Abstract
Selenite is a predominant form of selenium (Se) available to plants, especially in anaerobic soils, but the molecular mechanism of selenite uptake by plants is not well understood.
ltn1, a rice mutant previously shown to have increased phosphate (Pi) uptake, was found to exhibit higher selenite uptake than the wild-type in both concentration- and time-dependent selenite uptake assays. Respiratory inhibitors significantly inhibited selenite uptake in the wildtype and the ltn1 mutant, indicating that selenite uptake was coupled with H+ and energy-dependent. Selenite uptake was greatly enhanced under Pi-starvation conditions, suggesting that Pi transporters are involved in selenite uptake.
OsPT2, the most abundantly expressed Pi transporter in the roots, is also significantly up-regulated in ltn1 and dramatically induced by Pi starvation. OsPT2-overexpressing and knockdown plants displayed significantly increased and decreased rates of selenite uptake, respectively, suggesting that OsPT2 plays a crucial role in selenite uptake. Se content in rice grains also increased significantly in OsPT2-overexpressing plants.
These data strongly demonstrate that selenite and Pi share similar uptake mechanisms and that OsPT2 is involved in selenite uptake, which provides a potential strategy for breeding Se-enriched rice varieties.
Keywords: molecular mechanism, phosphate transporter, rice (Oryza sativa), selenite uptake, selenium (Se)
Introduction
Selenium (Se) is an essential micronutrient for humans and other animals (Schwarz & Foltz, 1957; Rotruck et al., 1973). It plays important roles in antioxidant function, thyroid hormone metabolism, male fertility, and immune responses (Stadtman, 1990; Kohrle, 2000; Rayman, 2002; Stranges et al., 2006). Several studies have shown that improvement of Se status can lower the risk of cancer (Clark et al., 1996; Combs, 2001; Rayman, 2005). People mainly acquire Se from plant foods through the food chain, especially cereal foods (Rayman, 2012). Because of the low Se concentrations in plant foods, the average daily Se intake is insufficient to meet the requirements of human health (Williams et al., 2009). Agronomic fortification was successfully performed with selenized fertilizers and resulted in an increase in Se intake, but it also increases the cost of agricultural products and brings potential environment risks (Hartikainen, 2005; Broadley et al., 2006). Genetic biofortification is a new and promising strategy which may increase Se accumulation in cereal crops and reduce the need for Se fertilizers (Broadley et al., 2006; Rayman, 2008). To this end, it is important to develop a comprehensive understanding of the mechanism of Se uptake in plants in order to improve the Se content in crops more efficiently.
Selenium occurs naturally as Se2−, Se0, Se2O32−, SeO32−, and SeO42− (Sors et al., 2005). The predominant forms of Se available to plants are selenate and selenite. Redox potential and pH can greatly affect the forms of Se present in soil. Selenate is the dominant species in solution at high redox (pE + pH > 15.0). In the medium redox range (7.5 < pE + pH < 15.0), either SeO32− or HSeO3− species is predominant (Elrashidi et al., 1987). Sulfate transporters are involved in selenate uptake of plants (Terry et al., 2000; White et al., 2004; Sors et al., 2005). AtSultr1;2, a high-affinity sulfate transporter, was first identified through the screening of selenate-resistant mutants in Arabidopsis (Shibagaki et al., 2002; El Kassis et al., 2007). Unlike that of selenate, the mechanism of selenite uptake in plants is not well understood. Previous studies have shown that selenite enters roots through passive diffusion (Shrift & Ulrich, 1969). This hypothesis is supported by the fact that selenite uptake is repressed only 20 or 30% by respiratory inhibitors (Arvy, 1989, 1993). However, it was shown that hydroxylamine and 2,4-dinitrophenol (DNP) inhibited selenite uptake by 72 and 86%, respectively, at pH 4.0 (Ulrich & Shrift, 1968). This discrepancy is mainly a result of the effect of pH on selenite species in the absorption solution (Zhang et al., 2010). Selenous acid is a diprotic weak acid with pKa1 and pKa2 values of 2.57 and 6.60, respectively. Selenite exists as H2SeO3, SeO32−, and HSeO3− at different pH values (Zhang et al., 2006, 2010; Zhao et al., 2010). The proportions of these Se aqueous species vary greatly with pH (Zhang et al., 2006, 2010; Zhao et al., 2010). Selenite could be absorbed by rice roots through aquaporins as the neutral H2SeO3 species (Zhang et al., 2006). Zhao et al. (2010) further demonstrated that a silicon (Si) influx transporter OsNIP2;1 (Lsi1), a nodulin 26-like intrinsic membrane protein (NIP) subfamily of aquaporins, is permeable to H2SeO3. However, how selenite enters roots as HSeO3− remains unclear. Earlier studies indicated that selenite uptake was repressed by an increase in the Pi concentration (Broyer et al., 1972; Hopper & Parker, 1999). Recent studies have suggested that selenite uptake is probably mediated by Pi transporters (Li et al., 2008). However, there is still no convincing molecular evidence to support this hypothesis.
An ideal approach to uncovering the mechanism of selenite uptake is to identify mutants that significantly alter this uptake. Previously, the ltn1 mutant was identified and found to be associated with leaf tip necrosis, predominantly in mature leaves, which is the result of Pi overaccumulation (Hu et al., 2011). Both concentration- and time-dependent selenite uptake assays were performed to determine whether ltn1 also has a higher uptake rate of selenite. ltn1 was indeed found to exhibit much higher rates of selenite uptake than wild-type plants. As the transcript abundances of OsPT1,OsPT2,OsPT4, and OsPT8 were significantly elevated in ltn1 roots, it is therefore reasonable to speculate that enhanced selenite uptake in ltn1 roots may be correlated with up-regulation of these Pi transporters. The present work demonstrated that OsPT2, a Pi transporter, is involved in selenite uptake, which provides direct evidence that this Pi transporter is also responsible for the active uptake of selenite. Se content in rice grains also increased dramatically in OsPT2-overexpressing plants, suggesting that this may be a suitable strategy for breeding Se-enriched rice varieties.
Materials and Methods
Plant materials and growth conditions
Rice (Oryza sativa L.) wild-type Nipponbare and its mutant (ltn1), wild-type Zhonghua11 and transgenic lines (Ox-1, Ox-3, Ri-2, and Ri-5) were used in this study. To evaluate selenite uptake in rice seedling, seeds were surfaced-sterilized, rinsed with deionized water, and then germinated on filter papers in an incubator at 35°C in darkness. When the radicals were 2 cm long, the seedlings were precultured in half-strength Kimura B nutrient solution (Ma et al., 2001) in an environmentally controlled growth chamber, with a light : dark cycle of 16 : 8 h (24 : 18°C). The light intensity was c. 300 μmol m−2 s−1. The humidity was controlled at 67%. After 3 d, seedlings were transferred to full-strength nutrient solution and grown for another 12 d, and then uniform seedlings were transplanted to 18 l containers containing the same nutrient solution. The pH was adjusted to 5.5 every day with 1 mM HCl or 1 mM NaOH. The solution was replenished every 4 d. Seedlings were harvested for the Se-uptake experiment after 8 d.
Kinetics of Se absorption
Roots of three individual plants were pretreated in Se-free solution (100 μM KH2PO4, 100 μM Ca(NO3)2 and 100 μM MgCl2, pH 5.5) for 0.5 h, then transferred to the absorption solution containing 5 mM MES (2-morpholinoethanesulphonic acid), 0.5 mM Ca(NO3)2 and different Se concentrations (0, 0.1, 0.2, 0.4, 0.8, 1.6, 3.2, 6.4, and 9.6 μM Na2SeO3, pH 5.5). After 3 h absorption, the roots were rinsed with an ice-cold desorption solution (5 mM MES, 0.5 mM Ca(NO3)2 and 0.5 mM K2SO4 (pH 5.5)) and then soaked in the same solution for 15 min to remove external Se. They were then blotted dry and used for Se analysis.
Similarly, roots of three individual plants were first pretreated in Se-free solution, then moved to absorption solution supplemented with 2 μM Na2SeO3 for 0.5, 1.0, 1.5, 2.0, 2.5, 3.0, 3.5, and 4.0 h. After the Se absorption, the roots were rinsed with the desorption solution and dried for Se analysis.
Effect of respiratory inhibitors on selenite uptake
Roots of three individual plants were pretreated in Se-free solution for 30 min and then transferred to an absorption solution containing 2.0 μM Na2SeO3 with or without 1 μM carbonyl cyanide m-chlorophenylhydrazone (CCCP) or 0.1 mM DNP. CCCP was initially dissolved in ethanol and added to the absorption solution with a final ethanol concentration of 0.01% (v/v) (Li et al., 2008). After 2 h absorption, the roots were rinsed with an ice-cold desorption solution and then immersed in the same solution for 15 min to remove external Se. Roots were blotted dry and analyzed for Se content. All reagents were purchased from Sigma.
Examination of selenite uptake
Seedlings of wild-type and ltn1 plants were washed thoroughly in deionized water and then transferred to normal, P-deficient or S-deficient medium. The medium was a modified version of Kimura B nutrient solution. The control was a normal nutrient solution. In the S-deficient and P-deficient solutions, KH2PO4, MgSO4, ZnSO4, and CuSO4 were substituted by an equimolar amount of corresponding chloride salts. After 3 d, seedlings were transferred to normal, S-deficient or P-deficient medium containing 2 μM Na2SeO3 for another 3 d, and then the roots were rinsed, dried, and analyzed for Se content.
Vector construction and rice transformation
For overexpression vector construction, the open reading frame (ORF) of OsPT2 was amplified and cloned into binary vector pCambia2300Actin between restriction sites XbaI and PstI. For RNAi vector construction, a 305 bp fragment of OsPT2 ORF was cloned in both orientations in pCambia2300Actin between restriction sites SalI and BamHI, separated by the first intron of GA20 oxidase of potato to form a hairpin structure (Luo et al., 2005). The resulting vectors were transformed into wild-type rice (Zhonghua11) using Agrobacterium tumefaciens-mediated transformation (Liu et al., 2006).
Selenite uptake
Roots of three individual plants were pretreated in Se-free solution for 30 min, then transferred to the absorption solution. After absorption for 3 h, the roots were rinsed with an ice-cold desorption solution, then soaked in it for 15 min to clean up the external Se, and then blotted dry before separating roots and shoots for Se analysis.
OsPT2 RNA in situ hybridization
RNA in situ hybridization was performed as previously described (Li et al., 2011). The primers used for probe amplification are GCACAAACTTCCTCGGTATG (ISPTZ-1F, sense probe) and AAGAGAAACCCCACAAATCC (ISPTZ-1R, antisense probe). Digoxigenin-labeled RNA probes were prepared using a DIG Northern Starter Kit (catalogue no. 2039672; Roche) according to the manufacturer's instructions. The hybridization signals were observed using bright field imaging with a microscope (Olympus BX51, Tokyo, Japan) and photographed with a Micro Color CCD camera (DVC Co., Austin, TX, USA).
Field experiment
Wild-type rice (Zhonghua11) and transgenic lines (Ox-1, Ox-3, Ri-2, and Ri-5) were planted in the field of the experimental station of the Institute of Genetics and Developmental Biology (IGDB), Chinese Academy of Sciences (CAS), in Lingshui County of Hainan Province, China. The area of the plot for wildtype rice and each transgenic line was 10 m2, with three replicates. The soil type is clay with pH 5.2. The concentrations of organic matter, available N, available P and available K were 19 620, 123.67, 32.64, and 137.23 mg kg−1, respectively. The concentrations of total Se and available Se in the soil were 0.18 mg kg−1 and 5.23 μg kg−1, respectively. The cultural management practices were the same for wild-type and transgenic rice. Upon ripening, the rice was harvested and the grains were dehusked by hand. The Se content was determined in brown rice grains.
Analysis of total Se
All glass vessels were initially soaked overnight in 10% HCl to eliminate Se contamination, then rinsed in double deionized water (> 18 MΩ) and oven-dried. Dried and homogenized samples (c. 100 mg) were weighed and put into 100 ml digestion tubes. Then 5 ml of an acid mixture of 4 ml HNO3 and 1 ml HClO4 were added. Samples were digested overnight at 25°C, then completely digested at 150°C in a digestion oven. After cooling, the digests were diluted with double deionized water to a final volume of 25 ml. Total Se in the digested samples was determined by inductively coupled plasma mass spectrometry (ICP-MS). All chemical reagents used were of a high-purity grade. A standard tea material (GSV-4, 0.072 mg Se kg−1, GBW07605; The Institute of Geophysical and Geochemical Exploration, Langfang City, China) and a blank were simultaneously digested with the test samples for quality control, with an average Se recovery of between 89 and 93% (Zhang et al., 2006).
Statistical analysis
Analysis of variance was performed using SPSS 13.0 for Windows (SPSS Inc., Chicago, IL, USA) to determine the significance of any differences between control and treatments. Statistical differences were assessed by Student's t-test. All data shown were the means of at least three samples.
Results
Selenite uptake is significantly increased in the ltn1 mutant
Both concentration- and time-dependent selenite uptake experiments were performed to evaluate whether ltn1 has a higher rate of uptake of selenite than the wild-type. Concentration-dependent kinetics suggested that selenite uptake by ltn1 increased in proportion to the Se concentration in the absorption solution (Fig.1a). A linear equation was fitted to the data with regression coefficients of 0.99. Selenite uptake by ltn1 became significantly higher than the wild-type as Se concentrations increased; however, selenite uptake by the wild-type followed saturation kinetics as Se concentrations increased. The data fitted a Michaelis–Menten saturation curve (R2 = 0.98). Vmax (maximum enzymatic reaction rate) was 4.45 μmol kg−1 DW h−1, and Km (substrate concentration at which the reaction rate is half maximum) was 3.77 μmol kg−1 DW. Time-dependent selenite uptake showed that ltn1 had significantly higher Se concentrations than the wild-type at all Se-treated time-points (Fig.1b). After 3 h exposure, Se concentrations in wild-type roots almost reached a plateau, whereas Se concentrations in ltn1 kept increasing with extending Se treatment. Given that ltn1 was characterized as a Pi overaccumulation mutant, it was reasonable to speculate that selenite uptake might be associated with the Pi uptake pathway.
Figure 1.
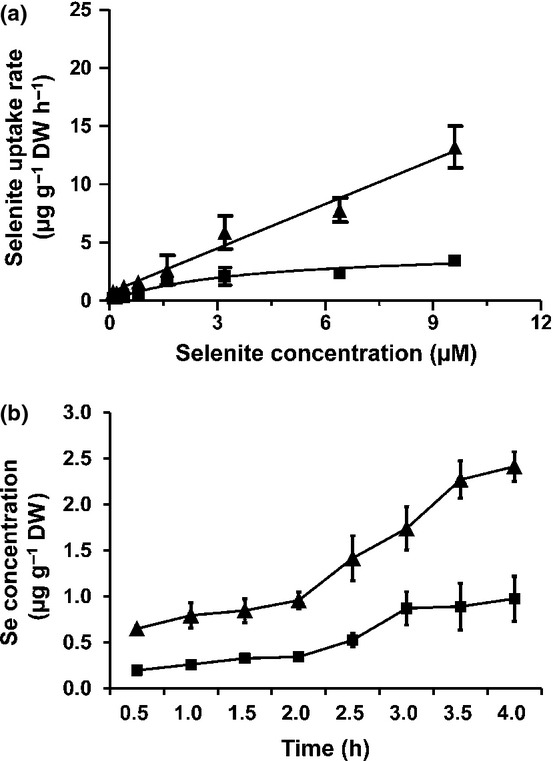
Difference in concentration- (a) and time-dependent (b) selenite uptake by roots of Nipponbare (Oryza sativa; squares) and its mutant ltn1 (triangles). Error bars indicate mean values ± SD (n = 5).
Selenite uptake is enhanced by Pi starvation
To investigate the effects of P or S starvation on selenite uptake, wild-type and ltn1 rice seedlings were grown in normal, P-deficient, or S-deficient medium. After 3 d, seedlings were then transferred, respectively, to normal, P-, or S-deficient medium containing 2 μM Na2SeO3 for another 3 d and the Se content was determined. The results showed that the Se content in the roots of wild-type plants and ltn1 mutants in P-deficient medium was significantly higher than that of the control, but S starvation had no effect on the Se content of either wild-type plants or ltn1 mutants (Fig.2). Under P-starvation conditions, the concentrations of Se in roots of the wild-type and ltn1 mutants were increased 2.58- and 3.81-fold relative to the control, respectively. These results showed that the selenite uptake capacity of wild-type and ltn1 plants was significantly increased under P-deficient conditions, indicated that Pi deficiency dramatically promotes selenite uptake.
Figure 2.
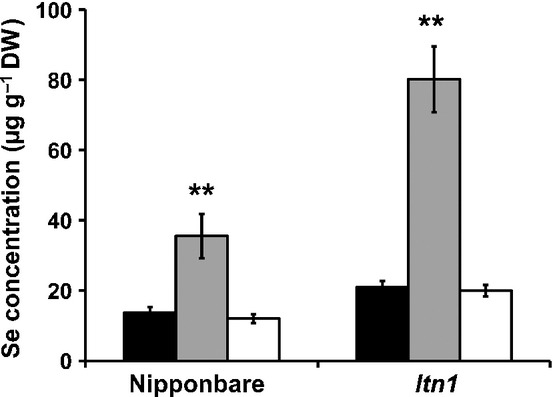
Effects of phosphorus (P) and sulfur (S) starvation on selenium (Se) concentration in roots of Nipponbare (the wild-type, Oryza sativa) and its mutant ltn1. Error bars indicate mean values ± SD (n = 5). Asterisks indicate the significant differences between the wild-type and ltn1 as determined by Student's t-test: **, P < 0.01. Control, black bars; no P, gray bars; no S, white bars.
Selenite uptake was mediated by symport with H+
To determine whether selenite uptake was mediated by symport of H+, selenite uptake of wild-type and ltn1 plants was measured after the roots had been exposed to 2 μM Na2SeO3 absorption solutions containing 1 μM CCCP or 20 μM DNP for 2 h. The control was supplied with the same absorption solutions without CCCP or DNP. Selenite uptake of both wild-type and ltn1 plants was significantly lower than in controls after addition of 1 μM CCCP or 20 μM DNP to the absorption solutions, and the rate of selenite uptake of ltn1 plants was reduced to the same value as the wild-type (Fig.3). Both CCCP and DNP are typical protonophores, which allow protons to freely transverse the membrane and inhibit anion uptake by depolarizing the electrical potential across the plasma membrane (Shioi & Taylor, 1984). These results indicated that selenite uptake was energy-dependent and mediated by symport of H+ and selenite anion, which is consistent with Pi uptake (Pao et al., 1998).
Figure 3.
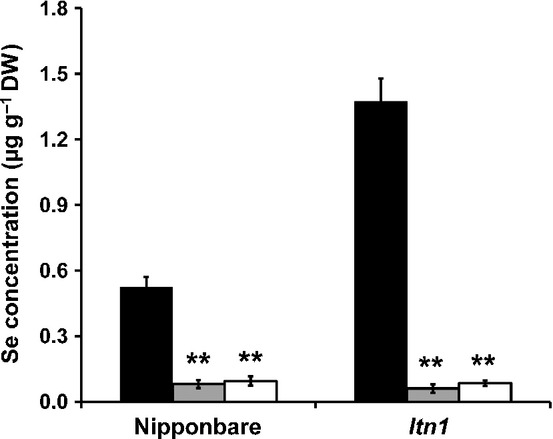
Effects of carbonyl cyanide m-chlorophenylhydrazone (CCCP) and 2,4-dinitrophenol (DNP) on selenite uptake by roots of Nipponbare (the wild-type, Oryza sativa) and ltn1. Error bars indicate mean values ± SD (n = 5). Asterisks indicate the significant differences between the wild-type and ltn1 as determined by Student's t-test: **, P < 0.01. Control, black bars; CCCP, gray bars; DNP, white bars.
OsPT2 was expressed most abundantly under Pi-deficient conditions
To examine the expression of Pi transporters, the wild-type and ltn1 plants were exposed to Pi-sufficient and Pi-deficient medium and the expression of the Pi transporters was determined by real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR). The results showed that the expression of OsPT1,OsPT2,OsPT4,OsPT6, and OsPT8 was substantially up-regulated in ltn1 relative to the wild-type under Pi-sufficient conditions, as in previous results (Fig.4a). Under Pi-deficient conditions, the expression of OsPT1,OsPT2,OsPT4,OsPT6, and OsPT8 was substantially up-regulated in both the wild-type and ltn1 (Fig.4b). The expression of OsPT2 was the most abundant and was far more highly expressed than other Pi transporters. These results indicate that OsPT2 probably plays a major role in either Pi or selenite uptake in rice.
Figure 4.
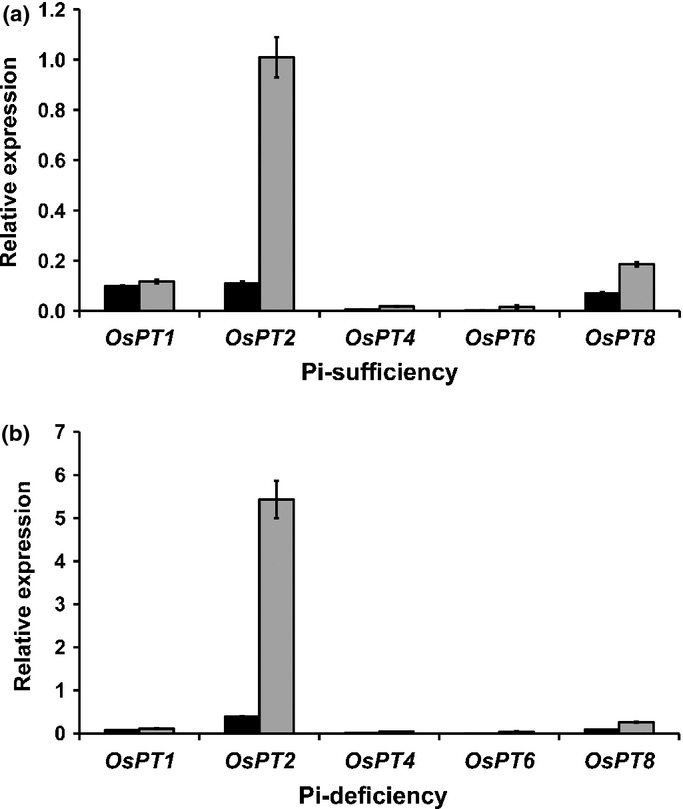
Effect of phosphorus (P) on gene expression of Pi transporters in roots of Nipponbare (the wild-type, Oryza sativa; black bars) and ltn1 (gray bars) under Pi-sufficient (a) and Pi-deficient (b) conditions. Error bars indicate mean values ± SD (n = 3).
Selenite uptake and selenium accumulation were correlated with OsPT2 expression
To further confirm the involvement of OsPT2 in selenite uptake, transgenic rice plants with overexpressing OsPT2 and knockdown OsPT2 by RNAi were generated. The results showed that the expression of OsPT2 was significantly higher in the overexpression lines but dramatically repressed in the RNAi lines (Fig.5a,b).
Figure 5.
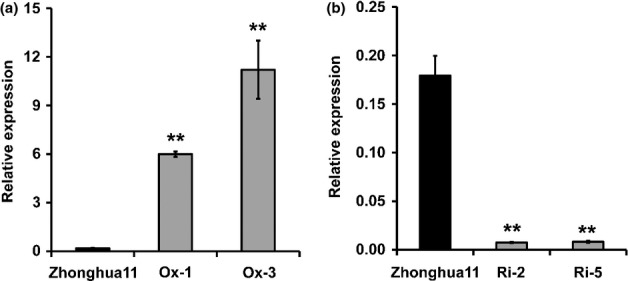
Expression of the OsPT2 gene in Zhonghua11 (Oryza sativa) and OsPT2 transgenic lines. Ox-1 and Ox-3 are two OsPT2-overexpressing lines; Ri-2 and Ri-5 are two OsPT2-RNAi lines. Error bars indicate mean values ± SD (n = 3). Asterisks indicate the significant differences between Zhonghua11 and OsPT2 transgenic lines as determined by Student's t-test: **, P < 0.01.
To investigate the relationship between selenite uptake and OsPT2 expression, the selenite uptake rate of the wild-type, OsPT2-overexpressing lines, and OsPT2 RNAi lines were compared in the presence of 2 μM Na2SeO3. After 3 h of incubation, the uptake rate of the two overexpression lines was significantly higher and that of the two RNAi lines was significantly lower than the wild-type (Fig.6a). After 4 d of incubation, Se content in these two overexpression lines was significantly higher than the wild-type in both roots and shoots. By contrast, the Se content of the RNAi lines was significantly lower than in the wild-type (Fig.6b). These results further demonstrated that OsPT2 played a crucial role in selenite uptake.
Figure 6.
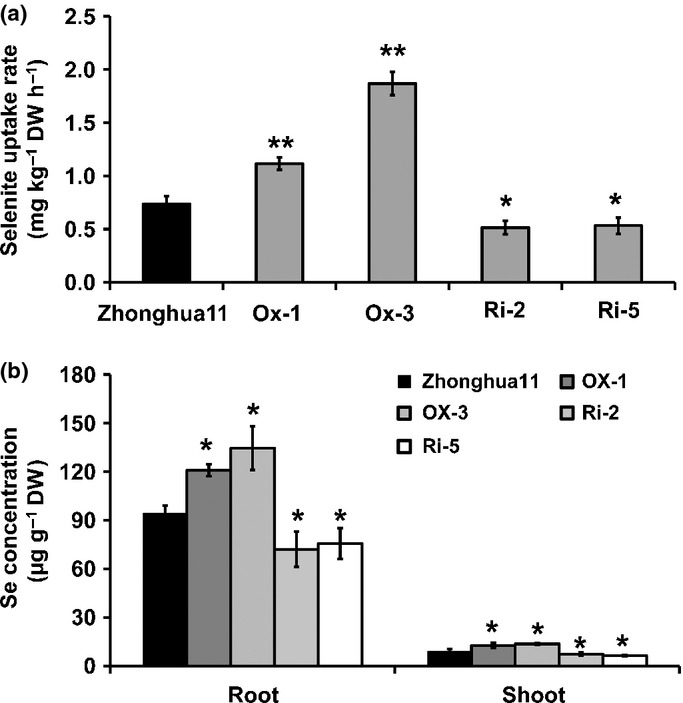
(a) Difference in selenite uptake by roots of Zhonghua11 (Oryza sativa) and OsPT2 transgenic lines. (b) Difference in selenium (Se) concentration in roots and shoots of Zhonghua11 and transgenic lines. Ox-1 and Ox-3 are two OsPT2-overexpressing lines; Ri-2 and Ri-5 are two OsPT2-RNAi lines. Error bars indicate mean values ± SD (n = 3). Asterisks indicate the significant differences between Zhonghua11 and OsPT2 transgenic lines as determined by Student's t-test: *, P < 0.05; **, P < 0.01.
OsPT2 was predominantly expressed in epidermal cells of primary roots
Previous studies have shown that OsPT2 can mediate Pi uptake from the external solution (Ai et al., 2009). It was therefore postulated that OsPT2 may be expressed in the epidermal cells of the primary root under P-deprived conditions. To confirm this hypothesis, RNA in situ hybridization was performed. Results showed that OsPT2 was highly expressed in the epidermal tissue of the primary roots with an OsPT2 antisense probe (Fig.7a). By contrast, hybridization with an OsPT2 sense probe showed no signal (Fig.7b). This location of the tissue strongly supported the conclusion that OsPT2 could mediate Pi uptake by the roots.
Figure 7.
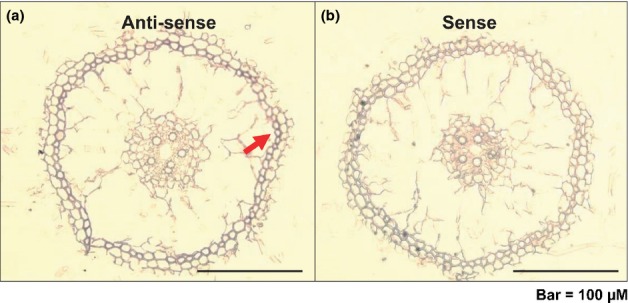
(a) OsPT2 tissue location in young root detected by mRNA in situ hybridization. The OsPT2 signal was detected in the epidermal tissue (indicated by a red arrow) of the primary roots of Zhonghua11 (Oryza sativa). (b) Negative control preparation made with an OsPT2 sense probe.
Se content was increased in rice grains of OsPT2-overexpressing plants
To investigate the effect of OsPT2 gene expression on Se content of rice grains, wild-type and transgenic rice plants were planted in a field and given the same cultural management. After harvest, the Se content of the grains was determined. The results showed that the Se content in rice grains of OsPT2-overexpressing plants (Ox-1 and Ox-3) was significantly higher than in the wild-type (Fig.8). Consistently, the Se content was significantly lower than the wild-type in the grains of OsPT2 RNAi lines (Ri-2 and Ri-5) (Fig.8) These results further demonstrated that Se content in seeds can be improved by enhanced Se uptake.
Figure 8.
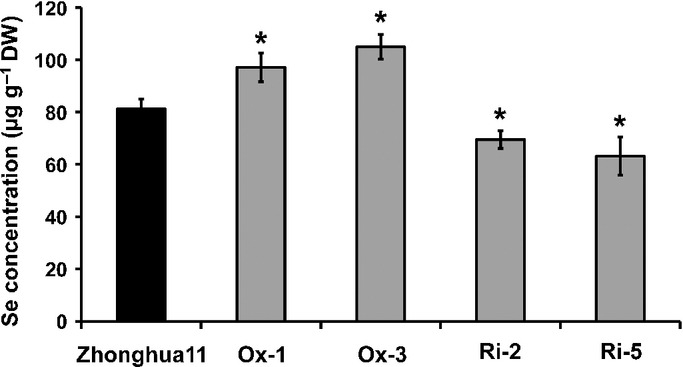
Selenium (Se) concentration in rice grains of Zhonghua11 (Oryza sativa) and OsPT2 transgenic lines. Ox-1 and Ox-3 are two OsPT2-overexpressing lines; Ri-2 and Ri-5 are two OsPT2-RNAi lines. Error bars indicate mean values ± SD (n = 3). Asterisks indicate the significant differences between Zhonghua11 (Oryza sativa) and OsPT2 transgenic lines as determined by Student's t-test: *, P < 0.05.
Discussion
Active uptake of selenite was mediated by Pi transporters
In the present study, it was shown that ltn1 exhibited a much higher selenite uptake rate than the wild-type. The ltn1 mutant was previously identified with Pi overaccumulation in the leaves (Hu et al., 2011). It has been strongly suggested that selenite uptake has a direct relationship with Pi uptake and may be mediated by Pi transporters. CCCP and DNP are typical protonophores, which allow protons to freely transverse the membrane and collapse the proton motive force, which results in the inhibition of anion uptake (Shioi & Taylor, 1984). Both CCCP and DNP significantly inhibited selenite uptake in wild-type and ltn1 plants, suggesting that the selenite uptake process is energy-dependent and involves symport of H+ and selenite anion. Previous studies have demonstrated that Pi uptake is strongly depressed by CCCP and DNP and mediated by H+/H2PO4− symport across the plasma membrane (Ullrich-Eberius et al., 1981; Leggewie et al., 1997; Daram et al., 1998; Liu et al., 1998; Raghothama, 1999). Selenite uptake was therefore postulated to be closely related to the H+/H2PO4− symporter.
OsPT2 is responsible for selenite uptake
A large number of the genes encoding Pi transporters have been identified in different plant species and are generally classified into Pht1, Pht2, Pht3, and PT gene families (Rausch & Bucher, 2002). The Pht1 family plays a crucial role in both Pi uptake from soil and translocation from roots to shoots. There are 13 members of the Pht1 family in the rice genome, of which 10 are expressed in roots; the transcripts of OsPT12 and OsPT13 were not detected in roots and the mRNA of OsPT7 was barely detectable (Paszkowski et al., 2002). Expression analysis revealed transcripts of OsPT1,OsPT2,OsPT4, and OsPT8 to be significantly up-regulated in ltn1 roots relative to the wild-type, with OsPT2 showing the highest expression. Under Pi-deficient conditions, the transcripts of OsPT1,OsPT2,OsPT4, and OsPT8 were significantly up-regulated in both the wild-type and ltn1 roots, and the expression of OsPT2 was the most abundant. Selenite uptake assays showed that ltn1 exhibits greater selenite uptake than the wild-type at different Se concentrations and time-points during Se treatment. Moreover, selenite uptake was much more pronounced in both wild-type and ltn1 plants under Pi-starvation conditions than under normal conditions, which can activate Pi transporters to increase Pi uptake. Therefore, it could be concluded that Pi transport is involved in selenite uptake. Among the Pi transporters, OsPT2 was found to be the most abundantly expressed one and most significantly up-regulated in ltn1, and was also more dramatically induced by Pi starvation. Therefore, it is most likely that enhanced selenite uptake in both the wildtype and ltn1 roots is mainly correlated with up-regulation of OsPT2 gene expression. The rate of selenite uptake was significantly higher in OsPT2 overexpression lines than in the wild-type and significantly lower in RNAi lines, strongly suggesting that OsPT2 was involved in selenite uptake.
Putative selenite-H+ symport process in the plant cell membrane
Plant Pht1 transporters belong to the major facilitator superfamily (MFS) transporters, which transduce electrochemical energy stored in the proton gradients into substrate concentration gradients (Saier et al., 1999; Kaback et al., 2001). The bacterial MFS transporters have 12 transmembrane domains, and both the N- and C-terminal halves surround a central hydrophilic cavity containing the substrate-binding sites determining specificity (Maiden et al., 1987; Pao et al., 1998; Hirai et al., 2002). Plant Pi transporters are predicted to have 12 transmembrane domains (TMs), containing two partially duplicated subdomains of six TM segments (Saier, 2000; Lagerstedt et al., 2004). To date, the process of phosphate-H+ symport in the membrane is still unclear. The putative mechanism of proton and Pi symport through a Pht1 transporter is consistent with the mechanism of proton and glycerol-3-phosphate symport in Escherichia coli (Abramson et al., 2003; Huang et al., 2003). Therefore, a possible symport process of proton and selenite through a phosphate-H+ symporter might be involved in protonation of OsPT2, with hydrophilic pores open outside and binding of the selenite anions, and then conformational change leading to opening of the pores inside, the release of selenite, OsPT2 deprotonation, and a return to the outward face conformation.
The process of phosphate-H+ symport was initially involved in Pi recognition. The specificity to Pi in substrate-binding sites in the transporter is determined by a few key amino acid residues. Arg45 and Arg269 are located at the proposed substrate-binding site and often participate in Pi recognition in proteins by forming hydrogen bonds with its oxygen atoms in glycerol-3-phosphate transporter from Escherichia coli (Huang et al., 2003). However, the Pi recognition site in plant Pi transporters is unclear. Previous studies have shown that a number of plant transporters do not exhibit high specificity for substrates. For example, an Arabidopsis sulfate transporter, Sultr1;2, identified from screening for mutants resistant to selenate, can also transport selenate, because of the structural and chemical similarity to sulfate (Shibagaki et al., 2002; El Kassis et al., 2007). A silicon influx transporter, OsNIP2;1, is permeable to arsenite, methylated arsenic, and selenite in rice roots (Ma et al., 2008; Zhao et al., 2010). In our work, OsPT2 was found to transport selenite as well as Pi, indicating that it does not have a highly specific binding site for Pi.
OsPT2 participates in selenite uptake but not root-to-shoot translocation
OsPT2's main role may be in Pi translocation, and up-regulation of OsPT2 may elevate Pi in rice shoots and result in Pi toxicity under normal conditions (Ai et al., 2009). However, the selenite that was taken up was poorly translocated to shoots and retained in the roots of plants overexpressing OsPT2. Previous studies have shown that selenite does not normally accumulate in the roots, and most of the Se in selenite-fed plants was converted to selenate or an unknown compound, postulated to be a selenotrisulfide (Asher et al., 1977). When treated with selenite for 1 h, Astragalus crotalariae contained c. 4% selenite, 2% selenate, and 92% as a neutral or basic form, most probably MeSeCys (Shrift & Ulrich, 1969). Brassica juncea treated with selenite showed 4.3% selenite, 51.2% selenomethionine Se-oxide hydrate, 1.2% S-(methylseleno) cysteine, and 34.2% selenomethionine accumulation in the shoots (Kahakachchi et al., 2004). In selenite-treated plants, selenomethionine, selenomethionine Se-oxide, Se-methyl-selenocysteine, and several other unidentified Se species were detected in the root extracts and xylem sap with limited translocation to shoots (Li et al., 2008). These results suggest that the major portion of selenite was readily reduced to organic Se compounds such as selenomethionine in roots. This explains why OsPT2 was not involved in selenite translocation from roots to shoots. However, the Se content was higher in the shoots of OsPT2-overexpressing plants than in the wild-type. This was predominantly the result of involvement of OsPT2 in selenite uptake and the resulting higher rate of Se accumulation in roots. Consequently, more selenite was speculated to be converted to organic Se and then transported to shoots, which resulted in greater Se accumulation in shoots. Therefore, increasing selenite uptake is a prerequisite to Se accumulation in shoots.
OsPT2 is predominantly expressed in roots. Under P-deprived conditions, it was strongly induced and expressed in the stele of primary roots and lateral roots, but not in epidermal or cortical cells (Ai et al., 2009). However, selenite was readily converted to other organic forms in epidermal and cortical cells before transportation to the stele. This raises the issue why OsPT2 could increase selenite uptake in roots if it could not transport organic Se. In this study, OsPT2 was also found to be highly expressed in the epidermal tissues of the primary roots, further supporting the idea of involvement of OsPT2 in selenite uptake. One reasonable explanation is that OsPT2 might also be expressed in root epidermal and cortical cells, depending on the developmental stages of root formation.
Overexpression of OsPT2 can improve Se content in rice grains
A global survey of Se in rice showed that Se concentrations in major rice-producing and rice-consuming countries are low and c. 75% of the grains from the production and export pools would fail to provide 70% of daily recommended Se intake (Williams et al., 2009). It is well documented that spraying foliage with Se was an effective way to increase Se content in rice grains and is widely used in agriculture (Cao et al., 2001; Hu et al., 2002). However, this practice is often limited by weather factors such as wind and rain. Moreover, the application of Se to foliage may pose some environmental risks. In the present study, OsPT2-overexpressing plants showed significantly higher Se content in rice grains than the wild-type, suggesting that genetic biofortification of Se in rice grains is a feasible means of increasing Se uptake, which may replace foliage spray or application of Se fertilizers in soils. When selenite is absorbed by plants overexpressing OsPT2, most of that Se is accumulated in the roots. The next target might be to facilitate the root-to-shoot movement of Se to further improve Se content in rice grains. It is therefore very important to identify transporters responsible for organic Se translocation from roots to shoots.
In conclusion, the Pi transporter OsPT2 is the first transporter identified in plants that is responsible for the active uptake of selenite. Overexpression of OsPT2 could significantly increase selenite uptake and Se accumulation in both shoots and roots, thus resulting in a higher Se content in rice grains, which could be a potential strategy for breeding Se-enriched rice varieties.
Acknowledgments
This work was supported by the 43rd China Postdoctoral Science Foundation (grant no. 20080430588), the talent foundation (grant no. 09001107) and the research foundation of the Henan University of Science and Technology (grant no. 13560036) and the Chinese Academy of Sciences (XDA08010401). We thank Hongzhi Zhang (Institute of Geographic Sciences and Natural Resources Research, Chinese Academy of Sciences) for carrying out selenium content analysis using ICP-MS.
References
- Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- Ai PH, Sun SB, Zhao JN, Fan XR, Xin WJ, Guo Q, Yu L, Shen QR, Wu P, Miller AJ, et al. Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. Plant Journal. 2009;57:798–809. doi: 10.1111/j.1365-313X.2008.03726.x. [DOI] [PubMed] [Google Scholar]
- Arvy MP. Some factors influencing the uptake and distribution of selenite in the bean plant (Phaseolus vulgaris. Plant and Soil. 1989;117:129–133. ) [Google Scholar]
- Arvy MP. Selenate and selenite uptake and translocation in bean plants (Phaseolus vulgaris. Journal of Experimental Botany. 1993;44:1083–1087. ) [Google Scholar]
- Asher CJ, Butler GW, Peterson PJ. Selenium transport in root systems of tomato. Journal of Experimental Botany. 1977;28:279–291. [Google Scholar]
- Broadley MR, White PJ, Bryson RJ. Biofortification of UK food crops with selenium. Proceedings of the Nutrition Society. 2006;65:169–181. doi: 10.1079/pns2006490. [DOI] [PubMed] [Google Scholar]
- Broyer TC, Johnson CM, Huston RP. Selenium and nutrition of Astragalus. II. Ionic sorption interactions among selenium, phosphate, and macronutrient and micronutrient cations. Plant and Soil. 1972;36:651–669. [Google Scholar]
- Cao ZH, Wang XC, Yao DH, Zhang XL, Wong MH. Selenium geochemistry of paddy soils in Yangtze River Delta. Environment International. 2001;26:335–339. doi: 10.1016/s0160-4120(01)00009-5. [DOI] [PubMed] [Google Scholar]
- Clark LC, Combs GF, Jr, Turnbull BW, Slate EH, Chalker DK, Chow J, Davis LS, Glover RA, Graham GF, Gross EG, et al. Effects of selenium supplementation for cancer prevention in patients with carcinoma of the skin. A randomized controlled trial. Nutritional Prevention of Cancer Study Group. Journal of the American Medical Association. 1996;276:1957–1963. [PubMed] [Google Scholar]
- Combs GF., Jr Impact of selenium and cancer-prevention findings on the nutrition-health paradigm. Nutrition and Cancer. 2001;40:6–11. doi: 10.1207/S15327914NC401_4. [DOI] [PubMed] [Google Scholar]
- Daram P, Brunner S, Persson BL, Amrhein N, Bucher M. Functional analysis and cell-specific expression of a phosphate transporter from tomato. Planta. 1998;206:225–233. doi: 10.1007/s004250050394. [DOI] [PubMed] [Google Scholar]
- El Kassis E, Cathala N, Rouached H, Fourcroy P, Berthomieu P, Terry N, Davidian JC. Characterization of a selenate-resistant Arabidopsis mutant: root growth as a potential target for selenate toxicity. Plant Physiology. 2007;143:1231–1241. doi: 10.1104/pp.106.091462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrashidi MA, Adriano DC, Workman SM, Lindsay WL. Chemical equilibria of selenium in soils: a theoretical development. Soil Science. 1987;144:141–152. [Google Scholar]
- Hartikainen H. Biogeochemistry of selenium and its impact on food chain quality and human health. Journal of Trace Elements in Medicine and Biology. 2005;18:309–318. doi: 10.1016/j.jtemb.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Hirai T, Heymann JA, Shi D, Sarker R, Maloney PC, Subramaniam S. Three-dimensional structure of a bacterial oxalate transporter. Nature Structural and Molecular Biology. 2002;9:597–600. doi: 10.1038/nsb821. [DOI] [PubMed] [Google Scholar]
- Hopper JL, Parker DR. Plant availability of selenite and selenate as influenced by the competing ions phosphate and sulfate. Plant and Soil. 1999;210:199–207. [Google Scholar]
- Hu B, Zhu C, Li F, Tang J, Wang Y, Lin A, Liu L, Che R, Chu C. LEAF TIP NECROSIS1 plays a pivotal role in regulation of multiple phosphate starvation responses in rice. Plant Physiology. 2011;156:1101–1115. doi: 10.1104/pp.110.170209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu QH, Chen LC, Xu J, Zhang Y, Pan GX. Determination of selenium concentration in rice and the effect of foliar application of Se-enriched fertilizer or sodium selenite on the selenium content of rice. Journal of the Science of Food and Agriculture. 2002;82:869–872. [Google Scholar]
- Huang YF, Lemieux MJ, Song JM, Auer M, Wang DN. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science. 2003;301:616–620. doi: 10.1126/science.1087619. [DOI] [PubMed] [Google Scholar]
- Kaback HR, Sahin-Tóth M, Weinglass AB. The kamikaze approach to membrane transport. Nature Reviews Molecular and Cell Biology. 2001;2:610–620. doi: 10.1038/35085077. [DOI] [PubMed] [Google Scholar]
- Kahakachchi C, Boakye HT, Uden PC, Tyson JF. Chromatographic speciation of anionic and neutral selenium compounds in Se-accumulating Brassica juncea (Indian mustard) and in selenized yeast. Journal of Chromatography A. 2004;1054:303–312. [PubMed] [Google Scholar]
- Kohrle J. The deiodinase family: selenoenzymes regulating thyroid hormone availability and action. Cellular and Molecular Life Sciences. 2000;57:1853–1863. doi: 10.1007/PL00000667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagerstedt JO, Voss JC, Wieslander A, Persson BL. Structural modeling of dual-affinity purified Pho84 phosphate transporter. FEBS Letters. 2004;578:262–268. doi: 10.1016/j.febslet.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Leggewie G, Willmitzer L, Riesmeier JW. Two cDNAs from potato are able to complement a phosphate uptake-deficient yeast mutant: identification of phosphate transporters from higher plants. Plant Cell. 1997;9:381–392. doi: 10.1105/tpc.9.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wang Y, Liu L, Hu Y, Zhang F, Mergen S, Wang G, Michael R, Chu C. A rice plastidial nucleotide sugar epimerase is involved in galactolipid biosynthesis and improves photosynthetic efficiency. PLoS Genetics. 2011;7:e1002196. doi: 10.1371/journal.pgen.1002196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HF, Steve PM, Zhao FJ. Selenium uptake, translocation and speciation in wheat supplied with selenate or selenite. New Phytologist. 2008;178:92–102. doi: 10.1111/j.1469-8137.2007.02343.x. [DOI] [PubMed] [Google Scholar]
- Liu H, Trieu AT, Blaylock LA, Harrison MJ. Cloning and characterization of two phosphate transporters from Medicago truncatula roots: regulation in response to phosphate and to colonization by Arbuscular mycorrhizal (AM) fungi. Molecular Plant-Microbe Interactions. 1998;11:14–22. doi: 10.1094/MPMI.1998.11.1.14. [DOI] [PubMed] [Google Scholar]
- Liu X, Bai X, Wang X, Chu C. OsWRKY71, a rice transcription factor, is involved in rice defense response. Journal of Plant Physiology. 2006;164:969–979. doi: 10.1016/j.jplph.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Luo A, Liu L, Tang Z, Bai X, Cao S, Chu C. Down-regulation of OsGRF1 gene in rice rhd1 mutant results in reduced heading date. Journal of Integrative Plant Biology. 2005;47:745–752. [Google Scholar]
- Ma JF, Goto S, Tamai K, Ichii M. Role of root hairs and lateral roots in silicon uptake by rice. Plant Physiology. 2001;127:1773–1780. [PMC free article] [PubMed] [Google Scholar]
- Ma JF, Yamaji N, Mitani N, Xu XY, Su YH, McGrath SP, Zhao FJ. Transporters of arsenite in rice and their role in arsenic accumulation in rice grain. Proceedings of the National Academy of Sciences, USA. 2008;105:9931–9935. doi: 10.1073/pnas.0802361105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiden MC, Davis EO, Baldwin SA, Moore DC, Henderson PJ. Mammalian and bacterial sugar transport proteins are homologous. Nature. 1987;325:641–643. doi: 10.1038/325641a0. [DOI] [PubMed] [Google Scholar]
- Pao SS, Paulsen IT, Saier MH., Jr Major facilitator superfamily. Microbiology and Molecular Biology Reviews. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszkowski U, Kroken S, Roux C, Briggs SP. Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proceedings of the National Academy of Sciences, USA. 2002;99:13324–13329. doi: 10.1073/pnas.202474599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama KG. Phosphate acquisition. Annual Review of Plant Physiology and Plant Molecular Biology. 1999;50:665–693. doi: 10.1146/annurev.arplant.50.1.665. [DOI] [PubMed] [Google Scholar]
- Rausch C, Bucher M. Molecular mechanisms of phosphate transport in plants. Planta. 2002;216:23–37. doi: 10.1007/s00425-002-0921-3. [DOI] [PubMed] [Google Scholar]
- Rayman MP. The argument for increasing selenium intake. Proceedings of the Nutrition Society. 2002;61:203–215. doi: 10.1079/PNS2002153. [DOI] [PubMed] [Google Scholar]
- Rayman MP. Selenium in cancer prevention: a review of the evidence and mechanism of action. Proceedings of the Nutrition Society. 2005;64:527–542. doi: 10.1079/pns2005467. [DOI] [PubMed] [Google Scholar]
- Rayman MP. Food-chain selenium and human health: emphasis on intake. British Journal of Nutrition. 2008;100:254–268. doi: 10.1017/S0007114508939830. [DOI] [PubMed] [Google Scholar]
- Rayman MP. Selenium and human health. Lancet. 2012;379:1256–1268. doi: 10.1016/S0140-6736(11)61452-9. [DOI] [PubMed] [Google Scholar]
- Rotruck JT, Pope AL, Ganther HE, Swanson AB, Hafeman DG, Hoekstra WG. Selenium: biological role as a component of glutathione peroxidase. Science. 1973;179:588–590. doi: 10.1126/science.179.4073.588. [DOI] [PubMed] [Google Scholar]
- Saier MH. Families of transmembrane transporters selective for amino acids and their derivatives. Microbiology. 2000;146:1775–1795. doi: 10.1099/00221287-146-8-1775. [DOI] [PubMed] [Google Scholar]
- Saier MH, Jr, Beatty JT, Goffeau A, Harley KT, Heijne WH, Huang SC, Jack DL, Jack PS, Lew K, Liu J, et al. The major facilitator superfamily. Journal of Molecular Microbiology and Biotechnology. 1999;1:257–279. [PubMed] [Google Scholar]
- Schwarz K, Foltz CM. Selenium as an integral part of Factor 3 against dietary necrotic liver degeneration. Journal of the American Chemical Society. 1957;79:3292–3293. [Google Scholar]
- Shibagaki N, Rose A, McDermott JP, Fujiwara T, Hayashi H, Yoneyama T, Davies JP. Selenate-resistant mutants of Arabidopsis thaliana identify Sultr1;2, a sulfate transporter required for efficient transport of sulfate into roots. Plant Journal. 2002;29:475–486. doi: 10.1046/j.0960-7412.2001.01232.x. [DOI] [PubMed] [Google Scholar]
- Shioi J, Taylor BL. Oxygen taxis and proton motive force in Salmonella typhimurium. Journal of Biological Chemistry. 1984;259:10983–10988. [PubMed] [Google Scholar]
- Shrift A, Ulrich J. Transport of selenate and selenite into Astragalus roots. Plant Physiology. 1969;44:893–896. doi: 10.1104/pp.44.6.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sors TG, Ellis DR, Salt DE. Selenium uptake, translocation, assimilation and metabolic fate in plants. Photosynthesis Research. 2005;86:373–389. doi: 10.1007/s11120-005-5222-9. [DOI] [PubMed] [Google Scholar]
- Stadtman TC. Selenium biochemistry. Annual Review of Biochemistry. 1990;59:111–127. doi: 10.1146/annurev.bi.59.070190.000551. [DOI] [PubMed] [Google Scholar]
- Stranges S, Marshall JR, Trevisan M, Natarajan R, Donahue RP, Combs GF, Farinaro E, Clark LC, Reid ME. Effects of selenium supplementation on cardiovascular disease incidence and mortality: secondary analyses in a randomized clinical trial. American Journal of Epidemiology. 2006;163:694–699. doi: 10.1093/aje/kwj097. [DOI] [PubMed] [Google Scholar]
- Terry N, Zayed AM, de Souza MP, Tarun AS. Selenium in higher plants. Annual Review of Plant Physiology and Plant Molecular Biology. 2000;51:401–432. doi: 10.1146/annurev.arplant.51.1.401. [DOI] [PubMed] [Google Scholar]
- Ullrich-Eberius CI, Novacky A, Fischer E, Lüttge U. Relationship between energy-dependent phosphate uptake and the electrical membrane potential in Lemna gibba G1. Plant Physiology. 1981;67:797–801. doi: 10.1104/pp.67.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich JM, Shrift A. Selenium absorption by excised Astragalus roots. Plant Physiology. 1968;43:14–20. doi: 10.1104/pp.43.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White PJ, Bowen HC, Parmaguru P, Fritz M, Spracklen WP, Spiby RE, Meacham MC, Mead A, Harriman M, Trueman LJ, et al. Interactions between selenium and sulphur nutrition in Arabidopsis thaliana. Journal of Experimental Botany. 2004;55:1927–1937. doi: 10.1093/jxb/erh192. [DOI] [PubMed] [Google Scholar]
- Williams PN, Lombi E, Sun GX, Schekel K, Zhu YG, Feng X, Zhu J, Carey AM, Adomako E, Lawgali Y, et al. Selenium characterization in the global rice supply chain. Environmental Science and Technology. 2009;43:6024–6030. doi: 10.1021/es900671m. [DOI] [PubMed] [Google Scholar]
- Zhang LH, Shi WM, Wang XC. Difference in selenite absorption between high- and low-selenium rice cultivars and its mechanism. Plant and Soil. 2006;282:183–193. [Google Scholar]
- Zhang LH, Yu FY, Shi WM, Li YJ, Miao YF. Physiological characteristics of selenite uptake by maize roots in response to different pH levels. Journal of Plant Nutrition and Soil Science. 2010;173:412–422. [Google Scholar]
- Zhao XQ, Mitani N, Yamaji N, Shen RF, Ma JF. Involvement of silicon influx transporter OsNIP2;1 in selenite uptake in rice. Plant Physiology. 2010;153:1871–1877. doi: 10.1104/pp.110.157867. [DOI] [PMC free article] [PubMed] [Google Scholar]


