SUMMARY
We have analyzed the yeast replicative lifespan of a large number of ORF deletions. Here we report that strains lacking genes SGF73, SGF11, and UBP8, encoding SAGA/SLIK complex histone deubiquitinase module (DUBm) components, are exceptionally long-lived. Strains lacking other SAGA/SALSA components, including the acetyltransferase encoded by GCN5, are not long-lived; however, these genes are required for the lifespan extension observed in DUBm deletions. Moreover, the SIR2-encoded histone deacetylase is required, and we document both a genetic and physical interaction between DUBm and Sir2. A series of studies, assessing Sir2-dependent functions, lead us to propose that DUBm strains are exceptionally long-lived because they promote multiple pro-longevity events, including reduced rDNA recombination and altered silencing of telomere-proximal genes. As ataxin-7, the human Sgf73 ortholog, causes the neurodegenerative disease spinocerebellar ataxia type 7, our findings indicate that the genetic and epigenetic interactions between DUBm and SIR2 will be relevant to neurodegeneration and aging.
INTRODUCTION
Aging is a fundamental biological process of great interest. Although yeast cells may seem an unlikely model organism to study human aging, insights from the study of yeast replicative lifespan (RLS), in which the number of daughter cells that one mother produces through budding is determined (Mortimer and Johnston, 1959), have advanced our understanding of eukaryotic and even mammalian aging (Steinkraus et al., 2008). Early studies led to the identification of the protein deacetylase encoded by SIR2 as a key modulator of aging, and subsequently Sirtuins have become a major focus of aging research (Guarente, 2011; Kaeberlein et al., 1999a; Kennedy et al., 1995). Additionally, studies in yeast, as well as other invertebrates, have highlighted the importance of the TOR pathway in aging (Kaeberlein et al., 2005b; Stanfel et al., 2009). Furthermore, a quantitative comparison of aging genes in yeast and worms has demonstrated with high statistical significance that longevity pathways are conserved between yeast and C. elegans (Smith et al., 2008), two organisms more divergent than worms are from humans. Hence, there is strong evidence to conclude that the orthologs of other yeast aging genes may influence mammalian aging.
Spinocerebellar ataxia type 7 (SCA7) is an autosomal dominant neurodegenerative disease resulting from the expansion of a polymorphic and unstable CAG tract in the ataxin-7 gene (David et al., 1997). In affected individuals, the CAG tract is translated into an abnormally long stretch of glutamine residues in the N-terminus of the ataxin-7 protein. When containing a track of 37 or more glutamine residues, polyglutamine-expanded ataxin-7 is not readily degraded, accumulates in protein aggregates, and causes neuronal dysfunction and neuronal cell death in the retina, cerebellum, and associated brainstem structures. This results in blindness, a severe loss of coordination, and ultimately premature death (Lebre and Brice, 2003). Ataxin-7 is a highly conserved member of the SPT3-TAFII31-GCN5L acetylase (STAGA) complex, one of the major transcriptional coactivator complexes in mammalian cells (Helmlinger et al., 2004; Martinez et al., 2001). More specifically, ataxin-7 is a component of the USP22 histone deubiquitinase module (DUBm) of the complex.
Based on limited sequence homology, SGF73 was proposed to be the yeast ortholog of ataxin-7 (Mushegian et al., 2000; Scheel et al., 2003), and later functional studies demonstrated this to be the case (Mal, 2006). In the DUBm of the yeast SAGA (Spt-Ada-Gcn5-Acetyltransferase) complex, which is analogous to the mammalian STAGA (SPT3-TAFII31-GCN5L acetylase) complex, Sgf73 serves to link the histone deubiquitinase Ubp8 to the rest of the complex (Lee et al., 2009). This function has also been proposed for ataxin-7 in mammals, where USP22, the ortholog of Ubp8, possesses H2B deubiquitinase activity (Zhang et al., 2008; Zhao et al., 2008). Ubp8 deubiquitinates histone H2B-K123 to confer large-scale SAGA-mediated transcriptional changes (Daniel et al., 2004; Henry et al., 2003).
In an ongoing genome-wide screen for long-lived yeast ORF deletions, we identified sgf73Δ as having a dramatically enhanced RLS, on par with the longest-lived single deletions that we found (Sutphin et al., 2012). Strains lacking other components of the DUBm also have exceptional RLS extension, but those lacking other SAGA components, including the histone acetyltransferase Gcn5, are not long-lived. Unexpectedly, lifespan extension in sgf73Δ and ubp8Δ depends entirely on SIR2, and further analysis indicates that lifespan extension in strains lacking components of the SAGA deubiquitinase complex occurs through coordinate control of multiple longevity pathways. Sgf73 can also physically interact with Sir2 and Ubp8, a finding replicated in mammals by interaction of USP22 with human SIRT1 (Armour et al., 2013; Lin et al., 2012), suggesting that altered chromatin-modifying activities in the central nervous system may contribute to SCA7 neurodegeneration.
RESULTS
SAGA DUBm components limit yeast replicative lifespan
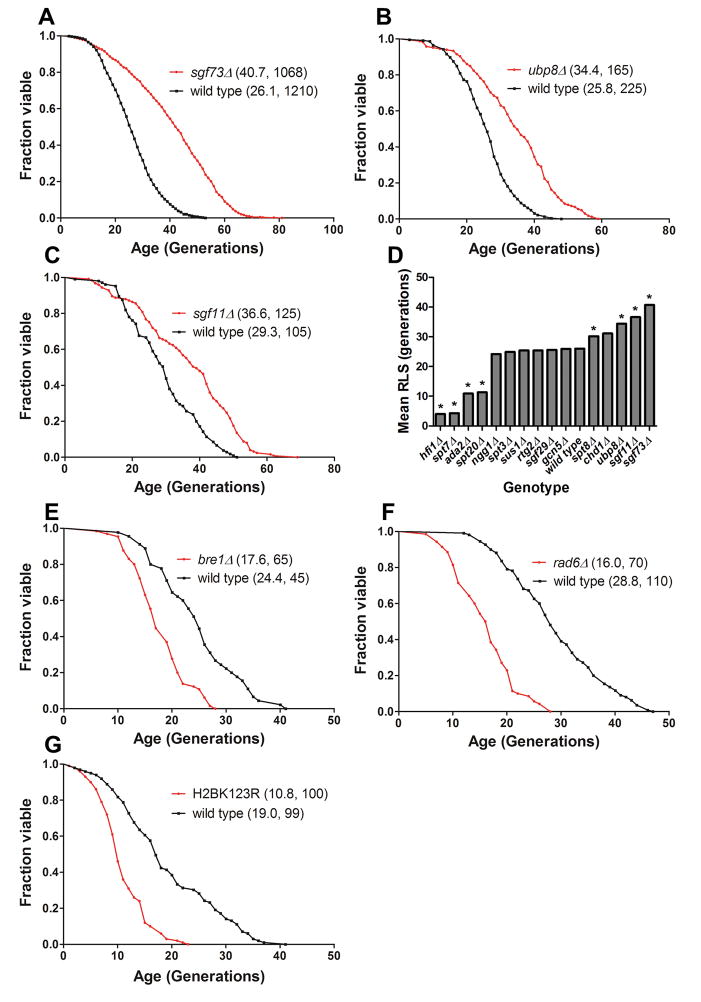
As part of an ongoing assessment of the effects of non-essential yeast genes on yeast replicative lifespan, we discovered that the sgf73Δ strain was one of the longest-lived strains yet identified, extending median and maximum lifespan by 65% and 53%, respectively (Figure 1A). SGF73 encodes a protein in the yeast SAGA and SLIK (SAGA-Like) complexes, chromatin modifying machines that control transcription of a large set of genes. These complexes contain at least two enzymatic activities: Gcn5 has histone acetyltransferase activity, and Ubp8 is a histone deubiquitinase that targets the histone H2B-K123 residue. A component of a four-protein DUBm, Sgf73 serves as a linking factor keeping the DUBm connected with the rest of the SAGA or SLIK complex (Lee et al., 2009). Therefore, we determined the lifespans of the other three components of the DUBm, and found that both sgf11Δ and ubp8Δ strains had robust lifespan extension (Figure 1B, C). Strains lacking the fourth component, SUS1, are not long-lived (Supplemental Figure S1A). Sus1 has other functions in addition to its activity in SAGA, which may make its role in yeast RLS unpredictable. Strains lacking both SGF73 and UBP8 have lifespans identical to the SGF73 single deletion, consistent with the prediction that both deletions enhance lifespan by a similar mechanism and cause increased levels of ubiquitinated H2B (Supplemental Figure S1B) (Kohler et al., 2008).
Figure 1.
Deletion of SAGA DUBm components SGF73, UBP8, and SGF11 significantly increases yeast RLS, and mutation of H2B-K123 or deletion of its monoubiquitinating enzymes shortens RLS. (A) sgf73Δ; (B)ubp8Δ ; (C) sgf11Δ; (D) summary of remaining viable SAGA component deletions; (E) bre1Δ; (F) rad6Δ; (G) H2B-K123R. Legends show (mean RLS, number of mother cells scored). See also Supplemental Figure S1, S2, and S3.
We then determined the lifespan of yeast deletions lacking other non-essential components of SAGA (Figure 1D, Supplemental Figure S2). For the most part, these strains were either not long-lived or had shortened lifespans. Exceptions were chd1Δ, which encodes a chromodomain protein involved in maintaining chromatin structure during transcription by preventing histone exchange (Smolle et al., 2012), and spt8Δ, which encodes a protein unique to the SAGA complex, influencing the TBP-TATA interaction at promoters (Belotserkovskaya et al., 2000). Interestingly, the gcn5Δ strain had no detectable effect on RLS. We conclude from these studies that enhanced RLS derives not from reduced SAGA function, but instead from a more specific effect linked to reduced DUBm function or to uncoupling of the acetyltransferase and deubiquitinase sub-complexes.
A primary target for Ubp8-mediated deubiquitination is histone H2B-K123. Monoubiquitination of this residue is mediated by a heterodimeric complex composed of Bre1, an E3 ubiquitin ligase, and Rad6, an E2 ubiquitin-conjugating enzyme. We determined the lifespan of yeast strains lacking BRE1 or RAD6, finding that they were both short-lived (Figure 1E, F). These data are consistent with the interpretation that replicative lifespan is mediated by H2B-K123 mono-ubiquitination and that higher levels of ubiquitination lead to enhanced lifespan. It should be noted that Rad6 serves as an E2 ubiquitin-conjugating enzyme for other E3s in addition to Bre1; hence, we cannot rule out the possibility that the short lifespan of rad6Δ could be attributable to other causes. To test the role of H2B-K123 mono-ubiquitination more directly, we determined the lifespan of a strain with one integrated copy of a H2BK123R mutation that is refractory to ubiquitination (and one wild-type H2B allele, since the gene is duplicated) (Figure 1G). Interestingly, this mutant was short-lived relative to a wild-type control, again consistent with an association of ubiquitination at H2B-K123 with enhanced longevity.
In addition to Ubp8, Ubp10 can also deubiquitinate histone H2B-K123 at alternative sites in the genome (Schulze et al., 2011). Therefore, we determined whether a ubp10Δmutant would also exhibit enhanced lifespan. Instead, lifespan was reduced in this strain (Supplemental Figure S3). This finding suggests that increased H2B-K123 ubiquitination at the sites normally regulated by Ubp8 and not Ubp10 mediates lifespan extension (Emre et al., 2005).
Lifespan extension by sgf73Δ or ubp8Δis SAGA-dependent but not SLIK-dependent
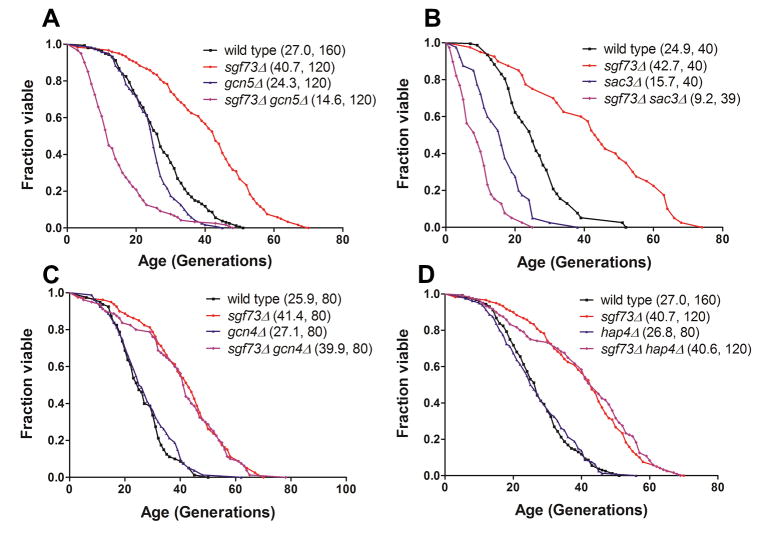
To further delineate the link between SAGA function and longevity, we performed epistasis studies to determine whether non-DUBm components of the SAGA complex are required for lifespan extension associated with sgf73Δand ubp8Δ. For genes encoding most components of the complex, including strains such as gcn5Δ( Figure 2A), loss of SGF73 not only fails to lengthen lifespan, but instead shortens it significantly. These findings are specific for SAGA components, as loss of other nuclear transcription factors, such as Gcn4 (Figure 2B), which blocks the lifespan extension of other long-lived mutants, and Hap4 (Figure 2C) have no impact on lifespan extension in sgf73Δ cells (Steffen et al., 2008).
Figure 2.
RLS epistasis to determine the role of SAGA components and other nuclear transcription factors in sgf73Δ life extension. Genes encoding the SAGA component GCN5 (A) is necessary for the extended RLS of sgf73Δ, while those encoding transcription factors GCN4 (B) and HAP4 (C) are not. Legends show (mean RLS, number of mother cells scored).
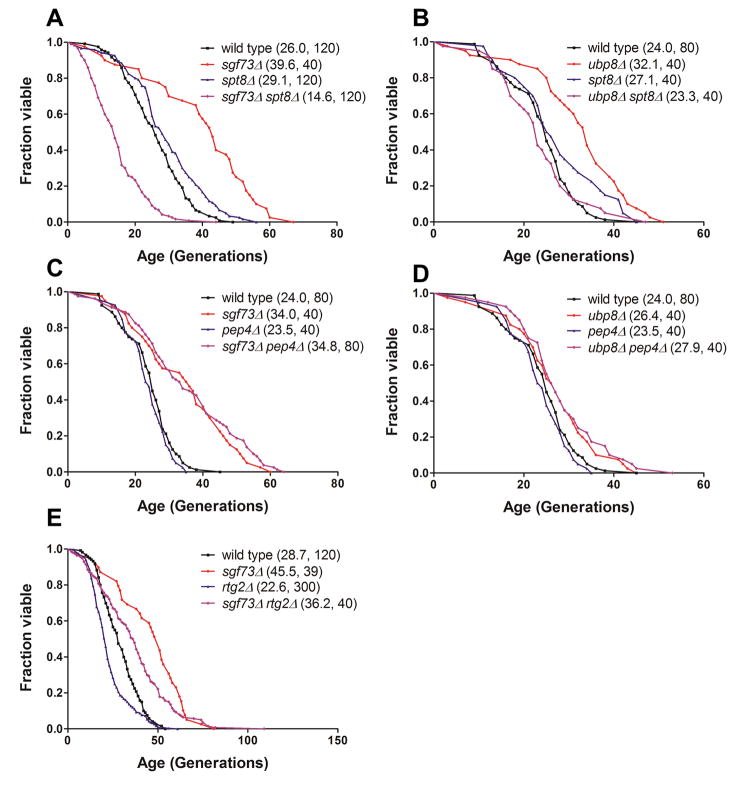
Our data indicate that loss of SAGA components reverses the longevity benefits of sgf73Δ; in other words, loss of DUBm components shortens the lifespan of strains lacking SAGA components rather than lengthening them. We wanted to determine whether this effect was specific to SAGA or SLIK, both of which contain Gcn5. Therefore, we conducted a series of experiments to distinguish between the two complexes by using genetic approaches known to specifically disrupt each complex. First, we determined whether Spt8, a component of SAGA and not SLIK, was required for lifespan extension in sgf73Δ or ubp8Δ, finding that lifespan effects were reversed in this genetic background in a manner similar to that of gcn5Δ (Figure 3A, B). Second, we determined the consequences of deletion of PEP4, which encodes an endopeptidase that cleaves full length Spt7 (associated with SAGA) into a smaller truncated form that associates specifically with SLIK (Spedale et al., 2010). In contrast to the findings with SPT8, both the sgf73Δ pep4Δ and ubp8Δ pep4Δ strains retained a long lifespan (Figure 3C, D). Together, these findings indicate that lifespan extension by loss of DUBm components requires an otherwise intact SAGA but not SLIK complex. To confirm this, we also checked the effects of deleting RTG2, another SLIK-specific component that is of particular interest, because it is linked to the retrograde response that communicates mitochondrial stress signals to the nucleus to evoke transcriptional responses. This pathway has been linked to RLS regulation in other contexts (Jazwinski, 2005). However, we found similar results to those in the pep4Δ strain: loss of SGF73 still resulted in lifespan extension to a comparable extent (Figure 3E). As expected, rtg2Δ alone was slightly short-lived in this strain background, as previously reported (Kaeberlein et al., 2005a). Two other components of retrograde signaling, RTG1 and RTG3, are also not required for lifespan extension by sgf73Δ( Supplemental Figure S4). These findings indicate that the integrity of the rest of the SAGA complex is required for lifespan extension by loss of deubiquitinase components.
Figure 3.
RLS epistasis to determine the relative roles of SAGA and SLIK in sgf73Δ and ubp8Δ lifespan extension. SAGA-specific component SPT8 is necessary for the extended RLS of (A) sgf73Δ and (B) ubp8Δ while neither PEP4, the protease necessary for processing of the SLIK-specific form of Spt7 (C, D), nor SLIK– specific component RTG2 (E) are necessary. Legends show (mean RLS, number of mother cells scored). See also Supplemental Figure S4.
The increased RLS of sgf73Δ and ubp8Δrequires SIR2
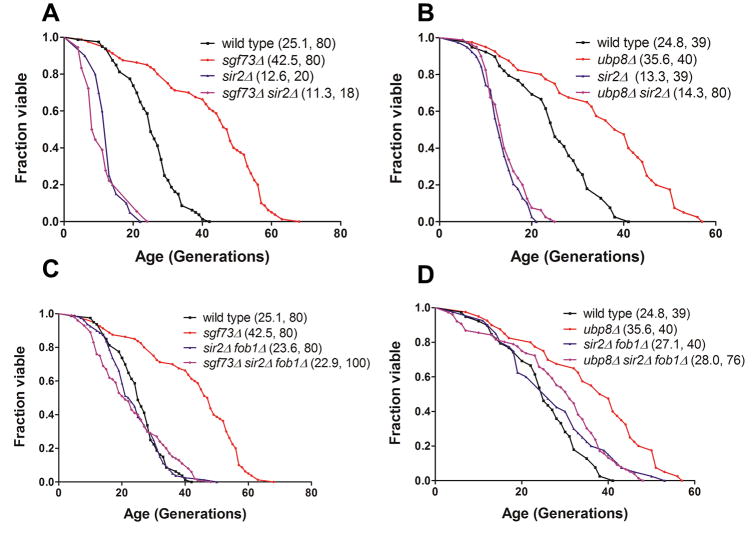
To explore the mechanism of DUBm-deficient lifespan extension, we performed epistasis analysis using several known lifespan-modulating genes. First, we determined whether sgf73Δ functions through the Sir2 longevity pathway by deleting SGF73 or UBP8 in a sir2Δ background. Strains lacking SIR2 are very short-lived, due to ERC accumulation and possibly other SIR2-dependent mechanisms (Kaeberlein et al., 1999a). Previous studies reported that other aging pathways do not make a significant contribution in the sir2 background; hence, genetic interventions that modulate these pathways no longer extend lifespan (Kaeberlein et al., 2004). We found a similar result for sgf73Δand ubp8Δ, with these deletions failing to extend thesir2Δ lifespan( Figure 4A, B). However, this finding is difficult to interpret. Whereas loss of SIR2 may indeed be epistatic to loss of SGF73 and UBP8, more often we have found that longevity interventions that fail to extend RLS in the sir2Δ background retain their ability to do so insir2 Δ fob1Δstrains ( Delaney et al., 2011; Kaeberlein et al., 2004), which have a RLS similar to wild-type strains, most likely due to the suppressive effect of fob1Δ on ERC formation (Defossez et al., 1999; Kaeberlein et al., 1999b). Thus it was possible that sgf73Δ and ubp8Δ strains would be long-lived in a sir2Δ fob1Δ background. However, neither loss of SGF73 nor loss of UBP8 affected longevity in the sir2Δ fob1Δbackground ( Figure 4C, D). These findings are consistent with a model in which deletion of components of the DUBm influences aging by enhancing or altering Sir2 function. One surprising facet of this series of experiments is that the replicative lifespan of either sgf73Δ or ubp8Δ far surpasses the extension observed by overexpression of SIR2 or deletion of FOB1 (Supplemental Figure S5) (Defossez et al., 1999; Kaeberlein et al., 1999a). A possibility is that overexpression of SIR2 and deletion of UBP8 might lead to lifespan extension by at least partially independent mechanisms, both of which are apparent in the sgf73Δ strain.
Figure 4.
SIR2 is necessary for the extended RLS of sgf73Δ (A) and ubp8Δ(B), even in strains lacking FOB1 (C, D). E, SIR2OE ubp8Δ double shows extended lifespan comparable to sgf73Δ. Legends show (mean RLS, number of mother cells scored). See also Supplemental Figure S5.
We also considered the alternate possibility that activity of the SAGA complex in the sir2Δ fob1Δstrain may counteract the longevity benefits associated with disrupting the deubiquitinase module. One way to think about this would be that unchecked Gcn5 acetyltransferase activity, when Sir2 is not there to compete, may have a negative impact on longevity that is independent of the benefits of the sgf73Δ strain. However, this appears not to be the case, as the sgf73Δ sir2Δ fob1Δ spt8Δ strain is not long-lived, as is the case with sgf73Δ sir2Δ fob1Δ (Supplemental Figure S6A). Therefore, we favor a model whereby loss of SGF73 leads to enhanced Sir2 activity.
Sgf73 regulates telomere silencing and rDNA recombination
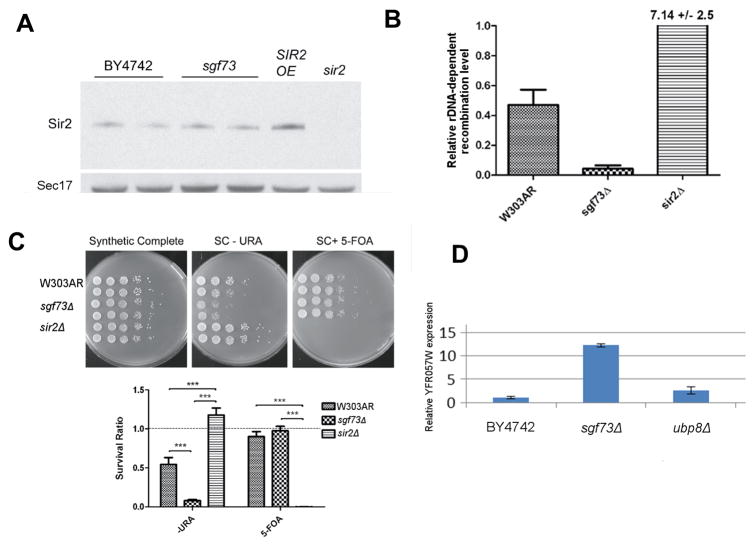
We reasoned that lifespan extension in strains lacking SGF73 could be due, at least in part, to enhanced Sir2 function at regions linked to longevity. Since increased Sir2 expression is linked to lifespan extension, one simple model would be that Sir2 levels are elevated in the sgf73Δ strain. However, we did not detect any difference in Sir2 expression in this strain (Figure 5A). Therefore, we turned to regions of the genome where Sir2 function is associated with longevity: rDNA and telomeres (Dang et al., 2009; Kaeberlein et al., 1999a). Interestingly, a subset of SAGA components, but not Gcn5, is required for Sir2-dependent silencing at both loci (Gottlieb and Esposito, 1989; Jacobson and Pillus, 2009).
Figure 5.
Sgf73 influences silencing at rDNA and telomeres. (A) Sir2 protein levels are unchanged in sgf73Δ; (B) rDNA-dependent recombination of an ADE2 locus is dramatically reduced in sgf73Δ. At telomeres, sgf73Δshows increased silencing of URA3 marker on VIIL (C), but decreased silencing of endogenous ORF YFR057W on VIR (D). Error bars indicate standard error of the mean. See also Supplemental Figure S6.
The rDNA locus contains between 100 and 200 nine-kb repeats. Recombination within the repeats leads to production of extrachromosomal rDNA circles (ERCs) (Sinclair and Guarente, 1997), which can replicate due to the presence of an ARS element, but fail to segregate to daughter cells because they lack centromeric sequences. Accumulation of ERCs in mother cells is one factor that promotes aging (Sinclair and Guarente, 1997), possibly through competition for replication factors in other regions of the genome (Kwan et al., 2013). Sir2 suppresses recombination within repeats, leading to fewer ERCs and longer lifespan (Gottlieb and Esposito, 1989; Kaeberlein et al., 1999a). Loss of the SAGA component Ada2 leads to reduced silencing at both telomeres and rDNA (Jacobson and Pillus, 2009), similar to loss of SIR2 (Aparicio et al., 1991; Smith and Boeke, 1997). In these studies, rDNA recombination was not tested and other SAGA components have not been assessed for rDNA silencing. We tested whether the sgf73Δ strain had reduced rDNA recombination using a standard assay whereby the ADE2 gene is inserted in one repeat, and recombination is tracked by the frequency of red/white half-sectored colonies – daughter cells do not receive the ADE2 locus when it is contained on an ERC. We found that rDNA-dependent recombination is dramatically reduced in the sgf73Δ strain, consistent with enhanced Sir2 function (Figure 5B). By examining double mutants lacking both SGF73 and SIR2, we were able to ascertain that rDNA recombination remains high, although perhaps slightly lower than in the sir2Δ strain alone (Supplemental Figure S6B). These findings indicate that whereas Ada2 is required for Sir2 function at the rDNA locus, Sgf73 counteracts Sir2 function and loss of Sgf73 enhances repression of rDNA recombination. We conclude that this phenotype contributes to the mechanism by which the sgf73Δ strain is long-lived.
Sir2 silences reporter genes placed near telomeres (Aparicio et al., 1991). However, its effect on endogenous genes in these locations has to date been evaluated most thoroughly for YFR057W, where there is a clear requirement for SIR2 (Vega-Palas et al., 2000). The deacetylase activity of Sir2 is required for silencing of this gene (Xu et al., 2007). Interestingly, disruption of SAGA components also affects expression of both reporter genes and YFR057W in a manner that indicates a complex interaction with Sir2. Loss of core SAGA components, ADA2 and ADA3, leads to disruption of silencing of both URA3 placed near the telomere of chromosome VIIL and ADE2 near the telomere of chromosome VR (Jacobson and Pillus, 2009); however, components of the deubiquitinase module have not been assessed. We tested silencing of both URA3 and YFR057w in a strain lacking SGF73, finding discordant results. Silencing of the URA3 locus occurs in a discrete and semi-heritable manner with some of the cells and their progeny expressing the gene, and other cells silencing it. We evaluated silencing of the URA3 reporter locus in a strain lacking SGF73 in two ways: (1) by spotting cell dilutions on plates either lacking uracil, where URA3 expression is required for growth, and (2) on plates containing 5-FOA, where URA3 expression is toxic. In wild-type cells, approximately half of the cells grow on each condition, and in the sir2Δ background, nearly all cells grow in the absence of uracil (Figure 5C). In contrast, sgf73Δ cells grow on 5-FOA but largely fail to grow on plates lacking uracil, indicating increased silencing. However, RT-PCR analysis of sgf73Δcells indicates reduced silencing of YFR057W (Figure 5D). Thus, our findings demonstrate that the effects of the sgf73Δ strain on silencing of the endogenous locus are distinct from those of core SAGA components with respect to the reporter gene. This discrepancy is not unprecedented, as prior studies indicate that mutants can have opposing effects on genes located near different telomeres and there can be complex effects with different reporter genes (Fourel et al., 1999; Pryde and Louis, 1999; Rossmann et al., 2011; Vega-Palas et al., 1997; Wyrick et al., 1999).
Sir2 interacts with Sgf73
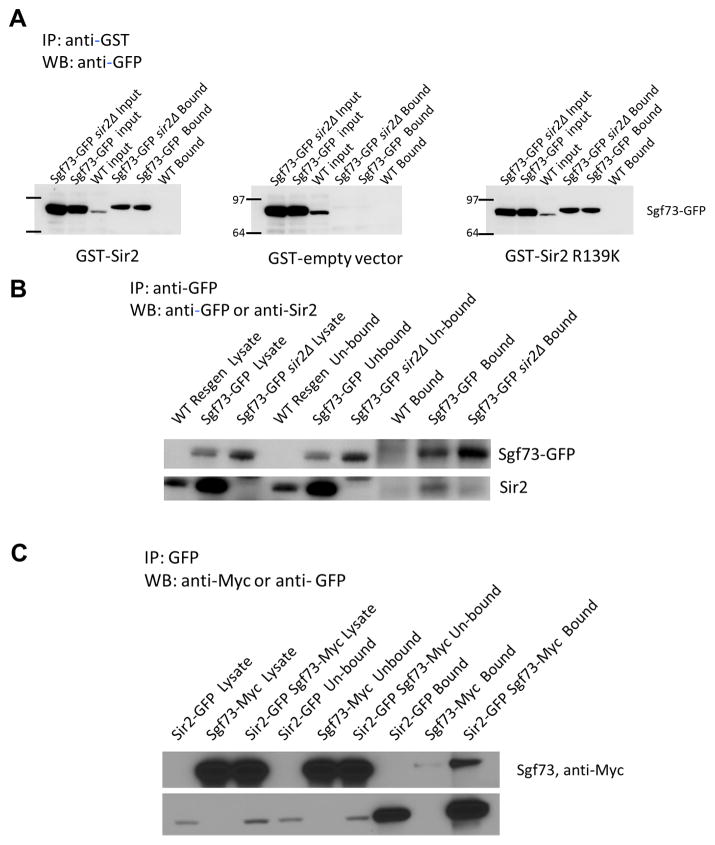
Based upon our findings of altered SIR2-dependent properties in the sgf73Δ yeast and the dependence on SIR2 for sgf73Δ RLS extension, we explored additional potential connections between Sgf73 and Sir2. Evidence exists in mammalian cells for an interaction between the ortholog of SIR2, SIRT1, and the ortholog of UBP8, USP22 (Armour et al., 2013; Lin et al., 2012). This suggests (1) that Sir2 may interact with DUBm components in yeast, and (2) that the functional interactions between the deacetylase and the deubiquitinase components may be deeply conserved. We tested whether Sir2 interactedwi th Sgf73 in yeast, finding evidence of an interaction by two different assays. First, we examined the interaction in vitro by mixing yeast lysates expressing an endogenously tagged Sgf73-GFP with purified recombinant GST-tagged Sir2 protein. Immunoprecipitation of Sir2 by glutathione sepharose was sufficient to recover Sgf73-GFP, upon immunoblot analysis of the precipitated material (Figure 6A). There is a non-specific band that is reactive to the anti-GFP antibody, which is present in the WT input lanes but is not immunoprecipitated. An enzymatically dead Sir2-R139K mutant protein also retained the capacity to interact with Sgf73-GFP, indicating that the deacetylase activity of Sir2 is not required for interaction. In a second experiment, we detected an in vivo interaction between Sgf73-GFP and endogenous Sir2 by co-immunoprecipitation of Sgf73-GFP, as well as between an endogenously tagged Sir2-GFP and endogenously tagged Sgf73-Myc by co-immunoprecipitation of Sir2-GFP (Figure 6B,C). Together, these experiments indicate an interaction between Sir2 and the DUBm of SAGA in yeast, and provide evidence for the deeply conserved nature of the interaction which has also been detected in mammalian cells.
Figure 6.
Sgf73 interacts with Sir2. (A), Interaction of recombinant Sir2-GST and Sgf73-GFP in the presence and absence of Sir2. Immunoprecipitation was done with glutathione beads coupled to either GST tagged Sir2, GST tagged enzymatically dead Sir2R319K, or GST, and western blot was probed with anti-GFP. (B), Interaction of endogenous Sir2 and Sgf73-GFP. Immunoprecipitation was done using anti-GFP antibody and western blots were probed using anti-Sir2 antibody.
DISCUSSION
In this study, we report that deletion of SGF73, the yeast ortholog of human ataxin-7, the causal gene in SCA7, results in one of the longest-lived mutant strains observed to date. Sgf73 is a transcriptional adaptor, serving as the linking factor that connects the core SAGA complex to the Ubp8 histone deubiquitinase module of the complex. Strains lacking the other DUBm components, encoded by UBP8 and SGF11, are also dramatically long-lived, whereas strains lacking GCN5 or other core SAGA components have either normal or shortened replicative lifespans. Strikingly, lifespan extension by loss of DUBm components requires SIR2, suggesting that DUBm mutants enhance Sir2 function. This view is somewhat nuanced by the observation that the sgf73Δ mutant has a much longer replicative lifespan than either strains lacking FOB1 or those overexpressing SIR2. From our studies and other reports describing their enzymatic activities, we propose that the extremely long lifespan of sgf73Δ is due to the contributions of multiple downstream pathways. The likelihood of involvement of each regulatory pathway is discussed below.
The foremost activity of Sir2 linked to extended RLS is repression of rDNA recombination. We find that a sgf73Δ strain has reduced rDNA recombination, consistent with enhanced longevity. A recent study indicates that polymorphisms in the ARS element, which is present in each rDNA repeat, can affect longevity (Kwan et al., 2013). A strong ARS element in the rDNA, possibly in combination with enhanced numbers of ERCs, competes with replication origins in other sites in the genome leading to replication stress. In support of this idea, Ubp8 has been identified as the deubiquitinase responsible for removing ubiquitin linkages on histone H2B-K123 in newly assembled histone octamers that are generated on replicated DNA. Strains lacking Bre1, the E3 ligase for H2B-K123, have reduced H2B-K123 ubiquitination. This leads to reduced assembly or stability of nucleosomes around newly replicated DNA, which slows fork progression, thereby resulting in replication stress (Trujillo and Osley, 2012). In contrast, ubp8Δ strains have increased nucleosome deposition and improved replication fork progression. In the context of old cells, where replication stress may be a key component contributing to aging, the ubp8Δ and sgf73Δ strains may be long-lived not only because they reduce ERC formation, but also because they promote replication fork progression.
Understanding the role of telomere-proximal genes is more complicated. Although Sir2 activity in these regions has also been linked to longevity, we find that the sgf73Δ strain has enhanced silencing of a telomere-proximal reporter gene at chromosome VIIL, but reduced repression of an endogenous gene clearly linked to Sir2-dependent silencing, YFR075W, located on chromosome VIR. This suggests that Sgf73 has a context-dependent effect on silencing of genes near telomeres, as observed for other genes at other natural telomeres (Fourel et al., 1999; Pryde and Louis, 1999; Vega-Palas et al., 2000; Vega-Palas et al., 1997; Wyrick et al., 1999). Future studies of telomeres with different genomic composition and structural features will clarify whether these effects are linked to aging. In addition, given that SAGA and Sgf73 have roles in heterochromatin-euchromatin barrier formation (Oki et al., 2004), with the C-terminus of the Sgf73 subunit of SAGA and SLIK important for retention of the larger complex and for heterochromatin boundary function (Kamata et al., 2013), it will be important to better understand the role of the DUBm in the context of the greater complex.
Finally, recent studies have linked SAGA activity to coordination of different metabolic phases of yeast growth in culture (Cai et al., 2011). This occurs through control of transcription, in part through H3K9 acetylation. Interestingly, SAGA has a direct role in regulating expression of ribosomal protein genes in a manner dependent on the transcription factor Ifh1 (Cai et al., 2013; Downey et al., 2013). Ribosomal protein gene deletions are highly enriched among long-lived mutants in yeast (Mehta et al., 2010). Thus, another potential link to enhanced longevity in DUBm mutants may be through reduced ribosome biogenesis. In summary, multiple mechanisms are likely to explain the extremely long lifespan of sgf73Δ and ubp8Δ strains, and further experimentation will be required to unravel the relative contribution of each component.
The discovery of the marked replicative lifespan extension in sgf73Δ mutants, together with the demonstration that the sgf73Δ strain lifespan extension phenotype stems from enhanced Sir2 function in combination with reduced Ubp8 function, may have important implications for the normal function of the mammalian Sgf73 ortholog, ataxin-7. Ataxin-7 was originally discovered as the causal gene in a dominantly inherited neurodegenerative disorder, known as spinocerebellar ataxia type 7 (SCA7) (David et al., 1997). Patients with SCA7 exhibit cerebellar and retinal degeneration, display CAG repeat expansions within the coding regions of their ataxin-7 genes, and produce mutant ataxin-7 protein containing expanded polyglutamine tracts ranging in size from 37 to >400 glutamines. SCA7 is thus one member of a family of nine CAG – polyglutamine diseases, and belongs to a large class of neurodegenerative disorders, including Alzheimer’s and Parkinson’s diseases. These “neurodegenerative proteinopathies” result from the production of misfolded proteins (La Spada and Taylor, 2010).
In the case of SCA7, polyglutamine-expanded ataxin-7 promotes transcriptional interference to produce neuropathology, as the mutant ataxin-7 protein localizes to the nucleus where it disrupts transcription factor function (Holmberg et al., 1998; Kaytor et al., 1999; La Spada et al., 2001). Unbiased mass spectrometry analysis of the STAGA complex yielded ataxin-7 as a core STAGA complex component, and further studies determined that polyglutamine-expanded ataxin-7 interferes with the GCN5-dependent histone acetyltransferase activity of the STAGA complex to impair STAGA-dependent transactivation (Palhan et al., 2005). Although polyglutamine-expanded ataxin-7 exhibits gain-of-function toxicity due to a proteotoxicity effect, the discovery of altered STAGA function in SCA7-mutant mice indicated a role for altered ataxin-7 protein function in SCA7 disease pathogenesis. Hence, SCA7-dependent neurodegeneration may result from divergent effects of mutant ataxin-7 protein on the activities of interacting proteins in the context of the STAGA complex, akin to polyglutamine-expanded ataxin-1 effects on its protein complex partners in SCA1 (Zoghbi and Orr, 2009).
As the Ubp8 ortholog USP22 was recently shown to interact with, deubiquitinate, and stabilize the Sir2 ortholog Sirt1 (Lin et al., 2012), and conversely Sirt1 has been shown to mediate deacetylation of USP22 and the SAGA coactivator complex (Armour et al., 2013), the discovery of genetic and functional relationships between Sgf73, Ubp8, and Sir2 in yeast strongly suggests that mutant ataxin-7 protein could be impairing the function of not only Gcn5 in the context of the STAGA complex, but also USP22 in the deubiquitinase module. Given the recent proposed linkage between USP22 and Sirt1, our findings predict that polyglutamine-expanded ataxin-7 could interfere with the normal function of both USP22 and SIRT1 in neurons. Indeed, whether and how ataxin-7 affects pathways of mammalian aging relevant to neural function and neuroprotection will be key questions for future studies, with implications for both SCA7 disease pathogenesis and therapy development and for a deeper understanding of the biology of aging in mammals.
MATERIALS AND METHODS
Strains and media
All yeast strains were derived from the parent strains of the haploid yeast ORF deletion collections (Winzeler et al., 1999), BY4742 (MATαhis3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 ) and BY4741 (MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ). sgf73Δ was reconstructed by deleting SGF73 via homologous recombination of a selectable URA3 marker in the deletion collection wild-type BY strain using standard PCR-mediated gene disruption, and this parent was used to generate all sgf73Δ containing double and triple mutants . SIR2OE (overexpression) strains contain a second integrated copy of SIR2 under the control of its endogenous promoter. RGY43 was a gift of Richard Gardner. Cells were grown on standard YPD containing 1% yeast extract, 2% peptone and 2% glucose.
Replicative Lifespan
RLS assays were performed as described previously (Steffen et al., 2009). Survival curves are pooled data from multiple experiments with accompanying experiment-matched controls, with mean RLS and number of mother cells scored shown for each curve. p-values for RLS survival curve comparisons were calculated using a Wilcoxon rank-sum test (Wilcoxon, 1946). Kaplan-Meier survival curves (Kaplan and Meier, 1958) were plotted using Prism (GraphPad, USA).
Yeast Interaction Studies
Yeast lysis
Standard yeast glass bead lysis protocol was followed. Briefly; Yeast were grown to an A600 of 0.8 and lysed in 50mM HEPES pH 7.5, 0.1M NaCl, 0.5% Np40, 10% glycerol, 1mM EDTA, with protease inhibitors including PMSF. Lysates were pre-cleared with 10μl of protein A/G (Dynabeads, Life Technologies) magnetic beads at 4°C for one hour.
Strains
All tagged protein strains used were endogenously tagged with either GFP or 13Myc. Tagged strains were demonstrated to be functional by mating for tagged Sir2 and by checking cycloheximide resistance for tagged Sgf73.
Recombinant protein purification
BL21 cells were transformed with one of three plasmids Sir2-GST, Sir2-R139K-GST, or GST empty vector. A 100ml culture was grown to A600 0.6 and IPTG was added to a final concentration of 0.5mM. The culture was then incubated with shaking at room temperature for 4 hours. Cells were collected and lysed (20mM Tris pH 8, 1mM EDTA, 1mM EGTA, 1% NP-40, 350mM NaCl, 10mM dithiothreitol, protease inhibitors (Roche 04693124001), PMSF, 200mg/ml lysozyme) for 30 min on ice, then sonicated 3x 1 min with 5 sec on 0.5 sec off cycles, and spun at 4°C for 20 minutes. Glutathione agarose was prepared and added to the bacterial lysates and incubated at 4°C for one hour.
Immunoprecipitation
In vitro: Conjugated beads were added to prepared yeast lysates and rotated at 4°C overnight. Beads were washed twice with yeast lysis buffer and twice with IP wash buffer (50mM HEPES pH 7.5, 150mM NaCl, 1mM EDTA). Proteins were analyzed by western blot, 2.5% of input was loaded, 12.5% of bound material was loaded. In vivo: Anti-GFP antibody (living colors JL8 632381) was added to yeast lysates and rotated at 4°C overnight, 50μl of protein A/G beads were then added and sample rotated for 1 hour at 4°C. Beads were washed twice with lysis buffer and twice with wash buffer. For Sgf73-GFP IP 2% of input and unbound, and 30% of bound material was loaded, for Sir2-GFP IP 1.25% of input and unbound, and 100% of bound material was loaded. Protein was analyzed via immunoblotting.
Immunoblot
Standard SDS-PAGE was used to separate proteins on 4–12% gels. After transfer to nitrocellulose, membrane was blocked with 5% milk and incubated with either anti-GFP (living colors JL8 632381) or anti-Sir2 (Garcia and Pillus, 2002).
Supplementary Material
HIGHLIGHTS.
S. cerevisiae strains lacking SAGA genes SGF73, SGF11, and UBP8 are long-lived.
Sir2 is required and shows genetic and physical interaction with SAGA.
Sgf73 ortholog causes neurodegenerative disease, suggesting potential human impact.
Acknowledgments
We thank members of the Kennedy, La Spada, and Pillus labs for technical assistance and helpful discussion. This study was supported by funds from the National Institutes of Health: R01 AG025549 and R01 AG043080 to B.K.K, GM090177, GM054778 to L.P., R01 EY014061 and R01 AG033082 to A.R.L. T32 AG000266 to M.M., T32 GM008666-14 to A.G.M., and F32 GM089101 to R.M.G. B.K.K. is an Ellison Medical Foundation Senior Scholar in Aging.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aparicio OM, Billington BL, Gottschling DE. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66:1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- Armour SM, Bennett EJ, Braun CR, Zhang XY, McMahon SB, Gygi SP, Harper JW, Sinclair DA. A high-confidence interaction map identifies SIRT1 as a mediator of acetylation of USP22 and the SAGA coactivator complex. Mol Cell Biol. 2013;33:1487–1502. doi: 10.1128/MCB.00971-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belotserkovskaya R, Sterner DE, Deng M, Sayre MH, Lieberman PM, Berger SL. Inhibition of TATA-binding protein function by SAGA subunits Spt3 and Spt8 at Gcn4-activated promoters. Mol Cell Biol. 2000;20:634–647. doi: 10.1128/mcb.20.2.634-647.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, McCormick MA, Kennedy BK, Tu BP. Integration of multiple nutrient cues and regulation of lifespan by ribosomal transcription factor ifh1. Cell reports. 2013;4:1063–1071. doi: 10.1016/j.celrep.2013.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Sutter BM, Li B, Tu BP. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Molecular cell. 2011;42:426–437. doi: 10.1016/j.molcel.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang W, Steffen KK, Perry R, Dorsey JA, Johnson FB, Shilatifard A, Kaeberlein M, Kennedy BK, Berger SL. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel JA, Torok MS, Sun ZW, Schieltz D, Allis CD, Yates JR, 3rd, Grant PA. Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. The Journal of biological chemistry. 2004;279:1867–1871. doi: 10.1074/jbc.C300494200. [DOI] [PubMed] [Google Scholar]
- David G, Abbas N, Stevanin G, Durr A, Yvert G, Cancel G, Weber C, Imbert G, Saudou F, Antoniou E, et al. Cloning of the SCA7 gene reveals a highly unstable CAG repeat expansion. Nature genetics. 1997;17:65–70. doi: 10.1038/ng0997-65. [DOI] [PubMed] [Google Scholar]
- Defossez PA, Prusty R, Kaeberlein M, Lin SJ, Ferrigno P, Silver PA, Keil RL, Guarente L. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Molecular cell. 1999;3:447–455. doi: 10.1016/s1097-2765(00)80472-4. [DOI] [PubMed] [Google Scholar]
- Delaney JR, Sutphin GL, Dulken B, Sim S, Kim JR, Robison B, Schleit J, Murakami CJ, Carr D, An EH, et al. Sir2 deletion prevents lifespan extension in 32 long-lived mutants. Aging Cell. 2011 doi: 10.1111/j.1474-9726.2011.00742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downey M, Knight B, Vashisht AA, Seller CA, Wohlschlegel JA, Shore D, Toczyski DP. Gcn5 and sirtuins regulate acetylation of the ribosomal protein transcription factor ifh1. Current biology : CB. 2013;23:1638–1648. doi: 10.1016/j.cub.2013.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emre NC, Ingvarsdottir K, Wyce A, Wood A, Krogan NJ, Henry KW, Li K, Marmorstein R, Greenblatt JF, Shilatifard A, et al. Maintenance of low histone ubiquitylation by Ubp10 correlates with telomere-proximal Sir2 association and gene silencing. Molecular cell. 2005;17:585–594. doi: 10.1016/j.molcel.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Fourel G, Revardel E, Koering CE, Gilson E. Cohabitation of insulators and silencing elements in yeast subtelomeric regions. The EMBO journal. 1999;18:2522–2537. doi: 10.1093/emboj/18.9.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia SN, Pillus L. A unique class of conditional sir2 mutants displays distinct silencing defects in Saccharomyces cerevisiae. Genetics. 2002;162:721–736. doi: 10.1093/genetics/162.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb S, Esposito RE. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 1989;56:771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- Guarente L. Cold Spring Harbor symposia on quantitative biology. 2011. Sirtuins, Aging, and Metabolism. [DOI] [PubMed] [Google Scholar]
- Helmlinger D, Hardy S, Sasorith S, Klein F, Robert F, Weber C, Miguet L, Potier N, Van-Dorsselaer A, Wurtz JM, et al. Ataxin-7 is a subunit of GCN5 histone acetyltransferase-containing complexes. Hum Mol Genet. 2004;13:1257–1265. doi: 10.1093/hmg/ddh139. [DOI] [PubMed] [Google Scholar]
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes & development. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg M, Duyckaerts C, Durr A, Cancel G, Gourfinkel-An I, Damier P, Faucheux B, Trottier Y, Hirsch EC, Agid Y, et al. Spinocerebellar ataxia type 7 (SCA7): a neurodegenerative disorder with neuronal intranuclear inclusions. Hum Mol Genet. 1998;7:913–918. doi: 10.1093/hmg/7.5.913. [DOI] [PubMed] [Google Scholar]
- Jacobson S, Pillus L. The SAGA subunit Ada2 functions in transcriptional silencing. Mol Cell Biol. 2009;29:6033–6045. doi: 10.1128/MCB.00542-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazwinski SM. Rtg2 protein: at the nexus of yeast longevity and aging. FEMS yeast research. 2005;5:1253–1259. doi: 10.1016/j.femsyr.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Sir2-independent life span extension by calorie restriction in yeast. PLOS Biology. 2004;2:1381–1387. doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Genes determining yeast replicative life span in a long-lived genetic background. Mech Ageing Dev. 2005a;126:491–504. doi: 10.1016/j.mad.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999a;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes & development. 1999b;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW, 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005b;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- Kamata K, Hatanaka A, Goswami G, Shinmyozu K, Nakayama J, Urano T, Hatashita M, Uchida H, Oki M. C-terminus of the Sgf73 subunit of SAGA and SLIK is important for retention in the larger complex and for heterochromatin boundary function. Genes to cells : devoted to molecular & cellular mechanisms. 2013;18:823–837. doi: 10.1111/gtc.12075. [DOI] [PubMed] [Google Scholar]
- Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Amer Statist Assn. 1958:53. [Google Scholar]
- Kaytor MD, Duvick LA, Skinner PJ, Koob MD, Ranum LP, Orr HT. Nuclear localization of the spinocerebellar ataxia type 7 protein, ataxin-7. Hum Mol Genet. 1999;8:1657–1664. doi: 10.1093/hmg/8.9.1657. [DOI] [PubMed] [Google Scholar]
- Kennedy BK, Austriaco NR, Zhang J, Guarente L. Mutation in the silencing gene SIR4 can delay aging in S. cerevisiae. Cell. 1995;80:485–496. doi: 10.1016/0092-8674(95)90499-9. [DOI] [PubMed] [Google Scholar]
- Kohler A, Schneider M, Cabal GG, Nehrbass U, Hurt E. Yeast Ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nature cell biology. 2008;10:707–715. doi: 10.1038/ncb1733. [DOI] [PubMed] [Google Scholar]
- Kwan EX, Foss EJ, Tsuchiyama S, Alvino GM, Kruglyak L, Kaeberlein M, Raghuraman MK, Brewer BJ, Kennedy BK, Bedalov A. A Natural Polymorphism in rDNA Replication Origins Links Origin Activation with Calorie Restriction and Lifespan. PLoS genetics. 2013;9:e1003329. doi: 10.1371/journal.pgen.1003329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Spada AR, Fu YH, Sopher BL, Libby RT, Wang X, Li LY, Einum DD, Huang J, Possin DE, Smith AC, et al. Polyglutamine-expanded ataxin-7 antagonizes CRX function and induces cone-rod dystrophy in a mouse model of SCA7. Neuron. 2001;31:913–927. doi: 10.1016/s0896-6273(01)00422-6. [DOI] [PubMed] [Google Scholar]
- La Spada AR, Taylor JP. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nature reviews Genetics. 2010;11:247–258. doi: 10.1038/nrg2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebre AS, Brice A. Spinocerebellar ataxia 7 (SCA7) Cytogenet Genome Res. 2003;100:154–163. doi: 10.1159/000072850. [DOI] [PubMed] [Google Scholar]
- Lee KK, Swanson SK, Florens L, Washburn MP, Workman JL. Yeast Sgf73/Ataxin-7 serves to anchor the deubiquitination module into both SAGA and Slik(SALSA) HAT complexes. Epigenetics & chromatin. 2009;2:2. doi: 10.1186/1756-8935-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Yang H, Kong Q, Li J, Lee SM, Gao B, Dong H, Wei J, Song J, Zhang DD, et al. USP22 antagonizes p53 transcriptional activation by deubiquitinating Sirt1 to suppress cell apoptosis and is required for mouse embryonic development. Molecular cell. 2012;46:484–494. doi: 10.1016/j.molcel.2012.03.024. [DOI] [PubMed] [Google Scholar]
- Mal AK. Histone methyltransferase Suv39h1 represses MyoD-stimulated myogenic differentiation. The EMBO journal. 2006;25:3323–3334. doi: 10.1038/sj.emboj.7601229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez E, Palhan VB, Tjernberg A, Lymar ES, Gamper AM, Kundu TK, Chait BT, Roeder RG. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol Cell Biol. 2001;21:6782–6795. doi: 10.1128/MCB.21.20.6782-6795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta R, Chandler-Brown D, Ramos FJ, Shamieh LS, Kaeberlein M. Regulation of mRNA translation as a conserved mechanism of longevity control. Advances in experimental medicine and biology. 2010;694:14–29. doi: 10.1007/978-1-4419-7002-2_2. [DOI] [PubMed] [Google Scholar]
- Mortimer RK, Johnston JR. Life span of individual yeast cells. Nature. 1959;183:1751–1752. doi: 10.1038/1831751a0. [DOI] [PubMed] [Google Scholar]
- Mushegian AR, Vishnivetskiy SA, Gurevich VV. Conserved phosphoprotein interaction motif is functionally interchangeable between ataxin-7 and arrestins. Biochemistry. 2000;39:6809–6813. doi: 10.1021/bi992694y. [DOI] [PubMed] [Google Scholar]
- Oki M, Valenzuela L, Chiba T, Ito T, Kamakaka RT. Barrier proteins remodel and modify chromatin to restrict silenced domains. Mol Cell Biol. 2004;24:1956–1967. doi: 10.1128/MCB.24.5.1956-1967.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palhan VB, Chen S, Peng GH, Tjernberg A, Gamper AM, Fan Y, Chait BT, La Spada AR, Roeder RG. Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:8472–8477. doi: 10.1073/pnas.0503505102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryde FE, Louis EJ. Limitations of silencing at native yeast telomeres. The EMBO journal. 1999;18:2538–2550. doi: 10.1093/emboj/18.9.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann MP, Luo W, Tsaponina O, Chabes A, Stillman B. A common telomeric gene silencing assay is affected by nucleotide metabolism. Molecular cell. 2011;42:127–136. doi: 10.1016/j.molcel.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel H, Tomiuk S, Hofmann K. Elucidation of ataxin-3 and ataxin-7 function by integrative bioinformatics. Hum Mol Genet. 2003;12:2845–2852. doi: 10.1093/hmg/ddg297. [DOI] [PubMed] [Google Scholar]
- Schulze JM, Hentrich T, Nakanishi S, Gupta A, Emberly E, Shilatifard A, Kobor MS. Splitting the task: Ubp8 and Ubp10 deubiquitinate different cellular pools of H2BK123. Genes & development. 2011;25:2242–2247. doi: 10.1101/gad.177220.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L. Extrachromosomal rDNA circles-a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- Smith ED, Tsuchiya M, Fox LA, Dang N, Hu D, Kerr EO, Johnston ED, Tchao BN, Pak DN, Welton KL, et al. Quantitative evidence for conserved longevity pathways between divergent eukaryotic species. Genome Res. 2008;18:564–570. doi: 10.1101/gr.074724.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Boeke JD. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes & development. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- Smolle M, Venkatesh S, Gogol MM, Li H, Zhang Y, Florens L, Washburn MP, Workman JL. Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nature structural & molecular biology. 2012;19:884–892. doi: 10.1038/nsmb.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spedale G, Mischerikow N, Heck AJ, Timmers HT, Pijnappel WW. Identification of Pep4p as the protease responsible for formation of the SAGA-related SLIK protein complex. The Journal of biological chemistry. 2010;285:22793–22799. doi: 10.1074/jbc.M110.108787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanfel MN, Shamieh LS, Kaeberlein M, Kennedy BK. The TOR pathway comes of age. Biochim Biophys Acta. 2009;1790:1067–1074. doi: 10.1016/j.bbagen.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen KK, Kennedy BK, Kaeberlein M. Measuring replicative life span in the budding yeast. J Vis Exp. 2009 doi: 10.3791/1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen KK, MacKay VL, Kerr EO, Tsuchiya M, Hu D, Fox LA, Dang N, Johnston ED, Oakes JA, Tchao BN, et al. Yeast lifespan extension by depletion of 60S ribosomal subunits is mediated by Gcn4. Cell. 2008;133:292–302. doi: 10.1016/j.cell.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinkraus KA, Kaeberlein M, Kennedy BK. Replicative aging in yeast: the means to the end. Annu Rev Cell Dev Biol. 2008;24:29–54. doi: 10.1146/annurev.cellbio.23.090506.123509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutphin GL, Olsen BA, Kennedy BK, Kaeberlein M. Genome-wide analysis of yeast aging. Sub-cellular biochemistry. 2012;57:251–289. doi: 10.1007/978-94-007-2561-4_12. [DOI] [PubMed] [Google Scholar]
- Trujillo KM, Osley MA. A role for H2B ubiquitylation in DNA replication. Molecular cell. 2012;48:734–746. doi: 10.1016/j.molcel.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega-Palas MA, Martin-Figueroa E, Florencio FJ. Telomeric silencing of a natural subtelomeric gene. Molecular & general genetics : MGG. 2000;263:287–291. doi: 10.1007/s004380051170. [DOI] [PubMed] [Google Scholar]
- Vega-Palas MA, Venditti S, Di Mauro E. Telomeric transcriptional silencing in a natural context. Nature genetics. 1997;15:232–233. doi: 10.1038/ng0397-232. [DOI] [PubMed] [Google Scholar]
- Wilcoxon F. Individual comparisons of grouped data by ranking methods. J Econ Entomol. 1946;39:269. doi: 10.1093/jee/39.2.269. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Wyrick JJ, Holstege FC, Jennings EG, Causton HC, Shore D, Grunstein M, Lander ES, Young RA. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature. 1999;402:418–421. doi: 10.1038/46567. [DOI] [PubMed] [Google Scholar]
- Xu F, Zhang Q, Zhang K, Xie W, Grunstein M. Sir2 deacetylates histone H3 lysine 56 to regulate telomeric heterochromatin structure in yeast. Molecular cell. 2007;27:890–900. doi: 10.1016/j.molcel.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XY, Varthi M, Sykes SM, Phillips C, Warzecha C, Zhu W, Wyce A, Thorne AW, Berger SL, McMahon SB. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Molecular cell. 2008;29:102–111. doi: 10.1016/j.molcel.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, Suzuki E, Le Guezennec X, Stunnenberg HG, Krasnov A, et al. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Molecular cell. 2008;29:92–101. doi: 10.1016/j.molcel.2007.12.011. [DOI] [PubMed] [Google Scholar]
- Zoghbi HY, Orr HT. Pathogenic mechanisms of a polyglutamine-mediated neurodegenerative disease, spinocerebellar ataxia type 1. The Journal of biological chemistry. 2009;284:7425–7429. doi: 10.1074/jbc.R800041200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.