Abstract
Tuberculosis has reemerged as a serious threat to human health because of the increasing prevalence of drug-resistant strains and synergetic infection with HIV, prompting an urgent need for new and more efficient treatments. The PhoP–PhoR two-component system of Mycobacterium tuberculosis plays an important role in the virulence of the pathogen and thus represents a potential drug target. To study the mechanism of gene transcription regulation by response regulator PhoP, we identified a high-affinity DNA sequence for PhoP binding using systematic evolution of ligands by exponential enrichment. The sequence contains a direct repeat of two 7 bp motifs separated by a 4 bp spacer, TCACAGC(N4)TCACAGC. The specificity of the direct-repeat sequence for PhoP binding was confirmed by isothermal titration calorimetry and electrophoretic mobility shift assays. PhoP binds to the direct repeat as a dimer in a highly cooperative manner. We found many genes previously identified to be regulated by PhoP that contain the direct-repeat motif in their promoter sequences. Synthetic DNA fragments at the putative promoter-binding sites bind PhoP with variable affinity, which is related to the number of mismatches in the 7 bp motifs, the positions of the mismatches, and the spacer and flanking sequences. Phosphorylation of PhoP increases the affinity but does not change the specificity of DNA binding. Overall, our results confirm the direct-repeat sequence as the consensus motif for PhoP binding and thus pave the way for identification of PhoP directly regulated genes in different mycobacterial genomes.
Mycobacterium tuberculosis (MTB), the etiologic agent of tuberculosis, is one of the leading causes of death worldwide among pathogens and is becoming a serious threat to public health because of the increasing emergence of drug-resistant strains and synergetic co-infection with HIV.1 The success of MTB as a pathogen relies on its ability to adapt to changing environmental conditions within the host through signal transduction systems, including two-component systems (TCS). TCS are major signaling systems in bacteria; they typically consist of a histidine kinase (HK) that senses external environmental signals and a response regulator (RR) that triggers the cellular response after being activated by its cognate HK.2
The MTB genome encodes 11 TCS,3 of which the PhoPR TCS plays a major role in virulence,4 although the signals it senses are still unknown. The phoPR knockout strains of MTB have a severe attenuation of virulence, and two studies comparing transcriptomes of phoP knockout strains to their corresponding wild-type parents have identified more 170 genes whose expression is affected by PhoP.5,6 The phoP mutant lacks complex mycobacterial lipids implicated in MTB virulence, including sulfolipids, polyacyltrehaloses, and diacyltrehaloses.6,7 Furthermore, a point mutation in phoP contributes to the avirulent phenotype of the MTB H37Ra strain, by preventing secretion of the ESAT-6 antigen, an important virulence factor and antigenic component of MTB.8−10 The important role of PhoPR in virulence makes this TCS an attractive target for developing anti-TB drugs11 and the phoPR-inactivated MTB strains ideal candidates for new TB vaccine development.12−14
The MTB PhoP protein belongs to the OmpR/PhoB subfamily, the largest of the response regulators.15 PhoP consists of two distinct domains: an N-terminal receiver domain with a conserved phosphorylation site that receives a phosphate group from the cognate HK PhoR and a C-terminal effector domain that harbors a winged helix–turn–helix DNA-binding motif.16,17 The effector domain binds to specific DNA sequences of the target promoters and interacts with the cellular transcription machinery. Most studies of the members of the OmpR/PhoB subfamily indicate that these RRs bind gene promoter DNA as dimers on direct-repeat sequences. The DNA sequence motif of the binding sites for PhoP from Streptomyces coelicolor is GTTCACC(N4)GTTCACC.18 The sequence of the pho box DNA for Escherichia coli PhoB binding is CTGTCAT(A/T)4CTGTCAT.19 The consensus sequence for PhoP of E. coli and Salmonella enterica is TGTTTA(N5)TGTTTA.20,21 Phosphorylation of PhoB from E. coli promotes dimerization, which enhances DNA binding.22,23 Phosphorylation of OmpR enhances its dimerization, and this dimerization enhancement is the energetic driving force for phosphorylation-mediated regulation of OmpR–DNA binding.24 However, KdpE, a member of the OmpR/PhoB family, binds independently to the half-sites of the target DNA sequences with equal affinity and no discernible cooperativity.25 The mechanism for the cooperativity in dimeric binding to DNA, or the lack of cooperativity in the case of KdpE, is currently unknown.
Despite an extensive number of publications about MTB PhoP and its DNA binding, the consensus DNA sequence and the mechanism of sequence recognition remained obscure, thus preventing identification of direct targets of PhoP. Sarkar and colleagues26−31 identified a direct repeat of two 9 bp motifs in the promoters of phoP (GGCAGACTGTTAGCAGACTACTGGCAACGAGC), pks2 (AGAACTAAAGAGCCACCAAAGACACAGCTACAT), and msl3 (also known as pks3) (CTGGTAGCGGCATGGCAACGGCCTGTGA), which they named DR1 and DR2 (underlined bases). The two motifs, DR1 and DR2, of the same gene promoter are somewhat similar, but they bear little resemblance among different gene promoters. Moreover, the direct-repeat motifs cannot be recognized in most of other gene promoters that bind PhoP. Cimino et al.32 studied the promoters of msl3, pks2, lipF, and fadD21, and they added a new DR3 located a variable distance from DR1 and DR2, which includes the same problem of inconsistency. Recently, two independent studies identified partial sequence motifs for PhoP binding in vivo, using results from chromatin immunoprecipitation sequencing (ChIP-seq). Solans et al.33 identified the motif as (C/T)(A/T)CAG(C/G)NNN(T/C)(T/A)CACAG, and Galagan et al.34 identified the motif as CTGNGNNNNNGCTG. Given the importance of PhoP and its target genes to MTB virulence, it is essential to confirm definitively the PhoP DNA-binding sequence.
In this study, we identified the PhoP-binding consensus sequence as a direct repeat of a 7 bp motif separated by a 4 bp spacer by using a method of systematic evolution of ligands by exponential enrichment (SELEX). We extended the search of the PhoP targets against the whole MTB genome with the consensus sequence. The direct interactions between PhoP and its identified target promoter sequences were confirmed by using isothermal titration calorimetry (ITC) and an electrophoretic mobility shift assay (EMSA). Furthermore, gel filtration chromatography, analytical ultracentrifugation (AUC), and ITC analyses showed that PhoP binds its target promoter sequences as a dimer in a cooperative manner.
Experimental Procedures
Protein Expression and Purification
Protein expression and purification were conducted as described previously.16 The phoP gene was cloned into a modified plasmid pET28a to generate the pET28-phoP plasmid, which encodes the PhoP protein with an N-terminal His tag that can be cleaved by the tobacco etch virus (TEV) protease. Plasmid pET28-phoP was transformed into E. coli strain BL21(DE3), and protein expression was induced by the addition of IPTG. For SELEX experiments (see below), the His tag was not removed; for all other experiments, the His tag was cleaved by the TEV protease, and the tag-free PhoP was separated from the His tag, uncleaved protein, and the TEV protease (His-tagged) by being passed through a His-Trap column (GE Life Sciences). All PhoP samples, with or without a His tag, were further purified and buffer exchanged with a Superdex 200 column (GE Life Sciences) for downstream applications.
Identification of the High-Affinity PhoP-Binding Sequence by SELEX
A random pool of oligonucleotides 5′-GGTGCAGGCATATGAAAG(N25)CTGGACCATATGCTCCAG-3′, where N25 represents 25 randomized nucleotides, was synthesized by equimolar incorporation of A, G, C, and T at each “N” position (Integrated DNA Technologies). The two sets of 18 nucleotides flanking the 25-nucleotide random core were designed for amplification by PCR. The double-stranded random DNA library was generated by a primer extension reaction, in which ∼20 μg of the random oligonucleotides was mixed with the reverse PCR primer complementary to the last 18 bases, T4 DNA polymerase (New England Biolabs), and dNTPs in a final volume of 50 μL. The reaction mixture was incubated at 37 °C for 30 min. The quality of the double-stranded random DNA was examined by agarose gel electrophoresis. To conduct the SELEX experiments, 10 μg of purified His-PhoP was bound to ∼10 μL of Ni-NTA Sepharose (Qiagen). This PhoP-Ni-NTA resin was washed twice with a binding buffer [20 mM Hepes (pH 7.5), 150 mM NaCl, 5 mM MgCl2, and 5% glycerol] and then resuspended in 200 μL of a binding buffer containing 50 μg/mL herring sperm DNA, 100 μg/mL poly(dI-dC), and 0.1 mg/mL BSA. The mixture was incubated for 30 min at room temperature. The primer extension product (50 μL) were then added and incubated for 1 h while being gently shaken. The resin was washed three times with 500 μL of a binding buffer and once with a binding buffer containing additional NaCl (concentration of 200 mM). The protein–DNA complex was eluted with 20 μL of elution buffer [25 mM phosphate (pH 7.4), 250 mM NaCl, and 300 mM imidazole]. The eluted DNA was amplified by 15 cycles of PCR with Taq DNA polymerase (Genscript, Piscataway, NJ). The PCR product was purified from a 6% native polyacrylamide gel with the QIAEX II gel extraction kit (QIAGEN). The purified PCR product was used in the second round of SELEX. After three or more serial selection rounds, the DNA was ligated into a TOPO vector using the TOPO TA cloning kit (Life Technologies) and subjected to DNA sequencing.
Electrophoretic Mobility Shift Assays
Double-stranded DNA fragments were prepared by mixing equimolar amounts of two complementary oligonucleotides in 10 mM Tris (pH 8.0) and 50 mM NaCl, heating the mixture at 90 °C for 10 min, and slowly cooling it to room temperature. The duplex DNA was purified from a 6% native polyacrylamide gel using the QIAEX II gel extraction kit. Purified DNA fragments were labeled with the biotin DNA labeling kit (Pierce). EMSA experiments were performed in a total volume of 10 μL containing 20 mM Hepes (pH 7.5), 50 mM NaCl, 5 mM MgCl2, 5% glycerol, 1 μg of poly(dI-dC), 0.12–0.15 μM labeled DNA, and 0.72–3.6 μM PhoP protein. The reaction mixtures were incubated at room temperature for 20 min and then loaded onto a 6% DNA retardation gel (Invitrogen). The gel was run at 100 V in 0.5× TBE buffer at 4 °C. The DNA was transferred to a nylon membrane by electroblotting and cross-linked to the membrane using a Stratalinker UV cross-linker on the autocrosslink setting. The blot was developed using the Pierce chemiluminescent nucleic acid detection kit.
To obtain phosphorylated PhoP, ∼18 μM protein was incubated with 50 mM acetyl phosphate (AcP) at room temperature in 500 μL of buffer containing 20 mM Hepes (pH 7.5), 100 mM NaCl, and 5 mM MgCl2. At certain time intervals, samples were taken, mixed with SDS sample buffer, and kept on ice. Samples were resolved on a 10% polyacrylamide gel containing 50 μM Phos-tag acrylamide35 to check the level of phosphorylation. The phosphorylated PhoP sample was used for an EMSA following the same procedure described above.
ITC Measurements
ITC experiments were conducted at 25 °C with a MicroCal iTC200 system in a buffer containing 20 mM Hepes (pH 7.5), 100 mM NaCl, and 5 mM MgCl2. The sample cell was stirred at 1000 rpm. The protein (10–20 μM) in the sample cell was titrated with 50–100 μM synthetic DNA duplex in the injection syringe. Titration was initiated by one 0.4 μL injection followed by 18 injections of 2 μL spaced by 120 s intervals. The data were analyzed using Origin 7.0 and fit with a one-set-of-sites binding model to obtain values of the stoichiometry (N), enthalpy change (ΔH), and association constant (Ka).
Size-Exclusion Chromatography
PhoP was mixed with double-stranded DNA fragments in a binding buffer [20 mM Hepes (pH 7.5), 100 mM NaCl, and 5 mM MgCl2] at room temperature for 20 min. The protein–DNA complexes were loaded onto a Superdex 200 HR 10/30 column (GE Life Sciences) equilibrated with the binding buffer and eluted at room temperature at a flow rate of 0.5 mL/min.
Analytical Ultracentrifugation
The AUC sedimentation velocity (SV) experiments were conducted in a Beckman Optima XL-A analytical ultracentrifuge using an An60Ti rotor and Epon charcoal standard double-sector centerpieces (12 mm optical path length). Samples containing DNA, protein, or the DNA–protein complex in a binding buffer [20 mM Hepes (pH 7.5), 100 mM NaCl, and 5 mM MgCl2] were centrifuged at 20 °C and 45000 rpm. Absorbance scans were taken at 260 nm for DNA alone and at 280 nm for protein and protein–DNA samples in continuous mode. SEDNTERP36 was used to calculate the buffer viscosity (η), buffer density (ρ), and protein partial specific volume values at 20 °C. The GC content for the DNA used in these experiments was ∼40%, and the partial specific volume was calculated to be 0.59 cm3 g–1. Sedimentation coefficient distributions were calculated with data from 300 SV scans by using SEDFIT.37 A resolution setting of 200 and a confidence interval of 0.8 were used. For phosphorylation samples, 26 μM PhoP was mixed with 50 mM AcP in a buffer identical to that for the PhoP alone sample, immediately prior to loading the sample into the AUC cell. The rotor with the sample was cooled and incubated at 20 °C for 2 h before centrifugation was started.
Results
PhoP Binds with High Affinity to DNA Sequences Containing a Direct Repeat of Two 7 bp Motifs Separated by a 4 bp Spacer
We used SELEX to identify high-affinity DNA sequences for PhoP binding. His-PhoP was immobilized on Ni-NTA Sepharose to enrich high-affinity DNA sequences from a pool of 25 bp randomized double-stranded DNA. After three rounds of SELEX, a single sequence, ctggagcatatggtccagTTAGTACCTCACAGCACTTTCAGAGctttcatatgcctgcacc (sequences of the flanking PCR priming sites are shown in lowercase letters), dominated the sequences obtained (Figure 1A). There were two identical 7 bp motifs (underlined residues) in a direct repeat with a 4 bp spacer. The last base of the second motif was from the 3′-PCR priming site sequence. This was the case for the majority of sequences derived from SELEX experiments, suggesting that the TTT sequence immediately following the second motif is likely to be favorable for PhoP binding. Some SELEX-derived sequences had mismatches in the direct repeat, but the motifs were easily recognizable. Seven sequences from the third round of SELEX contained only one motif or no recognizable motifs (each had a single occurrence); these sequences disappeared in later rounds of selection. The dominant sequence was further enriched with more cycles of SELEX (Figure 1A); at the seventh round, 12 of 16 clones sequenced had the identical sequence of the dominant picked up at the third round, suggesting that this sequence exhibits the highest affinity for PhoP among members of the selected DNA pool.
Figure 1.

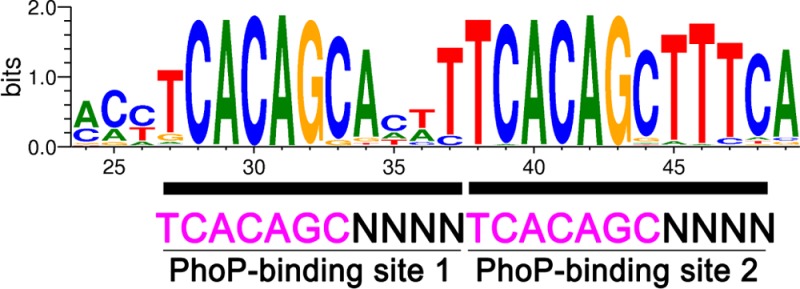
DNA sequences for PhoP binding determined by the SELEX experiments. (A) Sequence alignment of selected DNAs derived from the third, fourth, and seventh rounds of SELEX. The two direct-repeat motifs are colored red, with mismatches from the TCACAGC motif colored blue. The adjacent primer sequences (partial) are shown in lowercase letters. The right column shows the number of occurrences of each sequence. Seven sequences obtained from the third round of SELEX with only one recognizable motif or none are not shown. (B) Graphical representation of the consensus sequence generated with WebLogo. All SELEX-derived sequences shown in panel A, including adjacent primer sequences, were analyzed using WebLogo. The degree of conservation is indicated by the height of the letters. The direct repeat containing two 7 bp motifs separated by a 4 bp spacer can be easily recognized.
A figure generated by WebLogo38 from all SELEX-derived sequences containing the direct repeat showed clearly a direct repeat of the 7 bp motif, TCACAGC, with a 4 bp spacer (Figure 1B). In the first motif, the middle five bases were the best conserved, while the first base T was the least conserved. The last base position of the first motif was almost exclusively a cytosine, with a few sequences having a guanine. In the second motif, the first six bases were almost exclusively TCACAG, while the last base was less conserved with C strongly preferred. The sequences in the spacer are also strongly conserved, with an A and a T strongly preferred at the first and last positions, respectively. The spacer of most sequences was AT-rich (Figure 1A). The sequence immediately following the second motif had a high degree of conservation of TTT, which was higher than that of the last base C of the second motif. This could be partially biased because in most sequences these bases are not from the randomized sequence but derived from the PCR primer. However, the fact that the majority of selected sequences incorporated these bases right after the second motif suggests that the TTT sequence at this position is favorable for PhoP binding. In contrast, the sequence upstream of the 5′-end of the first motif is not well-conserved.
The Minimal Sequence for Optimal PhoP Binding Includes the Direct Repeat and Flanking Bases
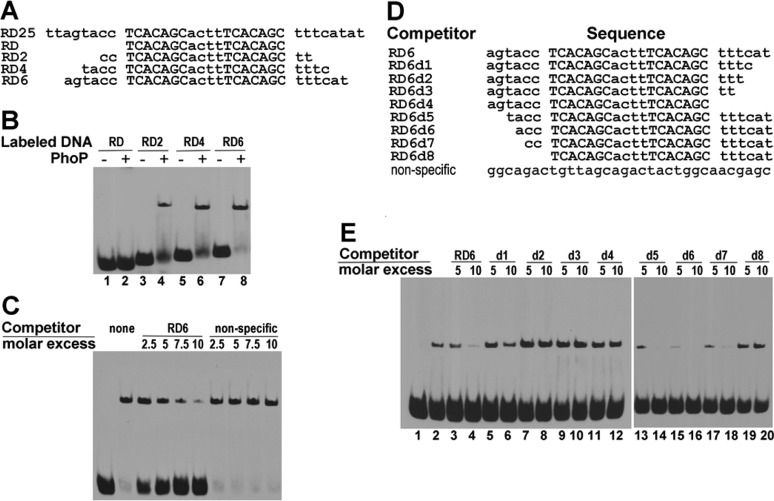
To confirm the role of the repeat motifs in binding specificity, we designed a series of duplex DNA sequences based on the dominant sequence identified from SELEX experiments and assayed their binding affinity for PhoP by an EMSA (Figure 2A). The shortest sequence (RD) contains only the direct repeat, while RD2, RD4, and RD6 contain two, four, and six flanking base pairs, respectively, at each end of the direct repeat. The sequence RD did not give a shifted band in the presence of PhoP under the experimental conditions, suggesting that the flanking sequences beyond the direct repeat are necessary for PhoP binding (lane 2 of Figure 2B). Sequences RD2, RD4, and RD6, to different degrees, were capable of forming a shifted band (Figure 2B), indicating that these sequences are able to form a stable PhoP–DNA complex. The relative intensity of the shifted bands suggested that, under the assay condition, RD6 has an affinity for PhoP higher than those of the rest of the sequences. Further increases in the length of the extension did not increase the binding affinity (data not shown).
Figure 2.
Characterization of interactions of PhoP with selected DNA by electrophoretic mobility shift assays. (A) Sequences of selected DNA used in the EMSA. Sequence RD25 is the dominant sequence derived from the SELEX assays. RD consists of only the two motifs; RD2, RD4, and RD6 contain two, four, and six flanking nucleotides at both ends of the motifs, respectively. (B) Identification of the minimal length of DNA required for binding PhoP. Double-stranded DNA shown in panel A was labeled at 3′-end with biotin and incubated without (lanes 1, 3, 5, and 7) or with 3.5 μM recombinant PhoP protein (lanes 2, 4, 6, and 8) at room temperature for 20 min. The EMSA was performed as described in Experimental Procedures. (C) Competition EMSA showing specificity of binding of RD6 to PhoP. Labeled RD6 was incubated without (lane 1) or with 0.7 μM PhoP (lanes 2–10). The presence of excess unlabeled RD6 reduced the retarded band, while an excess of a nonspecific DNA (sequence listed in panel D) did not have any effect. (D) DNA sequences used in the competition EMSAs. (E) Competition EMSA with various RD6 deletions. Biotin-labeled RD6 DNA was incubated without PhoP (lane 1) or with 0.7 μM PhoP protein (lane 2) and in the presence of 5- and 10-fold molar excesses of unlabeled wild-type RD6 (lanes 3 and 4, respectively) and RD6 deletions d1–d8 (lanes 5–20, respectively).
The 5′- and 3′-end flanking sequences are likely to have different roles in PhoP binding because they have different levels of conservation in the SELEX-derived sequences (Figure 1B). To define the minimal flanking sequence requirement at either end for optimal PhoP binding, a series of progressive deletions from either end of RD6 (Figure 2D) were used as competitors in the EMSA with labeled RD6. As shown in Figure 2E, the PhoP-retarded band was attenuated by the addition of excess unlabeled RD6 (lanes 3 and 4 vs lane 2). Removal of 2 bp from the 6 bp extension at the 3′-end (RD6d1) significantly reduced the binding affinity (Figure 2E, lanes 5 and 6). With a 10-fold molar excess of RD6d1, ∼7% of labeled RD6 DNA was in the retarded band, compared to <0.5% of labeled DNA retarded with a 10-fold excess of unlabeled RD6 (lane 4). Sequences with further deletions from the 3′-end (RD6d2, RD6d3, and RD6d4) could not effectively compete for binding with labeled RD6 (lanes 7–12). Deletion of up to 4 bp from the 5′-end of RD6, however, preserved efficient competition with RD6 (in Figure 2E, compare lanes 13–18 with lanes 3 and 4). Deletion of all flanking bases at the 5′-end (RD6d8) completely abolished the ability to compete (Figure 2E, lanes 19 and 20). As a negative control, a 32 bp DNA fragment bearing no direct-repeat motif was not able to compete with labeled RD6 (Figure 2C), confirming the specificity of the PhoP–RD6 binding interaction.
PhoP Specifically Binds the whiB6 Gene Promoter Region Containing the Direct-Repeat Motifs
We described previously that PhoP binds to the promoter region of whiB6,17 the gene encoding a transcription regulator WhiB-like protein that controls expression of genes including the ESX-1 secretion system.39 Binding of PhoP to the whiB6 promoter was confirmed in vivo by a recent ChIP-seq study,33 and PhoP upregulates transcription of whiB6 in clinical MTB strains but not in the common laboratory strain H37Rv.39
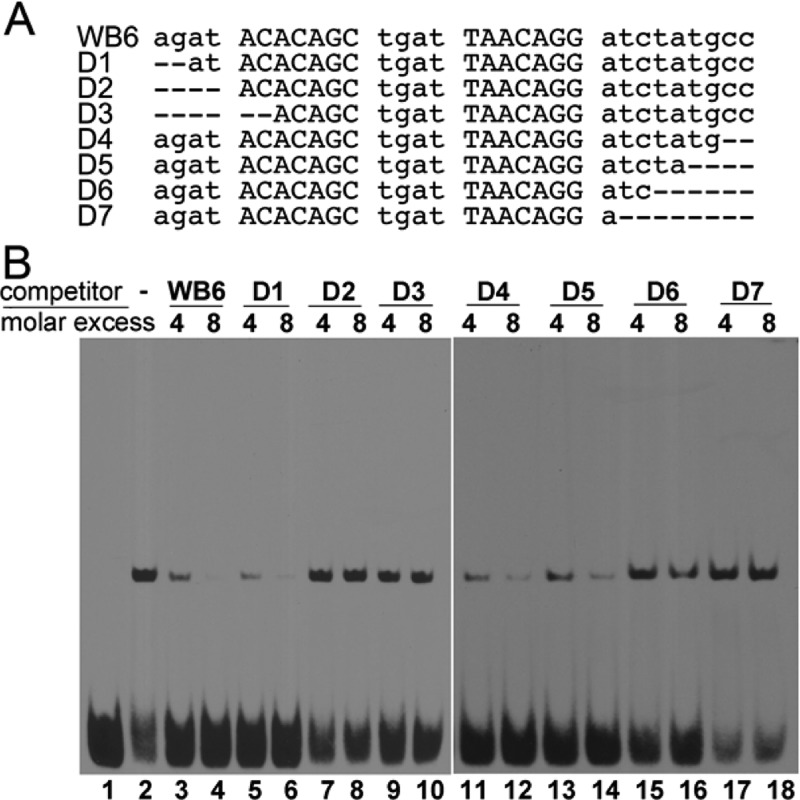
By trimming and walking along the whiB6 promoter sequence with an EMSA, we identified a 31 bp DNA fragment of the whiB6 promoter (WB6), agatACACAGCtgatTAACAGGatctatgcc, which bound PhoP with high affinity. The SELEX-derived direct repeat described above can be recognized in WB6 with three mismatches (motifs in uppercase letters with mismatches underlined). Unlabeled WB6 could competitively inhibit the binding of PhoP to labeled WB6 (Figure 3B, lanes 3 and 4 vs lane 2). Progressive deletions at the 5′- or 3′-end of WB6 showed effects on PhoP binding similar to those of RD6 (Figure 3). Deletions of two nucleotides at the 5′-end and four nucleotides at the 3′-end had no effects on the competition for the binding of PhoP with labeled WB6 (Figure 3B, D1, D4, and D5). Further deletions at either end reduced the competitiveness for PhoP binding (D2, D3, D6, and D7). These results indicate that 2 bp beyond the 5′-end of the first motif and 5 bp beyond the 3′-end of the second motif are required for optimal PhoP binding.
Figure 3.
Analysis of binding of PhoP to the whiB6 promoter sequence containing the direct repeat. (A) Sequence of WB6, the PhoP-binding site on the whiB6 promoter, and its truncated mutants used as competitors in the EMSA. (B) Competition EMSA studies with various truncated WB6 sequences shown in panel A. Biotin-labeled WB6 DNA was incubated without PhoP (lane 1) or with 1 μM PhoP protein in the absence (lane 2) or presence of 4- and 8-fold molar excesses of unlabeled wild-type WB6 (lanes 3 and 4, respectively) and its mutants D1–D7 (lanes 5–18).
PhoP Binds to the Direct Repeat as a Dimer in a Highly Cooperative Manner
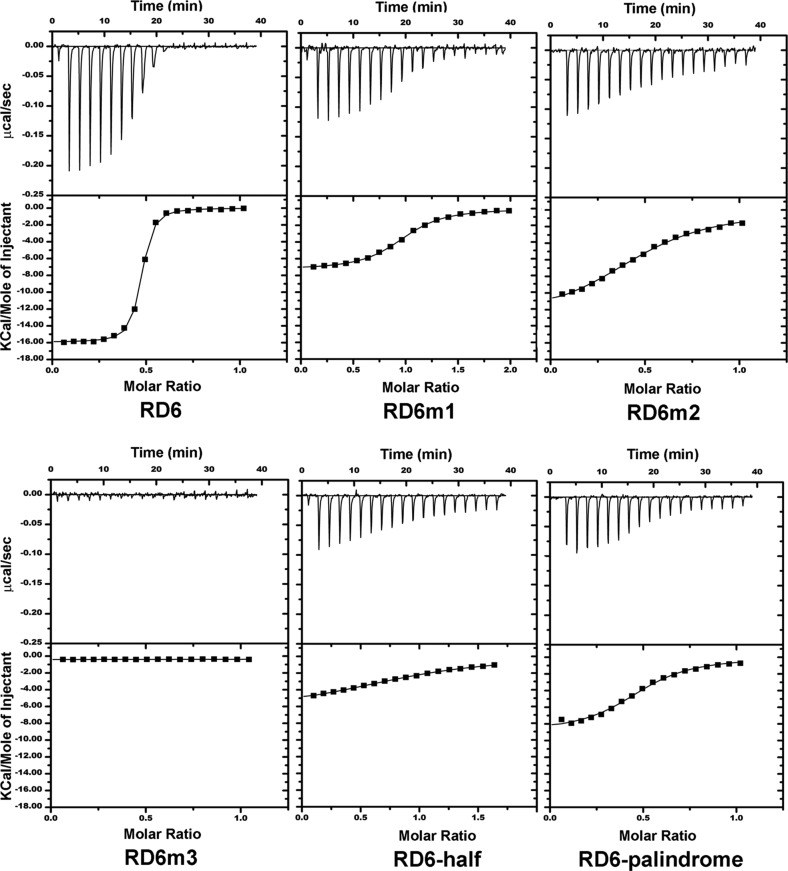
To further characterize the interaction of PhoP with DNA, we conducted ITC analysis of the variations of the RD6 DNA sequence listed in Table 1 for PhoP binding. Figure 4 shows representative ITC isotherms of binding of PhoP to DNA sequences. The titration data of RD6 could be best fit to a one-set-of-sites binding model that gave a Kd of ∼10 nM and a stoichiometry (N, ratio of RD6 to PhoP) of ∼0.5 (Table 1). A negative enthalpy change and a small negative entropy change exist, suggesting that the binding is enthalpy-driven. RD6-half, which contains the first half of the direct repeat and the spacer sequence ACTT, has an affinity for PhoP binding ∼500-fold lower than that of RD6 and a binding stoichiometry N of 1. The released binding heat was less than half of that of the perfect direct repeat.
Table 1. Thermodynamic Parameters Derived via ITC of Various DNA Sequences with the PhoP Protein.
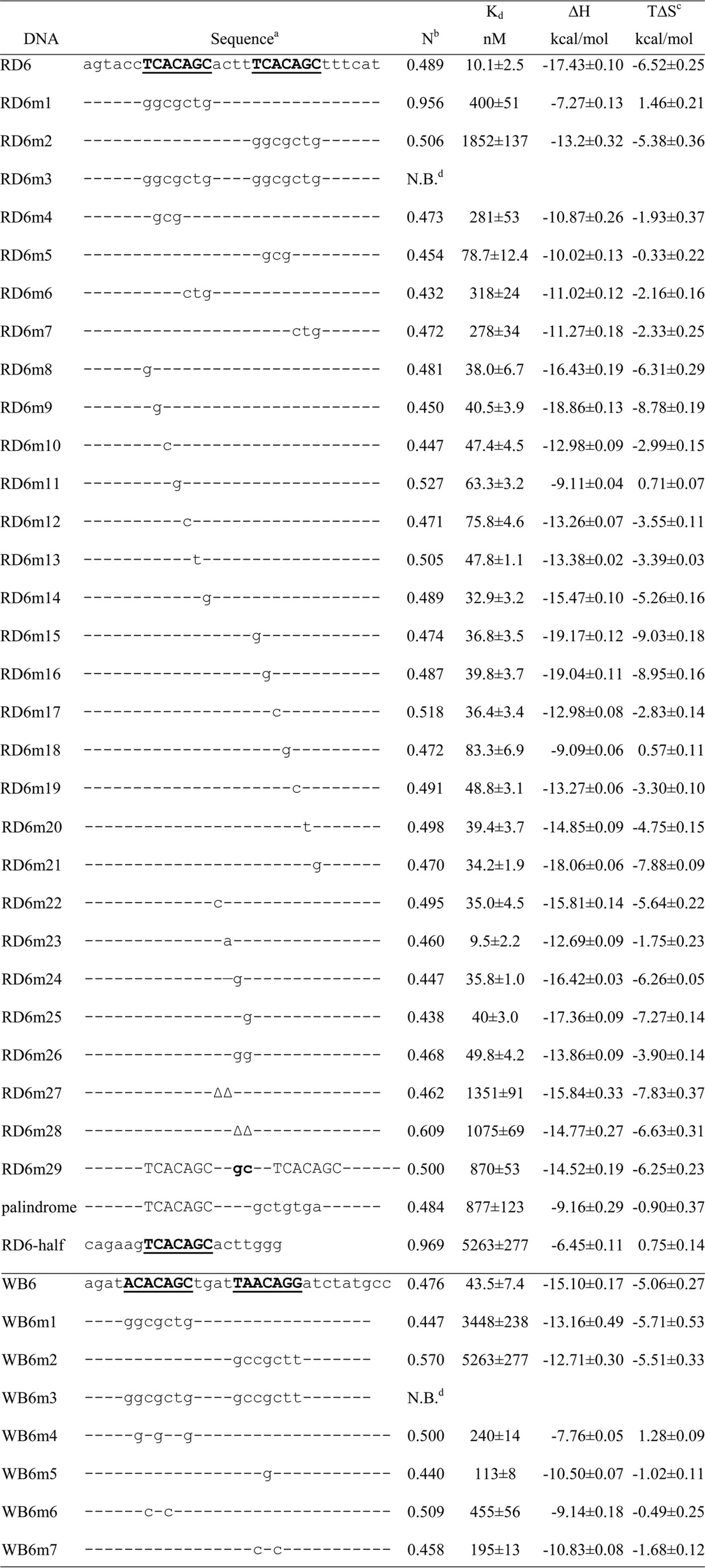
DNA sequences are aligned, except for RD6m29, in which insertion of two nucleotides between nucleotides c and t in the spacer causes a shift of the second motif. A hyphen represents an invariant base, and a delta (Δ) represents a deletion of the nucleotide at that position.
N is the stoichiometry, referring to the copy number of DNA per PhoP molecule.
Values of TΔS were calculated from the values of Kd and ΔH that were obtained from fitting the ITC titration data with Origin.
N.B. stands for no binding under the assay conditions, with a binding dissociation constant of >10 μM.
Figure 4.
Representative ITC isotherms for binding of PhoP to the RD6 DNA and its mutants. The isotherms could be best fit to the one-set-of-sites binding model. The top panels show raw data after baseline adjustments expressed as changes in thermal power with respect to time over the periods of titration. The bottom panels show the integrated heat of each titration. The DNA sequences used in ITC are listed in Table 1. All data are shown at the same scale for easy comparison.
Disruption of one motif of the direct repeat dramatically reduced the binding affinity for PhoP. When the first motif, T1CACAGC7 (superscripts indicate the positions of the base in the direct repeat), of RD6 was substituted with GGCGCTG (RD6m1 in Table 1), the binding affinity was reduced ∼40-fold with an N of ∼1. Mutation of the second motif T12CACAGC18 to the same random sequence (RD6m2) resulted in a greater reduction of the fitted Kd value. In this case, however, N was close to 0.5, suggesting that two copies of PhoP could bind to the DNA duplex. When both motifs were mutated (RD6m3), no binding was observed, demonstrating that the motifs are indeed the recognition sequence for PhoP. Replacing the second motif T12CACAGC18 with GCTGTGA to generate a palindromic sequence gave results similar to those of RD6m2 (palindrome in Table 1).
Analysis of the sequence derived from the whiB6 promoter (WB6) by ITC showed the requirements of two motifs for PhoP binding were the same as those of RD6. Binding of WB6 to PhoP gave a highly negative enthalpy change (Table 1). The ITC titration data could be best fit to a one-set-of-sites binding model that gave a Kd of ∼40 nM and a stoichiometry of 0.5. Disruption of either motif (WB6m1 and WB6m2) resulted in a dramatic decrease in binding affinity, and disruption of both motifs resulted in a complete loss of PhoP binding (WB6m3).
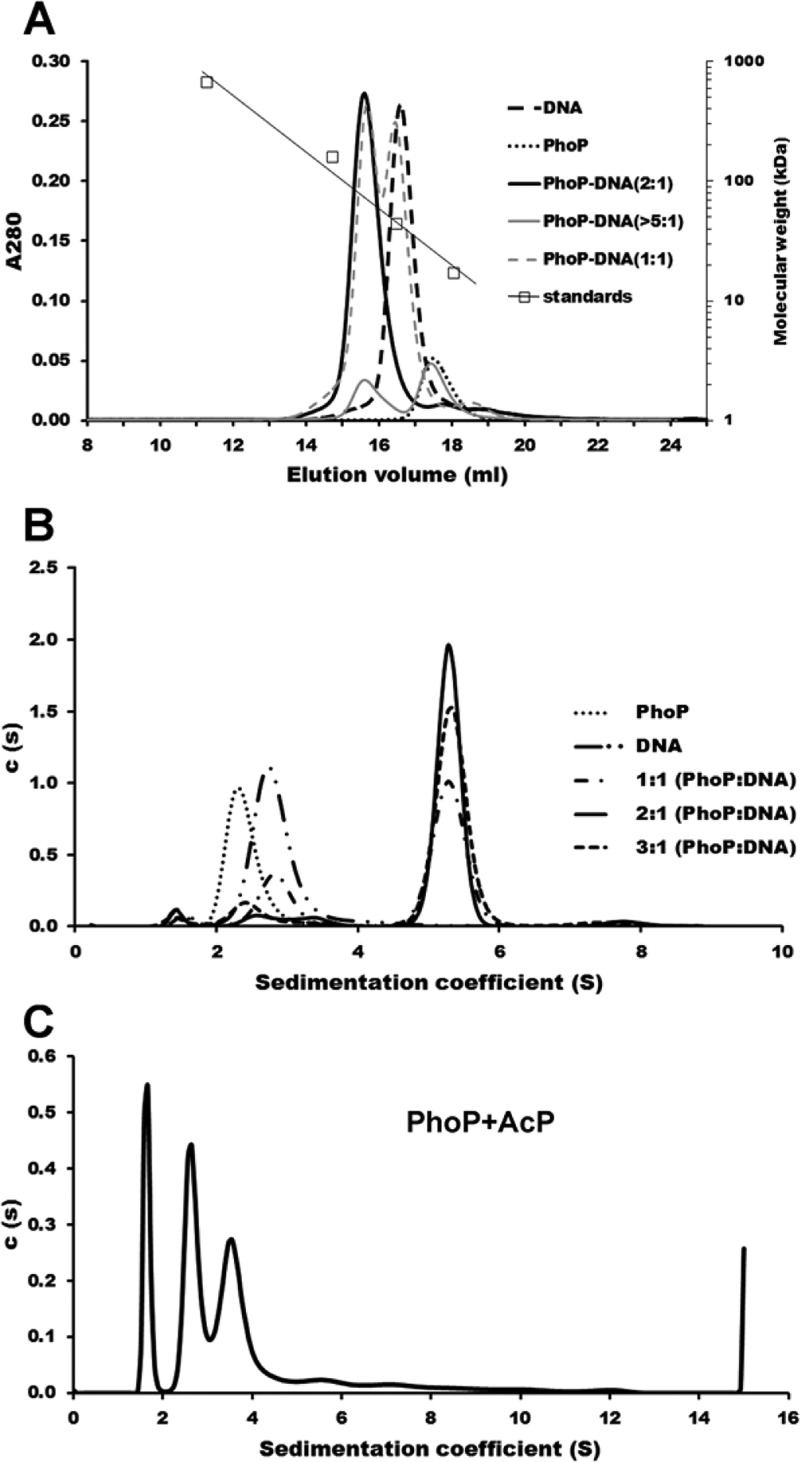
The results described above suggested that two PhoP molecules bind to RD6 in a cooperative manner. The binding stoichiometry of 0.5 (two copies of PhoP binding to one copy of the DNA duplex) was confirmed by size-exclusion chromatography experiments, in which PhoP and WB6 DNA formed a stable complex and co-eluted in a single peak (Figure 5A). PhoP was eluted from the size-exclusion column in a peak with an apparent molecular mass of ∼26.2 kDa (monomer molecular mass based on the sequence of 27.8 kDa), suggesting that it is a monomer in solution. The WB6 DNA duplex was eluted with an apparent molecular mass of 42.7 kDa in contrast to its actual molecular mass of 19.0 kDa, consistent with the long rod shape of double helix DNA. When the PhoP protein was mixed with WB6 at a molar ratio of 2:1, a single peak was detected at an apparent molecular mass of 73.6 kDa, consistent with a complex of two copies of PhoP binding to one copy of the WB6 duplex. This result also suggested that the PhoP–WB6 complex had a compact globular shape. To ascertain that only the 2:1 PhoP–WB6 complex exists, we ran the size-exclusion column with an excess of either DNA or PhoP in the mixture. A peak at the 2:1 complex was observed in all cases, with an additional peak corresponding to either unbound PhoP or WB6 DNA depending on which one was in excess (Figure 5A).
Figure 5.
Binding of PhoP to the whiB6 promoter as a dimer. (A) Size-exclusion chromatography analysis of the interaction of PhoP with the WB6 DNA sequence. The protein alone (17.6 μM), DNA alone (8.8 μM), and mixtures of PhoP and WB6 at molar ratios of 1:1 and 2:1 or with a large excess of PhoP were loaded onto a Superdex 200 gel filtration column equilibrated with the binding buffer. Calibration of the column is also shown with the molecular mass standards at 670, 158, 44, and 17 kDa. (B) Sedimentation velocity analysis of PhoP interacting with the WB6 DNA sequence. Sedimentation coefficient distributions c(s) of PhoP alone and the WB6 DNA alone gave single peaks at 2.33 and 2.71 S, respectively. The c(s) distributions of all mixtures containing PhoP and WB6 DNA at different molar ratios gave a major peak with a sedimentation coefficient of 5.276 S, corresponding to a PhoP–WB6 DNA complex of the same species; the slower peaks at 2.71 and 2.33 S correspond to the unbound DNA and free protein, respectively. (C) SV analysis of PhoP phosphorylation by acetyl phosphate. PhoP in the presence of AcP formed many oligomers, with three major peaks having fitted molecular masses of 28.2, 58.3, and 94.0 kDa, likely representing monomer, dimer, and trimer, respectively. The fitted frictional ratio is ∼1.9, resulting in the shift of the monomer peak to 1.7 S. The sample of PhoP alone (B, dotted line) has a frictional ratio of ∼1.3.
To further confirm the binding stoichiometry of PhoP with its target DNA, we performed AUC sedimentation velocity experiments on the mixture of PhoP with WB6. The sedimentation coefficient distribution [c(s)] of the WB6 DNA duplex had a single peak with a sedimentation coefficient of 2.71 S, compatible with a 31-mer WB6 DNA duplex (Figure 5B). The c(s) profile of PhoP had a peak at 2.33 S, compatible with the presence of a monomer in solution. In the presence of both PhoP and DNA, a biphasic c(s) distribution of two separate peaks was observed, with a faster peak at 5.276 S compatible with the PhoP–WB6 complex always present and an additional slower peak of free DNA or protein depending on the molar ratio of protein to DNA. At a 1:1 molar ratio, the slower peak is at 2.71 S, corresponding to unbound DNA. At a 2:1 protein:DNA molar ratio, the c(s) distribution showed a predominant species at 5.276 S. At a 3:1 protein:DNA molar ratio, the additional slower peak is at 2.33 S, corresponding to free PhoP. Taken together, these results support the formation of a 2:1 complex of the PhoP and its target DNA.
Roles of Individual Bases of the Direct-Repeat Sequence
To define the roles of individual bases of the direct-repeat motifs in binding PhoP, we designed a series of DNA sequences with mutations in the motifs and analyzed their binding affinity for PhoP by ITC (Table 1). We first assayed the effect of trinucleotide changes on PhoP binding. All trinucleotide changes significantly reduced binding affinity. Mutation of C2AC to GCG in the first motif resulted in a binding affinity lower than that after mutation of C13AC in the second motif to GCG (RD6m4 vs RD6m5 in Table 1), while mutation of A5GC in the first motif or A16GC in the second motif to CTG had a similar effect on PhoP binding (RD6m6 vs RD6m7).
To further define the sequence requirement for PhoP binding, we tested the effects of single-base substitutions on binding affinity. All single substitutions in the motifs reduced the binding affinity. Replacement of the last base C of either motif (C7 or C18) with a G had a relatively mild effect on binding affinity (RD6m14 and RD6m21 in Table 1). Substitutions of central positions of the motifs had a more significant effect, with those of the first motif having a more pronounced effect. Replacement of each base in A3CAG of the first motif and C15A of the second motif reduced the affinity more than replacement of other bases.
Base substitutions in the motifs of the WB6 sequence corroborated the results described above with regard to the direct-repeat motif sequence in RD6. Substitution of all C’s (C2, C4, and C7) in either motif with G’s resulted in a significantly reduced binding affinity compared to that of wild-type WB6 (Table 1, bottom section), with mutation of C15 (the only C in the second motif) to G having a milder effect. Mutation of A’s in either motif of WB6 that are also present in RD6 (consensus sequence) to C’s significantly impaired PhoP binding (WB6m6 and WB6m7), again with the second motif having a milder effect.
All SELEX-derived sequences have a 4 bp spacer between the two motifs, suggesting that there is a strict spacing between the two motifs for optimal PhoP binding. Insertion or deletion of bases from the spacer of RD6 resulted in a more than 85-fold reduction in binding affinity (Table 1, RD6m27–29). This is likely caused by disruption of the cooperativity of binding of the two PhoP molecules when the position of the second binding motif is shifted relative to the first motif. We next examined the effect of mutations of the spacer on PhoP binding. Replacement of A8CTT in RD6 with CCTT or ACGT modestly reduced the binding affinity, while replacement with ACTG or ACGG drastically reduced binding affinity (Table 1, RD6m22 and -24–26). Changing the spacer ACTT in RD6 to AATT did not affect the binding affinity (RD6m23). Taken together, these results suggest that an AT-rich spacer is favorable for PhoP binding.
Genomic Search of Promoters for Sequence Patterns Matching the Direct-Repeat Motif
The results described above established that the direct-repeat sequence TCACAGC(N4)TCACAGC is the consensus sequence for PhoP binding, thus making it possible to search for PhoP-binding sites on gene promoters to identify putative target genes of PhoP in the entire MTB genome. Gene promoter regions (from 600 bp upstream to 30 bp downstream of the translation initiation codon) of the MTB H37Rv genome were searched for the distribution of the consensus sequence. For a DNA sequence to bind PhoP, the motif can be on either the coding strand or the template strand, although the orientation of PhoP binding probably matters for activation of gene transcription. Because whiB6 and hsp, both of which have been shown to be regulated by PhoP,5,6 have the direct repeat of the TCACAGC motif located on the same strand as the coding strand, we describe this direction of the direct repeat as the forward direction. The reverse direction of the direct repeat thus has the motif sequence as GCTGTGA on the coding strand.
The number of hits of the in silico search depends on how many mismatches are allowed. The forward direction of the motifs has no hits with fewer than two mismatches for the direct repeat and 13 hits with two mismatches on 12 gene promoters (Table S1 of the Supporting Information). Four of these 12 genes, hsp, cfp2, Rv2633c, and PPE50, have been identified to be regulated by PhoP in transcriptome studies.5,6 The reverse direction has one site with a perfect match (pks16), one site with one mismatch (snoP), and seven sites with two mismatches (Table S1 of the Supporting Information). None of these genes were previously identified to be regulated by PhoP.
Analysis of Potential PhoP-Binding Sites on Gene Promoters for PhoP Binding Affinity
To validate the putative PhoP-binding sites derived from whole genome promoter search, we selected a list of sequences from the hits and analyzed their binding affinity for PhoP via ITC. The sequences are from 28 to 33 bp long, covering the direct-repeat motifs plus an extension of at least 4 bp at the 5′-end and 6 bp at the 3′-end. Table 2 lists some of the genes that gave a reasonable binding constant by ITC. Similar to that of the RD6 sequence, all ITC titration data fit well with a stoichiometry of two molecules of PhoP binding to one molecule of DNA duplex in a single binding event, suggesting that PhoP binds DNA as a dimer in a cooperative manner. The binding constants varied from ∼20 nM to >4 μM. The binding reactions are all exothermic with negative enthalpy changes (ΔH). Binding dissociation constants of >10 μM cannot be reliably measured, because of the small binding heat exacerbated by the limited solubility of PhoP.
Table 2. Results of ITC Titration of Selected Gene Promoter Sequences with the PhoP Proteina.
| gene | sequenceb | Pc | Nd | Kd (nM) | ΔH (kcal/mol) | TΔSe (kcal/mol) |
|---|---|---|---|---|---|---|
| cfp2 | tcgcTCACAGCtacgaCACAGacttgcc | –91 | 0.455 | 21.4 ± 1.6 | –14.62 ± 0.05 | –4.16 ± 0.10 |
| Rv2633c | tgatTCACAGCtaccTCACAttaatggg | –81 | 0.474 | 37.0 ± 3.7 | –16.37 ± 0.10 | –6.23 ± 0.16 |
| PPE50 | ctttTCACAGCaaagTCcCAGaaatggc | –379 | 0.375 | 45.7 ± 1.7 | –16.32 ± 0.04 | –6.31 ± 0.06 |
| fadD21 | cctgTttCAGCacatgCACAGCattgca | –111 | 0.437 | 50.3 ± 4.3 | –11.39 ± 0.07 | –1.43 ± 0.12 |
| hsp | ccggaCACAGCtaacTCACAaCgaagca | –37 | 0.434 | 51.5 ± 4.3 | –12.64 ± 0.07 | –2.70 ± 0.12 |
| fbpA | atacTgACAGCaagaTCACAattgagcc | –269 | 0.452 | 56.5 ± 2.6 | –13.9 ± 0.05 | –4.01 ± 0.07 |
| Rv3134c | gccatttCTGgGActttGCTGTGAaaagctg | –223 | 0.422 | 61.7 ± 5.3 | –15.28 ± 0.10 | –5.45 ± 0.15 |
| fadD9 | catcTCACAGCcgatcagCAGCaggctt | –122 | 0.459 | 82.0 ± 4.0 | –13.22 ± 0.05 | –3.55 ± 0.08 |
| phoP | agactacTggCAaCgagcTtTcAGgaattacac | –55 | 0.334 | 104 ± 10 | –18.49 ± 0.18 | –8.97 ± 0.24 |
| lipF | agacgtACAGCaaacTCcCAGtcataca | –572 | 0.434 | 115 ± 8 | –13.20 ± 0.08 | –3.73 ± 0.12 |
| pks3 | cgacgtcTggtAGCggcaTggCAaCggcctgtg | –236 | 0.316 | 130 ± 11 | –12.56 ± 0.12 | –3.17 ± 0.17 |
| Rv2331 | gtccTCgCAGCaagaaaACAGCgaaagc | –641 | 0.378 | 130 ± 6.1 | –6.62 ± 0.04 | 2.77 ± 0.06 |
| fas | cggcgtAgAGCgaatTCcCAGCataacg | –388 | 0.488 | 144 ± 6 | –3.18 ± 0.01 | 6.16 ± 0.17 |
| pks2 | aaagaCACAGCtacaTCgaAGgattgct | –50 | 0.409 | 154 ± 12 | –11.62 ± 0.09 | –2.33 ± 0.14 |
| Rv3312a | tgggTCACAGCgagtaatCAGCaagttc | –83 | 0.388 | 160 ± 8 | –11.08 ± 0.06 | –1.81 ± 0.09 |
| Rv1639c | cggaTCACAGgaaaccCcCAaCaaatca | –33 | 0.445 | 198 ± 12 | –10.24 ± 0.08 | –1.09 ± 0.12 |
| sirA | cgtcTCcCAGCggatTCcCgGgtcggcc | –313 | 0.363 | 332 ± 17 | –9.26 ± 0.07 | –0.42 ± 0.10 |
| cadI | cggtaCACAGCgcttgCAggGCttcagg | –431 | 0.360 | 403 ± 20 | –12.13 ± 0.11 | –3.41 ± 0.13 |
| Rv0520 | gcccgCACAGCcacgcCgtAGCaccggc | –200 | 0.542 | 472 ± 20 | –4.01 ± 0.03 | 4.62 ± 0.05 |
| Rv1217c | gaggTCgCAGCcgagcaACAGgtggcaa | –28 | 0.379 | 588 ± 17 | –10.21 ± 0.06 | –1.71 ± 0.08 |
| Rv2010 | gaacTCgtgGCcgccgCACAGCggatgt | –194 | 0.428 | 637 ± 16 | –7.96 ± 0.04 | 0.49 ± 0.06 |
| Rv3881c | atggTCACAGtcgggcCACAGttcgag | –336 | 0.509 | 714 ± 51 | –8.95 ± 0.11 | –0.57 ± 0.15 |
| cdh | gcgagCAggGCtgccgCACAGCgatctt | –246 | 0.461 | 935 ± 52 | –9.23 ± 0.13 | –1.00 ± 0.15 |
| ptrBa | ttgtcCgCAGCtggcTCACcGgctccga | –162 | 0.378 | 980 ± 77 | –11.98 ± 0.28 | –3.78 ± 0.32 |
| Rv3877 | ctcggCgCAGCgcgcgCtCAGCgacgcc | –470 | 0.365 | 1149 ± 66 | –5.41 ± 0.09 | 2.69 ± 0.12 |
| aroG | ttcgTCtCAtCacgtcCACAGacgatgc | –136 | 0.437 | 1163 ± 41 | –5.65 ± 0.06 | 2.45 ± 0.08 |
| umaA | tgacgCAagGCgagaTCACAGaccgaga | –105 | 0.423 | 1205 ± 58 | –9.23 ± 0.12 | –1.15 ± 0.15 |
| espA | cgcattgTCgCAGCgcagTtgCAGgagggcaa | –215 | 0.335 | 1587 ± 50 | –6.77 ± 0.08 | 1.14 ± 0.10 |
| Rv0964c | gcgcgCACgGCacacgCACcGCatcggc | –40 | 0.235 | 4348 ± 189 | –14.42 ± 0.98 | –7.11 ± 1.01 |
Entries are sorted by the binding constants.
The motifs are highlighted in bold, with mismatches shown in lowercase letters. Only the sequences of the coding strand are shown.
P represents the position relative to the translation initiation codon.
N is the stoichiometry, referring to the number of DNAs per PhoP molecule.
Values of TΔS were calculated from the values of Kd and ΔH that were obtained from fitting the ITC titration data with Origin.
The high-affinity promoter sequences with binding constants ranging from ∼20 to 100 nM (Table 2) have two to five mismatches compared with RD6 direct-repeat motifs. In the first motif, bases from the third to the last position, A3CAGC, are well-conserved, but the first two bases, T1C2, are much less conserved. In the second motif, the third base, A14, is not well-conserved and the last base, C18, is not conserved at all. The spacer and the sequence immediately following the second motif are AT-rich. When there were more than three G or C residues in the spacer, the binding affinity was significantly reduced (Table S1 of the Supporting Information). This is consistent with ITC studies in which mutating the spacer of RD6 to a GC-rich sequence resulted in a decrease in the affinity for PhoP binding (Table 1). Most sequences with high binding affinity have one to two G/C residues in the 4 bp spacer (Table 2, top half). Sequences right after the second motif were predominantly composed of A or T for the high-affinity sequences, while two or more G/C residues directly following the second motif were associated with low binding affinity. Examples are the promoter sequence of cyp123, agatcCACcGCcgcaTCACAtCggcgat, and that of papA4, agtaTCACtGCccgaTCACcGgtcgccc; both have highly conserved motif sequences with only three mismatches but had no measurable binding as determined by ITC (Table S1 of the Supporting Information).
Phosphorylation of PhoP Increased Its Binding Affinity for Direct-Repeat DNA Sequences but Did Not Alter Its Specificity
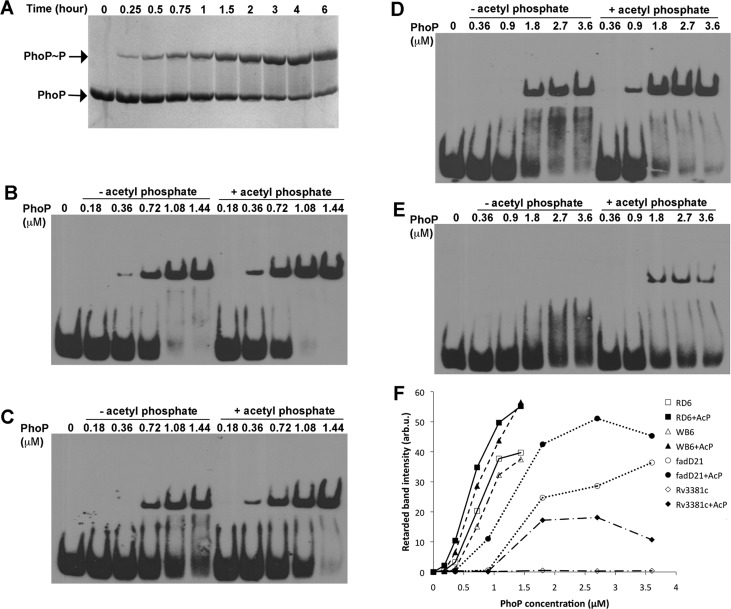
Phosphorylation of PhoP reached a steady state after incubation with acetyl phosphate for 2 h at room temperature, and more than 50% of PhoP was phosphorylated (Figure 6A). A significant increase in binding affinity of phosphorylated PhoP was observed for all sequences tested, ranging from the SELEX-derived highest-affinity RD6 (Figure 6B) and the high-affinity DNA sequence WB6 from the promoter of whiB6 (Figure 6C) to the lower-affinity sequences from the promoters of fadD21 (Figure 6D) and Rv3881c (Figure 6E). It is noteworthy that phosphorylation did not alter the sequence preference for PhoP binding. The high-affinity sequences maintained an affinity for phosphorylated PhoP higher than that of the lower-affinity sequences (Figure 6F). The promoter DNA of fadD21 was found to bind PhoP only when it is phosphorylated.32 However, our results showed that nonphosphorylated PhoP could bind a short sequence around the identified direct repeat on the fadD21 promoter, although a concentration higher than that of phosphorylated PhoP was necessary to form a stable complex. Nonphosphorylated PhoP did not shift the DNA sequence derived from the putative binding site of the Rv3881c promoter but showed only some smear at PhoP concentrations of ≤3.6 μM (Figure 6E). Phosphorylated PhoP was able to give a discrete shifted band at ∼1.8 μM protein.
Figure 6.
Effect of PhoP phosphorylation on the PhoP–DNA binding affinity. (A) Time course of PhoP phosphorylation by acetyl phosphate. The PhoP protein (15 μM) was incubated with 50 mM AcP at room temperature. At each specified time point, a sample was taken, mixed with SDS sample buffer, and kept on ice. The samples were run on a 10% acrylamide gel containing Phos-tag acrylamide, and the gel was stained with Coomassie blue. A slower-moving band appeared, and its intensity gradually increased over time (top band), corresponding to the phosphorylated PhoP protein. (B–E) EMSA results of binding of PhoP to RD6, WB6, the fadD21 promoter sequence, and the Rv3881c promoter sequence, respectively. The binding reactions were conducted with nonphosphorylated PhoP (lanes 2–6) or phosphorylated PhoP (lanes 7–11). The concentrations of PhoP used in binding reactions are as labeled. The DNA concentrations were 0.13 μM for RD6 and WB6 and 0.30 μM for the promoter DNA of fadD21 and Rv03881c. PhoP was phosphorylated as described above for 2 h prior to the binding reactions. (F) Intensity of retarded bands of EMSA gels in panel B–E were quantified and are plotted vs PhoP concentration. Data for nonphosphorylated PhoP and phosphorylated PhoP binding to the same DNA sequence are represented with the same type of line, but with empty and filled symbols, respectively.
Phosphorylation Promotes Dimerization and Oligomerization of PhoP
To check if phosphorylation increases the PhoP binding affinity of direct-repeat DNA by promoting PhoP dimerization, we conducted AUC SV experiments with PhoP in the presence of AcP (Figure 5C). In sharp contrast to the PhoP alone sample (Figure 5B, dotted line) that give a single peak of a monomer size, PhoP with AcP gave many peaks on the sedimentation profile, suggesting the presence of monomer, dimer, trimer, and other higher-order oligomers. The fitted frictional ratio for PhoP in the presence of AcP was ∼1.9, compared to a value of ∼1.3 for the PhoP alone sample. This high frictional ratio suggests that phosphorylation induces aggregation of PhoP into long chains, consistent with size-exclusion chromatography studies of the AcP phosphorylation of PhoP, in which most of the phosphorylated PhoP was lost on the prefiltration filter and the top filter of the column (data not shown). Binding of phosphorylated PhoP to direct-repeat DNA should promote dimerization and restrict further oligomerization.
Discussion
The SELEX-Derived Direct Repeat Is the Consensus Sequence for Binding of PhoP to Gene Promoters
First, the direct repeat has a pattern remarkably similar to that of all known DNA-binding consensus sequences for the OmpR/PhoB subfamily response regulators. The DNA sequence motif for PhoP from S. coelicolor,18 the pho box DNA for E. coli PhoB,19 and the consensus sequence for PhoP of both E. coli and Sa. enterica(20,21) all have 10 nucleotides between the equivalent bases of the two repeats, similar to that of the direct repeat identified in this study, TCACAGCN4TCACAGC. Second, this direct-repeat motif can be recognized in gene promoter sequences previously shown to bind PhoP (Table 2), such as those identified by an EMSA in the promoters of fadD21, phoP, and msl3.32 Third, as a general trend, synthetic oligo duplexes at the identified sites bind PhoP with a higher affinity for those having fewer mismatches (Table 2). In addition, for a few genes with available footprinting data, such as phoP, msl3, pks2, and lipF,28,32 the binding sites all fall within the PhoP-protected regions. Recently, two independent studies identified a partial sequence motif for PhoP binding using chromatin immunoprecipitation sequencing (ChIP-seq) techniques.33,34 Although the motifs are incomplete because of biological variability, the results are consistent with the direct-repeat consensus motif TCACAGC(N4)TCACAGC in the base positions and the spacing between the repeated motifs. Both works are performed in vivo and hence provide confidence to the results obtained in this work in vitro.
PhoP Binds Its Target Gene Promoters as a Dimer
As shown in the Results, the single-motif sequence has an affinity for PhoP drastically lower than that of the direct-repeat sequence with two motifs, suggesting that PhoP binds to gene promoters as a dimer. This observation is consistent with most results of other response regulators in the same subfamily. OmpR∼P is unable to make a stable complex with DNA containing only one binding site.40 PhoB of E. coli(41) and PhoP of Sa. enterica(21) are also found to bind to gene promoters as dimers. It was proposed that OmpR forms a dimer in solution prior to binding to DNA in a pairwise manner,24 although it is possible for monomer PhoP to assemble on the DNA direct repeat to form a dimer. In some gene promoters, there are multiple direct repeats, and it is possible that two or more PhoP dimers bind to these sites and form higher-order oligomers mediated by DNA. This type of cooperative binding of dimers has been demonstrated for binding of OmpR∼P to the subsites of the OmpF promoter.42
Phosphorylation of PhoP Increases Its Affinity for Target Promoters Likely by Enhancing PhoP Dimerization on Direct-Repeat DNA Sequences
PhoP has two distinct structural domains, a receiver domain that accepts a phosphoryl group from PhoR and an effector domain that is a DNA-binding domain with a winged helix–loop–helix fold.43,44 Evidence suggests that phosphorylation of the receiver domain affects the structure of the α4−β5−α5 face, thus altering its interaction with the DNA-binding domain or promoting dimerization of the receiver domain to modulate gene transcription.45 Structures of isolated receiver domains of the OmpR/PhoB subfamily response regulators reveal that they form dimers through the α4−β5−α5 face upon activation.46−48 The crystal structure of full-length MTB PhoP shows that the receiver domain forms a symmetric dimer through the α4−β5−α5 face, but the DNA-binding domain is merely tethered to the receiver domain through a flexible linker,16 allowing the DNA-binding domain the freedom to bind to direct-repeat DNA sequences as a tandem dimer. Because PhoP, as well as many other response regulators in the same subfamily, can bind DNA in the absence of phosphorylation,29,49 it is likely that phosphorylation activates DNA binding by modulating the α4−β5−α5 face and thus enhancing dimer formation.16,45 Recently, a crystal structure of KdpE in complex with DNA was reported, in which the effector domains form a tandem dimer binding to the DNA direct repeat while the receiver domain forms a symmetric dimer through the α4−β5−α5 face.50
Our EMSA results comparing phosphorylated to nonphosphorylated PhoP indicate that phosphorylation did not change the DNA sequence specificity. This is supported by the fact that a genomic promoter search with the SELEX-derived direct-repeat mapped to the same locations as that from the footprinting assays of genes phoP, msl3, pks2, and lipF,32 which were conducted with phosphorylated PhoP, although our SELEX experiments used nonphosphorylated PhoP. The list of potential PhoP-binding sites from gene promoter search overlaps well with transcriptome studies and recently published ChIP-seq results33,34 (Table S1 of the Supporting Information). The differences are due to the facts that PhoP might bind to some promoters exclusively in vivo through interacting with other DNA-binding proteins and some predicted binding sites might not be accessible to PhoP in vivo. Because ChIP-seq detects in vivo PhoP–DNA binding and potentially immunoprecipitates a mixture of phosphorylated and nonphosphorylated PhoP, the fact that the partial motif from those studies matches the SELEX-derived direct-repeat sequence further supports the validity of the in vitro results in this study.
Genomewide Transcription Regulation by PhoP Likely Occurs through Multiple Mechanisms
Gene transcription profiling studies comparing the wild-type and the phoP knockout MTB strains5,6 indicate that PhoP can either upregulate or downregulate its target genes. To function as a transcription activator, promoter-bound PhoP has to interact with other components of the transcription machinery to influence gene transcription. In this case, the position of binding is also important for efficient interaction with other components of the transcription machinery. However, binding to gene promoters with high affinity should serve well to block the initiation or progress of transcription. The hsp gene has one strong PhoP-binding site 37 bp upstream of the translation initiation codon (Table 2). PhoP binding to this site could block transcription if the transcription start site is near or upstream of this site.
While some genes are directly regulated by PhoP, many are likely to be regulated indirectly through PhoP-regulated transcription factors.17 All 173 genes that have been identified as being PhoP-regulated genes in two independent studies5,6 have sequences matching the direct repeat on their promoters, with most of them containing multiple potential binding sites. However, many of these putative PhoP-binding sites are likely to be false positives because many sequences containing the putative binding sites did not bind PhoP when they were assayed by ITC (Table S1 of the Supporting Information). Among known PhoP target genes that have strong PhoP-binding sites on their promoters, some encode transcription regulators, such as WhiB6 and DosR. These transcription regulators can in turn regulate many other genes whose expression is influenced by the PhoP–PhoR signaling system.
Some genes are likely to be regulated by clustering in an operon. As an example, the dormancy regulation genes, dosR and dosS,51 are regulated by PhoP.5,34 However, no PhoP-binding site was identified upstream of the genes. A close examination of the MTB genomic sequence revealed that Rv3134c is likely to be the first gene of the dosRS operon. Although Rv3134c was not identified to be regulated by PhoP in transcriptome studies, its promoter contains a strong PhoP-binding site that was confirmed by ITC (Table 2).
ITC results indicate that PhoP binds to gene promoters with a wide range of affinities, possibly reflecting the variable degree of regulation of each gene by PhoP. The binding affinity is related to the number of mismatches from the consensus, the positions of the mismatched bases in the motifs, and the sequences of the spacer and flanks. Consistent with the WebLogo analysis of the consensus sequence (Figure 1B) and analyses by an EMSA and ITC (Figure 2 and Table 1), mismatches at the edges of the motifs had a weaker impact than those in the middle of the motif on the binding affinity, and a GC-rich sequence in the spacer or immediately following the second motif significantly reduced the binding affinity.
Future Research Directions
With the PhoP-binding consensus motif available, it is possible to study the mechanism of PhoP function in gene regulation in a genomic scale. Genes directly regulated by PhoP can be identified from the list of potential sites of the gene promoter pattern matches of the consensus motif. However, as demonstrated in the results described above, establishing a relationship between the DNA sequence and PhoP binding affinity can be complicated, because a base at one position could influence the requirement of the base at another position. A systematic analysis of PhoP binding affinity of a representative subset of potential binding sites by ITC or an EMSA combined with a bioinformatic approach is necessary to establish a set of rules to identify true PhoP-binding sites from the results of a whole genome promoter search. Transcription start sites of potential PhoP-regulated genes will need to be mapped to understand the mechanism of interaction between PhoP and the rest of the transcription machinery. Gene promoter activity in relation to PhoP binding should be analyzed to verify PhoP regulation on gene transcription. A crystal structure of a PhoP–DNA complex will shed light on the atomic interactions between the protein and DNA and thus the mechanism of DNA sequence recognition.
Acknowledgments
We thank Zhao Zhang Li of the Biomedical Instrumentation Center, Uniformed Services University of the Health Sciences, for DNA sequencing, Sudipa Ghimire-Rijal for help with AUC and ITC experiments, Dr. John Burgner for help with AUC data processing and result interpretation, and Drs. Saibal Dey and Ernest Maynard for helpful comments and revisions of the manuscript. We are grateful to Dr. Issar Smith of the Public Health Research Institute for his continuous guidance throughout the project and his insightful comments and revisions of the manuscript. Dr. Smita Menon helped with the whiB6 gene promoter mapping experiments and participated in some SELEX experiments.
Glossary
Abbreviations
- AcP
acetyl phosphate
- AUC
analytical ultracentrifugation
- ChIP-seq
chromatin immunoprecipitation sequencing
- EMSA
electrophoretic mobility shift assay
- HK
histidine kinase
- IPTG
isopropyl β-d-1-thiogalactopyranoside
- ITC
isothermal titration calorimetry
- MTB
M. tuberculosis
- PMSF
phenylmethanesulfonyl fluoride
- PCR
polymerase chain reaction
- RR
response regulator
- SELEX
systematic evolution of ligands by exponential enrichment
- SV
sedimentation velocity
- TEV
tobacco etch virus
- TCS
two-component system.
Supporting Information Available
Some potential PhoP-binding sequences on the promoters of genes, along with available ITC titration data of weak or no detectable binding, and available transcription profile data comparing PhoP knockout and wild-type MTB strains (Table S1). This material is available free of charge via the Internet at http://pubs.acs.org.
Author Present Address
† X.H.: Department of Oncology, School of Medicine, Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC 20057.
Author Contributions
X.H. and S.W. designed the experiments and conducted the ITC, AUC, and SEC experiments. X.H. conducted the SELEX, EMSA, and phosphorylation experiments. X.H. and S.W. wrote the manuscript.
This work was supported by National Institutes of Health Grant R01GM079185 and Uniformed Services University of the Health Sciences Intramural Grant R071IR.
The opinions or assertions contained herein are the private ones of the authors and are not to be construed as official or reflecting the views of the Department of Defense or the Uniformed Services University of the Health Sciences.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Dalton T.; Cegielski P.; Akksilp S.; Asencios L.; Campos Caoili J.; Cho S. N.; Erokhin V. V.; Ershova J.; Gler M. T.; Kazennyy B. Y.; Kim H. J.; Kliiman K.; Kurbatova E.; Kvasnovsky C.; Leimane V.; van der Walt M.; Via L. E.; Volchenkov G. V.; Yagui M. A.; Kang H.; Akksilp R.; Sitti W.; Wattanaamornkiet W.; Andreevskaya S. N.; Chernousova L. N.; Demikhova O. V.; Larionova E. E.; Smirnova T. G.; Vasilieva I. A.; Vorobyeva A. V.; Barry C. E. III; Cai Y.; Shamputa I. C.; Bayona J.; Contreras C.; Bonilla C.; Jave O.; Brand J.; Lancaster J.; Odendaal R.; Chen M. P.; Diem L.; Metchock B.; Tan K.; Taylor A.; Wolfgang M.; Cho E.; Eum S. Y.; Kwak H. K.; Lee J.; Min S.; Degtyareva I.; Nemtsova E. S.; Khorosheva T.; Kyryanova E. V.; Egos G.; Perez M. T.; Tupasi T.; Hwang S. H.; Kim C. K.; Kim S. Y.; Lee H. J.; Kuksa L.; Norvaisha I.; Skenders G.; Sture I.; Kummik T.; Kuznetsova T.; Somova T.; Levina K.; Pariona G.; Yale G.; Suarez C.; Valencia E.; Viiklepp P. (2012) Prevalence of and risk factors for resistance to second-line drugs in people with multidrug-resistant tuberculosis in eight countries: A prospective cohort study. Lancet 380, 1406–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West A. H.; Stock A. M. (2001) Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26, 369–376. [DOI] [PubMed] [Google Scholar]
- Cole S. T.; Brosch R.; Parkhill J.; Garnier T.; Churcher C.; Harris D.; Gordon S. V.; Eiglmeier K.; Gas S.; Barry C. E. III; Tekaia F.; Badcock K.; Basham D.; Brown D.; Chillingworth T.; Connor R.; Davies R.; Devlin K.; Feltwell T.; Gentles S.; Hamlin N.; Holroyd S.; Hornsby T.; Jagels K.; Barrell B. G.; et al. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544. [DOI] [PubMed] [Google Scholar]
- Perez E.; Samper S.; Bordas Y.; Guilhot C.; Gicquel B.; Martin C. (2001) An essential role for phoP in Mycobacterium tuberculosis virulence. Mol. Microbiol. 41, 179–187. [DOI] [PubMed] [Google Scholar]
- Gonzalo-Asensio J.; Mostowy S.; Harders-Westerveen J.; Huygen K.; Hernandez-Pando R.; Thole J.; Behr M.; Gicquel B.; Martin C. (2008) PhoP: A missing piece in the intricate puzzle of Mycobacterium tuberculosis virulence. PLoS One 3, e3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters S. B.; Dubnau E.; Kolesnikova I.; Laval F.; Daffe M.; Smith I. (2006) The Mycobacterium tuberculosis PhoPR two-component system regulates genes essential for virulence and complex lipid biosynthesis. Mol. Microbiol. 60, 312–330. [DOI] [PubMed] [Google Scholar]
- Gonzalo Asensio J.; Maia C.; Ferrer N. L.; Barilone N.; Laval F.; Soto C. Y.; Winter N.; Daffe M.; Gicquel B.; Martin C.; Jackson M. (2006) The virulence-associated two-component PhoP-PhoR system controls the biosynthesis of polyketide-derived lipids in Mycobacterium tuberculosis. J. Biol. Chem. 281, 1313–1316. [DOI] [PubMed] [Google Scholar]
- Zheng H.; Lu L.; Wang B.; Pu S.; Zhang X.; Zhu G.; Shi W.; Zhang L.; Wang H.; Wang S.; Zhao G.; Zhang Y. (2008) Genetic basis of virulence attenuation revealed by comparative genomic analysis of Mycobacterium tuberculosis strain H37Ra versus H37Rv. PLoS One 3, e2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigui W.; Bottai D.; Majlessi L.; Monot M.; Josselin E.; Brodin P.; Garnier T.; Gicquel B.; Martin C.; Leclerc C.; Cole S. T.; Brosch R. (2008) Control of M. tuberculosis ESAT-6 secretion and specific T cell recognition by PhoP. PLoS Pathog. 4, e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. S.; Krause R.; Schreiber J.; Mollenkopf H. J.; Kowall J.; Stein R.; Jeon B. Y.; Kwak J. Y.; Song M. K.; Patron J. P.; Jorg S.; Roh K.; Cho S. N.; Kaufmann S. H. (2008) Mutation in the transcriptional regulator PhoP contributes to avirulence of Mycobacterium tuberculosis H37Ra strain. Cell Host Microbe 3, 97–103. [DOI] [PubMed] [Google Scholar]
- Ryndak M.; Wang S.; Smith I. (2008) PhoP, a key player in Mycobacterium tuberculosis virulence. Trends Microbiol. 16, 528–534. [DOI] [PubMed] [Google Scholar]
- Nambiar J. K.; Pinto R.; Aguilo J. I.; Takatsu K.; Martin C.; Britton W. J.; Triccas J. A. (2012) Protective immunity afforded by attenuated, PhoP-deficient Mycobacterium tuberculosis is associated with sustained generation of CD4+ T-cell memory. Eur. J. Immunol. 42, 385–392. [DOI] [PubMed] [Google Scholar]
- Aporta A.; Arbues A.; Aguilo J. I.; Monzon M.; Badiola J. J.; de Martino A.; Ferrer N.; Marinova D.; Anel A.; Martin C.; Pardo J. (2012) Attenuated Mycobacterium tuberculosis SO2 vaccine candidate is unable to induce cell death. PLoS One 7, e45213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbues A.; Aguilo J. I.; Gonzalo-Asensio J.; Marinova D.; Uranga S.; Puentes E.; Fernandez C.; Parra A.; Cardona P. J.; Vilaplana C.; Ausina V.; Williams A.; Clark S.; Malaga W.; Guilhot C.; Gicquel B.; Martin C. (2013) Construction, characterization and preclinical evaluation of MTBVAC, the first live-attenuated M. tuberculosis-based vaccine to enter clinical trials. Vaccine 31, 4867–4873. [DOI] [PubMed] [Google Scholar]
- Galperin M. Y. (2006) Structural classification of bacterial response regulators: Diversity of output domains and domain combinations. J. Bacteriol. 188, 4169–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon S.; Wang S. (2011) Structure of the response regulator PhoP from Mycobacterium tuberculosis reveals a dimer through the receiver domain. Biochemistry 50, 5948–5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.; Engohang-Ndong J.; Smith I. (2007) Structure of the DNA-binding domain of the response regulator PhoP from Mycobacterium tuberculosis. Biochemistry 46, 14751–14761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allenby N. E.; Laing E.; Bucca G.; Kierzek A. M.; Smith C. P. (2012) Diverse control of metabolism and other cellular processes in Streptomyces coelicolor by the PhoP transcription factor: Genome-wide identification of in vivo targets. Nucleic Acids Res. 40, 9543–9556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino K.; Shinagawa H.; Amemura M.; Nakata A. (1986) Nucleotide sequence of the phoB gene, the positive regulatory gene for the phosphate regulon of Escherichia coli K-12. J. Mol. Biol. 190, 37–44. [DOI] [PubMed] [Google Scholar]
- Kato A.; Tanabe H.; Utsumi R. (1999) Molecular characterization of the PhoP-PhoQ two-component system in Escherichia coli K-12: Identification of extracellular Mg2+-responsive promoters. J. Bacteriol. 181, 5516–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwir I.; Latifi T.; Perez J. C.; Huang H.; Groisman E. A. (2012) The promoter architectural landscape of the Salmonella PhoP regulon. Mol. Microbiol. 84, 463–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler U.; Weiss V. (1995) A common switch in activation of the response regulators NtrC and PhoB: Phosphorylation induces dimerization of the receiver modules. EMBO J. 14, 3696–3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack T. R.; Gao R.; Stock A. M. (2009) Probing the roles of the two different dimers mediated by the receiver domain of the response regulator PhoB. J. Mol. Biol. 389, 349–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri C. M.; Wu T.; Stock A. M. (2013) Comprehensive Analysis of OmpR Phosphorylation, Dimerization and DNA Binding Supports a Canonical Model for Activation. J. Mol. Biol. 425, 1612–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan A.; Paul L. N.; Tomar S.; Patil D. N.; Kumar P.; Yernool D. A. (2012) Structure-function studies of DNA binding domain of response regulator KdpE reveals equal affinity interactions at DNA half-sites. PLoS One 7, e30102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha A.; Gupta S.; Bhutani S.; Pathak A.; Sarkar D. (2008) PhoP-PhoP interaction at adjacent PhoP binding sites is influenced by protein phosphorylation. J. Bacteriol. 190, 1317–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A. K.; Pathak A.; Sinha A.; Datt M.; Singh B.; Karthikeyan S.; Sarkar D. (2010) A single-amino-acid substitution in the C terminus of PhoP determines DNA-binding specificity of the virulence-associated response regulator from Mycobacterium tuberculosis. J. Mol. Biol. 398, 647–656. [DOI] [PubMed] [Google Scholar]
- Goyal R.; Das A. K.; Singh R.; Singh P. K.; Korpole S.; Sarkar D. (2011) Phosphorylation of PhoP protein plays direct regulatory role in lipid biosynthesis of Mycobacterium tuberculosis. J. Biol. Chem. 286, 45197–45208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S.; Pathak A.; Sinha A.; Sarkar D. (2009) Mycobacterium tuberculosis PhoP recognizes two adjacent direct-repeat sequences to form head-to-head dimers. J. Bacteriol. 191, 7466–7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S.; Sinha A.; Sarkar D. (2006) Transcriptional autoregulation by Mycobacterium tuberculosis PhoP involves recognition of novel direct repeat sequences in the regulatory region of the promoter. FEBS Lett. 580, 5328–5338. [DOI] [PubMed] [Google Scholar]
- Pathak A.; Goyal R.; Sinha A.; Sarkar D. (2010) Domain structure of virulence-associated response regulator PhoP of Mycobacterium tuberculosis: Role of the linker region in regulator-promoter interaction(s). J. Biol. Chem. 285, 34309–34318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimino M.; Thomas C.; Namouchi A.; Dubrac S.; Gicquel B.; Gopaul D. N. (2012) Identification of DNA binding motifs of the Mycobacterium tuberculosis PhoP/PhoR two-component signal transduction system. PLoS One 7, e42876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solans L.; Gonzalo-Asensio J.; Sala C.; Benjak A.; Uplekar S.; Rougemont J.; Guilhot C.; Malaga W.; Martin C.; Cole S. T. (2014) The PhoP-dependent ncRNA Mcr7 modulates the TAT secretion system in Mycobacterium tuberculosis. PLoS Pathog. 10, e1004183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan J. E.; Minch K.; Peterson M.; Lyubetskaya A.; Azizi E.; Sweet L.; Gomes A.; Rustad T.; Dolganov G.; Glotova I.; Abeel T.; Mahwinney C.; Kennedy A. D.; Allard R.; Brabant W.; Krueger A.; Jaini S.; Honda B.; Yu W. H.; Hickey M. J.; Zucker J.; Garay C.; Weiner B.; Sisk P.; Stolte C.; Winkler J. K.; Van de Peer Y.; Iazzetti P.; Camacho D.; Dreyfuss J.; Liu Y.; Dorhoi A.; Mollenkopf H. J.; Drogaris P.; Lamontagne J.; Zhou Y.; Piquenot J.; Park S. T.; Raman S.; Kaufmann S. H.; Mohney R. P.; Chelsky D.; Moody D. B.; Sherman D. R.; Schoolnik G. K. (2013) The Mycobacterium tuberculosis regulatory network and hypoxia. Nature 499, 178–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri C. M.; Stock A. M. (2008) Universally applicable methods for monitoring response regulator aspartate phosphorylation both in vitro and in vivo using Phos-tag-based reagents. Anal. Biochem. 376, 73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laue M., Shah B. D., Ridgeway T. M., and Pelletier S. L. (1992) in Analytical Ultracentrifugation in Biochemistry and Polymer Science (Harding S., and Rowe A., Eds.) pp 90–125, Royal Society of Chemistry, London. [Google Scholar]
- Gabrielson J. P.; Randolph T. W.; Kendrick B. S.; Stoner M. R. (2007) Sedimentation velocity analytical ultracentrifugation and SEDFIT/c(s): Limits of quantitation for a monoclonal antibody system. Anal. Biochem. 361, 24–30. [DOI] [PubMed] [Google Scholar]
- Crooks G. E.; Hon G.; Chandonia J. M.; Brenner S. E. (2004) WebLogo: A sequence logo generator. Genome Res. 14, 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solans L.; Aguilo N.; Samper S.; Pawlik A.; Frigui W.; Martin C.; Brosch R.; Gonzalo-Asensio J. (2014) A Specific Polymorphism in Mycobacterium tuberculosis H37Rv Causes Differential ESAT-6 Expression and Identifies WhiB6 as a Novel ESX-1 Component. Infect. Immun. 82, 3446–3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlocker S. L.; Bergstrom L.; Inouye M. (1995) Tandem binding of six OmpR proteins to the ompF upstream regulatory sequence of Escherichia coli. J. Biol. Chem. 270, 26849–26856. [DOI] [PubMed] [Google Scholar]
- Blanco A. G.; Sola M.; Gomis-Ruth F. X.; Coll M. (2002) Tandem DNA recognition by PhoB, a two-component signal transduction transcriptional activator. Structure 10, 701–713. [DOI] [PubMed] [Google Scholar]
- Yoshida T.; Qin L.; Egger L. A.; Inouye M. (2006) Transcription regulation of ompF and ompC by a single transcription factor, OmpR. J. Biol. Chem. 281, 17114–17123. [DOI] [PubMed] [Google Scholar]
- Martinez-Hackert E.; Stock A. M. (1997) The DNA-binding domain of OmpR: Crystal structures of a winged helix transcription factor. Structure 5, 109–124. [DOI] [PubMed] [Google Scholar]
- Martinez-Hackert E.; Stock A. M. (1997) Structural relationships in the OmpR family of winged-helix transcription factors. J. Mol. Biol. 269, 301–312. [DOI] [PubMed] [Google Scholar]
- Wang S. (2012) Bacterial Two-Component Systems: Structures and Signaling Mechanisms. In Protein Phosphorylation in Human Health (Huang C., Ed.) pp 439–466, InTech, Rijeka, Croatia. [Google Scholar]
- Toro-Roman A.; Mack T. R.; Stock A. M. (2005) Structural analysis and solution studies of the activated regulatory domain of the response regulator ArcA: A symmetric dimer mediated by the α4-β5-α5 face. J. Mol. Biol. 349, 11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro-Roman A.; Wu T.; Stock A. M. (2005) A common dimerization interface in bacterial response regulators KdpE and TorR. Protein Sci. 14, 3077–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachhawat P.; Stock A. M. (2007) Crystal structures of the receiver domain of the response regulator PhoP from Escherichia coli in the absence and presence of the phosphoryl analog beryllofluoride. J. Bacteriol. 189, 5987–5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Zeng J.; Zhang H.; He Z. G. (2010) The characterization of conserved binding motifs and potential target genes for M. tuberculosis MtrAB reveals a link between the two-component system and the drug resistance of M. smegmatis. BMC Microbiol. 10, 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan A.; Kumar S.; Evrard A. N.; Paul L. N.; Yernool D. A. (2014) An asymmetric heterodomain interface stabilizes a response regulator-DNA complex. Nat. Commun. 5, 3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts D. M.; Liao R. P.; Wisedchaisri G.; Hol W. G.; Sherman D. R. (2004) Two sensor kinases contribute to the hypoxic response of Mycobacterium tuberculosis. J. Biol. Chem. 279, 23082–23087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.