Abstract
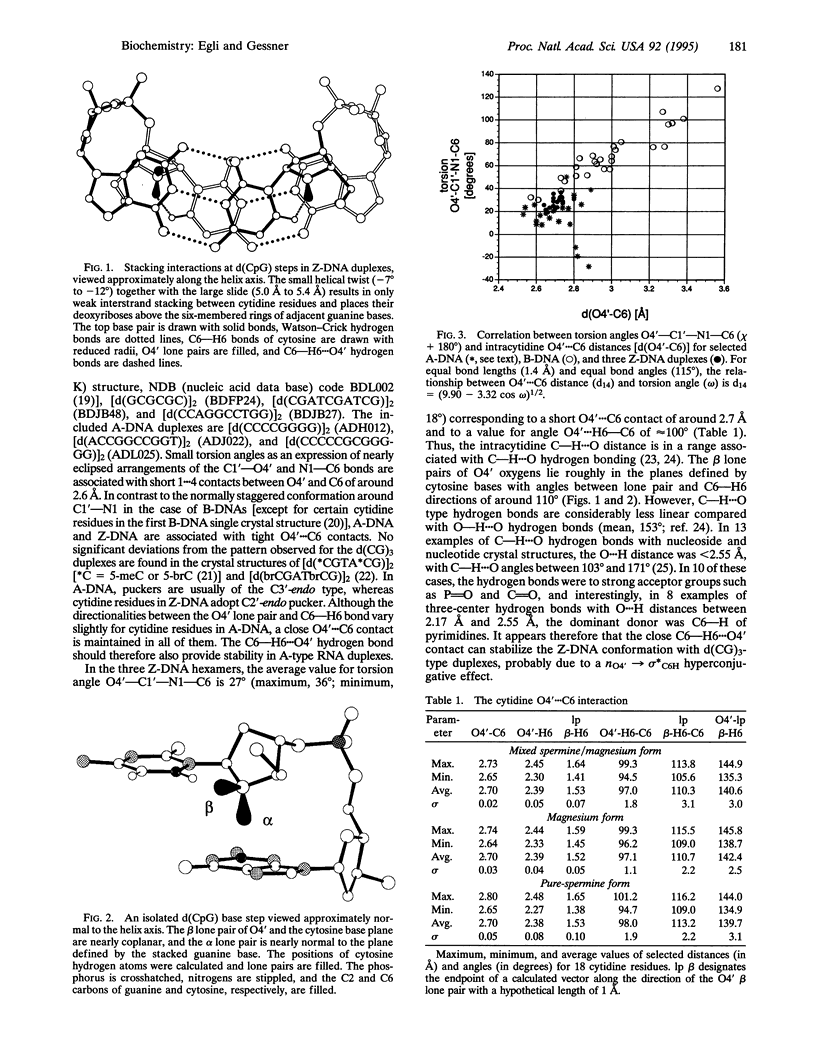
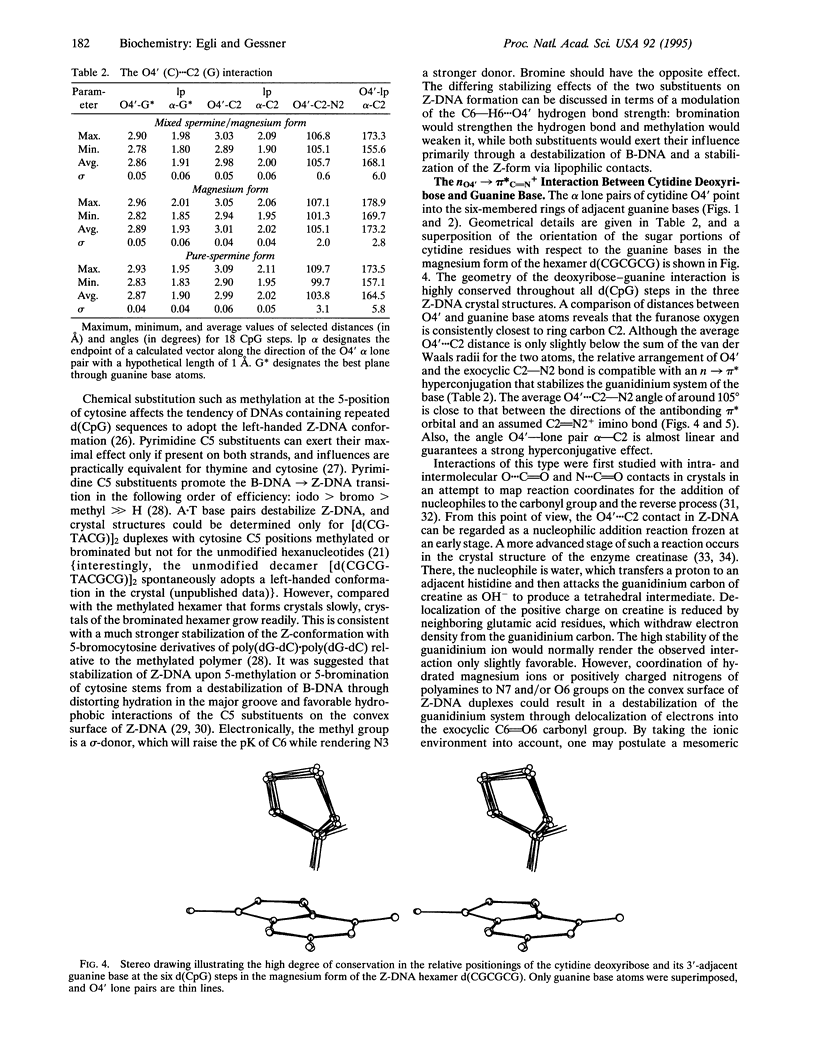
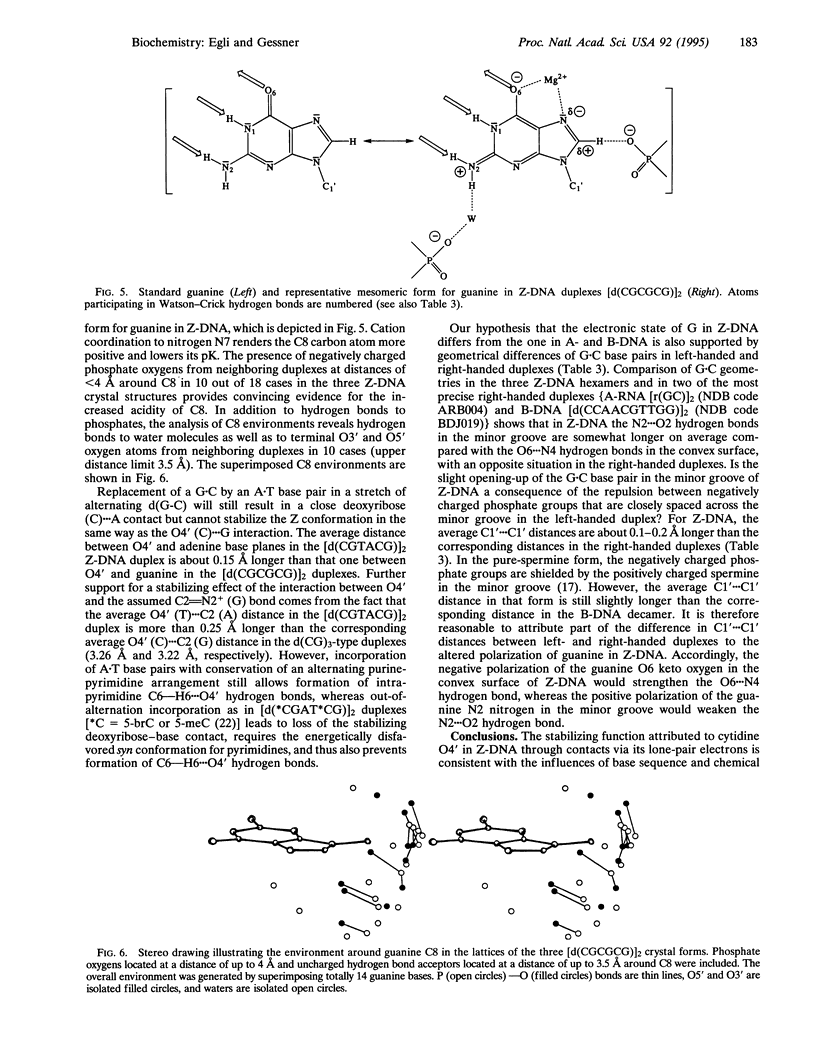
While B-DNA, the most common DNA conformation, displays rather regular twist angles and base stacking between successive base pairs, left-handed Z-DNA is characterized by the alternation of two different dinucleotide conformations with either a large twist and a small slide or a small twist and a large slide between adjacent base pairs. This results in poor stacking within the latter dinucleotide repeat that is in apparent contradiction to the rigidity and conformational stability of Z-DNA at high ionic strength. However, at d(CpG) steps the cytidine deoxyribose is situated such that its O4' sits directly over the six-membered ring of the guanine. We show here that the particular positionings of the two O4' lone-pair electrons provide stability through an intracytidine O4'...H6--C6 hydrogen bond and an n-->pi* interaction with the guanidinium system of the stacked base. Our model is based on the assumption of a strong polarization of the guanine bases in Z-DNA that is consistent with the Z-DNA-specific guanine O6 and N7 coordination to metal and organic cations and the proximity of its N2 and C8 positions to neighboring phosphate groups, as well as several other Z-DNA-specific conformational features.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bancroft D., Williams L. D., Rich A., Egli M. The low-temperature crystal structure of the pure-spermine form of Z-DNA reveals binding of a spermine molecule in the minor groove. Biochemistry. 1994 Feb 8;33(5):1073–1086. doi: 10.1021/bi00171a005. [DOI] [PubMed] [Google Scholar]
- Behe M., Felsenfeld G. Effects of methylation on a synthetic polynucleotide: the B--Z transition in poly(dG-m5dC).poly(dG-m5dC). Proc Natl Acad Sci U S A. 1981 Mar;78(3):1619–1623. doi: 10.1073/pnas.78.3.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H. M., Olson W. K., Beveridge D. L., Westbrook J., Gelbin A., Demeny T., Hsieh S. H., Srinivasan A. R., Schneider B. The nucleic acid database. A comprehensive relational database of three-dimensional structures of nucleic acids. Biophys J. 1992 Sep;63(3):751–759. doi: 10.1016/S0006-3495(92)81649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrondo M. A., Coll M., Aymami J., Wang A. H., van der Marel G. A., van Boom J. H., Rich A. Binding of a Hoechst dye to d(CGCGATATCGCG) and its influence on the conformation of the DNA fragment. Biochemistry. 1989 Sep 19;28(19):7849–7859. doi: 10.1021/bi00445a047. [DOI] [PubMed] [Google Scholar]
- Chevrier B., Dock A. C., Hartmann B., Leng M., Moras D., Thuong M. T., Westhof E. Solvation of the left-handed hexamer d(5BrC-G-5BrC-G-5 BrC-G) in crystals grown at two temperatures. J Mol Biol. 1986 Apr 20;188(4):707–719. doi: 10.1016/s0022-2836(86)80016-x. [DOI] [PubMed] [Google Scholar]
- Coll M., Knof S. H., Ohga Y., Messerschmidt A., Huber R., Moellering H., Rüssmann L., Schumacher G. Enzymatic mechanism of creatine amidinohydrolase as deduced from crystal structures. J Mol Biol. 1990 Jul 20;214(2):597–610. doi: 10.1016/0022-2836(90)90201-v. [DOI] [PubMed] [Google Scholar]
- Egli M., Usman N., Rich A. Conformational influence of the ribose 2'-hydroxyl group: crystal structures of DNA-RNA chimeric duplexes. Biochemistry. 1993 Apr 6;32(13):3221–3237. [PubMed] [Google Scholar]
- Egli M., Williams L. D., Gao Q., Rich A. Structure of the pure-spermine form of Z-DNA (magnesium free) at 1-A resolution. Biochemistry. 1991 Dec 3;30(48):11388–11402. doi: 10.1021/bi00112a005. [DOI] [PubMed] [Google Scholar]
- Frederick C. A., Coll M., van der Marel G. A., van Boom J. H., Wang A. H. Molecular structure of cyclic deoxydiadenylic acid at atomic resolution. Biochemistry. 1988 Nov 1;27(22):8350–8361. doi: 10.1021/bi00422a010. [DOI] [PubMed] [Google Scholar]
- Fujii S., Wang A. H., van der Marel G., van Boom J. H., Rich A. Molecular structure of (m5 dC-dG)3: the role of the methyl group on 5-methyl cytosine in stabilizing Z-DNA. Nucleic Acids Res. 1982 Dec 11;10(23):7879–7892. doi: 10.1093/nar/10.23.7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessner R. V., Frederick C. A., Quigley G. J., Rich A., Wang A. H. The molecular structure of the left-handed Z-DNA double helix at 1.0-A atomic resolution. Geometry, conformation, and ionic interactions of d(CGCGCG). J Biol Chem. 1989 May 15;264(14):7921–7935. doi: 10.2210/pdb1dcg/pdb. [DOI] [PubMed] [Google Scholar]
- Gessner R. V., Quigley G. J., Egli M. Comparative studies of high resolution Z-DNA crystal structures. Part 1: Common hydration patterns of alternating dC-dG. J Mol Biol. 1994 Mar 4;236(4):1154–1168. doi: 10.1016/0022-2836(94)90018-3. [DOI] [PubMed] [Google Scholar]
- Gessner R. V., Quigley G. J., Wang A. H., van der Marel G. A., van Boom J. H., Rich A. Structural basis for stabilization of Z-DNA by cobalt hexaammine and magnesium cations. Biochemistry. 1985 Jan 15;24(2):237–240. doi: 10.1021/bi00323a001. [DOI] [PubMed] [Google Scholar]
- Hoeffken H. W., Knof S. H., Bartlett P. A., Huber R., Moellering H., Schumacher G. Crystal structure determination, refinement and molecular model of creatine amidinohydrolase from Pseudomonas putida. J Mol Biol. 1988 Nov 20;204(2):417–433. doi: 10.1016/0022-2836(88)90586-4. [DOI] [PubMed] [Google Scholar]
- Jovin T. M., McIntosh L. P., Arndt-Jovin D. J., Zarling D. A., Robert-Nicoud M., van de Sande J. H., Jorgenson K. F., Eckstein F. Left-handed DNA: from synthetic polymers to chromosomes. J Biomol Struct Dyn. 1983 Oct;1(1):21–57. doi: 10.1080/07391102.1983.10507425. [DOI] [PubMed] [Google Scholar]
- Kopka M. L., Fratini A. V., Drew H. R., Dickerson R. E. Ordered water structure around a B-DNA dodecamer. A quantitative study. J Mol Biol. 1983 Jan 5;163(1):129–146. doi: 10.1016/0022-2836(83)90033-5. [DOI] [PubMed] [Google Scholar]
- Pjura P. E., Grzeskowiak K., Dickerson R. E. Binding of Hoechst 33258 to the minor groove of B-DNA. J Mol Biol. 1987 Sep 20;197(2):257–271. doi: 10.1016/0022-2836(87)90123-9. [DOI] [PubMed] [Google Scholar]
- Quintana J. R., Lipanov A. A., Dickerson R. E. Low-temperature crystallographic analyses of the binding of Hoechst 33258 to the double-helical DNA dodecamer C-G-C-G-A-A-T-T-C-G-C-G. Biochemistry. 1991 Oct 22;30(42):10294–10306. doi: 10.1021/bi00106a030. [DOI] [PubMed] [Google Scholar]
- Teng M. K., Usman N., Frederick C. A., Wang A. H. The molecular structure of the complex of Hoechst 33258 and the DNA dodecamer d(CGCGAATTCGCG). Nucleic Acids Res. 1988 Mar 25;16(6):2671–2690. doi: 10.1093/nar/16.6.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. H., Gessner R. V., van der Marel G. A., van Boom J. H., Rich A. Crystal structure of Z-DNA without an alternating purine-pyrimidine sequence. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3611–3615. doi: 10.1073/pnas.82.11.3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. H., Hakoshima T., van der Marel G., van Boom J. H., Rich A. AT base pairs are less stable than GC base pairs in Z-DNA: the crystal structure of d(m5CGTAm5CG). Cell. 1984 May;37(1):321–331. doi: 10.1016/0092-8674(84)90328-3. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Wing R., Drew H., Takano T., Broka C., Tanaka S., Itakura K., Dickerson R. E. Crystal structure analysis of a complete turn of B-DNA. Nature. 1980 Oct 23;287(5784):755–758. doi: 10.1038/287755a0. [DOI] [PubMed] [Google Scholar]



