluble O-Mad2 to C-Mad2. (B) Kinetochores catalyze production of the MCC. C-Mad2 generated through kinetochore Mad1/C-Mad2 binds to Cdc20, possibly through an intermediate conformation, I-Mad2 (not shown). Conserved motifs in Bub1 and Mad1 (stars) are necessary for checkpoint signaling (see text). Cdc20 and BubR1 localize to kinetochores in most organisms, although it is unresolved how this facilitates MCC formation. For instance, it remains to be determined whether kinetochore-bound or soluble BubR1 is incorporated into the MCC. The MCC inhibits APC/CCdc20 activity, and the critical mitotic targets of the APC/C (cyclin B and securin, see text) are thereby stabilized while the checkpoint is active.
Box 1: Conserved checkpoint kinases.
Original work showed that the checkpoint requires phosphorylation and that kinetochore phosphorylation is directly responsive to microtubule attachment state7,8 (see for review9). Considerable effort has since gone into investigating the role of kinases and identifying their substrates.
Cdk1/cyclin B:
Activity of Cdk1/cyclin B is required for mitotic entry, and anaphase progression depends on degradation of cyclin B and reversal of Cdk1 phosphorylation9,29. Cyclin B is a major target of the APC/C, and stabilizing it is a primary outcome of checkpoint activity. However, Cdk1 function is complex and it is involved both in activating and silencing the checkpoint, at least partly through Cdc20 phosphorylation9,140.
Aurora B:
Aurora B is critical for promoting chromosome biorientation by destabilizing incorrect attachments, a complementary function to the checkpoint. Aurora B is also critical for the checkpoint under certain circumstances, such as a lack of tension. Although this may largely be a consequence of the destabilizing function in yeast141, human Aurora B appears to also be important for loading or stabilizing Mps1 on kinetochores115-118,142.
Mps1:
Mps1 has a widely conserved role in the checkpoint (see for review143). Furthermore, Mps1 is unique in that its overexpression results in constitutive checkpoint activation, identifying it as the central checkpoint kinase144. Like Aurora B, Mps1 is important for promoting biorientation, although the mechanism for this is not clear115,122,145. Kinetochore phosphorylation by Mps1 signals the checkpoint through recruitment of the checkpoint proteins Bub1/3 and Mad1/248-50,78, and possibly through downstream substrates114,146,147.
Bub1:
Although Bub1 kinase activity is dispensable for a checkpoint response55, Bub1 phosphorylates the histone H2A which indirectly leads to Aurora B localization56. Bub1 kinase activity therefore has an indirect role in promoting biorientation and the checkpoint.
Pioneering laser ablation and cell biology studies7,18-20 determined that kinetochores activate the checkpoint by responding to missing or tensionless microtubule attachments. Because even a single unattached kinetochore is sufficient to arrest the cell cycle20, and because this is a minimal defect in the mitotic spindle, it led to the view that the checkpoint is switch-like, either completely on or off. However, cells appeared to arrest for different lengths of time depending on the severity of the attachment state defect21,22. Consistent with this, new reports now show that the checkpoint is graded, with more severe defects leading to a stronger response23,24. Furthermore, complementary findings about the kinetics of checkpoint signaling help explain why the checkpoint does not re-engage during anaphase when tension on kinetochores is lost25-28. Here, we will review recent results that elucidate the molecular basis for checkpoint activation at kinetochores as well as novel findings that suggest that nuclear pores can also generate a checkpoint signal. In Part II, we will relate these mechanistic findings to current reports on the dynamics and kinetics of checkpoint signaling. Overall, these findings resolve long-standing questions about checkpoint activation and redefine our view of the checkpoint response.
Part I - Kinetochore activation of the checkpoint
Mitotic progression is controlled by the Anaphase Promoting Complex/Cyclosome (APC/C), a ubiquitin ligase, and its coactivator Cdc20. Together, this complex activates anaphase onset and mitotic exit by targeting securin, a protein that protects sister chromatid cohesin, and cyclin B, the mitotic Cdk1 cofactor, for degradation29. The spindle checkpoint inhibits these APC/C functions by inactivating Cdc20 through the Mitotic Checkpoint Complex (MCC), which consists of Cdc20 in complex with Mad2, BubR1 (Mad3 in yeast), and Bub330. The MCC inactivates the APC/C through a variety of mechanisms31-35 and is the biochemical manifestation of the “wait anaphase” signal.
The key step in MCC formation is conformational activation of Mad2 from the free “open” form (O-Mad2) to the Cdc20-bound “closed” (C-Mad2) form (Fig. 2B) (see for review36,37). Studies employing FRAP to examine Mad2 localization dynamics implicated kinetochores as the site of Mad2 conversion38-40. Conversion is a catalytic process, occurring through the association of soluble O-Mad2 with kinetochore-bound CMad2. Mad1 is the receptor for C-Mad2 at the kinetochore (which is distinct from Cdc20-bound C-Mad2) and facilitates Mad2 conformational conversion (see for review36,37). A major function of the kinetochore, then, is to promote Mad2 conversion through a hierarchical recruitment of checkpoint proteins. This cascade appears to consist of the conserved kinases Aurora B and Mps1 at the top, followed by the recruitment of the Bub1 and Bub3 (Bub1/3) complex, then by the recruitment of BubR1 and Bub3 (BubR1/3), Mad1 and Mad2 (Mad1/2 heterotetramer)39,41-45. Cdc20 also localizes to kinetochores in most organisms except budding yeast, which also lacks Mad3 localization42. We will focus here on the newly elucidated mechanisms for Bub and Mad protein recruitment.
Bub protein kinetochore recruitment
Although kinetochore localization of checkpoint proteins is necessary for checkpoint signaling, the precise binding sites and mechanism regulating the binding of any checkpoint protein with the kinetochore remained unknown until recently. It has now been established that the core kinetochore protein Knl1 recruits the Bub1, BubR1 and Bub3 checkpoint proteins (Bub proteins)46-51, although the recruitment mechanisms are complex and not yet fully understood. Bub1 and BubR1 are related proteins that are universally required for the checkpoint and are important for kinetochore biorientation41,52,53 (see for review54). Bub1 is a protein kinase while BubR1 is a pseudokinase, and the catalytic domains of both proteins are apparently required for their biorientation functions but not for the spindle checkpoint53,55,56. Consistent with this, the yeast Mad3 checkpoint protein is related to BubR1 but lacks the pseudokinase domain and has no function in biorientation57 (Fig. 3A). Bub1 and BubR1 both form constitutive complexes with the Bub3 protein, which are mutually exclusive since they associate with the same surface of Bub358. Bub1 and BubR1 bind to Bub3 through a Bub3-binding or GLEBS domains (Fig. 3A).
Figure 3. Checkpoint protein regions and interactions.
(A) Interacting regions of kinetochore and checkpoint proteins are depicted. Lines between regions indicate established interactions, and phosphoregulated interactions are shown in red. BubR1/Mad3 domains vary by organism with the pseudokinase domain absent from some lineages57. Structures have been determined for interactions of Bub1-Bub3 and BubR1/Mad3-Bub358, Bub1/3-Knl151, BubR1/Mad3-Knl172, BubR1/Mad3-Mad232,148 and Mad1/2149. MELT-like repeats exhibit poor sequence-level conservation and repeat numbers vary from 2 in some fungi and plants to 27 in Xenopus tropicalis57,70. Note that a single MELT-like motif interacts with three WD40 domains51. C-helix, RLK, CD1, and Bub3-binding regions are all required for the checkpoint. RLK: A Mad1 motif required for Bub1 association86. C-helix: the Mad1 C-terminal helix. CD1: “conserved domain 1” of Bub190,93. TPR and WD40 are common protein domains. Diagrams are oriented with N-termini on the left. (B) Hypothetical BubR1 kinetochore binding mechanisms. (i) The KI2 motif of Knl1 interacts directly with BubR1, possibly facilitating recruitment of a single BubR1 molecule47,63,70. (ii) Bub1 may also recruit BubR1, possibly through a Bub1-BubR1 interaction. Bub3 that is associated with BubR1 at the kinetochore may therefore be available for other interactions63. Bub1:BubR1 stoichiometry at the kinetochore has not been systematically analyzed.
Although Knl1 is a relatively divergent protein, it contains several conserved motifs (see for review17,59,60). Two KNL1-specific motifs are the so-called “KI” motifs and the “MELT”-like ([M/I/L/V]-[E/D]-[M/I/L/V]-T) motifs47-50,57. Initial work characterized an interaction between a KI motif on Knl1 and the conserved TPR motif on Bub1 (Fig. 3A), and this was proposed to mediated Bub1/3 kinetochore recruitment46,47. Surprisingly, however, this interaction is dispensable for kinetochore targeting of Bub1 in vivo61. Additional work determined that Mps1 kinase activity stimulates Bub1 localization and checkpoint activation62, and a trio of studies ultimately determined that Mps1-mediated phosphorylation of the threonines on the MELT-like motifs of Knl1 is required for Bub1 kinetochore localization48-50. Crystallography and additional biochemistry revealed that the phosphorylation is required for a high affinity interaction between Bub3 and Knl151, indicating that Bub3 binding to Knl1 is the key step in localizing Bub1/3 to kinetochores. However, Bub1 contributes to stabilizing the Bub3-Knl1 interaction in vitro51,63, consistent with the observation that Bub1 is important for Bub3 localization in vivo44,64. Unexpectedly, the mechanism of BubR1 recruitment to the kinetochore is different from the related Bub1 protein and is still unresolved. BubR1 does not stabilize the Bub3-MELT interaction51,63, suggesting it may not localize through Bub3-Knl1 binding. BubR1 localization depends on Bub150,65-67, so one possibility is that Bub1 directly recruits BubR1 through dimerization68. This would explain why ectopically localized fission yeast Bub1 recruits Mad369 (Fig. 3B). Such a binding mode may leave BubR1-associated Bub3 available for interactions aside from MELT-like-motif binding63,61.
Quantitative analysis of Bub1 and BubR1 kinetochore recruitment has been investigated in human cell lines and in vitro63,70,71. MELT-like motifs within Knl1 exist in multiple copies (of varying number) in different organisms (Fig. 3A), suggesting multiple Bub protein binding sites. Indeed, in vivo analyses of Knl1-Bub protein binding indicated that the number of MELT-like motifs correlates with the amount of Bub1 binding, at least up to several motifs63,70,71. Interestingly, while a single MELT-like motif can recruit Bub1 and restore some checkpoint function, additional motifs are necessary to permit detectable BubR1 binding in vivo and promote chromosome congression, an effect attributed to BubR159,65,66. This raises the possibility that kinetochore-localized BubR1 is dispensable for the checkpoint and specifically regulates microtubule attachments.
Bub protein binding may also be enhanced by KI motifs through a distinct mechanism. In contrast to the MELT-like motifs, KI motifs are conserved only among some vertebrates57 and only two motifs are present in Knl1, although they may be specialized versions of more loosely defined repetitive motifs70. One KI motif (KI1) interacts only with Bub1, while the other (KI2) appears to interact only with BubR147,61,63,68,72. These motifs enhance Bub protein affinity for Knl1 over a MELT-like sequence alone63,70, but they are not strictly required for Bub protein kinetochore recruitment61. One possibility is that the KI motifs serve as a sensitized switch to recruit just enough Bub protein to signal the checkpoint without stabilizing microtubule attachments, a specific function of BubR1. This may be the case when kinetochores are attached to microtubules that lack tension and must subsequently be destabilized. In the case where kinetochores are completely unattached to microtubules, it would be desirable to both activate the checkpoint and stabilize newly formed microtubule attachments, which would be achieved by recruiting a complete complement of Bub proteins. As we discuss in Part II, attached but tensionless kinetochores appear to elicit a weaker checkpoint response than unattached kinetochores, consistent with the idea that they recruit less Bub proteins. KI motifs may therefore serve as modules to activate the checkpoint while preserving maximal error-correction capabilities at the kinetochore in response to a tension defect.
Mad1/2 kinetochore localization
While Bub1/3 localization is required for the checkpoint, its localization does not always correlate with checkpoint activation. For instance, some Bub1 is retained on early anaphase kinetochores39,44,73, and Bub1 but not Mad1 is present on kinetochores bound to the sides of microtubules, which do not signal the checkpoint74. In contrast, Mad1/2 kinetochore localization is strictly correlated with checkpoint signaling, implicating Mad1/2 kinetochore association as the deterministic step in checkpoint activation. This was elegantly demonstrated by tethering Mad1 to kinetochores, which resulted in constitutive checkpoint activation75,76. However, tethering Mad1 to chromosome arms did not signal the checkpoint, indicating that at least one additional kinetochore factor is required for activation. Identifying these factors has proved to be a major challenge due to the complexity of the kinetochore and the labile nature of Mad1 association.
Recent work showed that the Bub1 protein is a key component in localizing Mad1 to the kinetochore. In budding yeast, Mad1 interacts with Bub1 and requires an RLK motif present in Mad177 (Fig 3A). Mutating this motif or depleting Bub1 abolished kinetochore localization of Mad1 in budding yeast and C. elegans, and Mad1-Bub1 interactions were reconstituted in vitro78,79. Furthermore, a Mad1-kinetochore interaction was demonstrated in vitro with kinetochores and was found to require Mps1-mediated phosphorylation of Bub178. This phosphorylation is therefore a promising candidate for direct control of checkpoint activation in response to microtubule attachment.
Human Mad1 may require an additional receptor because the RLK mutation or a Bub1 depletion only reduced Mad1 levels at kinetochores by half in HeLa cells80. One candidate for an alternative Mad1 co-receptor is the Rod/Zwilch/Zw10 complex (RZZ), which is present mainly in metazoans57. Knl1 and its constitutive binding partner Zwint are required to localize RZZ81,82, and RZZ localization may be regulated through Aurora B phosphorylation of Zwint83. RZZ is required for stable Mad1 localization in human cells81, and the RZZ-associated protein Spindly interacts with Mad1 by coimmunoprecipitation in C. elegans84. However, RZZ/Spindly promotes dynein-mediated Mad1 stripping from kinetochores in metazoans, so it is difficult to discern a precise role in Mad1 recruitment (see for review85). The localization of Mad1 by super-resolution microscopy in human cells is consistent with binding to both Bub1/3 and RZZ, as both complexes localize within a few nanometers of Mad182. In budding yeast, Mad2 may fulfill this co-receptor function because it is required for Mad1 localization, in contrast to other organisms42,78,79,86.
While kinetochore binding of Mad1/C-Mad2 was thought to be sufficient for OMad2 conversion and checkpoint activation87,88, it now appears that there is an additional undefined Mad1 requirement. In budding yeast, Mad1 is constitutively associated with Mad289 while the Bub1-Mad1 interaction is checkpoint-specific86. This implies that the Mad1-Bub1 interaction may be necessary to stimulate Mad2 activation. Several recent studies have confirmed this idea, demonstrating that deletion of the Mad1 C-terminus prevents checkpoint signaling despite correct localization of Bub1 (and presumably Bub3), Mad1, and Mad290-92. Furthermore, tethering of C-Mad2 to kinetochores constitutively activates the checkpoint in a Mad1-dependent manner91. Both the Mad1 RLK motif and a conserved region of Bub1 are also required for the checkpoint with kinetochore-tethered Mad190,92,93. These findings point to the Bub1-Mad1 interaction as the key to activating Mad2 and transducing the checkpoint signal downstream of the kinetochore.
Silencing the kinetochore signal
Silencing the checkpoint signal is critical for mitotic progression and is regulated by multiple mechanisms, including MCC dissociation and regulation of metazoan Mad2 by the protein p31Comet. Here we will consider removal of checkpoint proteins from the kinetochore (for more detailed reviews, see85,94-96). Current models imply that loss of Mad1/2 from kinetochores halts Mad2 conversion and MCC formation, thereby extinguishing the checkpoint signal. At least two independent mechanisms lead to Mad1/2 kinetochore dissociation - dynein-mediated stripping of checkpoint proteins and reversal of activating phosphorylation through phosphatase activity (Fig. 4A,B). Dynein localizes to the kinetochore through RZZ/Spindly and microtubule association allows dynein to transport this module to the spindle pole. Mad1/2 and BubR1 are also removed by dynein during this process, coupling microtubule binding to stripping of kinetochore checkpoint proteins97,98-100. This elegant mechanism is not universal, however, because dynein does not localize to kinetochores in all eukaryotes. In addition, depletion of Spindly, the dynein receptor, does not completely prevent Mad1/2 removal, suggesting that Spindly may normally supersede an additional conserved removal pathway98,100. Interestingly, proteins of the centromere-proximal kinetochore subcomplex CCAN/COMA appear to have a role in opposing dynein stripping, as depletion of the CCAN protein CENP-I greatly enhances the rate of Mad1 dissociation101,102. Another possible silencing mechanism is direct displacement of Mad1/2 from kinetochores by microtubule binding, possibly by steric occlusion94 (Fig. 4C). However, Mps1 activity promotes Mad1 kinetochore localization despite microtubule attachment and biorientation103,104. Therefore, if this mechanism exists, it must be redundant or suppressed under these conditions.
Figure 4. Possible spindle checkpoint silencing mechanisms at the kinetochore.
Numerous mechanisms appear to be involved in silencing the checkpoint. Removal of Mad1/2 and inactivation of Mps1 may be the key silencing events at the kinetochore. (A) In human cells, the motor protein dynein transfers Mad1/2 and RZZ from kinetochores to microtubules, trafficking it towards the spindle pole. This is mediated by Spindly, which associates with RZZ and with dynein85. (B) The phosphatase PP1 associates with kinetochores and is important for checkpoint silencing71,105-109. PP1 activity promotes removal of Bub1/3, although it is not known whether it promotes dissociation of Bub1/3 from Mad1/248,66. Other phosphatases may also be important for silencing. (C) Microtubules may silence the checkpoint by physically displacing Mad1 or inducing conformational change at the kinetochore that reduces Mad1 affinity. (D) Mps1 kinase activity removes itself from the kinetochore57,107. The substrates that mediate removal are unknown. It is also unknown what drives Mps1 (re)association with kinetochores, although Aurora B and Checkpoint kinase 2 (Chk2) may be major factors103,115-118,150,151.
The most widely conserved silencing mechanism is the dephosphorylation of checkpoint proteins by protein phosphatase 1 (PP1/Glc7)66,105-109. PP1 localizes to the kinetochore through Knl1, but may also function through additional proteins, as is the case in fission yeast107,108,110. Preventing the PP1-Kn1l interaction results in constitutive checkpoint activation, and this may be caused by an attachment defect (due to a failure to reverse Aurora B phosphorylation110) or a checkpoint silencing defect107,108. PP1 removes Bub1/3 from kinetochores48,71, thereby promoting silencing upstream of Mad1/2 localization. PP1 may also target other checkpoint substrates. Downstream of the kinetochore, the phosphatase Cdc14 plays a critical role in maintaining checkpoint silencing during mitotic exit111,112.
Control of Mps1 localization may also be a key step to silencing the kinetochore signaling cascade, as Mps1 kinetochore levels are substantially reduced by the time of anaphase onset39,113 (Fig. 4D). Although the Mps1 recruitment mechanism remains incompletely defined, Mps1 turns over at kinetochores as determined by FRAP39, and its kinase activity promotes its own dissociation from kinetochores103,114. To retain checkpoint-signaling function, Mps1 must logically be re-recruited to the kinetochore. This re-recruitment may be a major regulatory step, and is likely mediated by Aurora B103,115-118. It is also possible that Knl1 is important for Mps1 regulation because Knl1 appears to have a role in checkpoint silencing that is independent of PP1 activity or dynein stripping119. After anaphase onset, degradation of checkpoint proteins Mps1 and Bub1 prevents checkpoint reactivation113,120.
The checkpoint and the nuclear pore
While kinetochores control the checkpoint throughout mitosis, the kinetochore does not appear to be necessary for all checkpoint signaling. For instance, although most cells do not assemble kinetochores outside of mitosis, they nonetheless harbor MCCs before mitotic entry, raising the possibility that kinetochores are not the only catalyst for MCC production. Additionally, Mad1 localizes to the nuclear pore complex (NPC), suggesting these large protein assemblies as alternative, pre-mitotic (interphase) checkpoint signaling scaffolds. A new study has validated this hypothesis121, demonstrating that NPC localization of Mad1 is required for a characteristic checkpoint-dependent delay in mitosis called the mitotic timer121-124. Because Bub1/3 has not been reported to localize to the NPC, it remains unclear whether NPCs use the same Mad2 activation mechanism as kinetochores or whether NPCs continuously activate Mad2 until they are disassembled at nuclear envelope breakdown. It is possible, for instance, that both NPCs and kinetochores stimulate checkpoint signaling by default, but that microtubule binding can only silence the kinetochore signal. Cells defective in Mad1 NPC targeting exhibit increased levels of lagging chromosomes at anaphase, indicative of merotelic attachments persisting through mitosis despite a functional kinetochore checkpoint121. Therefore, it appears that NPC-localized Mad1/2 assists the kinetochore-based mechanisms in promoting correct segregation. Presumably, this occurs by facilitating MCC production prior to mitotic entry, when kinetochores assemble, and by allowing more time for chromosomes to biorient121.
In addition to the mitotic timer, NPCs may have additional checkpoint-related functions. The conserved human nuclear pore protein Tpr helps to stabilize checkpoint proteins Mad1 and Mad2. Consistently, Tpr depletion has a moderate effect on the checkpoint response125. Mad1 also cycles between kinetochores and NPCs in checkpoint-active budding yeast, which retain their nuclear envelope during mitosis104. In these cells, NPC-localized Mad1 is proposed to facilitate checkpoint activity by blocking nuclear import of the checkpoint silencing phosphatase Glc7/PP1104 (see for review126).
II. The Checkpoint at the Limits
It is imperative to elucidate how the checkpoint functions at its limits to understand why failures occur that lead to missegregation and aneuploidy in a natural context. Because cancer cells are typically aneuploid and highly dependent upon checkpoint function, this may also shed light on ways to target the checkpoint for chemotherapy (see for review127,128). While classic studies established that both attachment and tension defects can lead to checkpoint arrest, these defects had not been systematically analyzed for their molecular output until recently. New studies have additionally expanded on previous work analyzing the timing from initial signaling to stabilization of APC/C targets.
Variation in checkpoint activation
Because a single unattached kinetochore can halt the cell cycle20, it appears that a minimal attachment defect is capable of a full checkpoint response. A logical conclusion is that the checkpoint response is binary – either completely on or off. However, hints that more complete attachment defects result in a stronger checkpoint emerged over time. For instance, it was realized that cells can undergo “mitotic slippage”, a bypass of cell-cycle arrest despite an active checkpoint, more quickly under certain circumstances21,22. Now, new reports have quantified variation in checkpoint response strength23,24,129 (Fig. 5A). Dick and Gerlich investigated this question using low doses of nocodazole, a drug that destabilizes microtubules, to generate unattached kinetochores. They found that a higher number of unattached kinetochores (as measured by chromosomes displaced from the metaphase plate) correlated with a slower degradation rate of securin, an APC/C substrate, indicating enhanced checkpoint function23. Another study measured destruction of cyclin A, which competes with checkpoint proteins for Cdc20 binding to be ubiquitylated and degraded, as a quantitative readout for checkpoint activity24,130. They found that different spindle poisons activate the checkpoint to different degrees. Taxol (paclitaxel), a microtubule-stabilizing drug that activates the checkpoint by generating tension defects, gave a significantly weaker response than nocodazole. Furthermore, checkpoint activity correlated with the amount of Mad2 localized to kinetochores, indicating the difference in duration is due to upstream signaling events at the kinetochore24.
Figure 5. The graded checkpoint response.
(A) Various treatments result in different degrees of checkpoint activity. Weak checkpoint activation results from minor defects, such as a lack of tension caused by Taxol treatment, which eliminates microtubule dynamics, or dimethyl anastrom (DMA) treatment, which collapses the mitotic spindle while preserving microtubule attachments. A weak checkpoint results in reduced mitotic duration (mitotic slippage), faster cyclin B degradation, and low kinetochore Mad2 levels. Complete disruption of microtubules results in a strong checkpoint response with an increased length of mitosis, slower cyclin B degradation, and increased Mad2 at kinetochores24. The rate of securin degradation, a readout for APC/C activity and, inversely, checkpoint strength, correlates with the number of unattached kinetochores23. (B) Hypothetical model for modulation of checkpoint strength through Mps1 activity. More severe perturbations may enhance or stabilize Mps1 phosphorylation on its targets, including Spc105 and Bub1. This in turn leads to greater kinetochore localization of Mad2 and a stronger checkpoint response indicated by greater MCC levels24 and less free Cdc20. Titration of Mps1 activity by inhibitor addition to checkpoint-arrested cells resulted in a proportionally attenuated checkpoint response, similarly to microtubule-directed drug treatments24. Activity of the opposing phosphatase PP1 may also be responsive to microtubule attachment.
Remarkably, the amount of Mad2 that localized to individual kinetochores, not just the number of kinetochores that bind to Mad2, was increased with greater microtubule destabilization24. This suggests that individual kinetochores can vary the strength of the checkpoint signal they generate. As was discussed in Part I, the Bub proteins have multiple binding sites at each kinetochore that could lead to variation in the checkpoint response, and the number of Bub proteins at the kinetochore likely correlates with the strength of the checkpoint signal63,70,71. The variation in Bub protein numbers could be caused by changes in upstream Mps1 activity (Fig. 5B). This is particularly likely in the case of Taxol treatment, as tensionless kinetochores may have a distinct or partial mechanism to activate the checkpoint relative to unattached kinetochores (see for review4). Different microtubule-binding modes of the kinetochore could conceivably trigger this variation, as microtubule binding can occur through the lattice (side) or end of a microtubule, and there are multiple microtubule-binding elements within the kinetochore131,132. In any case, elevated amounts of active Cdc20 may be the effector for reduced checkpoint efficiency. Stronger checkpoint response was correlated with greater levels of MCC-associated Cdc20, indicating more total Cdc20 was inactivated24. Furthermore, altering expression levels of Cdc20 and Mad2 by promoter swapping indicated that the ratio of Cdc20 to Mad2 in the cell must be carefully regulated for proper checkpoint function129.
It is presently unclear why the checkpoint response varies depending on the spindle defect, although it is reasonable for cells to risk undergoing segregation eventually because extended mitotic arrest can lead to inviability. Cells may therefore balance the risk of missegregation with that of extended arrest by modulating the strength of the checkpoint response. On the other hand, a weak checkpoint response is likely to be elicited early in mitosis, when kinetochores are newly formed (having had little time to accumulate MCCs) and microtubule attachments are present but not necessarily correct (leading to weak signaling). Therefore, cells may buffer this weak response by accumulating interphase MCCs through the mitotic timer121. This proposal is supported by the observations that mislocalizing Mad1 from nuclear pores sensitizes cells to weakened kinetochore signaling by Aurora B inhibition121, and that weak checkpoint activation is correlated with reduced levels of intact MCCs24. Thus, cells may prepare for weak signaling at mitotic onset, but become more sensitive to mitotic slippage as the risks due to prolonged arrest increase.
Checkpoint kinetics and the anaphase response
A critical feature of the checkpoint is that it must respond to low-tension kinetochores until metaphase but this must not continue through anaphase, when sister chromosomes are no longer held together by cohesin and kinetochores are therefore under little tension (Fig. 1). This is the so-called “anaphase problem”, and while multiple factors are involved in overcoming it (see for review112), we will focus here on recent studies that relate to checkpoint kinetics and activity of Cdk1 kinase, the mitotic master regulator.
Silencing during anaphase is mediated by phosphatase activity to reverse mitotic phosphorylation71,105-108,110 (see for review133) in conjunction with proteolysis of checkpoint proteins upon mitotic exit113,120 (see for review134). For instance, forced expression of Mps1 in an anaphase arrest was sufficient to induce checkpoint signaling113, suggesting that the checkpoint must stay silenced throughout anaphase. Additionally, the onset of cyclin B degradation, which indicates checkpoint silencing, correlates precisely with the completion of chromosome congression135. Recent temporal analysis of checkpoint reactivation has helped to clarify the anaphase checkpoint silencing process further. In vivo laser ablation of microtubules in metaphase human cells revealed that checkpoint signaling, measured by ensuing Mad2 recruitment, was intact throughout metaphase23. However, a subset of cells initiated anaphase within a brief window following Mad2 kinetochore recruitment, suggesting that cells already prepared for anaphase could not arrest. Consistent with this, the rate of securin degradation (indicating APC/C activity and an incomplete checkpoint response) was only slightly reduced within 0-5 minutes of checkpoint protein kinetochore recruitment, whereas it was reduced ~60 fold within 5-10 minutes23. Similar kinetics were also observed in fission yeast25, and in earlier studies that analyzed anaphase onset following the completion of chromosome congression38,136. Thus, a temporal gap exists between checkpoint protein recruitment and APC/C inactivation, meaning the checkpoint is functionally off during this time.
Several additional studies have analyzed the anaphase response further by using non-degradable cyclin B, the cofactor that activates Cdk1 towards its mitotic targets, in fission yeast25, mice27, and human cells26,28 Stabilizing cyclin B permitted securin degradation and sister chromatid separation with a corresponding loss of kinetochore tension25-28. Nonetheless, stabilized cyclin B allowed checkpoint proteins to relocalize to kinetochores upon anaphase onset, as had been previously observed25,26,28,29,111,134,137. This indicated that loss of tension at anaphase does indeed promote checkpoint signaling, but this response is normally repressed by Cdk1 inactivation. Since Cdk1 activity is required for the checkpoint (see for review9), its silencing by cyclin B degradation ensures the checkpoint is not reactivated26-28,129. It was further demonstrated that an anaphase checkpoint response is too slow to inhibit the APC/C, and this slow response may serve as a failsafe mechanism to prevent interruption of anaphase by checkpoint activation25,28. Therefore, transient satisfaction of the checkpoint is sufficient to activate the APC/C, and the fast kinetics of APC/C degradation of securin and cyclin B preclude functional checkpoint reactivation during anaphase (Fig. 6).
Figure 6. Timing of checkpoint signaling.
Interphase: Prior to mitosis, nuclear pores promote production of the MCC through Mad1/2, similarly to kinetochores, although the upstream activation signal(s), if any, are unknown121. Prometaphase/Metaphase: In most cell types, kinetochore-microtubule attachments are established following nuclear envelope breakdown and kinetochores, initially unattached, signal the checkpoint and thereby inhibit the APC/C. Metaphase: Upon biorientation at metaphase, kinetochore checkpoint signaling is silenced, leading to APC/C activity and the beginning of cyclin B degradation (arrow). Early Anaphase: At anaphase, attached kinetochores are no longer under tension. Rapid inactivation of Cdk1/cyclin B by the APC/C prevents these unattached kinetochores from signaling the checkpoint25-27,112. Late Anaphase: Following segregation, APC/C activity destroys checkpoint proteins, including Bub1 and Mps1, fully silencing checkpoint control over the APC/C113,120,134.
Conclusion/Perspective
Substantial advances have recently been made in the checkpoint field, including the identification and structural analyses of precise protein interactions between core kinetochore components and checkpoint proteins, the reconstitution of checkpoint activation steps in vitro, the characterization of NPCs as a generator of the MCC, and the description of the checkpoint as a graded and kinetically limited response. Given the complexity of the kinetochore, reconstituting checkpoint signaling events that occur there has been a major obstacle. As we describe, reconstitution of Bub and Mad protein binding to core kinetochore proteins has now been achieved, and it may soon be possible to entirely reconstitute the checkpoint in vitro to fully understand the signaling cascade. As discussed above, it appears that the canonical model for Mad1/2 activation is incomplete90-92, and it will therefore be critical to determine what additional event(s) are required for formation of the MCC downstream of Mad1 recruitment. Reconstitution advances will also permit structural studies that will be critical for detailed understanding of checkpoint mechanisms.
Cumulatively, recent work has established that Mps1 is the central checkpoint kinase at both kinetochores and NPCs121,122. The identification of NPCs as a mitotic timer raises new question about whether this timing mechanism is regulated and precisely why missegregation is increased in its absence. It is possible that initial kinetochore signaling is too weak or slow to delay the cell cycle in response to moderate defects without pre-formed MCCs derived from NPCs121. Since Bub1/3 has not been localized to the nuclear pore, an intriguing possibility is that the Rae1-Nup98 nuclear pore subcomplex functions analogously to Bub1/3 in activating Mad1/2. In addition to striking structural similarity to Bub1/351, Rae1-Nup98 is necessary for accurate segregation and securin stabilization138.
Regarding kinetochore checkpoint activation, a critical future question is what controls the kinase cascade. Ultimately, this activation signal must be opposed by tension-bearing microtubule attachments. Mps1 activity towards Bub1 may be specifically regulated because Bub1 and Mad1 kinetochore localization are separable (see for example74), and this may be the critical step controlled by microtubule attachment or tension. Kinetochore tethering of Mps1 can recruit checkpoint proteins and activate the checkpoint even while preserving bioriented attachments62,103,104, implying that checkpoint silencing requires either removal or inactivation of kinetochore-bound Mps1. Although PP1 opposes Mps1 activity48,71, phosphatase activity does not appear to be controlled by tension10, suggesting that another mechanism may regulate Mps1 activity towards Bub1. Mps1 actively cycles at kinetochores and promotes its own dissociation, so one possibility is that its re-recruitment may be controlled by microtubule attachments. Determining exactly how Mps1 output is connected to kinetochore-microtubule attachment is a major challenge for the future. Additional important questions to address are the underlying mechanism of BubR1 kinetochore recruitment, and the precise details of MCC formation.
The demonstration that the kinetochore response is variable in strength and kinetically limited raises new questions about aneuploidy and cancer. Weak activation and slow kinetics can possibly circumvent the checkpoint and lead to aneuploidy. It will be informative to learn how frequently missegregation results from mitotic slippage and late-arising attachment errors. Moreover, the checkpoint sensitivity to relative checkpoint protein abundance129 suggests that aneuploid cancer cells may evade the checkpoint in part through moderate perturbation of these levels (see for review139). A better understanding of quantitative protein ratios may therefore assist in targeting therapeutics to cancer cells. Additional knowledge about the graded checkpoint response will also entail identifying the underlying mechanisms that account for variability. Because Mad2 levels at individual kinetochores vary with the attachment defect24, upstream events likely tune the checkpoint response.
In the coming years, further advances will likely be made in understanding activation mechanisms, including the long-sought biochemical and structural basis for converting defects in forces at kinetochores into a phosphorylation signaling cascade. Structural studies will also inform how the Bub and Mad proteins interact at successive steps in the checkpoint. Advances in other areas, including checkpoint silencing and kinetics of individual checkpoint steps, will also likely be elucidated in the near future, enhancing our understanding of the dynamic limits of the checkpoint. Such work will lead to a greater understanding of fundamental questions of genome stability and improve our ability to target this critical pathway for improved cancer therapeutics.
Acknowledgments
We are grateful to the reviewers and Matt Miller for thoughtful comments on the manuscript and apologize to those who were not cited due to space limitations. NL was supported by an NIH center interdisciplinary training grant (T32 CA080416) and work in the Biggins lab is supported by NIH grants GM064386 and GM078069 to SB.
References
- 1.Gordon DJ, Resio B, Pellman D. Causes and consequences of aneuploidy in cancer. Nature Reviews Genetics. 2012;13(3):189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- 2.Ricke RM, van Deursen JM. Aneuploidy in health, disease, and aging. The Journal of cell biology. 2013;201:11–21. doi: 10.1083/jcb.201301061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McIntosh JR. Motors or dynamics: What really moves chromosomes? Nature cell biology. 2012;14:1234–1234. doi: 10.1038/ncb2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maresca TJ, Salmon ED. Welcome to a new kind of tension: translating kinetochore mechanics into a wait-anaphase signal. Journal of Cell Science. 2010;123:825–835. doi: 10.1242/jcs.064790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoyt MA, Totis L, Roberts BTS. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- 6.Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [5 and 6 established the existence of the spindle checkpoint and identified upstream checkpoint signaling genes through genetic screens.] [DOI] [PubMed] [Google Scholar]
- 7.Nicklas RB, Ward SC, Gorbsky GJ. Kinetochore chemistry is sensitive to tension and may link mitotic forces to a cell cycle checkpoint. The Journal of Cell Biology. 1995;130:929–939. doi: 10.1083/jcb.130.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Minshull J SH, Tonks NK, Murray AW. A MAP kinase-dependent spindle assembly checkpoint in Xenopus egg extracts. 1994;79:475–86. doi: 10.1016/0092-8674(94)90256-9. [DOI] [PubMed] [Google Scholar]
- 9.Funabiki H, Wynne DJ. Making an effective switch at the kinetochore by phosphorylation and dephosphorylation. Chromosoma. 2013;122:135–158. doi: 10.1007/s00412-013-0401-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X, Nicklas RB. Tension-sensitive kinetochore phosphorylation and the chromosome distribution checkpoint in praying mantid spermatocytes. Journal of cell science. 1997;110:537–545. doi: 10.1242/jcs.110.5.537. [DOI] [PubMed] [Google Scholar]
- 11.Nicklas RB, Campbell MS, Ward SC, Gorbsky GJ. Tension-sensitive kinetochore phosphorylation in vitro. Journal of cell science. 1998;111:3189–3196. doi: 10.1242/jcs.111.21.3189. [Classic studies that used innovative biophysical methods to conclusively demonstrate a role for tension in checkpoint satisfaction.] [DOI] [PubMed] [Google Scholar]
- 12.Gorbsky GJ, Ricketts WA. Differential expression of a phosphoepitope at the kinetochores of moving chromosomes. The Journal of cell biology. 1993;122:1311–1321. doi: 10.1083/jcb.122.6.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maresca TJ, Salmon ED. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. The Journal of Cell Biology. 2009;184:373–381. doi: 10.1083/jcb.200808130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foley EA, Kapoor TM. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nature Reviews Molecular Cell Biology. 2012;14:25–37. doi: 10.1038/nrm3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vader G, Maia AF, Lens SMA. The chromosomal passenger complex and the spindle assembly checkpoint: kinetochore-microtubule error correction and beyond. Cell Division. 2008;3:10. doi: 10.1186/1747-1028-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [Identification of the KMN network as a key microtubule-binding kinetochore element through biochemical reconstitution.] [DOI] [PubMed] [Google Scholar]
- 17.Varma D, Salmon ED. The KMN protein network - chief conductors of the kinetochore orchestra. Journal of Cell Science. 2013;125:5927–5936. doi: 10.1242/jcs.093724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen RH, Waters JC, Salmon ED, Murray AW. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science (New York, N.Y.) 1996;274:242–246. doi: 10.1126/science.274.5285.242. [First demonstration that a checkpoint protein localizes to the kinetochores, supporting the idea that the kinetochore generates the checkpoint signal.] [DOI] [PubMed] [Google Scholar]
- 19.Li X, Nicklas RB. Mitotic forces control a cell-cycle checkpoint. Nature. 1995;373:630–632. doi: 10.1038/373630a0. [DOI] [PubMed] [Google Scholar]
- 20.Rieder CL, Cole RW, Khodjakov A, Sluder G. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. The Journal of cell biology. 1995;130:941–948. doi: 10.1083/jcb.130.4.941. [Seminal study that, along with reference 19, established the kinetochore as central to the spindle checkpoint.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weaver BAA. Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. The Journal of Cell Biology. 2003;162:551–563. doi: 10.1083/jcb.200303167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rieder CL, Maiato H. Stuck in division or passing through: what happens when cells cannot satisfy the spindle assembly checkpoint. Developmental cell. 2004;7:637–651. doi: 10.1016/j.devcel.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 23.Dick AE, Gerlich DW. Kinetic framework of spindle assembly checkpoint signalling. Nature Cell Biology. 2013;15:1370–1377. doi: 10.1038/ncb2842. [A clever application of established optical methods to gain unprecedented time-resolution on specific checkpoint silencing events and measure the strength of the checkpoint response.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collin P, Nashchekina O, Walker R, Pines J. The spindle assembly checkpoint works like a rheostat rather than a toggle switch. Nature Cell Biology. 2013;15:1378–1385. doi: 10.1038/ncb2855. [Along with references 23 and 25 quantitatively established the variable limits of checkpoint kinetics and duration in response to different stimuli.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamenz J, Hauf S. Slow checkpoint activation kinetics as a safety device in anaphase. Current Biology. 2014;24(6):646–51. doi: 10.1016/j.cub.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 26.Vázquez-Novelle María D., et al. Cdk1 inactivation terminates mitotic checkpoint surveillance and stabilizes kinetochore attachments in anaphase. Current Biology. 2014;24(6):638–45. doi: 10.1016/j.cub.2014.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rattani A, et al. Dependency of the spindle assembly checkpoint on Cdk1 renders the anaphase transition irreversible. Current Biology. 2014 doi: 10.1016/j.cub.2014.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clijsters L, et al. Inefficient degradation of cyclin B1 re-activates the spindle checkpoint right after sister chromatid disjunction. Cell Cycle. 2014;13 doi: 10.4161/cc.29336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oliveira RA, Hamilton RS, Pauli A, Davis I, Nasmyth K. Cohesin cleavage and Cdk inhibition trigger formation of daughter nuclei. Nature Cell Biology. 2010;12:185–192. doi: 10.1038/ncb2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sudakin V. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. he Journal of Cell Biology. 2001;154:925–936. doi: 10.1083/jcb.200102093. [The original biochemical identification of the MCC.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burton JL, Solomon MJ. Mad3p, a pseudosubstrate inhibitor of APCCdc20 in the spindle assembly checkpoint. Genes & Development. 2007;21:655–667. doi: 10.1101/gad.1511107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chao WCH, Kulkarni K, Zhang Z, Kong EH, Barford D. Structure of the mitotic checkpoint complex. Nature. 2012;484:208–213. doi: 10.1038/nature10896. [Demonstrated the structural basis for Cdc20 inactivation by the MCC.] [DOI] [PubMed] [Google Scholar]
- 33.Han Joo S., et al. Catalytic assembly of the mitotic checkpoint inhibitor BubR1-Cdc20 by a Mad2-induced functional wwitch in Cdc20. Molecular Cell. 2013;51:92–104. doi: 10.1016/j.molcel.2013.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Izawa D, Pines J. Mad2 and the APC/C compete for the same site on Cdc20 to ensure proper chromosome segregation. The Journal of Cell Biology. 2012;199:27–37. doi: 10.1083/jcb.201205170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lau Derek T.C., Murray Andrew W. Mad2 and Mad3 cooperate to arrest budding yeast in mitosis. Current Biology. 2012;22:180–190. doi: 10.1016/j.cub.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo X, Yu H. Protein Metamorphosis: The two-state behavior of Mad2. Structure. 2008;16:1616–1625. doi: 10.1016/j.str.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skinner JJ, Wood S, Shorter J, Englander SW, Black BE. The Mad2 partial unfolding model: regulating mitosis through Mad2 conformational switching. The Journal of Cell Biology. 2008;183:761–768. doi: 10.1083/jcb.200808122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Howell BJ, Hoffman DB, Fang G, Murray AW, Salmon ED. Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. The Journal of cell biology. 2000;150:1233–1250. doi: 10.1083/jcb.150.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Howell BJ, et al. Spindle checkpoint protein dynamics at kinetochores in living cells. Current biology. 2004;14:953–964. doi: 10.1016/j.cub.2004.05.053. [In vivo analysis of checkpoint protein dynamics at kinetochores by FRAP that, together with references 38 and 40, have been instrumental in understanding how the kinetochore works as a catalytic scaffold.] [DOI] [PubMed] [Google Scholar]
- 40.Shah JV, et al. Dynamics of centromere and kinetochore proteins: implications for checkpoint signaling and silencing. Current biology. 2004;14:942–952. doi: 10.1016/j.cub.2004.05.046. [DOI] [PubMed] [Google Scholar]
- 41.Chen RH. BubR1 is essential for kinetochore localization of other spindle checkpoint proteins and its phosphorylation requires Mad1. The Journal of Cell Biology. 2002;158:487–496. doi: 10.1083/jcb.200204048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gillett ES. Spindle checkpoint proteins and chromosome-microtubule attachment in budding yeast. The Journal of Cell Biology. 2004;164:535–546. doi: 10.1083/jcb.200308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heinrich S, Windecker H, Hustedt N, Hauf S. Mph1 kinetochore localization is crucial and upstream in the hierarchy of spindle assembly checkpoint protein recruitment to kinetochores. Journal of Cell Science. 2012;125:4720–4727. doi: 10.1242/jcs.110387. [DOI] [PubMed] [Google Scholar]
- 44.Sharp-Baker H, Chen R-H. Spindle checkpoint protein Bub1 is required for kinetochore localization of Mad1, Mad2, Bub3, and CENP-E, independently of its kinase activity. The Journal of cell biology. 2001;153:1239–1250. doi: 10.1083/jcb.153.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vigneron S, et al. Kinetochore localization of spindle checkpoint proteins: who controls whom? Molecular biology of the cell. 2004;15:4584–4596. doi: 10.1091/mbc.E04-01-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kiyomitsu T, Obuse C, Yanagida M. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Developmental Cell. 2007;13:663–676. doi: 10.1016/j.devcel.2007.09.005. [Initial identification of Knl1 as the Bub1 and BubR1 kinetochore receptor that established the checkpoint requirement for Knl1.] [DOI] [PubMed] [Google Scholar]
- 47.Kiyomitsu T, Murakami H, Yanagida M. Protein interaction domain mapping of human kinetochore protein Blinkin reveals a consensus motif for binding of spindle assembly checkpoint proteins Bub1 and BubR1. Molecular and Cellular Biology. 2011;31:998–1011. doi: 10.1128/MCB.00815-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.London N, Ceto S, Ranish Jeffrey A., Biggins S. Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Current Biology. 2012;22:900–906. doi: 10.1016/j.cub.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shepperd Lindsey A., et al. Phosphodependent recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 kinase maintains the spindle checkpoint. Current Biology. 2012;22:891–899. doi: 10.1016/j.cub.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamagishi Y, Yang C-H, Tanno Y, Watanabe Y. MPS1/Mph1 phosphorylates the kinetochore protein KNL1/Spc7 to recruit SAC components. Nature Cell Biology. 2012;14:746–752. doi: 10.1038/ncb2515. [Along with references 48, 49 and 51 established the molecular basis for Bub1 and Bub3 localization to kinetochores, identifying the first core kinetochore phosphorylation event critical for the checkpoint.] [DOI] [PubMed] [Google Scholar]
- 51.Primorac I, et al. Bub3 reads phosphorylated MELT repeats to promote spindle assembly checkpoint signaling. Elife. 2013;2 doi: 10.7554/eLife.01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Campbell L. Analysis of Bub3 spindle checkpoint function in Xenopus egg extracts. Journal of Cell Science. 2003;116:617–628. doi: 10.1242/jcs.00255. [DOI] [PubMed] [Google Scholar]
- 53.Suijkerbuijk Saskia J.E., et al. The vertebrate mitotic checkpoint protein BUBR1 is an unusual pseudokinase. Developmental Cell. 2012;22:1321–1329. doi: 10.1016/j.devcel.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 54.Elowe S. Bub1 and BubR1: at the interface between chromosome attachment and the spindle checkpoint. Molecular and Cellular Biology. 2011;31:3085–3093. doi: 10.1128/MCB.05326-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fernius J, Hardwick KG. Bub1 kinase targets Sgo1 to ensure efficient chromosome biorientation in budding yeast mitosis. PLoS Genetics. 2007;3:e213. doi: 10.1371/journal.pgen.0030213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kawashima SA, Yamagishi Y, Honda T, Ishiguro K.i., Watanabe Y. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science. 2010;327:172–177. doi: 10.1126/science.1180189. [DOI] [PubMed] [Google Scholar]
- 57.Vleugel M, Hoogendoorn E, Snel B, Kops Geert J.P.L. Evolution and function of the mitotic checkpoint. Developmental Cell. 2012;23:239–250. doi: 10.1016/j.devcel.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 58.Larsen NA, Al-Bassam J, Wei RR, Harrison SC. Structural analysis of Bub3 interactions in the mitotic spindle checkpoint. Proceedings of the National Academy of Sciences. 2007;104:1201–1206. doi: 10.1073/pnas.0610358104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Caldas GV, DeLuca JG. KNL1: bringing order to the kinetochore. Chromosoma. 2013;123(3):169–81. doi: 10.1007/s00412-013-0446-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ghongane P, Kapanidou M, Asghar A, Elowe S, Bolanos-Garcia VM. The dynamic protein Knl1 - a kinetochore rendezvous. Journal of Cell Science. 2014;127:1–9. doi: 10.1242/jcs.149922. [DOI] [PubMed] [Google Scholar]
- 61.Krenn V, Wehenkel A, Li X, Santaguida S, Musacchio A. Structural analysis reveals features of the spindle checkpoint kinase Bub1-kinetochore subunit Knl1 interaction. The Journal of Cell Biology. 2012;196:451–467. doi: 10.1083/jcb.201110013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ito D, Saito Y, Matsumoto T. Centromere-tethered Mps1 pombe homolog (Mph1) kinase is a sufficient marker for recruitment of the spindle checkpoint protein Bub1, but not Mad1. Proceedings of the National Academy of Sciences. 2011;109:209–214. doi: 10.1073/pnas.1114647109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krenn V, Overlack K, Primorac I, van Gerwen S, Musacchio A. KI motifs of human Knl1 enhance assembly of comprehensive spindle checkpoint complexes around MELT Repeats. Current Biology. 2014;24:29–39. doi: 10.1016/j.cub.2013.11.046. [DOI] [PubMed] [Google Scholar]
- 64.Vanoosthuyse V, Valsdottir R, Javerzat JP, Hardwick KG. Kinetochore targeting of fission yeast Mad and Bub proteins is essential for spindle checkpoint function but not for all chromosome segregation roles of Bub1p. Molecular and Cellular Biology. 2004;24:9786–9801. doi: 10.1128/MCB.24.22.9786-9801.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johnson VL. Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression. Journal of Cell Science. 2004;117:1577–1589. doi: 10.1242/jcs.01006. [DOI] [PubMed] [Google Scholar]
- 66.Kadura S, He X, Vanoosthuyse V, Hardwick KG, Sazer S. The A78V mutation in the Mad3-like domain of Schizosaccharomyces pombe Bub1p perturbs nuclear accumulation and kinetochore targeting of Bub1p, Bub3p, and Mad3p and spindle assembly checkpoint function. Molecular biology of the cell. 2005;16:385–395. doi: 10.1091/mbc.E04-07-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Millband DN, Hardwick KG. Fission yeast Mad3p is required for Mad2p to inhibit the anaphase-promoting complex and localizes to kinetochores in a Bub1p-, Bub3p-, and Mph1p-dependent manner. Molecular and Cellular Biology. 2002;22:2728–2742. doi: 10.1128/MCB.22.8.2728-2742.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.D'Arcy S, Davies OR, Blundell TL, Bolanos-Garcia VM. Defining the molecular masis of BubR1 kinetochore interactions and APC/C-CDC20 inhibition. Journal of Biological Chemistry. 2010;285:14764–14776. doi: 10.1074/jbc.M109.082016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rischitor PE, May KM, Hardwick KG. Bub1 is a fission yeast kinetochore scaffold protein, and is sufficient to recruit other spindle checkpoint proteins to ectopic sites on chromosomes. PLoS ONE. 2007;2 doi: 10.1371/journal.pone.0001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vleugel M, et al. Arrayed BUB recruitment modules in the kinetochore scaffold KNL1 promote accurate chromosome segregation. The Journal of Cell Biology. 2013;203:943–955. doi: 10.1083/jcb.201307016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang G, Lischetti T, Nilsson J. A minimal number of MELT repeats supports all functions of KNL1 in chromosome segregation. Journal of cell science. 2013;127:871–84. doi: 10.1242/jcs.139725. [DOI] [PubMed] [Google Scholar]
- 72.Bolanos-Garcia Victor M., et al. Structure of a Blinkin-BUBR1 complex reveals an interaction crucial for kinetochore-mitotic checkpoint regulation via an unanticipated binding site. Structure. 2011;19:1691–1700. doi: 10.1016/j.str.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jablonski SA CG, Cooke CA, Earnshaw WC, Yen TJ. The hBUB1 and hBUBR1 kinases sequentially assemble onto kinetochores during prophase with hBUBR1 concentrating at the kinetochore plates in mitosis. Chromosoma. 1998;107:386–96. doi: 10.1007/s004120050322. [DOI] [PubMed] [Google Scholar]
- 74.Shimogawa MM, Wargacki MM, Muller E, Davis T. Laterally attached kinetochores recruit the checkpoint protein Bub1, but satisfy the spindle checkpoint. Cell Cycle. 2010;9:3619–3628. doi: 10.4161/cc.9.17.12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kuijt TEF, Omerzu M, Saurin AT, Kops GJPL. Conditional targeting of MAD1 to kinetochores is sufficient to reactivate the spindle assembly checkpoint in metaphase. Chromosoma. 2014 doi: 10.1007/s00412-014-0458-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Maldonado M, Kapoor TM. Constitutive Mad1 targeting to kinetochores uncouples checkpoint signalling from chromosome biorientation. Nature Cell Biology. 2011;13:475–482. doi: 10.1038/ncb2223. [Elegant fusion study demonstrating the pivotal role of Mad1 kinetochore localization in the checkpoint.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brady DM, Hardwick KG. Complex formation between Mad1p, Bub1p and Bub3p is crucial for spindle checkpoint function. Current biology: CB. 2000;10:675–678. doi: 10.1016/s0960-9822(00)00515-7. [DOI] [PubMed] [Google Scholar]
- 78.London N, Biggins S. Mad1 kinetochore recruitment by Mps1-mediated phosphorylation of Bub1 signals the spindle checkpoint. Genes & Development. 2014;28:140–152. doi: 10.1101/gad.233700.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Moyle MW, et al. A Bub1-Mad1 interaction targets the Mad1-Mad2 complex to unattached kinetochores to initiate the spindle checkpoint. The Journal of Cell Biology. 2014;204(5):647–57. doi: 10.1083/jcb.201311015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim S, Sun H, Tomchick DR, Yu H, Luo X. Structure of human Mad1 C-terminal domain reveals its involvement in kinetochore targeting. Proceedings of the National Academy of Sciences. 2012;109:6549–6554. doi: 10.1073/pnas.1118210109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kops GJPL. ZW10 links mitotic checkpoint signaling to the structural kinetochore. The Journal of Cell Biology. 2005;169:49–60. doi: 10.1083/jcb.200411118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Varma D, et al. Spindle assembly checkpoint proteins are positioned close to core microtubule attachment sites at kinetochores. The Journal of Cell Biology. 2013;202:735–746. doi: 10.1083/jcb.201304197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kasuboski JM, et al. Zwint-1 is a novel Aurora B substrate required for the assembly of a dynein-binding platform on kinetochores. Molecular biology of the cell. 2011;22:3318–3330. doi: 10.1091/mbc.E11-03-0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yamamoto TG, Watanabe S, Essex A, Kitagawa R. SPDL-1 functions as a kinetochore receptor for MDF-1 in Caenorhabditis elegans. The Journal of Cell Biology. 2008;183:187–194. doi: 10.1083/jcb.200805185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jia L, Kim S, Yu H. Tracking spindle checkpoint signals from kinetochores to APC/C. Trends in Biochemical Sciences. 2013;38:302–311. doi: 10.1016/j.tibs.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 86.Brady DM HK. Complex formation between Mad1p, Bub1p and Bub3p is crucial for spindle checkpoint function. Current Biology. 2000;10:675–8. doi: 10.1016/s0960-9822(00)00515-7. [DOI] [PubMed] [Google Scholar]
- 87.Vink M, et al. In vitro FRAP identifies the minimal requirements for Mad2 kinetochore dynamics. Current Biology. 2006;16:755–766. doi: 10.1016/j.cub.2006.03.057. [DOI] [PubMed] [Google Scholar]
- 88.Yang M, et al. Insights into Mad2 Regulation in the spindle checkpoint revealed by the crystal structure of the symmetric Mad2 dimer. PLoS Biology. 2008;6 doi: 10.1371/journal.pbio.0060050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen R-H, Brady DM, Smith D, Murray AW, Hardwick KG. The spindle checkpoint of budding yeast depends on a tight complex between the Mad1 and Mad2 proteins. Molecular biology of the cell. 1999;10:2607–2618. doi: 10.1091/mbc.10.8.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Heinrich S, et al. Mad1 contribution to spindle assembly checkpoint signalling goes beyond presenting Mad2 at kinetochores. EMBO reports. 2014 doi: 10.1002/embr.201338114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kruse T, et al. A direct role of Mad1 in the spindle assembly checkpoint beyond Mad2 kinetochore recruitment. EMBO reports. 2014;15(3):282–90. doi: 10.1002/embr.201338101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ballister ER, Riegman M, Lampson MA. Recruitment of Mad1 to metaphase kinetochores is sufficient to reactivate the mitotic checkpoint. The Journal of Cell Biology. 2014;204:901–908. doi: 10.1083/jcb.201311113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Klebig C, Korinth D, Meraldi P. Bub1 regulates chromosome segregation in a kinetochore-independent manner. The Journal of Cell Biology. 2009;185:841–858. doi: 10.1083/jcb.200902128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kops GJPL, Shah JV. Connecting up and clearing out: how kinetochore attachment silences the spindle assembly checkpoint. Chromosoma. 2012;121:509–525. doi: 10.1007/s00412-012-0378-5. [DOI] [PubMed] [Google Scholar]
- 95.Jin F, Wang Y. The signaling network that silences the spindle assembly checkpoint upon the establishment of chromosome bipolar attachment. Proceedings of the National Academy of Sciences. 2013;110:21036–21041. doi: 10.1073/pnas.1307595111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang Y, Jin F, Higgins R, McKnight K. The current view for the silencing of the spindle assembly checkpoint. Cell Cycle. 2014;13:0–7. doi: 10.4161/cc.29027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Howell BJ. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. The Journal of Cell Biology. 2001;155:1159–1172. doi: 10.1083/jcb.200105093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Barisic M, Geley S. Spindly switch controls anaphase: Spindly and RZZ functions in chromosome attachment and mitotic checkpoint control. Cell Cycle. 2011;10:449–456. doi: 10.4161/cc.10.3.14759. [DOI] [PubMed] [Google Scholar]
- 99.Gassmann R, et al. A new mechanism controlling kinetochore-microtubule interactions revealed by comparison of two dynein-targeting components: SPDL-1 and the Rod/Zwilch/Zw10 complex. Genes & Development. 2008;22:2385–2399. doi: 10.1101/gad.1687508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gassmann R, et al. Removal of Spindly from microtubule-attached kinetochores controls spindle checkpoint silencing in human cells. Genes & Development. 2010;24:957–971. doi: 10.1101/gad.1886810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Matson DR, Demirel PB, Stukenberg PT, Burke DJ. A conserved role for COMA/CENP-H/I/N kinetochore proteins in the spindle checkpoint. Genes & Development. 2012;26:542–547. doi: 10.1101/gad.184184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Matson DR, Stukenberg PT. CENP-I and Aurora B act as a molecular switch that ties RZZ/Mad1 recruitment to kinetochore attachment status. The Journal of Cell Biology. 2014;205:541–554. doi: 10.1083/jcb.201307137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jelluma N, Dansen TB, Sliedrecht T, Kwiatkowski NP, Kops GJPL. Release of Mps1 from kinetochores is crucial for timely anaphase onset. The Journal of Cell Biology. 2010;191:281–290. doi: 10.1083/jcb.201003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cairo Lucas V., Ptak C, Wozniak Richard W. Mitosis-specific regulation of nuclear transport by the spindle assembly checkpoint protein Mad1p. Molecular Cell. 2012;49(1):109–20. doi: 10.1016/j.molcel.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 105.Pinsky BA, Nelson CR, Biggins S. Protein Phosphatase 1 regulates exit from the spindle checkpoint in budding yeast. Current Biology. 2009;19:1182–1187. doi: 10.1016/j.cub.2009.06.043. [Along with reference 106 showed that PP1 activity is essential for checkpoint silencing.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Vanoosthuyse V, Hardwick KG. A novel Protein Phosphatase 1-dependent spindle checkpoint silencing mechanism. Current Biology. 2009;19:1176–1181. doi: 10.1016/j.cub.2009.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rosenberg Jessica S., Cross Frederick R., Funabiki H. KNL1/Spc105 recruits PP1 to silence the spindle assembly checkpoint. Current Biology. 2011;21:942–947. doi: 10.1016/j.cub.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Meadows John C., et al. Spindle checkpoint silencing requires association of PP1 to both Spc7 and Kinesin-8 motors. Developmental Cell. 2011;20:739–750. doi: 10.1016/j.devcel.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wei R, Ngo B, Wu G, Lee W-H. Phosphorylation of the Ndc80 complex protein, HEC1, by Nek2 kinase modulates chromosome alignment and signaling of the spindle assembly checkpoint. Molecular Biology of the Cell. 2011;22:3584–3594. doi: 10.1091/mbc.E11-01-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liu D, et al. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. The Journal of Cell Biology. 2010;188:809–820. doi: 10.1083/jcb.201001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mirchenko L, Uhlmann F. Sli15/INCENP dephosphorylation prevents mitotic checkpoint reengagement due to loss of tension at anaphase onset. Current Biology. 2010;20:1396–1401. doi: 10.1016/j.cub.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Vázquez-Novelle María D., Mirchenko L, Uhlmann F, Petronczki M. The ‘anaphase problem’: how to disable the mitotic checkpoint when sisters split. Biochemical Society Transactions. 2010;38 doi: 10.1042/BST0381660. [DOI] [PubMed] [Google Scholar]
- 113.Palframan WJ. Anaphase inactivation of the spindle checkpoint. Science. 2006;313:680–684. doi: 10.1126/science.1127205. [DOI] [PubMed] [Google Scholar]
- 114.Hewitt L, et al. Sustained Mps1 activity is required in mitosis to recruit O-Mad2 to the Mad1-C-Mad2 core complex. The Journal of Cell Biology. 2010;190:25–34. doi: 10.1083/jcb.201002133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Santaguida S, Tighe A, D'Alise AM, Taylor SS, Musacchio A. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. The Journal of Cell Biology. 2010;190:73–87. doi: 10.1083/jcb.201001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Saurin AT, van der Waal MS, Medema RH, Lens SMA, Kops GJPL. Aurora B potentiates Mps1 activation to ensure rapid checkpoint establishment at the onset of mitosis. Nature Communications. 2011 doi: 10.1038/ncomms1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nijenhuis W, et al. A TPR domain-containing N-terminal module of MPS1 is required for its kinetochore localization by Aurora B. The Journal of Cell Biology. 2013;201:217–231. doi: 10.1083/jcb.201210033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhu T, et al. Phosphorylation of microtubule-binding protein Hec1 by mitotic kinase Aurora B specifies spindle checkpoint kinase Mps1 signaling at the kinetochore. Journal of Biological Chemistry. 2013;288:36149–36159. doi: 10.1074/jbc.M113.507970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Espeut J, Cheerambathur DK, Krenning L, Oegema K, Desai A. Microtubule binding by KNL-1 contributes to spindle checkpoint silencing at the kinetochore. The Journal of Cell Biology. 2012;196:469–482. doi: 10.1083/jcb.201111107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Qi W, Yu H. KEN-Box-dependent degradation of the Bub1 spindle checkpoint kinase by the Anaphase-promoting Complex/Cyclosome. Journal of Biological Chemistry. 2006;282:3672–3679. doi: 10.1074/jbc.M609376200. [DOI] [PubMed] [Google Scholar]
- 121.Rodriguez-Bravo V, et al. Nuclear pores protect genome integrity by assembling a premitotic and Mad1-dependent anaphase inhibitor. Cell. 2014;156:1017–1031. doi: 10.1016/j.cell.2014.01.010. [Identified nuclear pore-associated Mad1 as the source of the kinetochore-independent mitotic timer.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Maciejowski J, et al. Mps1 directs the assembly of Cdc20 inhibitory complexes during interphase and mitosis to control M phase timing and spindle checkpoint signaling. The Journal of Cell Biology. 2010;190:89–100. doi: 10.1083/jcb.201001050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Malureanu LA, et al. BubR1 N terminus acts as a soluble inhibitor of Cyclin B degradation by APC/CCdc20 in interphase. Developmental Cell. 2009;16:118–131. doi: 10.1016/j.devcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Meraldi P, Draviam VM, Sorger PK. Timing and checkpoints in the regulation of mitotic progression. Developmental cell. 2004;7:45–60. doi: 10.1016/j.devcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 125.Schweizer N, et al. Spindle assembly checkpoint robustness requires Tpr-mediated regulation of Mad1/Mad2 proteostasis. The Journal of Cell Biology. 2013;203:883–893. doi: 10.1083/jcb.201309076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lucas V Cairo CP, Wozniak Richard W. Dual personality of Mad1: Regulation of nuclear import by a spindle assembly checkpoint protein. Nucleus. 2013;4:367–73. doi: 10.4161/nucl.26573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kops GJPL, Weaver BAA, Cleveland DW. On the road to cancer: aneuploidy and the mitotic checkpoint. Nature Reviews Cancer. 2005;5:773–785. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- 128.Manchado E, Guillamot M, Malumbres M. Killing cells by targeting mitosis. Cell Death & Differentiation. 2012;19:369–377. doi: 10.1038/cdd.2011.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Heinrich S, et al. Determinants of robustness in spindle assembly checkpoint signalling. Nature Cell Biology. 2013;15:1328–1339. doi: 10.1038/ncb2864. [DOI] [PubMed] [Google Scholar]
- 130.Di Fiore B, Pines J. How cyclin A destruction escapes the spindle assembly checkpoint. The Journal of Cell Biology. 2010;190:501–509. doi: 10.1083/jcb.201001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Asbury CL, Tien JF, Davis TN. Kinetochores’ gripping feat: conformational wave or biased diffusion? Trends in Cell Biology. 2011;21:38–46. doi: 10.1016/j.tcb.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tanaka TU. Kinetochore–microtubule interactions: steps towards bi-orientation. The EMBO Journal. 2010;29:4070–4082. doi: 10.1038/emboj.2010.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lesage B, Qian J, Bollen M. Spindle checkpoint silencing: PP1 rips the balance. Current Biology. 2011;21:R898–R903. doi: 10.1016/j.cub.2011.08.063. [DOI] [PubMed] [Google Scholar]
- 134.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nature Reviews Molecular Cell Biology. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 135.Clute P, Pines J. Temporal and spatial control of cyclin B1 destruction in metaphase. Nature cell biology. 1999;1:82–87. doi: 10.1038/10049. [DOI] [PubMed] [Google Scholar]
- 136.Rieder CL, et al. Mitosis in vertebrate somatic cells with two spindles: implications for the metaphase/anaphase transition checkpoint and cleavage. Proceedings of the National Academy of Sciences. 1997;94:5107–5112. doi: 10.1073/pnas.94.10.5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Parry DH, Hickson GRX, O'Farrell PH. Cyclin B destruction triggers changes in kinetochore behavior essential for successful anaphase. Current biology. 2003;13:647–653. doi: 10.1016/s0960-9822(03)00242-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jeganathan KB, Malureanu L, van Deursen JM. The Rae1–Nup98 complex prevents aneuploidy by inhibiting securin degradation. Nature. 2005;438:1036–1039. doi: 10.1038/nature04221. [DOI] [PubMed] [Google Scholar]
- 139.Schuyler SC, Wu YF, Kuan VJW. The Mad1-Mad2 balancing act - a damaged spindle checkpoint in chromosome instability and cancer. Journal of Cell Science. 2012;125:4197–4206. doi: 10.1242/jcs.107037. [DOI] [PubMed] [Google Scholar]
- 140.Miniowitz-Shemtov S, et al. Role of phosphorylation of Cdc20 in p31comet-stimulated disassembly of the mitotic checkpoint complex. Proceedings of the National Academy of Sciences. 2012;109:8056–8060. doi: 10.1073/pnas.1204081109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Pinsky BA, Kung C, Shokat KM, Biggins S. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nature Cell Biology. 2006;8:78–83. doi: 10.1038/ncb1341. [DOI] [PubMed] [Google Scholar]
- 142.van der Waal MS, et al. Mps1 promotes rapid centromere accumulation of Aurora B. EMBO reports. 2012;13:847–854. doi: 10.1038/embor.2012.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Liu X, Winey M. The MPS1 family of protein kinases. Annual Review of Biochemistry. 2012;81:561–585. doi: 10.1146/annurev-biochem-061611-090435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hardwick KG WE, Luca FC, Winey M, Murray AW. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. 1996;273:953–6. doi: 10.1126/science.273.5277.953. [Demonstrated that Mps1 kinase activity is sufficient for checkpoint activation, indicating it is the pivotal kinase in the signaling cascade.] [DOI] [PubMed] [Google Scholar]
- 145.Maure J-F, Kitamura E, Tanaka TU. Mps1 kinase promotes sister-kinetochore bi-orientation by a tension-dependent mechanism. Current Biology. 2007;17:2175–2182. doi: 10.1016/j.cub.2007.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tipton AR, et al. Monopolar Spindle 1 (MPS1) kinase promotes production of closed MAD2 (C-MAD2) conformer and assembly of the mitotic checkpoint complex. Journal of Biological Chemistry. 2013;288:35149–35158. doi: 10.1074/jbc.M113.522375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zich J, et al. Kinase activity of fission yeast Mph1 is required for Mad2 and Mad3 to stably bind the Anaphase Promoting Complex. Current Biology. 2012;22:296–301. doi: 10.1016/j.cub.2011.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tipton AR, et al. BUBR1 and Closed MAD2 (C-MAD2) interact directly to assemble a functional mitotic checkpoint complex. Journal of Biological Chemistry. 2011;286:21173–21179. doi: 10.1074/jbc.M111.238543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sironi L MM, Knapp S, De Antoni A, Jeang KT, Musacchio A. Crystal structure of the tetrameric Mad1-Mad2 core complex: implications of a ‘safety belt’ binding mechanism for the spindle checkpoint. EMBO J. 2002;21:2496–506. doi: 10.1093/emboj/21.10.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Petsalaki E, Zachos G. Chk2 prevents mitotic exit when the majority of kinetochores are unattached. J Cell Biol. 2014;205(3):339–56. doi: 10.1083/jcb.201310071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yeh CW, Yu ZC, Chen PH, Cheng YC, Shieh SY. Phosphorylation at threonine 288 by cell cycle checkpoint kinase 2 (CHK2) controls human monopolar spindle 1 (Mps1) kinetochore localization. Journal of Biological Chemistry. 2014;289(22):15319–27. doi: 10.1074/jbc.M114.552273. [DOI] [PMC free article] [PubMed] [Google Scholar]