Abstract
We previously identified the NS5A/HSP70 binding site to be a hairpin moiety at C-terminus of NS5A domain I and showed a corresponding cyclized polyarginine-tagged synthetic peptide (HCV4) significantly blocks virus production. Here, sequence comparison confirmed five residues to be conserved. Based on NS5A domain I crystal structure, Phe171, Val173, and Tyr178 were predicted to form the binding interface. Substitution of Phe171 and Val173 with more hydrophobic unusual amino acids improved peptide antiviral activity and HSP70 binding, while similar substitutions at Tyr178 had a negative effect. Substitution of non-conserved residues with arginines maintained antiviral activity and HSP70 binding and dispensed with polyarginine tag for cellular entry. Peptide cyclization improved antiviral activity and HSP70 binding. The cyclic retro-inverso analog displayed the best antiviral properties. FTIR spectroscopy confirmed a secondary structure consisting of an N-terminal beta-sheet followed by a turn and a C-terminal beta-sheet. These peptides constitute a new class of anti-HCV compounds.
Keywords: Peptide, HSP70, NS5A, HCV, Protein binding, IRES
Introduction
The hepatitis C virus (HCV) infects 3% of the world population and is mainly responsible for liver transplantation in patients with cirrhosis in developed countries (Shepard et al., 2005). Furthermore, HCV is the most common chronic blood borne pathogen in the United States (U.S.) affecting 1.8% of the population and is the major etiologic factor responsible for the recent doubling of hepatocellular carcinoma (HCC) (El-Serag, 2002).
Recently HCV infection has been treated with a combination of pegylated interferon-α (PEG-IFN), ribavirin (RBV) and NS3/4A protease inhibitors which have improved sustained virological response (SVR) from PEG-IFN and RBV alone, but also increased adverse side effects including anemia and gastrointestinal symptoms (Ciesek and Manns, 2011). In December of 2013 Sofosbuvir which targets the viral polymerase was FDA approved in combination with PEG-IFN and RBV and showed very high efficacy (at least 90% in clinical trials) but at great cost $84,000 to $168,000. Efficacy and adverse events outside clinical trials remain to be evaluated. There is optimism that a combination of direct acting antivirals (DAAs) may eliminate the need for PEG-IFN and RBV, the major culprits responsible for significant side effects. However, DAAs can result in resistant virus as targeting viral proteins puts direct selective pressure for resistant mutants. Therefore, there is still a need for new compounds to use in drug cocktails to improve efficacy, avert resistance, reduce cost, and reduce adverse events.
HCV is an RNA virus classified in the genus Hepacivirus in the Flaviviridae family. It possesses an approximately 9.6kb positive sense RNA genome that is translated as a single polypeptide approximately 3000 amino acids in length (Baron, 1996; Lindenbach and Rice, 2005). It is subsequently proteolytically cleaved into 10 viral proteins including the structural proteins Core, E1, E2, and the integral membrane ion channel p7, as well as the non-structural (NS) proteins NS2, NS3, NS4A, NS4B, NS5A and NS5B (Lindenbach and Rice, 2005). The 5’ non-coding region (NCR) of the viral genome possesses an internal ribosomal entry site (IRES) (Wang et al., 1993), a cis-acting element found in some host RNA transcripts as well as in viruses that allows ribosomal translation initiation to occur internally within a transcript in lieu of 5’ cap dependent translation (Pacheco and Martinez-Salas, 2010). The HCV viral life cycle in a cell can be divided into six phases: 1) binding and internalization, 2) cytoplasmic release and uncoating 3) viral polyprotein translation and processing, 4) RNA genome replication, 5) packaging and assembly, and 6) virus morphogenesis and secretion (Moradpour et al., 2007). NS5A, a 56-59 kDa multi-functional phosphoprotein, is a component of the viral replicase complex and has been implicated in regulation of HCV genome replication, IRES-mediated viral protein translation, virion assembly, and infectious virion secretion (He et al., 2003; Hughes et al., 2009; Khachatoorian et al., 2012a; Khachatoorian et al., 2012b; Tellinghuisen et al., 2008).
Interestingly, cellular heat-shock proteins (HSPs) also play an important role in the replication of RNA viruses (Vasconcelos et al., 1998; Weeks and Miller, 2008; Zheng et al., 2010). HSPs, which normally assist unfolded or mis-folded polypeptide chains to (re)fold into functional proteins, are crucial for cell survival during stressful conditions (Mayer and Bukau, 2005). We have previously identified, through co-immunoprecipitation and subsequent mass spectrometric analyses, an NS5A/HSP complex composed of NS5A, HSP70, and HSP40 (cofactor of HSP70) and demonstrated their co-localization in Huh-7 cells (Gonzalez et al., 2009). We further showed that both NS5A-augmented IRES-mediated translation and virus production are blocked by 1) HSP70 knockdown, 2) the HSP synthesis inhibitor quercetin, a bioflavonoid, and 3) a small hairpin peptide (HCV4) from NS5A domain I that is capable of blocking NS5A/HSP70 interaction, with no associated cytotoxicity (Gonzalez et al., 2009; Khachatoorian et al., 2012a; Khachatoorian et al., 2012b). These findings support our hypothesis that HCV utilizes an NS5A/HSP complex for the IRES-mediated translation of its genome.
The HCV4 hairpin peptide described previously (Khachatoorian et al., 2012b) is a potent small molecule inhibitor of virus specific NS5A-driven IRES-mediated translation of the viral genome and is also capable of disrupting the NS5A/HSP70 complex formation in vitro. This peptide was identified by deletion analyses of NS5A which narrowed down the site of NS5A/HSP70 interaction to the C terminal 34 amino acids of NS5A domain I (C34) followed by alanine scanning mutagenesis on the C34 region. These analyses, identified the NS5A/HSP70 binding region on NS5A to be the hairpin moiety at C34 hereafter referred to as the HSP binding domain (HBD). Subsequently, we sought to further characterize the HBD site and identify the specific amino acid residues involved in this interaction, with the goal of generating peptides that would be even more potent than the HCV4 analog. We refer to these peptides as HSP interfering peptides (HIPs).
Results
Five amino acid residues are conserved in the HSP binding domain (HBD) of NS5A
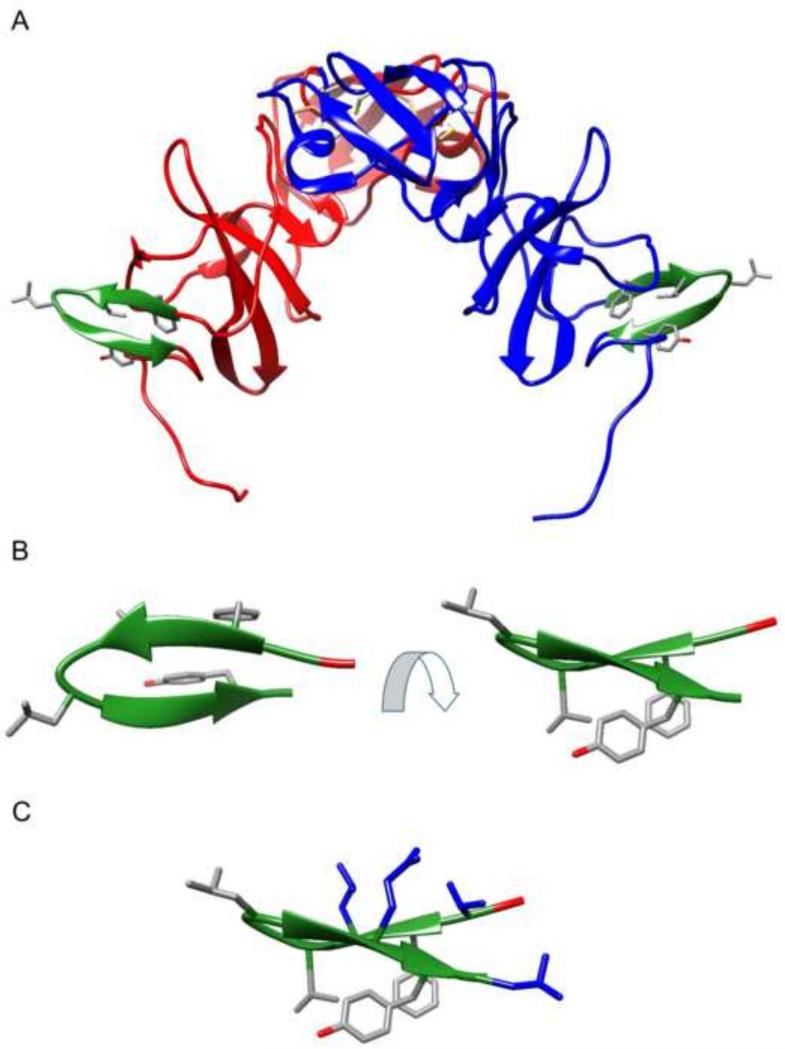
HBD consists of ten amino acids that form a beta-sheet structure at the C terminus of NS5A domain I (Figure 1). Comparison of the H77 and Con1 sequences at the site of HBD revealed five conserved residues: Phe171, Val173, Gly174, Leu175, and Tyr178 (Table 1, highlighted in green). We hypothesized that these five residues may be critical for the NS5A/HSP70 interaction.
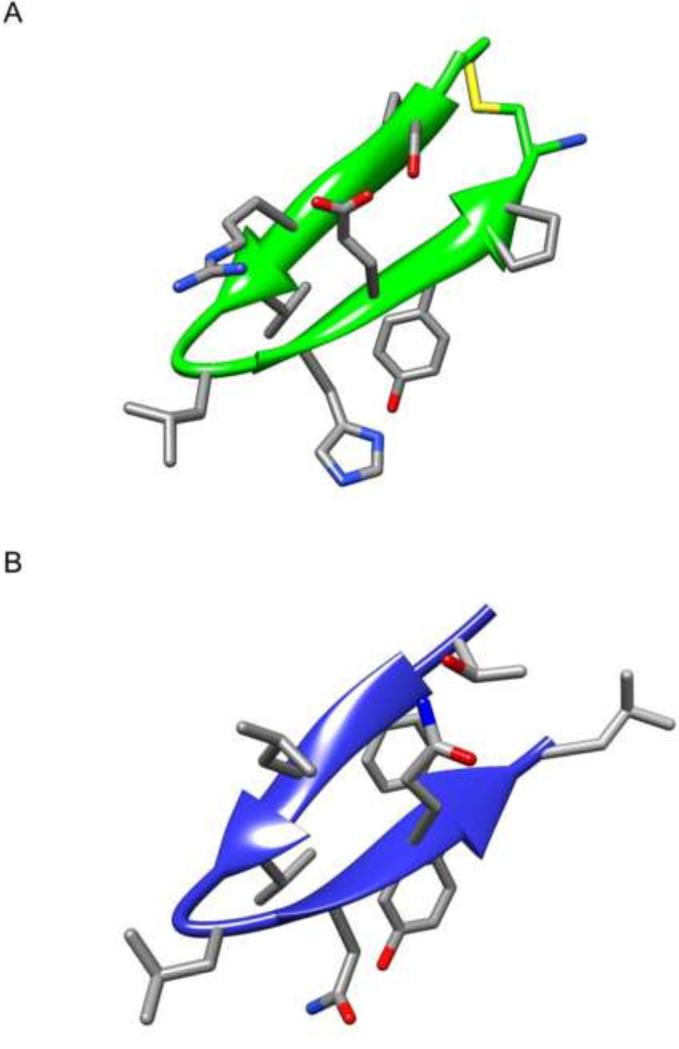
Figure 1.
Schematic representation of the HBD region (green) based on the previously reported crystal structure of dimeric NS5A domain I (Tellinghuisen et al., 2005). A. NS5A domain I dimer. B. The HBD hairpin moiety shown at two different angles. The side chains of four conserved residues (Phe171, Val173, Leu175, and Tyr178) are also shown. C. The HBD hairpin moiety showing four additional residues in blue (Thr170, Leu172, Gln177 and Leu179) that were substituted with arginine residues to enhance cell permeability of HCV peptides.
Table 1.
Sequence and characterization of all peptides generated.
| Sequence | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H77 Amino Acid Position | 169 | 170 | 171 | 172 | 173 | 174 | 175 | 176 | 177 | 178 | 179 | 180 | |||||
| H77 | Val | Ser | Phe | Arg | Val | Gly | Leu | His | Glu | Tyr | Pro | Val | |||||
| Con1 (Crystal Structure) | Val | Thr | Phe | Leu | Val | Gly | Leu | Asn | Gln | Tyr | Leu | Val | |||||
| Peptide | Cyclisation ? | ||||||||||||||||
| HCV1 | Val | Ser | Phe | Arg | Val | Gly | Leu | His | Glu | Tyr | Pro | Val | -CON H2 | No | |||
| HCV2 | (scrambled sequence) | Tyr | Phe | Val | Pro | His | Glu | Ser | Gly | Arg | Val | Val | Leu | -CON H2 | No | ||
| HCV3 | CysS- | Ser | Phe | Arg | Val | Gly | Leu | His | Glu | Tyr | Pro | Cyss | -CON H2 | -S-S- Bridge | |||
| HCV4 | Arg-(Ahx-Arg)6-Ahx-Ahx- | CysS- | Ser | Phe | Arg | Val | D-Pro | Cha | His | Glu | Tyr | Pro | Cyss | -CON H2 | -S-S- Bridge | ||
| HCV5 | Arg | CysS- | Arg | Phe | Arg | Val | D-Pro | Cha | His | Arg | Tyr | Arg | Cyss | Arg | -CON H2 | -S-S- Bridge | |
| HCV6 | Pal-Ahx-Ahx- | Arg | CysS- | Arg | Phe | Arg | Val | D-Pro | Cha | His | Arg | Tyr | Arg | CysS- | Arg | -CON H2 | -S-S- Bridge |
| HCV7 | Arg | CysS- | Arg | Phe | Arg | Val | D-Pro | Cha | His | Arg | Bip | Arg | CysS- | Arg | -CON H2 | -S-S- Bridge | |
| HCV8 | Arg | CysS- | Arg | Phe | Arg | Val | D-Pro | Cha | His | Arg | Dpa | Arg | CysS- | Arg | -CON H2 | -S-S- Bridge | |
| HCV9 | Arg | CysS- | Arg | Phe | Arg | Chg | D-Pro | Cha | His | Arg | Tyr | Arg | CysS- | Arg | -CON H2 | -S-S- Bridge | |
| HCV10 | Arg | CysS- | Arg | Phe | Arg | CystBu | D-Pro | Cha | His | Arg | Tyr | Arg | CysS- | Arg | -CON H2 | -S-S- Bridge | |
| HCV11 | Arg | CysS- | Arg | 2Nal | Arg | Val | D-Pro | Cha | His | Arg | Tyr | Arg | CysS- | Arg | -CON H2 | -S-S- Bridge | |
| HCV12 | Arg | CysS- | Arg | Dpa | Arg | Val | D-Pro | Cha | His | Arg | Tyr | Arg | CysS- | Arg | -CON H2 | -S-S- Bridge | |
| HCV13 | Arg | CysS- | Arg | Trp | Arg | Val | D-Pro | Cha | His | Arg | Dpa | Arg | CysS- | Arg | -CON H2 | -S-S- Bridge | |
| HCV14 | Arg | CysS- | Arg | 2Nal | Arg | Chg | D-Pro | Cha | His | Arg | Bip | Arg | CysS- | Arg | -CON H2 | -S-S- Bridge | |
| HCV15 | CysS- | Arg | 2Nal | Arg | Val | D-Pro | Cha | His | Arg | Tyr | Arg | CysS- | -CON H2 | -S-S- Bridge | |||
| HCV15R | CysSH | Arg | 2Na | Arg | Val | D-Pro | Cha | His | Arg | Tyr | Arg | CysSH | -CON H2 | No (reduced form) | |||
| HCV16 | -Ac | Arg | 2Nal | Arg | Val | D-Pro | Cha | His | Arg | Tyr | Arg | CysS- | -CON H2 | -S-CH2-(thioether) | |||
| HCV18 | CysS- | Arg | 2Nal | Arg | Chg | D-Pro | Cha | His | Arg | Tyr | Arg | CysS- | -CON H2 | -S-S- Bridge | |||
| HCV18RI | H2NOC- | D-CysS- | D-Arg | D-2Nal | D-Arg | D-Chg | L-Pro | D-Cha | D-His | D-Arg | D-Tyr | D-Arg | D-CysS- | -S-S- Bridge | |||
The green highlights indicate the conserved amino acids. The red highlights indicate amino acid substitutions that resulted in peptides with better efficacy at blocking HCV proliferation and higher HSP70 binding affinity compared to HCV4, while the blue highlights indicate lower antiviral activity and HSP70 binding affinity. Cyclization of the peptides is also emphasized by gray highlights at both the N and C termini. The symbols to left of the table refer to specific modifications to peptides: ▲ indicates substitution of Phe171 and/or Val173 with more hydrophobic residues; △ indicates substitution of Tyr178 with more hydrophobic residues; ■ indicates addition of a lipid tag to the peptide; * indicates the retro-inverso analog; ◇ indicates peptide linearization; and □ indicates decreasing the length of cyclizing linker achieved by a thioether linkage. Solid symbols indicate a beneficial modification (i.e. improved antiviral activity and HSP70 binding), while open symbols indicate a negative effect. (2Nal = 2-naphthylalanine; -Ac = acetyl group; Ahx = 6-aminohexanoic acid; Bip = L-4,4’-biphenylalanine; Cha = cyclohexylalanine; Chg = cyclohexylglycine; CysSH = reduced cysteine; CystBu = S-tert-butyl-L-cysteine; Dpa = L-3,3′-diphenylalanine; Pal = palmitoic acid).
A structural analysis of the hairpin region in the crystal structure of dimeric NS5A domain I (Tellinghuisen et al., 2005) revealed that Phe171, Val173, and Tyr178 occur within and on one side of the plane of the two beta-sheets, while Leu175 extends on the opposite face and is located on the linker region between the two beta-sheets (Figure 1). The spatial orientation of Phe171, Val173, and Tyr178 supports their role in forming a binding interface that interacts with HSP70. We reasoned that Gly174 and Leu175 are necessary for a turn between the two beta-sheets of the hairpin and may, therefore, play a stabilizing role in the NS5A/HSP70 interaction consistent with the role of glycine and leucine in facilitating beta-turns.
Based on these observations, we hypothesized that Phe171, Val173, and Tyr178 form the main binding motif and that certain substitutions (e.g. more hydrophobic) in these positions might affect peptide antiviral activity. Through computer assisted molecular modeling, we also reasoned that Gly174 may be substituted with D-Pro to facilitate a turn between the hairpin's two beta-sheets in order to help maintaining a hairpin-like structure. Thus, all the peptides generated for this study bear D-Pro instead of Gly at position 174 (except for the retro-inverso analog HCV18RI which bears an L-Pro at this position) (Table 1). Also Leu175 was substituted with the more hydrophobic cyclohexylalanine (Cha) (Table 1).
We proceeded to generate groups of peptide derivatives of HCV4 with different modifications as described below, and all peptides were tested for their antiviral activity. Representative peptides from each group were further assayed to determine their dose response as well as HSP70 binding affinity. HCV1-3 were not tested for their antiviral activity in this study as they do not possess an arginine tag or internal arginine residues and would, therefore, be incapable of cellular entry. However, we have previously reported an effective liposome-mediated delivery of HCV2 and HCV3 (Khachatoorian et al., 2012b). HCV1-3 were, nonetheless, tested for their structural conformation.
Substitution of Phe171 and Val173 with more hydrophobic residues increases peptide antiviral activity and HSP70 binding affinity
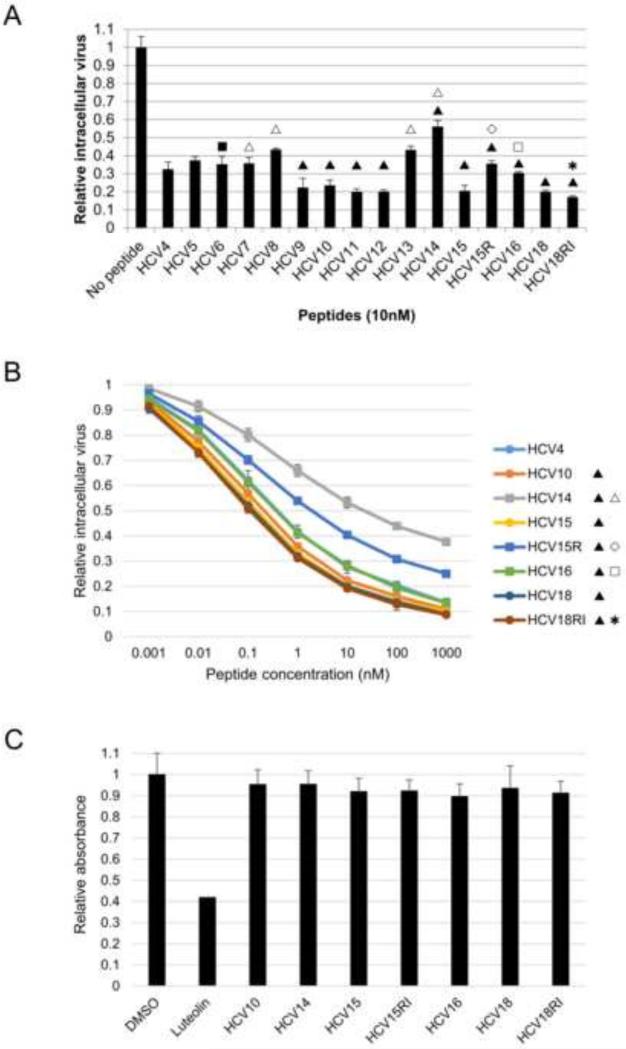
Based on the spatial arrangement of the Phe171, Val173, and Tyr178 residues, we hypothesized that substitutions within the synthetic hairpin peptides at these residues would affect their binding to HSP70 and, therefore, alter the antiviral activity of these peptides. All amino acid substitutions used were intended to increase the hydrophobicity of these residues as they possess larger hydrophobic side chains. To this end, we used L-2-naphthylalanine (2Nal) and L-3,3’-diphenylalanine (Dpa) as substituents for Phe171 and L-cyclohexylglycine (Chg) and S-tert-butyl-L-cysteine (CystBu) as substituents for Val173. All peptides bearing any of these modifications at Phe171 and/or Val173 are marked with the solid triangle symbol ▲ in Table 1 and Figure 2. HCV9, HCV10, HCV11, HCV12, HCV15, and HCV18 indeed showed improved antiviral activity compared with the HCV4 peptide in the HCV cell culture (HCVcc) system (Figure 2A).
Figure 2.
Antiviral activity of peptides and their cytotoxicity profile. A. Antiviral activity of peptides. Intracellular virus production assay was performed on cells treated with the indicated peptides at a concentration of 10 nM. The y axis reflects relative Renilla luciferase levels. B. Dose response of peptides. Intracellular virus production assay was performed on cells treated with the indicated peptides at a concentration range of 0.001 nM to 1000 nM. The y axis reflects relative Renilla luciferase levels. C. Peptides are not cytotoxic. MTT assay was performed on cells treated with 10 nM to 10 μM of indicated peptides. Assay was performed every 24 hours up to 72 hours after treatment. The data for 10 μM and 72 hour treatment is shown. The symbols in panels A and B refer to specific modifications to peptides: ▲ indicates substitution of Phe171 and/or Val173 with more hydrophobic residues; △ indicates substitution of Tyr178 with more hydrophobic residues; ■ indicates addition of a lipid tag to the peptide; ✱ indicates the retro-inverso analog; ◇ indicates peptide linearization; and □ indicates decreasing the length of cyclizing linker achieved by a thioether linkage. Solid symbols indicate a beneficial modification (i.e. improved antiviral activity and HSP70 binding), while open symbols indicate a negative effect.
We also determined the dose response of HCV10, HCV15, and HCV18 and showed that the IC50 of these peptides is lower compared with HCV4 (Table 2). In addition, we determined the HSP70 binding affinity of these peptides through surface plasmon resonance (SPR) analyses utilizing recombinant full-length HSP70 in fusion with N-terminal maltose binding protein (MBP) immobilized on the SPR chip. The HSP70 binding affinity of HCV10, HCV15, and HCV18 were found to be higher than that of HCV4 (Table 3).
Table 2.
The IC50 values for the indicated peptides.
| Peptide | IC50 |
|---|---|
| (nM) | |
| HCV4 | 0.452 |
| HCV10 | 0.234 |
| HCV14 | 31 |
| HCV15 | 0.192 |
| HCV15R | 3 |
| HCV16 | 0.345 |
| HCV18 | 0.048 |
| HCV18RI | 0.029 |
Intracellular virus production assay was performed on cells treated with the indicated peptides at a concentration range of 0.001 nM to 1000 nM. The symbols to left of the table refer to specific modifications to peptides: ▲ indicates substitution of Phe171 and/or Val173 with more hydrophobic residues; △ indicates substitution of Tyr178 with more hydrophobic residues; * indicates the retro-inverso analog; ◇ indicates peptide linearization; and □ indicates decreasing the length of cyclizing linker achieved by a thioether linkage. Solid symbols indicate a beneficial modification (i.e. improved antiviral activity and HSP70 binding), while open symbols indicate a negative effect.
Table 3.
The dissociation constants for the indicated peptides.
| Peptide | Kd |
|---|---|
| (M) | |
| NS5A domain I | 3.16 × 10–7 |
| HCV4 | 7.03 × 10–6 |
| HCV10 | 4.23 × 10–7 |
| HCV14 | 2.76 × 10–5 |
| HCV15 | 2.86 × 10–7 |
| HCV15R | 1.13 × 10–5 |
| HCV16 | 5.19 × 10–6 |
| HCV18 | 8.36 × 10–7 |
| HCV18RI | 6.57 × 10–7 |
Constants were calculated using SPR assays conducted by injecting the indicated peptides at different concentrations onto the HSP70-bound chip. The symbols to left of the table refer to specific modifications to peptides: ▲ indicates substitution of Phe171 and/or Val173 with more hydrophobic residues; △ indicates substitution of Tyr178 with more hydrophobic residues; * indicates the retro-inverso analog; ◇ indicates peptide linearization; and □ indicates decreasing the length of cyclizing linker achieved by a thioether linkage. Solid symbols indicate a beneficial modification (i.e. improved antiviral activity and HSP70 binding), while open symbols indicate a negative effect.
Substitution of Tyr178 with more hydrophobic residues decreases peptide antiviral activity and HSP70 binding affinity
Tyr178 is a polar residue, and we hypothesized that substituting it with highly hydrophobic residues may impair the antiviral activity of the peptides. We utilized L-4,4’-biphenylalanine (Bip) or L-Dpa as substituents for Tyr178, and peptides bearing these modifications at Tyr178 are marked with the open triangle symbol △. As shown in Figure 2A, these analogs (HCV7, HCV8, HCV13, and HCV14) showed decreased efficacy in blocking virus production in comparison with the HCV4 peptide. The dose response of HCV14 which bears the Tyr178Bip substitution (IC50 ~ 31 nM) was also much lower compared with HCV4 (IC50 ~ 0.452 nM) (Table 2). Furthermore, SPR analyses demonstrated that the HSP70 binding affinity of HCV14 decreased compared with HCV4 (Table 3).
Substitution of the non-conserved amino acids with arginines enables membrane penetration without a polyarginine tag and maintains peptide antiviral activity
The HCV4 peptide consists of a polyarginine tag at its N-terminus which allows for cellular internalization (Table 1). Addition of this tag significantly increases the molecular weight of the peptide and may lead to cellular toxicity. The two beta-sheets of the hairpin structure include five non-conserved residues: Thr170, Leu172, Asn176, Gln177, and Leu179 (Table 1). We hypothesized that these non-conserved residues can be substituted with arginines without significantly altering the antiviral activity of the peptides, and we substituted Thr170, Leu172, Gln177, and Leu179 with arginines (Figure 1C, blue residues). As shown in Figure 2A, peptides bearing arginines at these positions (HCV5-HCV15, HCV15R, HCV16, HCV18, and HCV18RI) maintained their antiviral activity compared to the HCV4 peptide, which indicates that they were capable of penetrating the plasma membrane. Furthermore, these peptides have the benefit of being significantly smaller. All the peptides in this study were generated without the polyarginine tag.
Addition of a lipid tag to the peptide does not interfere with its antiviral activity
We also sought to determine whether conjugation of a lipid (e.g. palmitic acid) to the HCV5 peptide would interfere with its antiviral activity, as lipidation may be beneficial for in vivo studies (targeting the peptide to the liver and increasing the plasma half-life). Therefore, we generated the HCV6 peptide (Table 1), the palmitoylated analog of HCV5 (marked with the solid square symbol ■), and demonstrated that its antiviral activity is comparable to that of HCV5 (Table 1 and Figure 2A).
The retro-inverso form of the hairpin peptide maintains its antiviral activity and HSP70 binding affinity
As peptides are subject to degradation in the intracellular environment and in vivo, we sought to determine whether the retro-inverso form of the hairpin peptide would be feasible in terms of antiviral activity. We generated a retro-inverso analog of HCV18 (HCV18RI) where the peptide was synthesized in reverse order and all the L-amino acids were substituted with their D-counterparts, while the DPro174 was changed to L-Pro. HCV18RI is represented with the star ✱ symbol. In comparison with HCV18, HCV18RI maintained antiviral activity (Figure 2A), dose response (Figure 2B and Table 2), and HSP70 binding affinity (Table 3).
Cyclization of the peptides improves their antiviral activity
We reasoned that the characteristic anti-parallel beta-sheet structure of the HBD hairpin may be important for the antiviral activity of the analog peptides. Therefore, we generated the HCV15R peptide, represented with the open diamond ◇ symbol, which is linear due to the reduced disulfide bond that cyclizes the peptide (Table 1). As shown in Figure 2A, linearization of the peptide reduced its antiviral activity with the cyclized form being more effective than the linear form. Furthermore, the dose response of HCV15R (IC50 ~ 3 nM) was significantly reduced compared to HCV15 (IC50 ~ 0.192 nM) (Figure 2B and Table 2). Linearization of the peptide also decreased the binding affinity of HCV15R to HSP70 compared with HCV15 and HCV4 (Table 3).
Linearization of the peptide may reduce the efficacy of the peptide by eliminating the optimal hairpin-like conformation of the peptide and/or decreasing its half-life as linear peptides may be more susceptible to enzymatic degradation. We synthesized all peptide analogs used in this study (except HCV15R) with a cyclizing linker.
Decreasing the length of the cyclizing linker in peptides lowers their antiviral activity
The majority of the peptide analogs used in this study were cyclized by a disulfide linker to achieve the hairpin-like structural conformation observed in the crystal structure (Tellinghuisen et al., 2005) (Figure 1). To determine if altering the length of the linker would affect the efficacy of the peptide, we synthesized an analog of the HCV15 peptide (HCV16, marked with the open square □ symbol) in which the disulfide linker is replaced by the shorter S-carboxymethyl-linker, obtained by substituting the N-terminal cysteine with a chloroacetyl group and subsequent intramolecular S-alkylation (Table 1). Decreasing the linker length negatively impacted the antiviral efficacy of the peptide (Figure 2A). The IC50 of the peptide also increased from 0.192 nM (HCV15) to 0.345 nM (HCV16) (Table 2). This may indicate that the binding of the HBD hairpin is highly optimized in the binding pocket and that minute structural changes may affect its conformation effectively decreasing the peptide binding affinity. Thus, we tested the HSP70 binding affinity of HCV16 and found that its dissociation constant was higher than that of HCV15 (Table 3).
Hairpin peptide analogs do not display cytotoxic effects
To ensure that the antiviral activity of the peptides in the HCVcc system is not affected by their potential cytotoxic effects, MTT assays were performed at a concentration range of 10 nM to 10 μM for 72 hours of treatment. As shown in Figure 2C, no significant cytotoxicity was observed at the 10 μM concentration after 72 hours of peptide treatment.
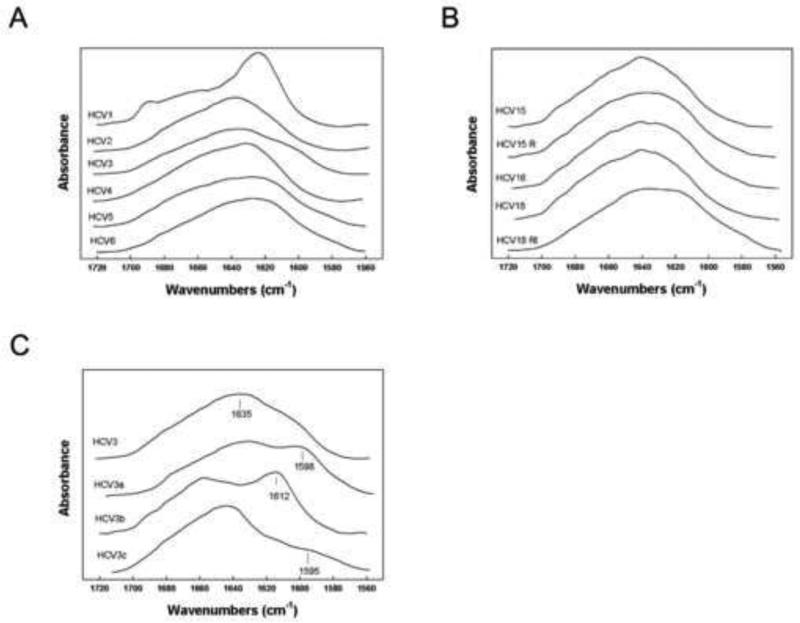
Secondary structure analysis of HCV peptides by Fourier transform infrared (FTIR) spectroscopy demonstrates a predominant beta-sheet structure
We proceeded to determine the secondary structure of the HCV suite of peptides in aqueous solvent systems using FTIR spectroscopy. The results of FTIR spectral analyses are shown in Figure 3A and 3B, and the results of the deconvolution of the spectra are shown in Table 4. In aqueous buffer, all of the HCV peptides have a dominant beta-sheet structure (Table 4). The parental peptide HCV1 has a secondary structure in aqueous buffer that is anti-parallel beta-sheet with a major peek centered around 1625 cm−1 and a minor signature band at 1690 cm−1 (Figure 3A) that is similar to the conformation observed in the crystal structure of NS5A domain I (Tellinghuisen et al., 2005). In contrast to the spectrum of HCV1, all other HCV variants have a broader spectrum centered from 1630 to 1638 cm−1, and that is more typical of peptides assuming mostly parallel beta-sheet conformations (Figure 3A and 3B). Overall, the deconvolution results suggest that all peptides have largely beta-sheet structure followed by turn and disordered conformations in aqueous environments. There were only minor contributions for alpha helical conformations with the most notable amount for the reduced form of the peptide (HCV15R) (Table 4). Further, the contribution of turns is lowest for HCV15R. These observations are consistent with our findings above that the HCV15R peptide (with reduced disulfide bond) displays a significant decrease in antiviral activity and HSP70 binding affinity (Figure 2A and Table 2 and 3).
Figure 3.
FTIR spectroscopy analyses of HCV peptides. A and B. FTIR spectral analysis of unlabeled HCV peptide secondary structure in deuterated 10 mM phosphate buffer pD 5.6. C. FTIR spectral analysis of HCV3 13C isotope enhanced peptide secondary structure in deuterated 10 mM phosphate buffer pD 5.6. HCV3 is unlabeled peptide; HCV3a is isotopically enhanced at residues Phe3 and Val5; HCV3b is isotopically enhanced at Gly6 and Leu7; and HCV3c is isotopically enhanced at Tyr10.
Table 4.
Proportions of secondary structurea for HCV peptidesb in deuterated phosphate buffer as estimated from Fourier self-deconvolution of the FTIR spectrac of the peptide mide I band.
| Peptide | % Conformation |
|||
|---|---|---|---|---|
| α-Helix | β-Sheet | Turn | Disordered | |
| HCV1 | 2.10 | 43.11 | 31.05 | 23.74 |
| HCV2 | 4.66 | 53.56 | 28.94 | 17.84 |
| HCV3 | 11.60 | 54.34 | 21.96 | 12.04 |
| HCV4 | 11.86 | 41.48 | 29.21 | 17.45 |
| HCV5 | 10.55 | 50.90 | 25.72 | 12.83 |
| HCV6 | 12.34 | 52.87 | 20.39 | 14.40 |
| HCV15 | 11.05 | 39.52 | 31.46 | 17.97 |
| HCV15R | 24.77 | 49.12 | 12.97 | 13.14 |
| HCV16 | 14.68 | 40.36 | 18.71 | 26.25 |
| HCV18 | 5.88 | 47.91 | 29.74 | 16.47 |
| HCV18RI | 7.82 | 53.88 | 22.55 | 15.75 |
Data are the means of four separate determinations and have an SE ± 5% or better.
HCV Peptide [i.e., 2 mM HCV] desolvated in deuterated 10 mM phosphate buffer pH 5.6 (Figure 3).
FTIR spectra were deconvoluted as described in Materials and Methods.
13C-enhanced FTIR spectroscopy of HCV3 peptide indicates a secondary structure consisting of an N terminal beta-sheet followed by a turn and a C terminal beta-sheet
To gain further insights into the residue specific structure of the HCV variants, we isotopically enhanced HCV3 residues with 13C carbonyl labels for selective amino acids in the N-terminal, mid-sequence, and C-terminal domains (Dwivedi and Krimm, 1984; Tadesse et al., 1991). The spectra for the selectively labeled sequences are shown in Figure 3C. When HCV3 peptide was enhanced with 13C labels at Phe at position 3 (Phe 171) and Val at position 5 (Val173), there was a characteristic shift of thirty-seven wavenumbers from 1635 cm−1 to 1598 cm−1 indicative of these residues assuming parallel beta-sheet conformations (Figure 3C, spectrum HCV3a). Spectra of the mid-sequence 13C carbonyl labels at Gly at position 6 (Gly174) and Leu at position 7 (Leu175) also showed a spectral down shift from 1649 cm−1 to 1612 cm−1 typical of residues participating in Type III beta-turn (310 helix) conformations when in deuterated buffer (Byler et al., 1986) (Figure 3C, spectrum HCV3b). When the Tyr at position 10 (Tyr178) was enhanced with 13C, there was also a shift from the beta-sheet region of the unlabeled parent FTIR spectra centered in the 1632 cm−1 absorbance to 1695 cm−1 suggesting that this residue was also assuming mostly parallel beta-sheet conformations (Figure 3C, spectrum HCV3c).
Molecular simulation confirms a beta-hairpin secondary structure for HCV3 peptide
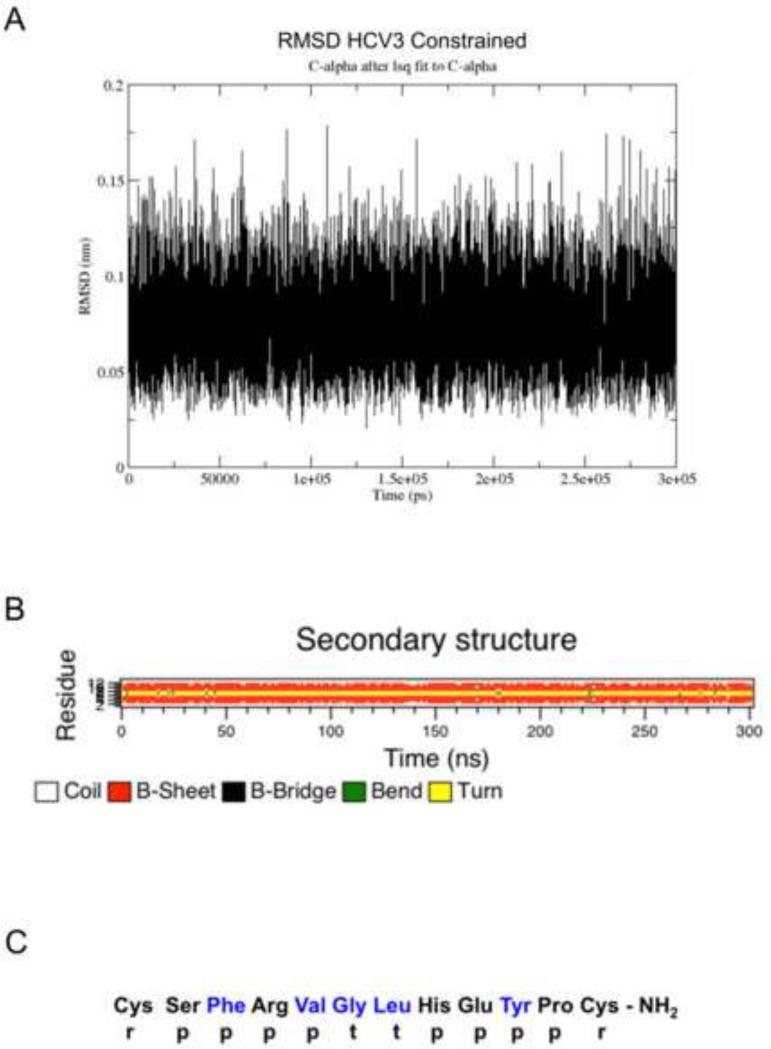
Based on the above 13C labeled residue assignments obtained by isotope enhanced FTIR measurements, a highly accurate model of HCV3 in water was then determined using molecular dynamics refinement. This was facilitated by conformationally constraining the labeled residue while allowing the remaining residues to assume conformations indicative of an equilibrium conformation in a simulated water environment. As shown in Figure 4A and 4B, the HCV3 peptide backbone rapidly reached a stable equilibrium secondary conformation within the first few nanoseconds of simulation. Based on the molecular dynamics refinement, the residue specific secondary structure of HCV3 is diagramed in Figure 4C. The sequential map indicates that the disulfide linked Cys1 and Cys12 have a random or disordered conformation while the beta-sheet components flank a well-defined turn element that includes Gly-6 and Leu-7. There are two beta-sheet components one from Ser2 to Val5 and the other from His8 to Pro11. The beta-sheets assume mostly parallel conformations and are configured with the turn to form a beta-hairpin structure (Figure 5A) which is remarkably similar to the HDB moiety from the crystal structure (Tellinghuisen et al., 2005) (Figure 5B). This overall structure has an amphipathic character with amino acids on one side of the beta-hairpin with polar residues and the other side with amino acids having hydrophobic side chains. There are also two intra-chain components that stabilize the parallel beta-hairpin format along with the disulfide linkage between Cys1 to Cys12. One element is the salt bridge between Arg4 and Glu9. The existence of this ion-lock is confirmed by the fact that the distance between the side-chain charged group centroids is maintained during the simulation to be within the 4 angstrom distance typical of most salt-bridges in proteins (Kumar and Nussinov, 1999). A second non-covalent side chain interaction that potentially adds to the stability of the beta-hairpin structure is the interaction between Phe3 and Tyr10. This Phe-Tyr aromatic pair maintains Cα - Cα distances over the time course of the molecular dynamics run that favor π-stacking and T-shape orientations for possible interactions that enhance intra-chain stability of the hair-pin structure (Chelli et al., 2002; McGaughey et al., 1998).
Figure 4.
Molecular simulation of the HCV3 secondary structure. A. Root Mean Square Deviation (RMSD) of the c-alpha backbone for HCV as a function of simulation time. B. Evolution of HCV3 secondary structure as a function of simulation time in aqueous periodic solvent box. C. Residue specific assignments for HCV3 derived from FTIR measurements of isotopically enhanced peptides and refined by constrained molecular dynamics in aqueous buffer. Residues in blue typeface identify those labeled with 13C at their carbonyl atoms. Conformational codes are as follows: r = random – disordered structures; p = parallel beta-sheet; and t = turn conformations.
Figure 5.
Structure of HCV3 in aqueous buffer. A. The HCV3 beta-sheet domains are in ribbon-arrow format; Arg4 – Glu9 ion pair are in stick format; and the disulfide linkage between Cys1 and Cys13 is indicated by yellow stick connectivity. B. Crystal structure of HBD (Tellinghuisen et al., 2005) displayed in the same orientation as the HCV3 peptide in panel A.
Discussion
In this study, we have characterized the interaction of the NS5A HBD with HSP70, identified the amino acid residues involved in the NS5A/HSP70 interaction, and developed peptides that are more efficacious than HCV4 in binding HSP70 and in blocking virus production.
As reported previously, the HBD-mediated NS5A/HSP70 interaction is involved in the NS5A-driven IRES-mediated translation of the viral genome, and treatment of cells with the peptides corresponding to HBD significantly inhibits IRES-mediated translation (Khachatoorian et al., 2012b). Furthermore, knockdown of HSP70 was shown by our group and others to significantly reduce viral IRES-mediated translation and protein production (Gonzalez et al., 2009; Lim et al., 2012). These and our current results indicate that the NS5A/HSP70 interaction is critical for viral proliferation. Recently, we also analyzed the binding of heat shock cognate protein (HSC) 70 with NS5A and found that NS5A and HSC70 directly bind each other as well (Khachatoorian et al., 2014). Nonetheless, our data as well as another report indicated that HSC70 is involved in infectious virion assembly rather than IRES-mediated translation (Khachatoorian et al., 2014; Parent et al., 2009). We also showed that the compensatory upregulation of HSP70 due to knockdown of HSC70 fails to improve infectious virion assembly and that simultaneous knockdown of HSP70 and HSC70 leads to a similar decrease in viral translation as knockdown of HSP70 alone further supporting the distinct roles that HSP70 and HSC70 play in the viral life cycle. We are currently investigating the NS5A/HSC70 interaction biochemically to identify the precise site(s) of interaction on both proteins. It may be possible that HSC70 also utilizes the HBD hairpin to bind to NS5A.
We demonstrated that cyclization of the peptide improves antiviral activity and HSP70 binding affinity potentially by locking the peptide in a more native hairpin-like structure and/or by rendering the peptide less likely to be proteolytically cleaved. Furthermore, cyclization of the peptide significantly decreases the alpha-helical contributions to the peptide secondary structure which is only observed in the linearized peptide (HCV15R). In addition, we showed that the length of the cyclizing linker is also important for its HSP70 binding affinity and for the function of the peptide in blocking virus production.
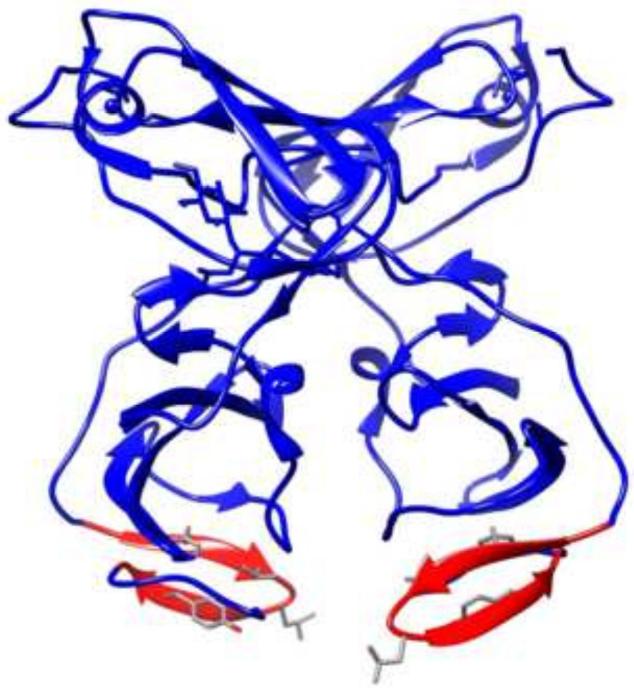
We hypothesized that the amino acid residues within the HBD hairpin that are critical for the NS5A/HSP70 interaction should most likely be conserved, and sequence comparison confirmed the existence of five conserved residues in the hairpin: Phe171, Val173, Gly174, Leu175, and Tyr178. Analysis of the previously reported crystal structure of dimeric NS5A domain I (Tellinghuisen et al., 2005) readily revealed that Phe171, Val173, and Tyr178 occur on one side of the plane of the hairpin beta-sheets suggesting that they could form a binding surface with HSP70. In addition, the crystal structure also shows the HBD and these three residues to be on the surface of the dimer and exposed supporting their role as the NS5A/HSP70 binding interface. In an alternate crystal structure of dimeric NS5A domain I (Love et al., 2009), the two subunits appear as more compact closing the proposed RNA binding cleft (Figure 6). In this structure, while the two HBD moieties appear to be still exposed, their location changes from the side of the molecule to the bottom (Figure 6). It is possible that the former crystal structure provides for a better conformation for the HBD to bind HSP70. Nevertheless, NS5A likely needs to undergo a conformational change to allow HBD to form a tight binding interface with HSP70, and this may be possible in both crystal structure conformations.
Figure 6.
Schematic representation of the previously reported alternate crystal structure of dimeric NS5A domain I (Love et al., 2009). The HBD hairpins are highlighted in red. The side chains of the four conserved residues (Phe171, Val173, Leu175, and Tyr178) are also shown.
To verify that these three conserved residues are involved in the NS5A/HSP70 interaction, we substituted them with artificial amino acids with large hydrophobic side chains. Substitutions at Phe171 and Val173 improved the antiviral activity and binding affinity of the peptide, whereas substitution of Tyr178 had the opposite effect. This is expected as tyrosine is a polar amino acid. In addition substitution of the other non-conserved amino acids within the hairpin did not affect cellular entry, antiviral activity, and binding affinity of the peptide. Thus, we conclude that the binding of NS5A with HSP70 is mediated by these three conserved amino acids within the C34 hairpin.
To confirm our structural predictions of the peptide in aqueous environments, we performed FTIR spectroscopy on the majority of the peptides generated and observed that all peptides primarily consist of beta-sheets and beta-turns consistent with the crystal structure of the hairpin in the NS5A domain I (Tellinghuisen et al., 2005). Alpha-helical contributions were minor except for the linearized HCV15R peptide which also shows significantly reduced antiviral activity and HSP70 binding affinity. FTIR spectroscopy also indicated the existence of turn element in the peptide consistent with our analysis of the crystal structures. Furthermore, 13C-enhanced spectroscopy of the HCV3 peptide clearly demonstrated that Gly174 and Leu175 form a turn element between the two beta-sheets of the hairpin confirming our original predictions. Molecular simulation of the HCV3 FTIR spectral analyses further confirmed the existence of all the predicted elements of the peptide secondary structure including the N and C terminal beta-sheets flanking a beta-turn.
Thus, we have confirmed the antiviral activity of a number of HIPs that bind HSP70 and block the NS5A/HSP70 interaction. Considering that most available therapeutic agents target the viral proteins and induce the generation of resistant viral strains, HIPs may serve as a potential new class of therapeutics for HCV infection by targeting the host HSP70 as opposed to viral proteins in order to minimize the development of resistance. Our data indicates that the antiviral activity of HIPs is comparable to that of sofosbuvir and ledipasvir in the HCVcc system (Lawitz et al., 2012; Sofia et al., 2010). In combination with DAAs, HIPs may further be able to dispense with the need for PEG-IFN and RBV that are currently utilized in combination therapies. Further, insight provided here may also allow for development of therapies for others viruses that utilize HSP70 recruitment.
Materials and methods
Plasmid constructs
NS5A domain I was cloned in the pET-28b plasmid (Novagen, 69865-3). The HCV IRES reporter plasmid has been previously described (Gonzalez et al., 2009). An intra-genotype 2 chimeric monocistronic reporter virus, pNRLFC based on pJ6/JFH-C parental virus has been described previously (Arumugaswami et al., 2008). For the current study, we have used a chemically synthesized plasmid pFNX-RLuc (having similar sequences to pNRLFC) for construction of recombinant virus.
Peptide synthesis and characterization
Peptides were synthesized by the solid phase method using CEM Liberty automatic microwave peptide synthesizer (CEM Corporation), applying 9-fluorenylmethyloxycarbonyl (Fmoc) chemistry (Fields and Noble, 1990) and standard, commercially available amino acid derivatives and reagents (EMD Biosciences and Chem-Impex International). Rink Amide MBHA resin (EMD Biosciences) was used as a solid support. Peptides were cleaved from resin using modified reagent K (TFA 94% (v/v); phenol, 2% (w/v); water, 2% (v/v); TIS, 1% (v/v); EDT, 1% (v/v); 2 hours) and precipitated by addition of ice-cold diethyl ether. Reduced peptides were purified by preparative reverse-phase high performance liquid chromatography (RP-HPLC) and their purity evaluated by matrix-assisted laser desorption ionization spectrometry (MALDI-MS) as well as by analytical RP-HPLC). Disulfide bond formation: Peptides were dissolved at a final concentration of 0.25 mg/ml in 50% DMSO:H2O and stirred overnight at room temperature. Subsequently peptides were lyophilized and re-purified on a preparative C18 SymmetryShield™ RP-HPLC column to >95% homogeneity. Their purity was evaluated by MALDI-MS as well as by analytical RP-HPLC. Analytical RP-HPLC: Analytical RP-HPLC was performed on a Varian ProStar 210 HPLC system equipped with ProStar 325 Dual Wavelength UV-Vis detector with wavelengths set at 220 nm and 280 nm (Varian Inc.). Mobile phases consisted of solvent A, 0.1% TFA in water, and solvent B, 0.1% TFA in acetonitrile. Analyses of peptides were performed with an analytical reversed-phase C18 SymmetryShield™ RP18 column, 4.6[x1]250 mm, 5μm (Waters Corp.) applying linear gradient of solvent B from 0 to 100% over 100 min (flow rate: 1 ml/min).
Cell culture
The cell line huh-7.5 was maintained in a humidified atmosphere containing 5% CO2 at 37°C in Dulbecco's Modified Eagle Medium (Mediatech, 10-013-CM) supplemented with 10% fetal bovine serum (Omega Scientific, FB-01) and 2 mM L-Gln (Life Technologies, 25030). Huh-7.5 cells were a kind gift from Charles.
Cell viability
Cell viability was determined using MTT Cell Proliferation assay (ATCC, 30-1010K) according to manufacturer's instructions.
Infectious virus production
pFNX-RLuc was in vitro transcribed, and the purified RNA was electroporated into huh-7.5 cells to generate infectious viral supernatant as previously described (Arumugaswami et al., 2008).
Viral assays
The HCV reporter virus was utilized as described previously (Khachatoorian et al., 2012a). All infection assays were performed with a viral titer of 103 focus forming units and a multiplicity of infection (MOI) of 0.1. Intracellular virus production: Huh-7.5 cells were infected for three hours at which point the medium was replaced, indicated peptides were added, and infection was allowed to proceed. Cells were harvested 48 hours later, and luciferase activity was measured using the Renilla Luciferase Assay System (Promega, E2820).
Production and purification of recombinant proteins
NS5A domain I was expressed and purified as described previously (Wheatley et al., 2013). Briefly, protein expression was induced with 1 mM IPTG in BL21(DE3) E. coli cells, shaking at 250 rpm, for 5 hours at 37°C. Cells were centrifuged for 5 minutes at 6000rpm and stored at −20°C. Cells were resuspended in 50 mM Tris-HCl pH 7.6, 300 mM NaCl, and 20 mM imidazole, with Protease Inhibitor Cocktail (Sigma-Aldrich, P8849-5ml) and lysed by sonication. Cells were spun down in rotor SS-34 at 16,500 rpm for 30 min, filtered through a 0.2μm filter, and applied to a Hi-trap Nickel column by syringe at room temperature. Proteins were eluted in one step with of 50 mM Tris-HCl pH 7.6, 300 mM NaCl, and 400 mM imidazole.
Surface plasmon resonance (SPR)
Binding studies were performed on a Biacore 3000 instrument (Biacore AB, Uppsala, Sweden). Proteins were immobilized on CM5 sensor chips by amine coupling. The solution phase analytes were dissolved in HBS-EP buffer, which contained 0.15M NaCl, 10 mM HEPES, pH 7.4, 3 μM EDTA, and 0.005% polysorbate 20. The solutions traversed the sensors at a flow rate of 50 μl/minute. Costar low-retention polypropylene tubes (Corning, 3207) were used throughout. Binding results were expressed in resonance units. Kinetics and dissociation constants were analyzed and calculated with BIAevaluation Software Version 4.1.
Fourier transform infrared (FTIR) spectroscopy
Infrared spectra were recorded at 25°C using a Bruker Vector 22™ FTIR spectrometer with a deuterated triglycine sulfate (DTGS) detector, and averaged over 256 scans at a gain of 4 with a resolution of 2 cm−1. Peptide samples were initially freeze-dried several times from 10 mM HCl in D2O to remove any interfering counter ions and residual H2O. Solution spectra of peptides were made in deuterated 10 mM phosphate buffer, pD 5.6 (pD = pH+0.4). The spectra were acquired at a peptide concentration of 2 mM in deuterium buffer using a temperature controlled, demountable liquid cell with calcium fluoride windows fitted with a 50 μm thick Mylar spacer (Harrick Scientific, Pleasantville, NY). The relative proportions of α-helix, turn, β-sheet, and disordered conformations of solution and multilayer IR spectra were determined by Fourier self-deconvolution for band narrowing and area calculations of component peaks of the FTIR spectra using curve-fitting software supplied by Galactic Software (GRAMS/AI, version 8.0; Thermo Electron Corp., Waltham, MA). The frequency limits for the different structures were: α-helix (1662-1645 cm−1), β-sheet (1637-1613 and 1710-1682 cm−1), turns (1682-1662 cm−1), and disordered or random (1650-1637 cm−1) (Byler and Susi, 1986).
Molecular Structure Simulation of HCV3 in Aqueous Solution
A starting structure for the HCV3 peptide amino acid sequence was approximated by using Hyperchem 8.0 (http://www.hyper.com) to template the backbone structure of the beta-hairpin conformation that was derived from the homologous segment of the crystal structure of NS5A domain I (PDB 1ZH1, residues 173 - 184) (Tellinghuisen et al., 2005). This preliminary structure was placed in a periodic 56Å3 box of TIP4P water and the ensemble was neutralized with counter ions to simulate the environment used for the FTIR isotope enhanced experimental measurements. The peptide in the solution box was conjugate-gradient minimized using the Polak-Ribiere approach implemented in Hyperchem. This minimized HCV3 aqueous ensemble was then ported to the Gromacs program suite, version 4.5.5 (http://www.gromacs.org), and subjected the steepest descent method using the Amber03 force field option (Hess et al., 2008).
The steepest descent minimized system was further subjected to 40 picoseconds of pre-run molecular dynamics at 300°K to allow the solvent to equilibrate while restraining the peptide. After pre-run solvent equilibration the peptide structure was further refined by 300 nsec of MD simulation at 300°K with residue specific constraints based on FTIR isotope enhanced experimental observations. The Berendsen protocol was used for temperature and pressure coupling and the Particle Mesh Ewald methods employed for evaluating long-range electrostatic interactions. The time-dependent evolution of the peptide secondary structure (i.e., analyzed using the DSSP criteria (Kabsch and Sander, 1983) for the peptide in the water environment indicated when equilibrium was reached. Molecular model illustrations were rendered using PyMOL v0.99 (http://www.pymol.org).
Statistical analysis
Error bars reflect the standard deviation. P values were determined by student t test. All viral assays were done with six biological replicates for each individual assay.
IC50 calculation
IC50 values were calculated based on the formula derived from a third degree polynomial curve fit in Microsoft™ Excel™.
NS5A binds HSP70 via three conserved residues in HSP binding domain (HBD) of NS5A.
Arginine substitution of non-conserved residues allows cellular uptake of peptides.
Addition of a lipid tag to peptide does not interfere with its antiviral activity.
The retro-inverso peptide maintains antiviral activity and HSP70 binding affinity.
FTIR spectroscopy confirms peptide secondary structure as in crystal structures
Acknowledgements
The authors thank David Dawson for helpful discussions and Charles Rice for providing huh-7.5 cells.
Financial support:
NIH R01DK090794, SWF.
NIH R21AI084090, AD.
List of abbreviations
- Ac
acetyl group
- 2Nal
2-naphthylalanine
- Ahx
6-aminohexanoic acid
- Bip
L-4,4’-biphenylalanine
- C34
C terminal 34 amino acids of NS5A domain I
- Cha
cyclohexylalanine
- Chg
cyclohexylglycine
- CysSH
reduced cysteine
- CystBu
S-tert-butyl-L-cysteine
- DAA
direct acting antiviral
- Dpa
L-3,3 -diphenylalanine
- DMSO
dimethyl sulfoxide
- EDT
ethanedithiol
- Fmoc
9-fluorenylmethyloxycarbonyl
- FTIR
Fourier transform infrared (spectroscopy)
- HBD
HSP binding domain
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- HCVcc
HCV cell culture
- HIP
HSP interfering peptide
- HSP
heat shock protein
- IC50
half-maximal inhibition concentration
- IRES
internal ribosomal entry site
- MALDI-MS
matrix-assisted laser desorption ionization mass spectrometry
- MBP
maltose binding protein
- NBD
nucleotide binding domain
- NCR
noncoding region
- NS
nonstructural
- NS5A
nonstructural protein 5A
- Pal
palmitic acid
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- PEG-IFN
pegylated interferon
- RBV
ribavirin
- RLuc
Renilla luciferase
- RP-HPLC
reverse-phase high performance liquid chromatography
- SBD
substrate binding domain
- SPR
surface plasmon resonance
- SVR
sustained virological response
- TIS
thioanisole
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
California Center for Antiviral Drug Discovery grant, University of California Office of the President, MRPI # 143226, AD.
References
- Arumugaswami V, Remenyi R, Kanagavel V, Sue EY, Ngoc Ho T, Liu C, Fontanes V, Dasgupta A, Sun R. High-resolution functional profiling of hepatitis C virus genome. PLoS Pathog. 2008;4:e1000182. doi: 10.1371/journal.ppat.1000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron S. Medical microbiology. 4th ed. University of Texas Medical Branch at Galveston; Galveston, Tex.: 1996. [PubMed] [Google Scholar]
- Byler DM, Susi H. Examination of the secondary structure of proteins by deconvolved FTIR spectra. Biopolymers. 1986;25:469–487. doi: 10.1002/bip.360250307. [DOI] [PubMed] [Google Scholar]
- Chelli R, Gervasio FL, Procacci P, Schettino V. Stacking and T-shape competition in aromatic-aromatic amino acid interactions. J Am Chem Soc. 2002;124:6133–6143. doi: 10.1021/ja0121639. [DOI] [PubMed] [Google Scholar]
- Ciesek S, Manns MP. Hepatitis in 2010: the dawn of a new era in HCV therapy. Nat Rev Gastroenterol Hepatol. 2011;8:69–71. doi: 10.1038/nrgastro.2010.219. [DOI] [PubMed] [Google Scholar]
- Dwivedi AM, Krimm S. Vibrational analysis of peptides, polypeptides, and proteins. XVIII. Conformational sensitivity of the alpha-helix spectrum: alpha I- and alpha II-poly(L-alanine). Biopolymers. 1984;23:923–943. doi: 10.1002/bip.360230509. [DOI] [PubMed] [Google Scholar]
- El-Serag HB. Hepatocellular carcinoma: an epidemiologic view. J Clin Gastroenterol. 2002;35:S72–78. doi: 10.1097/00004836-200211002-00002. [DOI] [PubMed] [Google Scholar]
- Fields GB, Noble RL. Solid phase peptide synthesis utilizing 9-fluorenylmethoxycarbonyl amino acids. Int J Pept Protein Res. 1990;35:161–214. doi: 10.1111/j.1399-3011.1990.tb00939.x. [DOI] [PubMed] [Google Scholar]
- Gonzalez O, Fontanes V, Raychaudhuri S, Loo R, Loo J, Arumugaswami V, Sun R, Dasgupta A, French SW. The heat shock protein inhibitor Quercetin attenuates hepatitis C virus production. Hepatology. 2009;50:1756–1764. doi: 10.1002/hep.23232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Yan W, Coito C, Li Y, Gale M, Jr., Katze MG. The regulation of hepatitis C virus (HCV) internal ribosome-entry site-mediated translation by HCV replicons and nonstructural proteins. J Gen Virol. 2003;84:535–543. doi: 10.1099/vir.0.18658-0. [DOI] [PubMed] [Google Scholar]
- Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4:[uni2009] Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. Journal of Chemical Theory and Computation. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- Hughes M, Griffin S, Harris M. Domain III of NS5A contributes to both RNA replication and assembly of hepatitis C virus particles. J Gen Virol. 2009;90:1329–1334. doi: 10.1099/vir.0.009332-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Khachatoorian R, Arumugaswami V, Raychaudhuri S, Yeh GK, Maloney EM, Wang J, Dasgupta A, French SW. Divergent antiviral effects of bioflavonoids on the hepatitis C virus life cycle. Virology. 2012a;433:346–355. doi: 10.1016/j.virol.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khachatoorian R, Arumugaswami V, Ruchala P, Raychaudhuri S, Maloney EM, Miao E, Dasgupta A, French SW. A cell-permeable hairpin peptide inhibits hepatitis C viral nonstructural protein 5A-mediated translation and virus production. Hepatology. 2012b;55:1662–1672. doi: 10.1002/hep.25533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khachatoorian R, Ganapthy E, Ahmadieh Y, Wheatley N, Sundberg C, Jung C-L, Arumugaswami V, Raychaudhuri S, Dasgupta A, French SW. The NS5A-binding heat shock proteins HSC70 and HSP70 play distinct roles in the hepatitis C viral life cycle. Virology. 2014 doi: 10.1016/j.virol.2014.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Nussinov R. Salt bridge stability in monomeric proteins. J Mol Biol. 1999;293:1241–1255. doi: 10.1006/jmbi.1999.3218. [DOI] [PubMed] [Google Scholar]
- Lawitz EJ, Gruener D, Hill JM, Marbury T, Moorehead L, Mathias A, Cheng G, Link JO, Wong KA, Mo H, McHutchison JG, Brainard DM. A phase 1, randomized, placebo-controlled, 3-day, dose-ranging study of GS-5885, an NS5A inhibitor, in patients with genotype 1 hepatitis C. J Hepatol. 2012;57:24–31. doi: 10.1016/j.jhep.2011.12.029. [DOI] [PubMed] [Google Scholar]
- Lim YS, Shin KS, Oh SH, Kang SM, Won SJ, Hwang SB. Nonstructural 5A protein of hepatitis C virus regulates heat shock protein 72 for its own propagation. J Viral Hepat. 2012;19:353–363. doi: 10.1111/j.1365-2893.2011.01556.x. [DOI] [PubMed] [Google Scholar]
- Lindenbach BD, Rice CM. Unravelling hepatitis C virus replication from genome to function. Nature. 2005;436:933–938. doi: 10.1038/nature04077. [DOI] [PubMed] [Google Scholar]
- Love RA, Brodsky O, Hickey MJ, Wells PA, Cronin CN. Crystal structure of a novel dimeric form of NS5A domain I protein from hepatitis C virus. J Virol. 2009;83:4395–4403. doi: 10.1128/JVI.02352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaughey GB, Gagne M, Rappe AK. pi-Stacking interactions. Alive and well in proteins. J Biol Chem. 1998;273:15458–15463. doi: 10.1074/jbc.273.25.15458. [DOI] [PubMed] [Google Scholar]
- Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- Pacheco A, Martinez-Salas E. Insights into the biology of IRES elements through riboproteomic approaches. J Biomed Biotechnol. 2010;2010:458927. doi: 10.1155/2010/458927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent R, Qu X, Petit MA, Beretta L. The heat shock cognate protein 70 is associated with hepatitis C virus particles and modulates virus infectivity. Hepatology. 2009;49:1798–1809. doi: 10.1002/hep.22852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005;5:558–567. doi: 10.1016/S1473-3099(05)70216-4. [DOI] [PubMed] [Google Scholar]
- Sofia MJ, Bao D, Chang W, Du J, Nagarathnam D, Rachakonda S, Reddy PG, Ross BS, Wang P, Zhang HR, Bansal S, Espiritu C, Keilman M, Lam AM, Steuer HM, Niu C, Otto MJ, Furman PA. Discovery of a beta-d-2′-deoxy-2′-alpha-fluoro-2′-beta-C-methyluridine nucleotide prodrug (PSI-7977) for the treatment of hepatitis C virus. Journal of medicinal chemistry. 2010;53:7202–7218. doi: 10.1021/jm100863x. [DOI] [PubMed] [Google Scholar]
- Tadesse L, Nazarbaghi R, Walters L. Isotopically enhanced infrared spectroscopy: a novel method for examining secondary structure at specific sites in conformationally heterogeneous peptides. Journal of The American Chemical Society. 1991;113:7036–7037. [Google Scholar]
- Tellinghuisen TL, Foss KL, Treadaway JC, Rice CM. Identification of residues required for RNA replication in domains II and III of the hepatitis C virus NS5A protein. J Virol. 2008;82:1073–1083. doi: 10.1128/JVI.00328-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellinghuisen TL, Marcotrigiano J, Rice CM. Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature. 2005;435:374–379. doi: 10.1038/nature03580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasconcelos DY, Cai XH, Oglesbee MJ. Constitutive overexpression of the major inducible 70 kDa heat shock protein mediates large plaque formation by measles virus. J Gen Virol. 1998;79(Pt 9):2239–2247. doi: 10.1099/0022-1317-79-9-2239. [DOI] [PubMed] [Google Scholar]
- Wang C, Sarnow P, Siddiqui A. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J Virol. 1993;67:3338–3344. doi: 10.1128/jvi.67.6.3338-3344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks SA, Miller DJ. The heat shock protein 70 cochaperone YDJ1 is required for efficient membrane-specific flock house virus RNA replication complex assembly and function in Saccharomyces cerevisiae. J Virol. 2008;82:2004–2012. doi: 10.1128/JVI.02017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley NM, Gidaniyan SD, Liu Y, Cascio D, Yeates TO. Bacterial microcompartment shells of diverse functional types possess pentameric vertex proteins. Protein science : a publication of the Protein Society. 2013;22:660–665. doi: 10.1002/pro.2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng ZZ, Miao J, Zhao M, Tang M, Yeo AE, Yu H, Zhang J, Xia NS. Role of heat-shock protein 90 in hepatitis E virus capsid trafficking. J Gen Virol. 2010;91:1728–1736. doi: 10.1099/vir.0.019323-0. [DOI] [PubMed] [Google Scholar]