Abstract
To identify novel genetic bases of early-onset epithelial ovarian tumors, we used the trio exome sequencing strategy in a patient without familial history of cancer who presented metastatic serous ovarian adenocarcinomas at 21 years of age. We identified a single de novo mutation (c.1157A>G/p.Asn386Ser) within the INHBA gene encoding the βA-subunit of inhibins/activins, which play a key role in ovarian development. In vitro, this mutation alters the ratio of secreted activins and inhibins. In a second patient with early-onset serous borderline papillary cystadenoma, we identified an unreported germline mutation (c.179G>T/p.Arg60Leu) of the INHA gene encoding the α-subunit, the partner of the βA-subunit. This mutation also alters the secreted activin/inhibin ratio, by disrupting both inhibin A and inhibin B biosynthesis. In a cohort of 62 cases, we detected an additional unreported germline mutation of the INHBA gene (c.839G>A/p.Gly280Glu). Our results strongly suggest that inhibin mutations contribute to the genetic determinism of epithelial ovarian tumors.
Keywords: cancer, ovary, exome, inhibin, activin, INHA, INHBA
In many families suspected to present an inherited form of cancer, analyses focused on genes known to be involved in Mendelian predisposition to cancer often remain negative, despite a personal and/or familial history strongly suggestive of a genetic determinism. Therefore, other pathways involved in cancer genetics remain to be identified. We considered that the comparative index case-parents exome sequencing strategy, originally developed to detect de novo mutation in early-onset severe conditions such as mental retardation [Vissers et al., 2010], could be a powerful method to identify novel molecular bases of cancer. Indeed, given the numerous examples of de novo mutations involved in genetic conditions recently described in humans [Veltman and Brunner, 2012], a fraction of early-onset sporadic cancer cases could result from de novo germline mutations affecting unknown cancer predisposing genes. The genes affected by these de novo mutations may represent novel candidate genes for familial forms of cancer.
For this study, we used as a paradigm sporadic early-onset epithelial ovarian cancer. In the general population, epithelial ovarian tumors usually occur after 50 years of age and the development of epithelial ovarian tumors before the age of 40 is therefore suggestive, in clinical practice, of a genetic determinism. Early-onset epithelial ovarian tumors occur in patients with germline mutations of BRCA1, BRCA2, and the MMR genes [Lynch et al., 2009], and recently, germline mutations of other genes known to be involved in cancer such as BARD1, BRIP1, CHEK2, MRE11A, NBN, PALB2, RAD50, RAD51C, RAD51D, or TP53 have been reported in patients with early-onset epithelial ovarian cancers [Loveday et al., 2011; Walsh et al., 2011]. Nevertheless, in a fraction of patients developing early-onset epithelial ovarian tumors, genetic analyses of these genes remain negative, suggesting the existence of uncharacterized deleterious mutations in other genes. We report here the identification by exome sequencing of a de novo germline mutation affecting the βA-subunit of inhibins/activins in a sporadic case of early-onset serous ovarian cancer and provide arguments indicating that germline inhibin mutations contribute to the genetic determinism of epithelial ovarian tumors by altering the inhibin/activin production.
The patient is a French woman of Caucasian origin who had developed at 21 years of age large bilateral ovarian tumors corresponding to serous low-grade ovarian adenocarcinomas (FIGO IV). These tumors were complicated by numerous peritoneal, uterine, colorectal, and pancreatic metastases that lead to a radical surgery followed by chemotherapy. Her father and mother, at 56 and 52 years of age, respectively, had neither personal nor familial history of cancer. In cancer genetics, the development of a bilateral ovarian adenocarcinoma at 21 years of age is strongly suggestive of a genetic predisposition. Screening for BRCA1, BRCA2 and TP53 point mutations and genomic rearrangements in the index case peripheral blood DNA had revealed no germline mutation. Considering the absence of cancer in the family, we speculated that a germline de novo mutation could be responsible for the early-onset of the tumors observed in this patient. To identify such mutations, we performed a comparative exome analysis of the patient and her healthy parents (Supp. Materials and Methods). For the parents, exome sequencing was performed on peripheral blood DNA. For the index case, the exome was performed on genomic DNA extracted from nontumoral ovarian tissue to be able to detect also postzygotic de novo mutations that may be present only in the ovarian tissue and could be missed by analyzing the peripheral blood DNA. Across the three exomes, we obtained an average of 8 Gb with 98% of mappable sequences, a mean Read Depth of 69x, 88% of bases were covered to a minimum depth of 10x and 89% of the read bases had a Qscore above 30. On average, 17,395 exonic variants were identified per exome. First, variants with a read coverage of less than 10× and a Qscore <30 were filtered out. We first checked in the index case exome the absence of deleterious mutations within the different genes known to be involved in the genetic determinism of ovarian cancer [Lynch et al., 2009; Loveday et al., 2011; Walsh et al., 2011]. Then, using the Exome Variation Analyser software [Coutant et al., 2012], we filtered the variations against the 1000 Genomes Project data set (May 2011, 20101123 release) and the ESP cohort data set (ESP6500), using an allelic frequency filter of 0.001. Variations present in our in-house database including 72 exomes were also filtered out. With these additional filters, an average of 295 novel variations was retained per individual. When we subtracted the variants detected in the parents from those detected in the index case to detect de novo mutations, 46 variations remained and were analyzed in more detail. Visualization of the BAM files revealed that 37/46 variations were in fact present in a low fraction of the reads obtained in the parents. Among the remaining variations, eight were detected in less than 20% of the reads and Sanger sequencing performed on such variations did not confirm their existence. These variations likely correspond to sequencing artifacts. After this filtering scheme, only one de novo mutation was detected in the patient exome: the c.1157A>G/p.Asn386Ser mutation, within the INHBA gene (NM_002192.2; MIM #147290; www.lovd.nl/inhba) encoding the βA-subunit of inhibin/activin proteins that play a key role in ovary [Knight et al., 2012].
The Asn386 residue is located within the carboxy-terminal mature domain of the inhibin βA-subunit and is conserved from human to chicken. This previously unreported mutation is absent from the 1000 Genomes Project data set (May 2011, 20101123 release) and from the Exome Sequencing Project cohort (ESP6500), which includes 4,300 European American individuals. Targeted Sanger sequencing confirmed the presence of this heterozygous mutation in the nontumoral and tumoral ovarian tissues of the patient and also in her peripheral blood. Its absence was confirmed in the peripheral blood DNA of the parents. These results indicate that the c.1157A>G, p.Asn386Ser INHBA mutation is a bona fide germline de novo mutation that had probably occurred at the prezygotic level.
INHBA is a member of the TGF-β super family and encodes the βA-subunit of the activins/inhibins. Homodimerization of the βA-subunit results in activin A and heterodimerization of the βA-subunit with the inhibin α-subunit, encoded by the INHA gene (MIM #147380), results in inhibin A [Walton et al., 2012]. Activins/inhibins play a crucial role in ovarian development and, in ovaries, the different subunits are mostly secreted by granulosa cells and the targeted cells include epithelial cells [Knight et al., 2012]. The average rate of de novo mutations per exome per generation is only 0–1 [Sanders et al., 2012; Veltman and Brunner, 2012]. This rate and the key role of inhibin in ovaries make very unlikely that this INHBA mutation, identified as the single de novo mutation in a sporadic bilateral epithelial ovarian cancer occurring at 21 years of age, is not related to ovarian cancer and that more relevant mutations were missed in the 12% fraction analyzed at lower coverage in the trio exomes.
To determine the impact of the detected INHBA mutation, we performed functional assays in HEK-293F cells (Supp. Materials and Methods). After transient cotransfection of cells with plasmids expressing wild-type α-subunit and wild-type or mutant βA-subunit, we determined the quantitative effect of the mutation on the activin A and inhibin A production in conditioned media, using an E4/E4 and an E4/R1 immunoassay, respectively, as previously described [Makanji et al., 2008]. As shown in Figure1, we observed for the p.Asn386Ser mutant as compared with the wild-type βA-subunit, a 28% decrease in activin A production and a 44% increase in inhibin A production (P = 0.0007 and P = 0.048, respectively, two-tailed t-test). Activin A has been shown to inhibit the growth of epithelial cells including breast cancer cells and ovarian cancer cells through Smads and CDK inhibitor activation [Burdette et al., 2005; Ramachandran et al., 2009] suggesting therefore that a reduction of activin A, such as the one observed with the INHBA p.Asn386Ser mutation, could favor the growth of epithelial ovarian cells. Therefore, the identification in an early-onset sporadic bilateral ovarian adenocarcinoma case of a single de novo mutation of a gene playing a pivotal role in ovarian development and the functional impact of this mutation are two key arguments indicating that germline INHBA mutation can cause, alone or in combination with other transmitted genetic variations, early-onset epithelial ovarian cancer.
Figure 1.
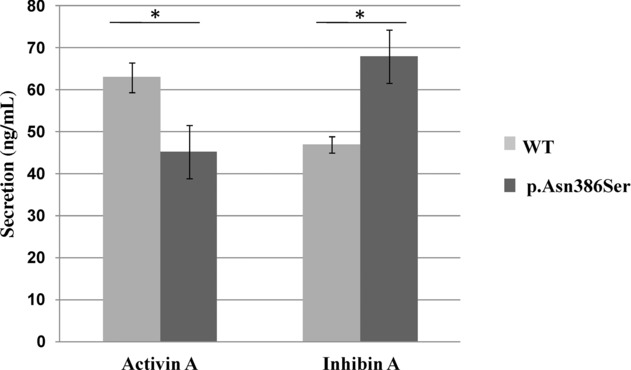
Impact of the INHBA p.Asn386Ser mutation on the amount of secreted activin A and inhibin A. Activin A and inhibin A concentrations in the conditioned media of HEK-293F cells, 2 days after cotransfection of plasmids expressing the inhibin α-subunit and the wild-type or mutant (p.Asn356Ser) βA-subunits, were measured using an E4/E4 immunoassay and an E4/R1 immunoassay, respectively. The results correspond to three independent transfections. *, Two-tailed t-test at P < 0.05.
To determine the prevalence of germline INHBA mutations in early-onset ovarian tumors, we then screened for germline INHBA mutations by Sanger sequencing 43 other patients (14 originated from France and 29 from USA), who had developed ovarian epithelial tumors before 40 years of age and in whom analysis of BRCA1 and BRCA2 genes had revealed no deleterious mutation. We observed no additional mutation of the INHBA gene in these 43 patients. Since the identified de novo mutation of INHBA alters the ratio of inhibin A versus activin A and the βA-subunit heterodimerizes with the α-subunit to form inhibin A, we also screened the 14 French patients for germline mutations of the INHA gene encoding the α-subunit. This analysis revealed, in a Caucasian patient who presented at 29 years of age a serous borderline papillary cystadenoma, a germline heterozygous mutation c.179G>T/p.Arg60Leu (NM_002191.3; www.lovd.nl/INHA), which had not been previously reported in the different databases. DNA from the unaffected parents was not available. The presence of this heterozygous INHA mutation was confirmed in the ovarian tumor of the patient. Arg60 is a very conserved residue that forms part of a secondary furin cleavage site located at the amino terminus of the inhibin α prodomain [Walton et al., 2012]. The measurement, using immunoassays, of inhibin A and B production in conditioned media from HEK-293F cells cotransfected with plasmids expressing wild-type or mutant α-subunit and wild-type βA- or βB-subunit (Fig. 2), revealed that the p.Arg60Leu INHA mutation disrupts both inhibin A biosynthesis (75% decrease, P = 0.00006) and inhibin B biosynthesis (55% decrease, P = 0.002). Remarkably, INHA −/− female mice specifically develop mixed ovarian tumors indicating that INHA could act as a tumor suppressor gene with a gonadal specificity [Matzuk et al., 1992], which is in agreement with the possible oncogenic effect of the p.Arg60Leu INHA gene detected in this study.
Figure 2.
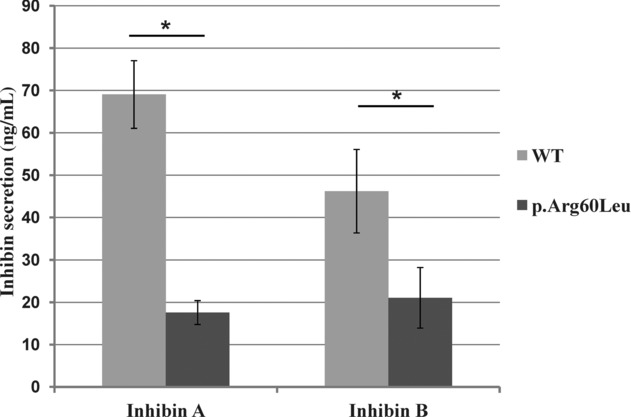
The impact of the INHA p.Arg60Leu mutation on the amount of secreted inhibins A and B. Activin A and B concentrations in the conditioned media of HEK-293F cells, 2 days after cotransfection of plasmids expressing the wild-type or mutant (p.Arg60Leu) inhibin α-subunit and the wild-type βA- or βB-subunits, were measured using an E4/R1 immunoassay and a C5/R1 immunoassay, respectively. The results correspond to six independent transfections. *, Two-tailed t-test at P < 0.05.
Exome sequencing data are available for a cohort of 316 high-grade serous ovarian tumors [The Cancer Genome Atlas Network, 2011]. Somatic alterations of the INHA or INHBA genes are reported in 5% of the cases but no germline mutation of these genes is reported. Because this TCGA cohort is not enriched in early-onset cases but rather reflects the overall age distribution of serous ovarian cancer, we decided to complement the INHBA and INHA mutation screening by Sanger sequencing a second series of 62 other French early-onset ovarian epithelial cases (<40 years), without detectable BRCA mutation. We detected within the INHBA gene an additional new germline heterozygous variation, c.839G>A, p.Gly280Glu (NM_002192.2). This variation was detected in a patient who developed a serous papillary ovarian adenocarcinoma at 27 years of age. DNA sample from the unaffected parents was not available. The Gly280 residue lies within the prodomain of the α-subunit in a region involved in the interaction with the extracellular matrix [Li et al., 2010]. Interestingly, we also detected in this cohort several nonsynonymous variants of the INHA and INHBA that are reported in the databases associated with a low allele frequency (Supp. Table S1). To check for a possible enrichment of rare nonsynonymous variants in our patient cohort compared with the general population, we compared, using five statistical methods dedicated to rare variants, the overall load of rare nonsynonymous variants within INHBA and INHA between the total replication cohort of 76 French early-onset ovarian epithelial tumor cases (corresponding to the 14 French early-onset ovarian tumor cases extended to 62 additional other French cases also analyzed for INHBA and INHA mutations) and two control sequencing datasets, respectively, extracted from the 1000 Genomes Project and from the EVS6500 project (Supp. Materials and Methods). Despite limitations due to both potential population stratification bias and heterogeneity in sequencing technologies, all of the five statistical methods used, including burden and bidirectional tests [Stitziel et al., 2011], agree on a significant enrichment (Supp. Table S2) providing an additional argument in favor of the implication of inhibin mutations in ovarian cancer.
In conclusion, we think that this study provides the following strong arguments showing that alteration of the inhibin/activin pathway may contribute to the development of early-onset epithelial ovarian tumors by the alteration of the inhibin/activin ratio and, therefore, alteration of the cross-talk between granulosa and epithelial cells: (1) the identification, in a bilateral epithelial ovarian cancer developed at 21 years of age, of only one de novo mutation affecting the INHBA that encodes the βA-subunit of inhibin/activin, (2) the role of the inhibin pathway in ovaries, (3) the impact of the INHBA mutation on inhibin/activin production, (4) the subsequent identification of a mutation affecting INHA, which encodes the partner of the βA-subunit, also disrupting in vitro the inhibin/activin production, (5) the existence of an animal model implicating this gene as a tumor suppressor gene with gonadal specificity, and (6) the enrichment of rare inhibin variants in patients with early-onset ovarian cancers, as compared with control sequencing datasets.
More generally, this study highlights the power of comparative trio exome analysis for identifying new molecular pathways involved in the genetic determinism of cancer, by the detection of de novo mutations in patients presenting early-onset single or multiple primary tumors.
Acknowledgments
The authors thank Prof. Mario Tosi for reading the manuscript and commenting on it.
Disclosure statement: The authors declare no conflict of interest.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
References
- Burdette JE, Jeruss JS, Kurley SJ, Lee EJ, Woodruff TK. Activin A mediates growth inhibition and cell cycle arrest through Smads in human breast cancer cells. Cancer Res. 2005;65:7968–7975. doi: 10.1158/0008-5472.CAN-04-3553. [DOI] [PubMed] [Google Scholar]
- Coutant S, Cabot C, Lefebvre A, Léonard M, Prieur-Gaston E, Campion D, Lecroq T, Dauchel H. EVAExome Variation Analyzer, an efficient and versatile tool for filtering strategies in medical genomics. BMC Bioinformatics. 2012;13(Suppl 14):S9. doi: 10.1186/1471-2105-13-S14-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilissen C, Hoischen A, Brunner HG, Veltman JA. Disease gene identification strategies for exome sequencing. Eur J Hum Genet. 2012;20:490–497. doi: 10.1038/ejhg.2011.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight PG, Satchell L, Glister C. Intra-ovarian roles of activins and inhibins. Mol Cell Endocrinol. 2012;359:53–65. doi: 10.1016/j.mce.2011.04.024. [DOI] [PubMed] [Google Scholar]
- Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Shimono C, Norioka N, Nakano I, Okubo T, Yagi Y, Hayashi M, Sato Y, Fujisaki H, Hattori S, Sugiura N, Kimata K, et al. Activin A binds to perlecan through its pro-region that has heparin/heparan sulfate binding activity. J Biol Chem. 2010;285:36645–36655. doi: 10.1074/jbc.M110.177865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loveday C, Turnbull C, Ramsay E, Hughes D, Ruark E, Frankum JR, Bowden G, Kalmyrzaev B, Warren-Perry M, Snape K, Adlard JW, Barwell J, et al. Germline mutations in RAD51D confer susceptibility to ovarian cancer. Nat Genet. 2011;43:879–882. doi: 10.1038/ng.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch HT, Casey MJ, Snyder CL, Bewtra C, Lynch JF, Butts M, Godwin AK. Hereditary ovarian carcinoma: heterogeneity, molecular genetics, pathology, and management. Mol Oncol. 2009;3:97–137. doi: 10.1016/j.molonc.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen BE, Browning SR. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 2009;5:e1000384. doi: 10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makanji Y, Walton KL, Wilce MC, Chan KL, Robertson DM, Harrison CA. Suppression of inhibin A biological activity by alterations in the binding site for betaglycan. J Biol Chem. 2008;283:16743–16751. doi: 10.1074/jbc.M801045200. [DOI] [PubMed] [Google Scholar]
- Matzuk MM, Finegold MJ, Su JG, Hsueh AJ, Bradley A. Alpha-inhibin is a tumour-suppressor gene with gonadal specificity in mice. Nature. 1992;360:313–319. doi: 10.1038/360313a0. [DOI] [PubMed] [Google Scholar]
- Morgenthaler S, Thilly WG. A strategy to discover genes that carry multi-allelic or mono-allelic risk for common diseases: a cohort allelic sums test (CAST. Mutat Res. 2007;615:28–56. doi: 10.1016/j.mrfmmm.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Neale BM, Rivas MA, Voight BF, Altshuler D, Devlin B, Orho-Melander M, Kathiresan S, Purcell SM, Roeder K, Daly MJ. Testing for an unusual distribution of rare variants. PLoS Genet. 2011;7:e1001322. doi: 10.1371/journal.pgen.1001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran A, Marshall ES, Love DR, Baguley BC, Shelling AN. Activin is a potent growth suppressor of epithelial ovarian cancer cells. Cancer Lett. 2009;285:157–165. doi: 10.1016/j.canlet.2009.05.010. [DOI] [PubMed] [Google Scholar]
- Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, Ercan-Senciek AG, DiLullo NM, Parikshak NN, Stein JL, Walker MF, Ober GT, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitziel NO, Kiezun A, Sunyaev S. Computational and statistical approaches to analyzing variants identified by exome sequencing. Genome Biol. 2011;12:227. doi: 10.1186/gb-2011-12-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltman JA, Brunner HG. De novo mutations in human genetic disease. Nat Rev Genet. 2012;13:565–575. doi: 10.1038/nrg3241. [DOI] [PubMed] [Google Scholar]
- Vissers LE, de Ligt J, Gilissen C, Janssen I, Steehouwer M, de Vries P, van Lier B, Arts P, Wieskamp N, del Rosario M, van Bon BW, Hoischen A, et al. A de novo paradigm for mental retardation. Nat Genet. 2010;42:1109–1112. doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- Walsh T, Casadei S, Lee MK, Pennil CC, Nord AS, Thornton AM, Roeb W, Agnew KJ, Stray SM, Wickramanayake A, Norquist B, Pennington KP, et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc Natl Acad Sci USA. 2011;108:18032–18037. doi: 10.1073/pnas.1115052108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton KL, Makanji Y, Harrison CA. New insights into the mechanisms of activin action and inhibition. Mol Cell Endocrinol. 2012;359:2–12. doi: 10.1016/j.mce.2011.06.030. [DOI] [PubMed] [Google Scholar]
- Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article.


