Abstract
Objective
Nonsense mutations account for 5–70% of all genetic disorders. In the United States, nonsense mutations in the CLN1/PPT1 gene underlie >40% of the patients with infantile neuronal ceroid lipofuscinosis (INCL), a devastating neurodegenerative lysosomal storage disease. We sought to generate a reliable mouse model of INCL carrying the most common Ppt1 nonsense mutation (c.451C>T) found in the United States patient population to provide a platform for evaluating nonsense suppressors in vivo.
Methods
We knocked-in c.451C>T nonsense mutation in the Ppt1 gene in C57 embryonic stem (ES) cells using a targeting vector in which LoxP flanked the Neo cassette, which was removed from targeted ES cells by electroporating Cre. Two independently targeted ES clones were injected into blastocysts to generate syngenic C57 knock-in mice, obviating the necessity for extensive backcrossing.
Results
Generation of Ppt1-KI mice was confirmed by DNA sequencing, which showed the presence of c.451C>T mutation in the Ppt1 gene. These mice are viable and fertile, although they developed spasticity (a “clasping” phenotype) at a median age of 6 months. Autofluorescent storage materials accumulated throughout the brain regions and in visceral organs. Electron microscopic analysis of the brain and the spleen showed granular osmiophilic deposits. Increased neuronal apoptosis was particularly evident in cerebral cortex and abnormal histopathological and electroretinographic (ERG) analyses attested striking retinal degeneration. Progressive deterioration of motor coordination and behavioral parameters continued until eventual death.
Interpretation
Our findings show that Ppt1-KI mice reliably recapitulate INCL phenotype providing a platform for testing the efficacy of existing and novel nonsense suppressors in vivo.
Introduction
Neuronal ceroid lipofuscinoses (NCLs), commonly known as Batten disease,1–3 constitute a group of the most common (1 in 12,500 births)4 neurodegenerative lysosomal storage disorders (LSDs) for which there is no effective treatment. Mutations in more than 14 different genes (called CLNs)5,6 underlie various types of NCLs. Despite the age variability in disease onset, the clinical features common to all NCLs include psychomotor retardation, seizures, visual loss, and premature death.1–7 Pathological findings include rapidly progressive brain atrophy resulting from destruction of mostly cerebrocortical neurons and the presence of auto fluorescent storage material both in neurons and in other cell types.1–7
Among all NCLs, the infantile NCL (INCL), caused by mutations in the CLN1 gene,8 encoding palmitoyl-protein thioesterase-1 (PPT1), is one of the most devastating diseases. The isolation of PPT1 from bovine brain, its purification to homogeneity, and the characterization of its thioesterase activity9 significantly advanced the field of INCL research. Subsequent molecular cloning of bovine and rat Ppt1-cDNAs and expression10 allowed further characterization, including the demonstration that PPT1 is a lysosomal enzyme.11,12 Moreover, previous linkage analyses localized an INCL candidate locus on human chromosome 1p32, a region in which the CLN1/PPT1 gene was colocalized.13 The identification of inactivating mutations in the CLN1/PPT1 gene in patients with INCL clearly established the molecular basis underlying this devastating lysosomal storage disease (LSD).8 However, the precise molecular mechanism(s) of INCL pathogenesis remains largely unclear and there is no effective treatment for this predominantly childhood neurodegenerative disease.
Palmitoylation (also called S-acylation) is a posttranslational modification in which a long-chain, saturated fatty acid (predominantly palmitate) is attached to cysteine residues in polypeptides via thioester linkage.14 While palmitoylation plays an important role in protein function, depalmitoylation of these proteins is equally critical for recycling or degradation by lysosomal hydrolases. Thus, dynamic palmitoylation (palmitoylation–depalmitoylation), like phosphorylation–dephosphorylation, has emerged as a regulatory mechanism for the function of many important proteins.15 Furthermore, fatty-acylated proteins are refractory to degradation by lysosomal hydrolases.16 The inactivating mutations in the Cln1/Ppt1gene impair the degradation of these lipid-modified proteins (constituents of ceroid) causing their accumulation in lysosomes, which leads to INCL. In time, increased accumulation of ceroids in lysosomes of Ppt1-deficient cells organize to form the so called granular osmiophilic deposits (GRODs).2
Although infantile NCL (INCL) is rare (1 in >100,000 births),2,3,17 it is one of the most rapidly progressive and uniformly fatal neurodegenerative LSDs. The children afflicted with INCL are phenotypically normal at birth but by 11–18 months of age they manifest psychomotor retardation. By 2 years of age, these children undergo complete retinal degeneration and blindness. Around 4 years of age, an isoelectric electroencephalogram (EEG) attests to the lack of brain function and these patients remain in a vegetative state for several more years before eventual death.2,3,17,18 These grim facts underscore an urgent need for the development of effective therapeutic strategies for this devastating disease. While various strategies are being used to develop effective therapies, currently, there is no effective treatment for INCL or any of the other NCL types.19
Nonsense mutations in a gene cause premature translational termination in 5–70% of most inherited disorders. These mutations give rise to in-frame UAA, UAG, or UGA codons in the messenger RNA coding region leading to premature translational termination and truncated protein products. Nonsense mutations also promote destabilization of mRNAs by nonsense-mediated mRNA decay (NMD). Studies on genotype-phenotype correlation studies of patients with INCL have indicated that worldwide, 25% of these patients harbor nonsense mutations in the CLN1/PPT1 gene whereas in the United States, 40% of the INCL patient population carries Ppt1-nonsense mutations.20 Thus, drugs capable of suppressing nonsense mutations in a gene that prevents premature termination by selectively inducing ribosomal read through without affecting the normal termination codons are being sought. Indeed, several compounds that suppress disease causing nonsense mutations, including PTC124 (Ataluren),21 are now being tested.22 The effects of compounds with nonsense suppressor activity can be tested in vitro using cultured cells from patients with genetic diseases caused by nonsense mutations. However, the results obtained from such in vitro studies may not be directly extrapolated in vivo. Thus, a reliable animal model of a disease carrying nonsense mutations may provide a platform for in vivo validation of the results derived from in vitro experiments.
In this study, we engineered a mouse model in which the nonsense mutation, c.451C>T (p.R151X), in the Cln1/Ppt1 gene, most frequently found in the INCL patients in the United States, was knocked-in using C57 embryonic stem (ES) cells. We used ES cells derived from C57 mice in order to generate syngenic C57 mice carrying the c.451C>T nonsense mutation. This strategy obviated the necessity of extensive backcrossing, which would have been required for converting the genetic background of mice generated by targeting the ES cells from 129 mouse strain. Our results showed that homozygous mutant (c.451C>T/c.451C>T) mice (hereafter called Ppt1-KI mice) are viable and fertile. Importantly, these mice manifest virtually all clinical and pathological features of INCL, providing a platform for the in vivo evaluation of nonsense suppressors alone or in combination with other agents for their potential therapeutic efficacy.
Materials and Methods
Animals
Mice lacking Ppt1 (Ppt1−/− mice),23 which recapitulate INCL phenotype,24 were a generous gift from Dr. Sandra L. Hofmann, University of Texas Southwestern Medical Center, Dallas, TX. These mice were used for comparison of the phenotype in our Ppt1-KI mice. All animal experiments were conducted after approval of an animal protocol (ASP#10-012) by the Animal Care and Use Committee of the Eunice Kennedy-Shriver National Institute of Child Health and Human Development, National Institutes of Health.
Generation of Ppt1 c.451C>T (p.R151X) targeting vector
To construct the targeting vector, a 5′ homology arm (∼1.9 kb) and a 3′ homology arm (∼5.2 kb) of the mouse Ppt1 gene (GenBank accession# NP_032943.2) were generated by PCR using mouse genomic DNA as the template and high fidelity Taq DNA polymerase. The primers used to generate the 5′ arm fragment were as follows: Forward: 5′- acg ttc gc GAC TGC TCT TCT GAA GGT CCT GAG TTC A-3′, Reverse: 5′- acg tgg cgc gcc ata TGA GAG AGC CAG GTC CTA TGG AGC T-3′. The primers used to generate the 3′ arm fragment were as follows: Forward: 5′-acg tcc gc GGG ACA CAG ACA AGC ATG TAG TCA AAA C-3′, Reverse: 5′- acg tgc ggc cg CAG AAT GTC CCA AAC CAT GCT C-3′. The amplified PCR fragments were subcloned into a cloning vector (FV vector, Taconic, Cranbury, NJ). The c.451C>T (R151X) mutation was introduced into the 5′ arm of exon 5 of the mouse Ppt1 gene using a site-directed mutagenesis kit (Stratagene, La Jolla, CA). The primers used for site-directed mutagenesis were as follows: Forward: 5′-GGT GTC TTT GGA CTC CCC TGA TGC CCA GGA GAG AGT TCT-3′, Reverse: 5′-AGA ACT CTC TCC TGG GCA TCA GGG GAG TCC AAA GAC ACC-3′. The recombinant plasmids were confirmed by restriction digestion and DNA sequencing. In addition to the homology arms, the targeting vector also contained LoxP flanking the neomycin (Neo) resistance cassette and a DTA expression cassette. The complete nucleotide sequence of the final targeting vector was confirmed by DNA sequencing.
The targeting vector was linearized by NotI and electroporated into C57BL/6J ES cells (Taconic, Cranbury, NJ). The transfected ES cells resistant to G418 were screened for homologous recombination by PCR and the correctly targeted ES cells were identified by Southern blot analysis. Two independent clones of recombinant ES cells were electroporated with a Cre-expression plasmid. The resulting deletion of the Neo cassette in recombinant, Cre-transfected clones was confirmed by PCR analysis. The correctly targeted ES clones, without the Neo cassette, were injected into BALB/C blastocysts and implanted into pseudopregnant mice. The male chimeras were mated with C57BL/6J females to generate offspring. The male and female offspring with WT/c.451C>T mutation were identified by PCR. Germline transmission of the mutation was confirmed by PCR-based genotyping and further verified by direct DNA sequencing. The breeding of the chimeric founders to generate WT/c.451C>T mice was performed by Taconic (Cranbury, NJ). Further breeding of the male and female progeny carrying the WT/c.451C>T mutation was carried out yielding WT, WT/c.451C>T, and c.451C>T/c.451C>T mice.
RNA isolation
Total RNA from cerebral cortices of WT, heterozygous, and Ppt1-KI mice were isolated using TRIzol reagent (Life Technologies, Carlsbad, CA, Lot# 543602). The RNA samples were further purified using a QIAGEN RNeasy Mini Kit and treated with DNAse I (30 U/μg total RNA) (QIAGEN, Gaithersburg, MD) to eliminate any traces of DNA. The purity and amount of the RNA present in 1 μL of the sample was assessed by using a nanodrop spectrometer (NanoDrop Technologies Thermoscientific Wilmington, DE). The ratio of absorbance at 260 and 280 nm was measured. A ratio of ∼2.0 or greater attested the purity of the RNA samples.
Quantitative analysis of RNA
The RNA samples were reverse transcribed and converted to cDNA using the SuperScript III First-Strand Synthesis Kit (Invitrogen, Carlsbad, CA). Expression of mRNA was quantitated using a SYBR Green PCR Master Mix and cDNA equivalent to 8–10 ng of total RNA. A C-1000 Touch Thermo Cycler CFX96 Real-Time System (Bio-Rad Laboratories, Inc., Hercules, CA) was used to amplify the cDNA and the results were analyzed using CFX Manager Software Version 3.0 (Bio-Rad Laboratories, Inc., Hercules, CA). The primers used were as follows: Ppt1 (Forward: 5′-AGT GGC TCA GAG ATG CCC AAC-3′, Reverse: 5′-CCT GAT GAA GTC GCA GAT GTG AG -3′); GFAP (Forward: 5′-CAC GAA CGA GTC CCT AGA GC-3′, Reverse: 5′- ATG GTG ATG CGG TTT TCT TC-3′); Iba-1 (Forward: 5′-TCT GCC GTC CAA ACT TGA AGC C-3′, Reverse: 5′-CTC TCC AGC TCT AGG TGG GTC T-3′). The relative amounts of mRNA were calculated from the cycle threshold (Ct) values using GAPDH for normalization. The mRNA levels in brain tissues from the heterozygous, Ppt1-KI, and Ppt1−/− mice are presented as fold change compared with those of their WT littermates. At least three independent experiments were performed for each genotype and the results are expressed as the mean (n = 3) ±SD.
Western blot analyses
The cortical tissues from the brains of WT, heterozygous, Ppt1-KI and the Ppt1−/− mice were homogenized in 1 mL of RIPA Buffer (Thermo Scientific, Rockford, IL, Lot# OC183166) containing 10 μL of protease-inhibitor cocktail (Thermo Scientific, Rockford, IL, Cat # 1861278). Total proteins (80 μg) from each sample were resolved by electrophoresis using 4–12% NuPage Bis-tris gels (Invitrogen, Carlsbad, CA, Cat # NP0335BOX) under denaturing and reducing conditions. Proteins were then electrotransferred to PVDF Polyvinylidene difluoride membranes (Novex Life Technologies, Carlsbad, CA, Lot: PRO8083-01). The membranes were blocked with 5% nonfat dry milk (Bio-Rad, Hercules, CA, Cat# 170-6404) in 1x TBST and then incubated with the primary antibodies overnight at 4°C with gentle shaking. Primary antibodies used were against Ppt1, glial fibrillary acidic protein (GFAP), Iba-1 and cleaved Caspase-3. Western blot analysis was performed using an ImageQuant LAS 4000 mini machine (General Electric, Piscataway, NJ). Chemiluminescence was detected using SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific, Rockford, IL, Lot # MK166442) according to the manufacturer's specifications. Rat anti-Ppt1 (1:1000) was a generous gift from Dr. S.L. Hofmann (University of Texas, SW Medical Center, Dallas, TX).
Ppt1 enzyme activity assay
Cerebrocortical tissues were homogenized in 1 mL of cold RIPA buffer (Thermo Scientific, Rockford, IL, Lot # OC183166) and centrifuged at 10,000g for 10 min at 4°C to remove the nuclei. The protein quantification was performed using a 96-well plate and a Micro BCA Protein Assay Kit (Thermo Scientific, Rockford, IL, Lot # NF170026). The experiment was performed using a protocol provided by the manufacturer of Ppt1-substrate (Moscerdam Substrates, 2341 KS Oegstgeest, the Netherlands). Briefly, each well in a 96-well plate contained a combination of either 10 μL of diluted cortical tissue homogenate or 10 μL of 0.2% BSA (control) mixed with 20 μL of substrate (3.4 mmol/L MU-6s-paml-βGlc in a 2:1 Chloroform: Methanol mixture). The reaction mixture was incubated at 37°C for an hour and the reaction was terminated by adding 200 μL of the stop buffer (0.5 mol/L NaHCO3/0.5 mol/L Na2CO3 buffer + 0.025% Triton X-100, pH 10). Ppt1 enzyme activity was measured using a FlexStation II spectrofluorometer (Molecular Devices, Sunnyvale, CA) at absorbance wavelengths of 355 nm, λex and 460 nm, λem. Activities were calculated according to the instructions provided by the substrate manufacturer (Moscerdam Substrates) and the results were expressed as nmol/h per mg protein.
Autofluorescence
The method for fluorescence microscopy has been described previously.25 Briefly, the tissues were fixed in 4.0% paraformaldehyde, embedded in paraffin, and processed for histological sectioning (American Histolab, Germantown, MD). The slides with the tissue sections were dipped in xylene for 5 min to remove paraffin and then successively passed through decreasing concentrations of ethanol (100–0%) in water. The sections were then mounted with Vectashield Mounting Medium (Vector Laboratories, Burlingame, CA, Lot #R0712) and covered with cover slips. Autofluorescence was visualized using the dark field in an Axioskop2 plus fluorescence microscope (Carl Zeiss Thornwood, NY, USA) and the images were captured with a Nikon DS-Fi1 digital camera (Nikon Melville, NY, USA).
Histological and immunohistological analyses
Whole brain sections were fixed in 4.0% paraformaldehyde at room temperature after perfusion with cold 1x PBS. The samples were sectioned and the staining process began by first incubating in Citrate Buffer (pH 6) overnight at 95°C for antigen retrieval. After washing five times in 1x PBS, a 0.3% H2O2 in 70% methanol mixture was added to the slides followed by 30 min of incubation in the dark at room temperature. The slides were washed in PBS and permeabilized by incubating in 0.4% Triton X-100 in PBS for 1 h at room temperature. After washing in PBS, the non-specific binding of primary antibodies to tissues were blocked using an Avidin/Biotin Blocking Kit (Vector Laboratories, Burlingame, CA, Cat #SP-20001). Immediately after blocking, the tissue sections were layered over with anti-GFAP (Cell Signaling, Cat # H36705, 1:500 Danvers, MA, USA) or anti-IbA-1 (WAKO, Lot # 5TJ5648, 1:200 Irvine, CA, USA) and incubated overnight in a humidified chamber at 4°C. Sections were then washed three times with 1x PBS, and incubated with an anti-mouse or an anti-rabbit biotinylated secondary antibody (Vector Laboratories, Burlingame, CA, 1:1000 dilution, Lot #X1103, Y0515), respectively. A VECTASTAIN Elite ABC kit (Vector Laboratory, Burlingame, CA, PK-6100) was used to facilitate in the detection of biotinylated molecules prior to the color staining. Color staining was achieved using a protocol provided in the 3,3′-diaminobenzidine (DAB) peroxidase substrate kit (Vector Laboratories, Inc., Burlingame, CA, SK-4100).
TUNEL assay
To determine the level of apoptosis in the brain, we performed TUNEL assay using the TACS 2 TdT-DAB in situ Apoptosis Detection Kit (Trevigen, Gaithersburg, MD, Cat #4810-30-K) as per manufacturer's instructions. Briefly, the paraffin embedded sections of brain tissues from the WT mice and their Ppt1-KI littermates as well as those from Ppt1−/− mice were placed on glass slides. The slides were then dipped in xylene for 5 min to remove the paraffin. They were then passed through serially decreasing concentrations of ethanol (100–30%) followed by washing three times in 1x PBS (10 min each). The slides were then placed in the proteinase-K solution from the TUNEL assay kit for 30 min at room temperature, immersed in Quenching Solution (5 min), and finally, in 1x TdT labeling buffer for 5 min at room temperature. The labeling reaction mixture was added and incubated in a humidified chamber at 37°C for 1 h. To stop the reaction, slides were immersed in stop buffer for 5 min at room temperature and washed twice with deionized water (5 min each) before covering with Strep-HRP solution for 10 min at 37°C. After washing twice in 1x PBS (2 min/wash) to remove the Strep-HRP, the samples were immersed in DAB solution for 6 min at room temperature. The tissue sections were then counterstained with Methyl Green for 2 min at room temperature. After applying coverslips, images were acquired using a Nikon DS-Fi1 digital camera (Nikon).
Transmission electron microscopy
Transmission electron microsocopic analyses of the cortical tissues from WT, heterozygous, Ppt1-KI, and those of the Ppt1−/− mice were performed as previously described.25 Briefly, thin sections of perfused brains were fixed in 2.5% glutaradlehyde in 0.1 mol/L sodium cacodylate buffer, pH 7.4, followed by three washings in 0.1 mol/L sodium cacodylate buffer at room temperature. Ultrathin sections of the brain tissues were stained with lead citrate and uranyl acetate and examined with an LEO 912 electron microscope (JFE Enterprises, University of Maryland, College Park, MD).
Histological analysis of the retina
Eyes from the WT, Ppt1-KI, and Ppt1−/− mice were extracted and fixed in 4% paraformaldehyde at room temperature after perfusion. The samples were sectioned and stained with hematoxylin and eosin. The stained sections were examined using an Axioskop2 plus fluorescence microscope (Carl Zeiss). Images were captured using a Nikon DS-Fi1 digital camera (Nikon).
ERG analysis of retinal function
For electroretinogram (ERG) recordings were performed as previously reported.26 Briefly, the mice were dark-adapted overnight and anesthetized with intraperitoneal injection of a mixture containing ketamine (100 mg/kg) and xylazine (6 mg/kg). Pupils were dilated with 2.5% phenylephrine and 1% tropicamide, and the cornea was anesthetized with 1% proparacaine. Animals were placed on a heating pad to maintain body temperature. All procedures were performed under dim red light. ERGs were recorded from both eyes using gold wire loops with a commercial Espion E2 system (Diagnosys LLC, Lowell, MA). A gold wire loop placed in the mouth was used as a reference electrode. LEDs build in colordome provide ganzfeld light stimulus. The intensity of light stimulus was calibrated with IL 1700 Research Radiometer (International Light Technologies, Inc., Peabody, MA). Responses were amplified with a bandwidth of 0.3–300 Hz and sampled at 1 kHz. Light-adapted ERG responses were recorded in the presence of a rod-saturating background of 20 sc cd/m2. The a-wave amplitude was measured from the baseline to the negative peak and the b-wave was measured from the a-wave trough to the maximum positive peak. ERG recordings were performed on eight mice per group and data were presented as mean ± SEM.
Rotarod test for evaluating motor coordination
Motor coordination of the Ppt1-KI mice and their WT littermates was assessed using Rotarod (UGO Basile, Varese, Italy) performance test27 at a fixed speed of 12 rpm while rotating in a single direction. Animals were trained twice daily at 1 min intervals for three consecutive days. They were allowed to rest for 1 min between the two trials. Rotarod experiments were performed for 180 sec on day 4 and the amount of time a mouse was on the rotarod before falling from the rotating rod was recorded. This test was performed by an independent investigator who had no knowledge of the genotype of the animals.
Kaplan–Meier plot for longevity
The Kaplan–Meier Longevity Plot28 was used to estimate the longevity of the Ppt1-knock-in mice (n = 62). When the mice could no longer reach for food and water, this was considered the endpoint and the mice were killed. For each mouse, the age (in days) at which they were killed was recorded and then plotted as the probability on a line graph. For the Ppt1-KI mice the probability of survival steadily declined as the number of mice in the study decreased.
Statistical analysis
Statistical analysis was performed with Microsoft Excel 2003–3007. Information on specific statistical test and mouse numbers is provided in the figure legends. Statistical significance was tested using Student's t-test and the results were considered statistically significant when P < 0.05.
Results
Generation of knock-in mice carrying c.451C>T nonsense mutation in the Cln1/Ppt1 gene
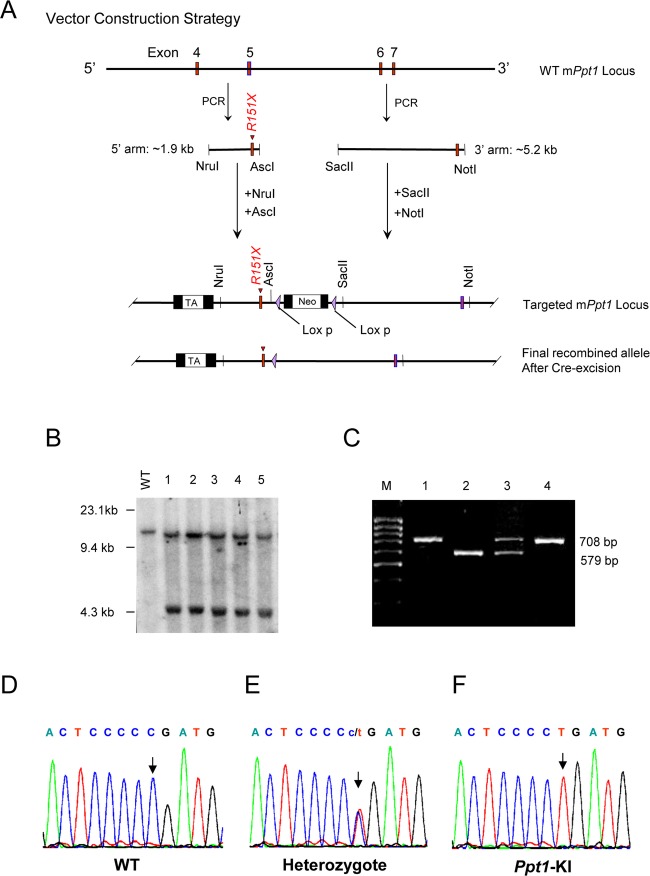
To generate the Ppt1 knock-in mice (hereafter called Ppt1-KI mice), we first engineered a targeting construct in which c.451C>T nonsense mutation was incorporated into the 5′ arm of exon 5 of mouse Ppt1 gene by site-directed mutagenesis. Our strategy for the construction of the targeting verctor used the Cre-Lox recombination system in which the Neo cassette was flanked by two 34-bp LoxP sequence (Fig.1A). The removal of the neo casette from the targeted ES cells was achieved by electroporating Cre. The knock-in of the c.451C>T Ppt1 nonsense mutation in the targeted vector was confirmed by DNA sequencing (Fig. S1A). The final targeting vector was electroporated into C57BL/6 ES cells. The correctly targeted ES cells were identified by Southern blot analysis (Fig.1B). The chimeric founder mice were mated with C57BL/6 mice to generate the F1 offspring. Male and female F1 heterozygous Ppt1-KI littermates were mated to generate Ppt1-KI mice. Germline transmission of the Ppt1 nonsense mutation was confirmed by PCR genotyping (Fig. S1B and Fig.1C). Sequencing of the genomic DNA confirmed the identity of the WT (Fig.1D), heterozygous (Fig.1E), and Ppt1-KI (Fig.1F) littermates.
Figure 1.
Targeting vector used for targeting C57 ES cells and generation of Ppt1-KI mice. (A) Structure of the targeting vector, which was generated with a C>T mutation in exon 5 of mouse Ppt1 gene. Cre was excised from the final recombined allele of the correctly targeted recombinant clones. (B) Southern blot analysis of WT and recombinant clones. Lane 1, WT; Lanes 2–6 recombinant clones. (C) The genotyping results of representative F2 mice: the 708 bp band corresponds to the constitutive knock-in allele and the 579 bp band corresponds to the endogenous wild type allele. Lanes 1 and 4: Ppt1-KI; Lane 2: WT; Lane 3: heterozygous. Electropherograms of genomic DNAs from F2 mice. (D) WT; (E) heterozygous and (F) Ppt1-KI. The arrows indicate the mutation sites (C>T).
Biochemical and molecular characterizations
The brain tissues of Ppt1-KI mice lack Ppt1-mRNA, Ppt1-protein, and Ppt1-enzyme activity
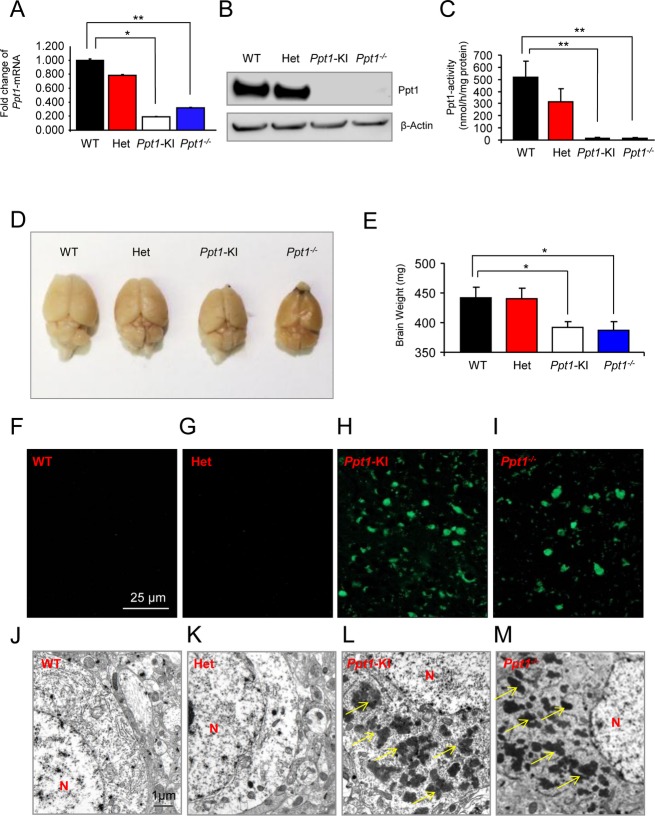
To determine whether the homozygous mice carrying c.451C>T/c.451C>T mutations (hereafter called Ppt1-KI mice) lack PPT1 enzyme activity, we first measured the levels of Ppt1-mRNA and Ppt1-protein in the brain by real-time RT-PCR using total RNA and Western blot analyses, respectively. The results showed that compared with their WT littermates the cortical tissues of Ppt1-KI mice and those of their Ppt1−/− counterparts showed significantly less (P < 0.02) Ppt1-mRNA (Fig.2A). While there was no apparent difference in the Ppt1-protein levels in the brain of heterozygous mice and those of their WT littermates, there was no detectable Ppt1-protein in the brain of Ppt1-KI mice (Fig.2B). As expected, the brain tissues of Ppt1−/− mice, which were used for comparison, had no detectable Ppt1-protein. To determine whether the Ppt1-proteins translated to Ppt1 enzymatic activity, we performed Ppt1-enzyme assay. The results showed that compared with the WT mice the brain tissues of the Ppt1-KI littermates showed no detectable Ppt1 activity. The Ppt1-activity in the heterozygous mice was readily detectable, albeit lower than that in WT mice (Fig.2C). As expected, the brain tissues of the Ppt1−/− mice, used as a comparison, showed no Ppt1-enzyme activity. Taken together, these results suggested that the Ppt1-KI mice carrying c.451C>T/c.451C>T mutations are deficient in Ppt1-protein as well as Ppt1 enzyme activity.
Figure 2.
Macroscopic, biochemical, and pathological characterization of Ppt1-KI mice. The Ppt1-mRNA levels (A) in the brain tissues of heterozygous and Ppt1-KI mice as well as those of their WT littermates. The Ppt1-mRNA levels in Ppt1−/− mice were used for comparison. Western blot analysis (B) of brain tissues from heterozygous and Ppt1-KI mice and those of their WT littermates. Brain tissues from Ppt1−/− mice were used as control. β-actin was used as a loading control. The Ppt1 enzyme activities (C) in the brain tissue lysates from heterozygous and Ppt1-KI mice and those of their WT littermates. Brain tissue lysates from Ppt1−/− mice served as a control. Morphology of the brain (D) from representative heterozygous and Ppt1-KI mice and that of their WT littermate. The brain from a Ppt1−/− mouse was used for comparison. Note that the reduced brain size in a representative Ppt1-KI mouse and that of a Ppt1−/− counterpart is visually appreciable compared with that of the heterozygous and WT mouse. Brain weights (E) for heterozygous and Ppt1-KI mice and those of their WT littermates. Brain weights of Ppt1−/− mice are used for comparison. The results are presented as the mean (n = 12) ±SD. Autofluorescent storage material in brain tissues from representative WT (F), heterozygous (G), and Ppt1-KI (H) mice compared with that in the Ppt1−/− mouse (I). Note that while intense autofluorescence was readily detectable in the brains of Ppt1-KI and Ppt1−/− mice, those of the heterozygous mice and that in their WT littermates showed no detectable autofluorescence. Transmission electron microscopic analyses of granular osmiophilic deposits (GRODs) in the brain tissues from WT (J), heterozygous (K), and Ppt1-KI (L) mice compared with those in Ppt1−/− mouse brain (M). Note the heavy accumulation of GRODs (arrows) in the brains of Ppt1-KI and Ppt1−/− mice while the GRODs were not detectable in the brains of heterozygous mice and in those of their WT littermate. N, nucleus. *P < 0.05, **P < 0.01, P < 0.001.
Brain morphology, weight, and accumulation of autofluorescent material in Ppt1-KI mice
Because of rapid neurodegeneration and accumulation of intracellular autofluorescent material are characteristic features of INCL pathology, we evaluated the morphology, weight, and the accumulation of autofluorescent material in the brain and in the spleen of WT, heterozygous, and Ppt1-KI mice. In addition, we compared these parameters with those of the Ppt1−/− mice. Our results showed that while the brain morphology (Fig.2D) as well as brain weights (Fig.2E) of heterozygous mice were comparable to those of the WT mice, the Ppt1-KI mice had a significantly smaller brain size and appreciably reduced brain weights (Fig.2E). Notably, these parameters in Ppt1-KI mice were comparable to those in Ppt1−/− mice (Fig.2D and E). We also determined the levels of autofluorescence in the brains of WT, heterozygous, and Ppt1-KI mice as well as those of Ppt1−/− counterparts. The brains of Ppt1−/− mice were used for comparison. The results showed that while autofluorescence in the brains from WT mice (Fig.2F) and those of the heterozygous littermates (Fig.2G) was virtually undetectable, it was readily detectable in the brain of Ppt1-KI littermates (Fig.2H). As expected, the brains of Ppt1−/− mice also had high levels of autofluorescence (Fig.2I). We then evaluated the brains of WT, heterozygous, and Ppt1-KI littermates for GRODs by transmission electron microscopy. For comparison, we analyzed the brains of Ppt1−/− mice. The results showed that while the WT mouse brain (Fig.2J) and that of their heterozygous littermate (Fig.2K) contained no detectable GRODs, those from both the Ppt1-KI (Fig.2L) and the Ppt1−/− mice (Fig.2M) contained numerous GRODs. These results showed that the Ppt1-KI mice, like their Ppt1−/− counterparts, recapitulate several characteristic pathological features of INCL.
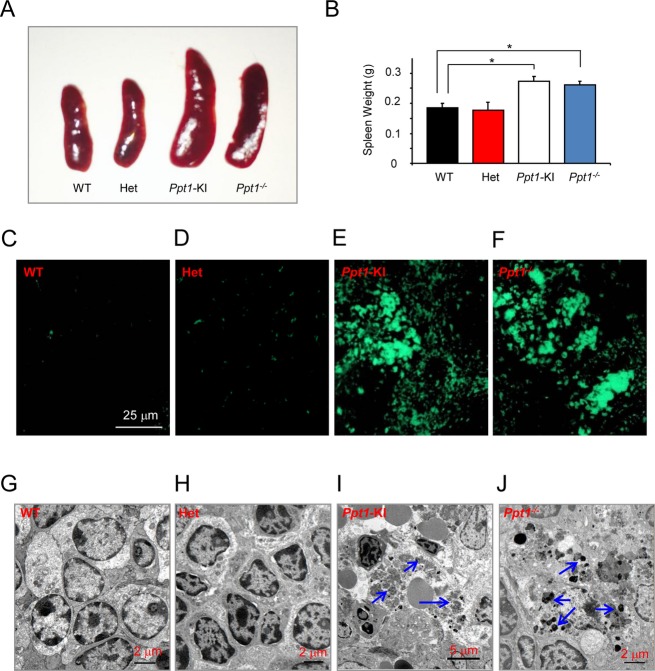
The Ppt1-KI mice show enlargement of spleen with autofluorescence and GRODs
The deficiency of Ppt1 causes lipopigment deposition in visceral organs, but it appears to have limited functional impairment compared to the devastating dysfunction it causes in the CNS Central Nervous System. The deficiency of Ppt1 has deleterious effects on the central nervous system but it adversely affects the visceral organs. Accordingly, we investigated the spleens from WT, heterozygous, and Ppt1-KI mice to determine the morphology, weight, the presence or absence of autofluorescence, and GRODs. For comparison, we also examined the spleens of Ppt1−/− mice. The results showed that the size of the spleens from Ppt1-KI and Ppt1−/− mice were substantially larger compared with that of the WT and heterozygous mice (Fig.3A). Consistent with this finding, the weights of the spleens from Ppt1-KI and Ppt1−/− mice were significantly higher (P < 0.05) compared with those of the Ppt1-KI and WT littermates (Fig.3B). We also determined the levels of autofluorescence in the spleens from WT, heterozygous, and Ppt1-KI littermates as well as those of the Ppt1−/− mice. The results showed that while autofluorescence in the spleens of WT (Fig.3C) and heterozygous mice (Fig.3D) was virtually undetectable, whereas those of the Ppt1-KI (Fig.3E) and Ppt1−/− mice (Fig.3F) was highly intense and readily detectable. We also examined the spleen tissues by TEM Transmission electron microscopy for the presence of GRODs. The results showed that while the GRODs were not detectable in the spleen of WT (Fig.3G) and heterozygous mice (Fig.3H), those from the Ppt1-KI (Fig.3I) and Ppt1−/− mice (Fig.3J) contained large macrophage-like phagocytic cells in which there were numerous GRODs. Taken together, these results demonstrated that the spleens of Ppt1-KI mice, like those of the Ppt1−/− counterparts, were morphologically larger, heavier, and contained both autofluorescent material as well as the GRODs. These results also suggested that both the brains and the visceral organs of Ppt1-KI mice, like those of their Ppt1−/− counterparts, recapitulate characteristic pathological features of INCL.
Figure 3.
Macroscopic, histopathological, and ultrastructural analyses of the spleen. (A) Morphology of the spleen from representative heterozygous, Ppt1-KI mice and their WT littermate. The spleen from a Ppt1−/− mouse was used for comparison. Note that the increased spleen sizes in Ppt1-KI and Ppt1−/− mice are visually appreciable compared to those of the heterozygous and WT mice. (B) Spleen weights from heterozygous and Ppt1-KI mice and those of their WT littermates. Spleen weights of Ppt1−/− mice served as controls. The results are presented as the mean (n = 16) ±SD. Autofluorescent storage material in the spleen from WT (C), heterozygous (D), and Ppt1-KI (E) mice compared with those of the Ppt1−/− mouse (F), which served as a control. Note that while intense autofluorescence was readily detectable in the spleens of Ppt1-KI and Ppt1−/− mice, those of the heterozygous and WT littermates of the Ppt1-KI mice showed virtually no autofluorescence. Transmission electron microscopic analyses of GRODs in the spleen of a representative WT (G), heterozygous (H), and Ppt1-KI mice (I) compared with those of a representative Ppt1−/− mouse (J), which served as a control. Note the heavy accumulation of GRODs (arrows) in the spleens of Ppt1-KI and Ppt1−/− mice but the GRODs were not detectable in the spleen of heterozygous mice and their WT littermate. *P < 0.05, P < 0.01, P < 0.001.
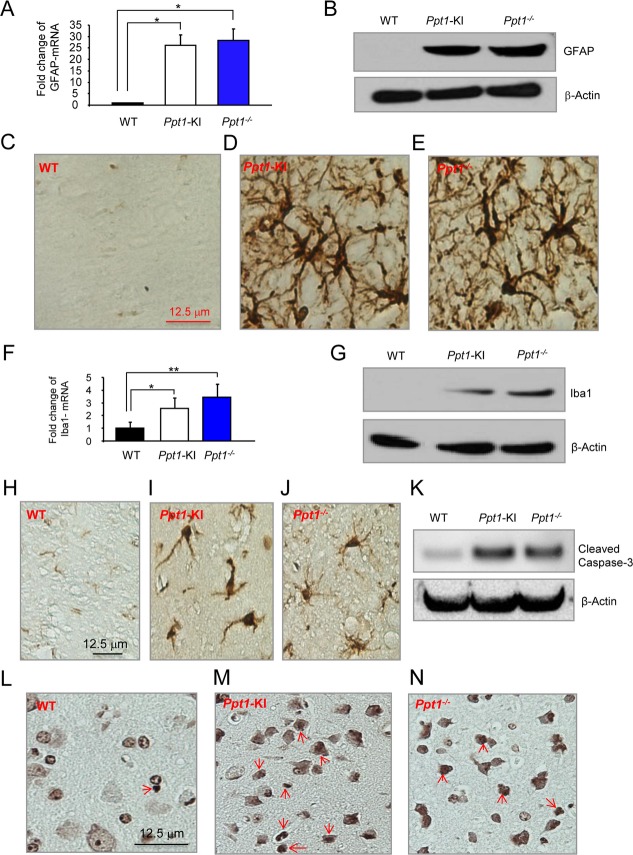
Increased levels of activated astrocytes and microglia in Ppt1-KI mouse brain
It has been reported that in INCL, the activation of astrocytes and infiltration of microglia in the brain occur as the degeneration of the neurons progresses.25 Thus, we performed real-time RT-PCR and Western blot analyses, respectively, to determine the mRNA levels of GFAP, a marker for activated astrocytes, and Iba1, a marker of microglia. The results showed that compared with the GFAP-mRNA levels in the brain tissues of WT mice, those in the Ppt1-KI littermates and Ppt1−/− mice were substantially elevated (Fig.4A). Consistent with these findings, the results of Western blot analyses showed that although GFAP-protein was undetectable in the brain of WT mice (Fig.4B, left lave) those of the Ppt1-KI (Fig.4B, middle lane) and Ppt1−/− mice (Fig.4B, right lane) showed readily detectable GFAP-protein bands. To confirm these results we performed immunohistochemical analysis of the brain tissues from WT and Ppt1-KI mice as well as those of their Ppt1−/− counterparts. The results confirmed that compared with WT mouse brain (Fig.4C), high levels of GFAP immunoreactivity in the brains of Ppt1-KI (Fig.4D) and in the Ppt1−/− mice (Fig.4E) were detectable. Similarly, compared with WT mice, the levels of Iba1-mRNA (Fig.4F) and Iba1-protein (Fig.4G) in the brain tissues of Ppt1-KI mice and Ppt1−/− counterparts were markedly higher. To confirm these results, we also performed immunohistochemical analyses of brain tissue sections using antibodies against microglial marker, Iba1. The results showed that compared with brain tissues of WT mice (Fig.4H), which contained virtually no Iba1-positive cells, those of the Ppt1-KI littermates contained readily identifiable Iba1-positive microglia (Fig.4I). For comparison, we stained the Ppt1−/− mouse brain sections using Iba1-antibody, which were also positive for microglia (Fig.4J). Since increased levels of apoptosis have been suggested to cause neuronal degeneration in INCL patients as well as in Ppt1−/− mice, we sought to determine the levels of cleaved caspase-3 protein, which is a marker for cells undergoing apoptosis. The results of the Western blot analysis showed that compared with the WT mice, the brains of the Ppt1-KI and Ppt1−/− mice contained substantially higher levels of cleaved caspase-3 protein (Fig.4K). We also performed the TUNEL assay to detect the apoptotic cells in the brains of WT (Fig.4L), Ppt1-KI (Fig.4M), and Ppt1−/− mice. The results showed that compared with the brains from WT mice those of the Ppt1-KI and Ppt1−/− (Fig.4N) mice showed numerous TUNEL-positive cells. Because oxidative stress causes neuronal death by apoptosis in INCL and in Ppt1−/− mouse brain, we sought to determine the oxidative stress levels in brain tissues of Ppt1-KI mice we evaluated catalase-mRNA as well as catalase-protein levels by quantative RT-PCR and Western blot analysis, respectively. The results showed that both catalase-mRNA (Fig. S2A) as well as catalase-protein levels (Fig. S2B) were substantially higher in Ppt1-KI mouse brain compared to those of their WT littermates. Moreover, we determined the mRNA and protein levels of the neuron marker, NeuN to determine whether neuron levels have declined in Ppt1-KI mouse brain due to increased apoptosis. The results showed that both NeuN-mRNA (Fig. S2C) and NeuN-protein (Fig. S2D) levels are significantly decreased in Ppt1-KI mouse brain compared with those of their WT littermates. Taken together, these results provided evidence that the Ppt1-KI mice reliably recapitulate the characteristic pathology reported in INCL patients and in Ppt1−/− mice.
Figure 4.
Biochemical and histochemical analysis of GFAP and Iba1 levels. The GFAP-mRNA levels in the cortical brain tissues of Ppt1-KI mice as well as those of their WT littermates. The GFAP-mRNA levels in Ppt1−/− mice were used for comparison. The results are presented as the mean (n = 12) ±SD. (B) Western blot analysis of cortical tissues from for Ppt1-KI mice and their WT littermates. Cortical tissues from Ppt1−/− mice were used as a control. β-actin was used as a loading standard. Immunohistochemical staining of GFAP of the brain tissues from WT mice (C) as well as those of their Ppt1-KI littermates (D). Brain sections from Ppt1−/− mice (E) were used for comparison. The Iba1-mRNA levels (F) in the cortical tissues of Ppt1-KI as well as those of their WT littermates. The Iba1-mRNA levels in Ppt1−/− mice were used as a control. The results are presented as the mean (n = 12) ±SD. (G) Western blot analysis of cortical tissues from for Ppt1-KI mice and those of their WT littermates. Cortical tissues from Ppt1−/− mice were used as a control. β-actin was used as a loading control. Iba-1 immunohistochemical color stain in the paraffin brain sections of WT mice (H) as well as those of their Ppt1-KI littermates (I). Brain sections from Ppt1−/− mice (J) were used for comparison. (K) Western blot analysis of cleaved caspase-3 in cortical brain tissues from for Ppt1-KI mice and those of their WT littermates. Cortical brain tissues from Ppt1−/− mice were used as a control. β-actin was used as a loading control. TUNEL Stain in the paraffin brain sections of WT mice (L) as well as those of their Ppt1-KI littermates (M). Brain sections from Ppt1−/− mice (N) were used for comparison. *P < 0.05, **P < 0.01, P < 0.001.
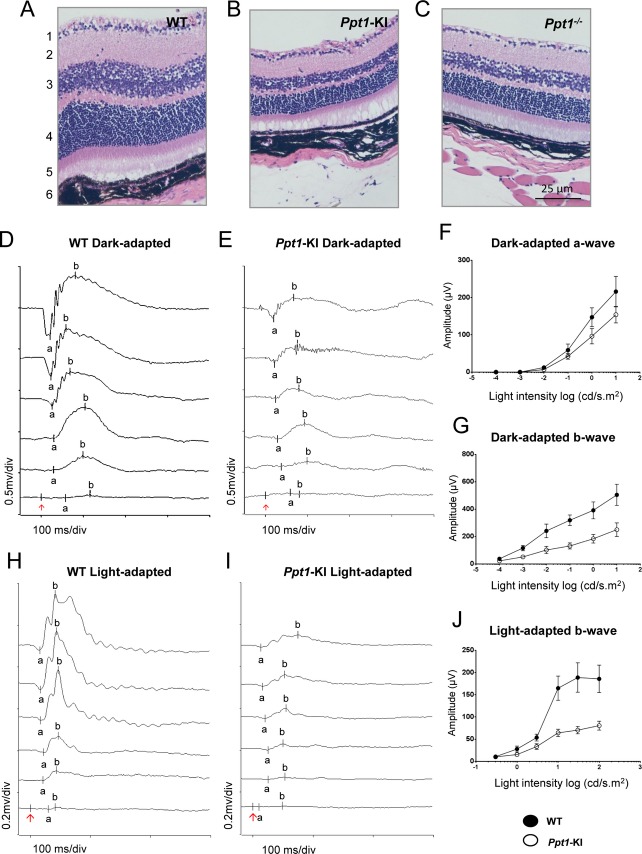
Structural abnormalities in the retina of Ppt1-KI mice
One of the early clinical signs of INCL is retinal degeneration and blindness.1–7,17,18 Thus, we sought to determine whether the Ppt1-KI mice also undergo retinal degeneration and consequent blindness. We first performed histological analyses of the retina from WT mice and from their Ppt1-KI littermates. The results showed that compared with the widths of the retinal layers in WT mice (Fig.5A), those in the Ppt1-KI littermates (Fig.5B) were appreciably thinner. Similar results were found in the retina of the Ppt1−/− mice (Fig.5C), which were used for comparison. More specifically, these changes were clearly appreciable in the inner and outer nuclear layers of the retina. These results provided evidence that like the Ppt1−/− mice, the Ppt1-KI mice also show structural deterioration of the retinal layers at around 6 months of age.
Figure 5.
Histopathological analysis of retinal layers and electroretinography. Representative hematoxylin- and eosin-stained retinas from (A) WT, (B) Ppt1-KI, and (C) Ppt1−/− mice are shown. Note that compared with the WT mice, reduced thickness of the retinal layers three (inner nuclear layer) and four (outer nuclear layer) in Ppt1-KI and Ppt1−/− mice are readily appreciable. In addition, the number of cells in retinal ganglion cell layer is also reduced in Ppt1-KI and Ppt1−/− mice compared with that of the WT control. Representative dark-adapted ERG waveforms from WT (D) and Ppt1-KI mice (E), respectively, in response to a series of light flashes with intensities of 10−4, 10−3, 0.01, 0.1, 1, and 10 cd.s/m2 (from bottom to the top traces). (F) Averaged a-wave amplitudes of dark-adapted ERG from WT and Ppt1-KI mice. (G) Averaged b-wave amplitudes of dark-adapted ERG from WT and Ppt1-KI mice. Representative light-adapted ERG waveforms from WT (H) and KI mice (I), respectively, were in response to flash intensities of 0.3, 1, 3, 10, 30, and 100 cd.s/m2 (from the bottom to the top traces). (J) Averaged b-wave amplitudes of light-adapted ERG from WT (n = 8) and Ppt1-KI (n = 8) mice.
Abnormal ERG findings attest to deterioration of retinal function
As the comparison of the various layers of the retina in Ppt1-KI mice with those of their WT littermates clearly showed signs of degeneration, we sought to determine whether the structural abnormalities correlated with the functional deterioration of the retina. Accordingly, we performed ERG studies using 6-month-old WT mice and their Ppt1-KI littermates. Representative ERG waveforms are shown from a WT mouse (Fig.5D) in response to a series of light flashes of intensity 10−4 cd.s/m2 for bottom trace, and 10-fold brighter for each consecutive trace and until 10 cd.s/m2 (top trace) was reached. Example of ERG responses recorded from an eye of a representative Ppt1-KI littermate is presented (Fig.5E). These responses were recorded using a series of flashes of same intensity as used for the WT mice. The responses from Ppt1-KI mice were much weaker than those from the WT littermates, indicating functional deterioration of the retina in the mutant animals. Amplitudes of a-wave (Fig.5F) and b-wave (Fig.5G) for dark-adapted ERG responses were measured from WT mice (n = 8) and Ppt1-KI littermates (n = 8). Although statistically not significant, a-wave amplitudes recorded from Ppt1-KI mice were consistently lower than those of the WT mice for each of the light intensities tested (Fig.5F). Even larger reduction in b-wave amplitudes were noticed for Ppt1-KI mice when compared with those of the WT littermates (Fig.5G). While the a-waves of dark-adapted ERG responses reflect the activity of rod photoreceptors, the b-waves predominantly represent the activities of the second-order neurons (bipolar cells) in the retina. These results are consistent with the observation of nonspecific deterioration of all neuron types in the retina of Ppt1-KI mice.
Light-adapted ERG responses elicited by a series of flashes with intensity of 0.3–100 cd.s/m2 were recorded from a representative WT mouse (Fig.5H) and a Ppt1-KI mouse (Fig.5I). These results provided additional evidence for functional deterioration in mutant animals. Averaged b-wave amplitudes (Fig.5J) indicated substantial reductions in ERG responses recorded from Ppt1-KI mice. Because light-adapted ERG responses are mediated by cone photoreceptor pathways in the retina, these data provide further evidence for general degeneration of neurons in the Ppt1-KI animals. Taken together, these results demonstrated that in Ppt1-KI mice the structural deterioration of the retina correlated well with the functional deterioration revealed by the ERG results.
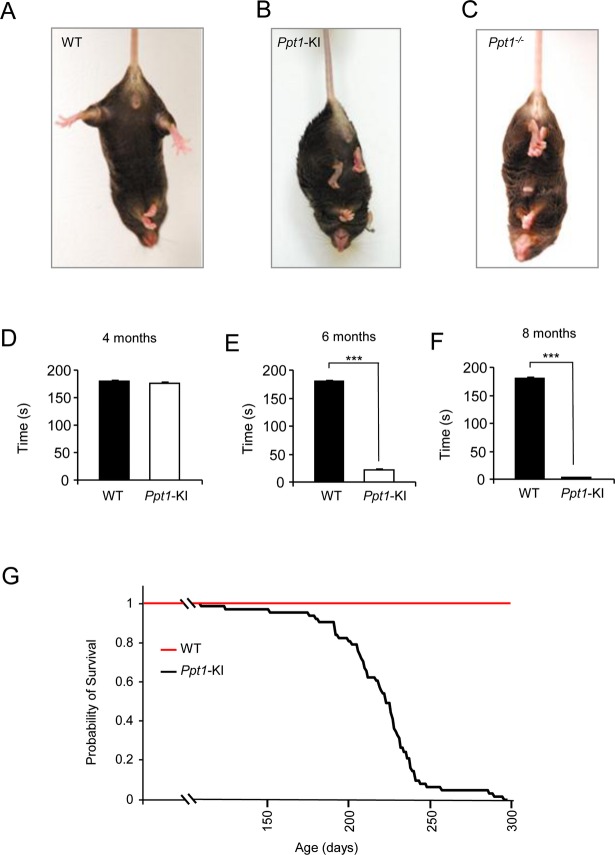
Deterioration of motor coordination in Ppt1-KI mice
To determine the onset of neurological deterioration, we tested the Ppt1-KI mice for the manifestation of clasping behavior. The results showed that at 6 month of age the WT mice showed no clasping (Fig.6A), whereas both the Ppt1-KI (Fig.6B) as well as Ppt1−/− mice (Fig.6C) readily manifested this behavior. In contrast, the heterozygous Ppt1-KI mice, like their WT littermates, did not manifest clasping behavior (data not shown). The loss of motor coordination is one of the cardinal signs of neurodegeration. Accordingly, the rotarod test27 was performed to assess the motor coordination of the WT and Ppt1-KI mice. The results of the rotarod test in Ppt1−/− mice, used for comparison, were virtually identical. The WT mice and their Ppt1-KI littermates were first evaluated when they were 4 months old, when they manifest no clasping behavior and at 6 months of age when they readily manifest clasping. The results of endurance test for motor coordination of 4-month-old mice showed that at a rotarod speed of 12 rpm there were virtually no difference between the WT mice and that of their Ppt1-KI littermates (Fig.6D). However, when the same test was performed using 6-month-old WT and Ppt1-KI littermates, striking deterioration in the rotarod performance of the Ppt1-KI mice was clearly evident (Fig.6E). Finally, we performed the rotarod test using the same mice when they were 8 months old. The results showed that not only were the Ppt1-KI mice unable to maintain the motor coordination, but they were also unable to hold on to the rotarod for more than a few seconds (Fig.6F). These data clearly demonstrated that Ppt1-KI mice undergo progressive deterioration in motor coordination in an age-dependent manner.
Figure 6.
Clasping behavior, motor coordination by rotarod and lifespan assessment. The development of clasping behavior, which attests neurological deterioration, is not manifested in WT mice (A), but can be readily seen in their 6-month-old Ppt1-KI littermates (B). The Ppt1−/− mouse (C), which also manifest clasping behavior, was used for comparison. Graphic representation of the results of rotarod test for motor coordination of WT mice and their Ppt1-KI littermates at 4 months (D), 6 months (E), and 8 months (F) of age. The rotarod performance results are presented as the mean (n = 12) ±SD. Note that while the rotarod test results were not appreciably different between 4-month-old WT and Ppt1-KI littermates, these results were highly significant (***P < 0.001) when the rotarod performances of WT and Ppt1-KI mice at 6 and 8 months of age, respectively, were compared. Kaplan–Meier survival curve (G) of Ppt1-KI mice (n = 62). The median lifespan of the Ppt1-KI mice was 227 days.
Lifespan estimate of Ppt1-KI mice by Kaplan–Meier plot
Two of the most devastating manifestations of INCL are the rapidly progressive neurodegeneration and markedly reduced lifespan. The manifestation of the clasping behavior as well as the declining motor coordination in rotarod test attested to neurodegeneration in the Ppt1-KI mice. Next, we sought to estimate their longevity. Accordingly, we allowed Ppt1-KI mice to live until they were unable to reach for food and water, at which time they were killed and the age of the mice was recorded. The results showed that median longevity of the Ppt1-KI mice was around 227 days (Fig.6G), which is considerably shorter than that of the WT littermates. Our results, taken together, showed that the Ppt1-KI mice not only manifest progressive neurological and retinal deterioration but also demonstrate a shortened lifespan demonstrating that these mice represent a reliable animal model of INCL.
Discussion
In this study, we have described the generation and phenotypic characterization of mice carrying the nonsense mutation in the Ppt1 gene (c.451C>T), commonly found in the US INCL patient population.20 We knocked-in this mutation into the genome of C57 ES cells in order to generate syngenic C57 mice without requiring extensive backcrossing needed to obtain homogenous C57 genetic background if 129 ES cells were used. The resulting c.451C>T/c.451C>T (Ppt1-KI) mice, like the Ppt1−/− counterparts,23,24 are normal at birth and continue to develop and reproduce normally. However, by 6 months of age they began to manifest neurological deficits attested by the clasping phenotype. These mice also had a shortened median lifespan of 227 days, which is considerably shorter than that of the WT mice. The Ppt1-mRNA as well as Ppt1-protein levels were virtually undetectable in brain tissues of the Ppt1-KI mice. Consistent with these findings, Ppt1 enzymatic activity was virtually undetectable in the brain of these mice. Evaluation of these mice by rotarod performance test showed progressive impairment of motor coordination at around 6 months of age. Histopathological analyses showed accumulation of autofluorescent material in the brain and spleen. Activated astrocytes and microglia in the brain were readily detected and histopathological analysis of the eye revealed degeneration of the retinal layers. Evaluation of retinal function by ERG showed progressive deterioration. In Ppt1-KI mice, a-wave of dark-adapted ERG was smaller, indicating a reduced rod photoreceptor function. There were even larger reductions in b-waves of dark-adapted ERG responses, which indicated additional loss of function of the bipolar cells in the retina of the mutant mice. Moreover, decreased light-adapted ERG responses revealed the loss of cone-pathway signals. Taken together, these results showed that the Ppt1-KI mice recapitulate virtually all characteristic clinical and pathological features of INCL.
As neomycin resistance gene (Neo) is used in our targeting construct and its expression is driven by a strong promoter, insertion of the neo cassette in an intron may affect endogenous gene expression and may result in the inactivation or reduction of the floxed genes.29–31 To circumvent this problem, Cre/LoxP technology has been employed for generating mutant organisms by conditional gene knockout.32 In this study, we removed the Neo cassette in the targeted ES cells by transient expression of Cre recombinase. This was carried out in order to eliminate any potential effects of the Neo gene affecting the phenotype of the Ppt1-KI mice. We used the C57 ES cells for targeting the nonsense mutation in to obviate the necessity of extensive backcrossing to obtain syngenic C57 Ppt1-KI mice had we used 129 ES cells.
It has been reported that out of the 64 possible triplet codons, 61 encode specific amino acids, and the remaining three codons, UAA, UAG, and UGA, are noncoding or nonsense codons, which cause translation termination.33 Emerging evidence suggests that premature translation termination may differ from the normal translation termination process. The mRNAs with a premature termination codon (PTC) undergo accelerated degradation, a process commonly known as NMD.22 Investigations of NMD led to the concept that a single small-molecule suppressing PTCs in a subset of patients with genetic diseases caused by nonsense mutations may be an effective therapeutic strategy.
Earlier experiments to suppress NMD recognized the effectiveness of streptomycin and other aminoglycoside antibiotics in suppressing nonsense mutations.34 Aminoglycosides in general inhibit bacterial protein synthesis. The clinical use of these drugs to suppress nonsense mutations is precluded because of toxicity of these drugs at high dose, required for a low level of read through of premature nonsense codons.35,36 Recently, investigations have been directed to generate nontoxic small molecules to suppressing PTCs. One such compound, PTC124 also known Ataluran, has shown promise in suppressing nonsense mutations in several genetic diseases including cystic fibrosis and Duchenne muscular dystrophy.22 Previously, we reported the effects of treating cultured cells from INCL patients carrying nonsense mutations in the CLN1/PPT1 gene, used to generate our Ppt1-KI mice, with PTC124, which induced a modest increase in Ppt1 enzyme activity with some beneficial effects.37 However, to be clinically relevant, it is important to determine if the results derived from in vitro experiments are reproducible in vivo. Thus, we generated the Ppt1-KI mice, which provide a platform for evaluating not only PTC124, but also other nonsense suppressors alone or in combination with a recently characterized thioesterase-mimetic small molecule, N-tert (Butyl) hydroxylamine (NtBuHA).38 Moreover, our Ppt1-KI mice may also be used as a platform for evaluating novel treatment strategies for INCL patients carrying nonsense mutations in the PPT1 gene including a combination of NtBuHA and nonsense suppressors.24 Our ongoing experiments are designed to evaluate this and other potential strategies using these mice.
While our manuscript was in review, we found that Miller et al.39 reported the generation of a Ppt1 R151X mouse model. The phenotypes described in our model are in general agreement with those of Miller et al.
Acknowledgments
We thank M. B. Bagh, G. Chandra, and S. Peng for valuable suggestions and advice on carrying out some of the experiments. We also thank Sandra Hofmann for a generous gift of the Rat anti-PPT1 antibody used for the detection of Ppt1 protein levels by Western blotting. This research was supported in full by the intramural program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, The National Institutes of Health (NIH).
Author Contribution
The project was conceived by A. B. M. and Z. Z. who also provided the design of the targeting construct used to generate the Ppt1-KI mice. The characterization of the Ppt1-KI mice including genotyping, mating of heterozygous (WT/c.451C>T) mice to generate Ppt1-KI mice and their WT littermates as well as all the experiments were conducted by A. B., who also analyzed the results. Illustrations were prepared jointly by A. B. and Z. Z. The results of ERG were analyzed by Y. L. and H. Q. All authors contributed to the preparation of the manuscript and in the revisions that followed to generate the final version for publication.
Conflict of Interest
None declared.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1. Confirmation of c.451 C>T mutation in targeted ES clones. (A) The c.451 C>T (R151X) mutations in targeted ES clones were confirmed by direct DNA sequencing. The arrow shows the mutation site (C>T). (B) Schematic of the PCR-based genotype analysis. The sequences of primers were listed in the Materials and Methods.
Figure S2. Biochemical analysis of Catalase and NeuN levels. (A) The Catalase-mRNA levels in the brain tissues of Ppt1-KI as well as those of their WT littermates. The Catalase-mRNA levels in Ppt1−/− mice were used as a control. The results are presented as the mean (n = 12) +SD. (B) Western blot analysis of brain tissues from Ppt1-KI mice and those of their WT littermates. Brain tissues from Ppt1−/− mice were used for comparison. β-actin was used as a loading control. (C) The NeuN-mRNA levels in the brain tissues of Ppt1-KI as well as in those of their WT littermates. The NeuN-mRNA levels in Ppt1−/− mice were used as control. The results are presented as the mean (n = 12) +SD. (D) Western blot analysis of brain tissues from Ppt1-KI mice and those of their WT littermates. Brain tissues from Ppt1−/− mice were used as a control. β-actin was used as a loading control. *P < 0.05, **P < 0.01, ***P < 0.001.
References
- 1.Anderson GW, Goebel HH, Simonati A. Human pathology in NCL. Biochim Biophys Acta. 2013;1832:1807–1826. doi: 10.1016/j.bbadis.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 2.Goebel HH, Wisniewski KE. Current state of clinical and morphological features in human NCL. Brain Pathol. 2004;14:61–69. doi: 10.1111/j.1750-3639.2004.tb00499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haltia M. The neuronal ceroid lipofuscinoses. Biochim Biophys Acta. 2006;1762:850–856. doi: 10.1016/j.bbadis.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 4.Rider JA, Rider DL. Batten disease: past present and future. Am J Med Genet. 1988;5(suppl):21–26. doi: 10.1002/ajmg.1320310606. [DOI] [PubMed] [Google Scholar]
- 5.Kousi M, Lehesjoki AE, Mole SE. Update of the mutation spectrum and clinical correlations of over 360 mutations in eight genes that underlie the neuronal ceroid lipofuscinoses. Hum Mutat. 2012;33:42–46. doi: 10.1002/humu.21624. [DOI] [PubMed] [Google Scholar]
- 6.Siintola E, Lehesjoki AE, Mole SE. Molecular genetics of the NCLs—status and perspectives. Biochim Biophys Acta. 2006;1762:857–864. doi: 10.1016/j.bbadis.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 7.Mole SE, Williams RE, Goebel HH. Correlations between genotype, ultrastructural morphology and clinical phenotype in the neuronal ceroid lipofuscinoses. Neurogenetics. 2005;6:107–126. doi: 10.1007/s10048-005-0218-3. [DOI] [PubMed] [Google Scholar]
- 8.Vesa J, Hellsten E, Verkruyse LA, et al. Mutations in the palmitoyl protein thioesterase gene causing infantile neuronal ceroid lipofuscinosis. Nature. 1995;376:584–587. doi: 10.1038/376584a0. [DOI] [PubMed] [Google Scholar]
- 9.Camp LA, Hofmann SL. Purification and properties of a palmitoyl-protein thioesterase that cleaves palmitate from H-Ras. J Biol Chem. 1993;268:22566–22574. [PubMed] [Google Scholar]
- 10.Camp LA, Verkruyse LA, Afendis SJ, et al. Molecular cloning and expression of palmitoyl-protein thioesterase. J Biol Chem. 1994;269:23212–23219. [PubMed] [Google Scholar]
- 11.Hellsten E, Vesa J, Olkkonen VM, et al. Human palmitoyl-protein thioesterase: evidence for lysosomal targeting of the enzyme and disturbed cellular routing in infantile neuronal ceroid lipofuscinosis. EMBO J. 1996;15:5240–5245. [PMC free article] [PubMed] [Google Scholar]
- 12.Verkruyse LA, Hofmann SL. Lysosomal targeting of palmitoyl-protein thioesterase. J Biol Chem. 1996;271:15831–15836. doi: 10.1074/jbc.271.26.15831. [DOI] [PubMed] [Google Scholar]
- 13.Hellsten E, Vesa J, Heiskanen M, et al. Identification of YAC clones for human chromosome 1p32 and physical mapping of the infantile neuronal ceroid lipofuscinoses (INCL) locus. Genomics. 1995;25:404–412. doi: 10.1016/0888-7543(95)80040-s. [DOI] [PubMed] [Google Scholar]
- 14.Linder ME, Deschenes RJ. Palmitoylation: policing protein stability and traffic. Nat Rev Mol Cell Biol. 2007;8:74–84. doi: 10.1038/nrm2084. [DOI] [PubMed] [Google Scholar]
- 15.Salaun C, Greaves J, Chamberlain LH. The intracellular dynamic of protein palmitoylation. J Cell Sci. 2010;191:1229–1238. doi: 10.1083/jcb.201008160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu JY, Verkruyse LA, Hofmann SL. Lipid thioesters derived from acylated proteins accumulate in infantile neuronal ceroid lipofuscinosis: correction of the defect in lymphoblasts by recombinant palmitoyl-protein thioesterase. Proc Natl Acad Sci USA. 1996;93:10046–10050. doi: 10.1073/pnas.93.19.10046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofmann SL, Peltonen L. The neuronal ceroid lipofuscinosis. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The metabolic and molecular basis of inherited disease. 8th ed. Vol. 3. New York, NY: McGraw–Hill; 2001. pp. 3877–3894. [Google Scholar]
- 18.Santavuori P, Vanhanen SL, Sainio K, et al. Infantile neuronal ceroid-lipofuscinosis (INCL): diagnosis criteria. J Inherit Metab Dis. 1993;16:227–229. doi: 10.1007/BF00710250. [DOI] [PubMed] [Google Scholar]
- 19.Kohan R, Cismondi IA, Oller-Ramirez AM, et al. Therapeutic approaches to the challenge of neuronal ceroid lipofuscinoses. Curr Pharm Biotechnol. 2011;12:867–883. doi: 10.2174/138920111795542633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Das AK, Becerra CH, Yi W, et al. Molecular genetics of palmitoyl-protein thioesterase deficiency in the U.S. J Clin Invest. 1998;102:361–370. doi: 10.1172/JCI3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Welch EM, Barton ER, Zhuo J, et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447:87–91. doi: 10.1038/nature05756. [DOI] [PubMed] [Google Scholar]
- 22.Peltz SW, Morsy M, Welch EM, Jacobson A. Ataluren as an agent for therapeutic nonsense suppression. Annu Rev Med. 2013;64:407–425. doi: 10.1146/annurev-med-120611-144851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta P, Soyombo AA, Atashband A, et al. Disruption of PPT1 or PPT2 causes neuronal ceroid lipofuscinoses in knockout mice. Proc Natl Acad Sci USA. 2001;98:13566–13571. doi: 10.1073/pnas.251485198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bible E, Gupta P, Hofmann SL, et al. Regional and cellular neuropathology in the palmitoyl- protein thioesterase-1 null mutant mouse model of infantile neuronal ceroid lipofuscinoses. Neurobiol Dis. 2004;16:346–359. doi: 10.1016/j.nbd.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Z, Lee YC, Kim SJ, et al. Palmitoyl-protein thioesterase-1 deficiency mediates the activation of the unfolded protein response and neuronal apoptosis in INCL. Hum Mol Genet. 2006;15:337–346. doi: 10.1093/hmg/ddi451. [DOI] [PubMed] [Google Scholar]
- 26.Qian H, Shah MR, Alexander KR, Ripps H. Two distinct processes are evident in rat cone flicker ERG responses at low and high temporal frequencies. Exp Eye Res. 2008;87:71–75. doi: 10.1016/j.exer.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamm RJ, Pike BR, O'Dell DM, et al. The rotarod test: an evaluation of its effectiveness in assessing motor deficits following traumatic brain injury. J Neurotrauma. 1994;11:187–196. doi: 10.1089/neu.1994.11.187. [DOI] [PubMed] [Google Scholar]
- 28.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- 29.Hirotsune S, Fleck MW, Gambello MJ, et al. Graded reduction of Pafah1b1 (Lis1) activity results in neuronal migration defects and early embryonic lethality. Nat Genet. 1998;19:333–339. doi: 10.1038/1221. [DOI] [PubMed] [Google Scholar]
- 30.Chen L, Adar R, Yang X, et al. Gly369Cys mutation in mouse FGFR3 causes achondroplasia by affecting both chondrogenesis and osteogenesis. J Clin Invest. 1999;104:1517–1525. doi: 10.1172/JCI6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iwata T, Chen L, Li C, et al. A neonatal lethal mutation in FGFR3 uncouples proliferation and differentiation of growth plate chondrocytes in embryos. Hum Mol Genet. 2000;9:1603–1613. doi: 10.1093/hmg/9.11.1603. [DOI] [PubMed] [Google Scholar]
- 32.Gu H, Marth JD, Orban PC, et al. Deletion of the DNA polymerase beta gene in T cells using tissue-specific gene targeting. Science. 1994;265:103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- 33.Nirenberg M, Leder P, Bernfield M, et al. RNA codewords and protein synthesis, VII. On the general nature of the RNA code. Proc Natl Acad Sci USA. 1965;53:1161–1168. doi: 10.1073/pnas.53.5.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hermann T. Aminoglycoside antibiotics: old drugs and new therapeutic approaches. Cell Mol Life Sci. 2007;64:1841–1852. doi: 10.1007/s00018-007-7034-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manuvakhova M, Keeling K, Bedwell DM. Aminoglycoside antibiotics mediate context-dependent suppression of termination codons in a mammalian translation system. RNA. 2000;6:1044–1055. doi: 10.1017/s1355838200000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burke JF, Mogg AE. Suppression of a nonsense mutation in mammalian cells in vivo by the aminoglycoside antibiotics G-418 and paromomycin. Nucleic Acids Res. 1985;13:6265–6272. doi: 10.1093/nar/13.17.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sarkar C, Zhang Z, Mukherjee AB. Stop codon read-through with PTC124 induces palmitoyl- protein thioesterase-1 activity, reduces thioester load and suppresses apoptosis in cultured cells from INCL patients. Mol Genet Metab. 2011;104:338–345. doi: 10.1016/j.ymgme.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarkar C, Chandra G, Peng S, et al. Neuroprotection and lifespan extension in Ppt1(−/−) mice by NtBuHA: therapeutic implications for INCL. Nat Neurosci. 2013;16:1608–1617. doi: 10.1038/nn.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller JN, Kovács AD, Pearce DA. The novel Cln1R151X mouse model of infantile neuronal ceroid lipofuscinoses (INCL) for testing nonsense suppression therapy. Hum Mol Genet. 2014 doi: 10.1093/hmg/ddu428. September 8 pii: ddu428. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Confirmation of c.451 C>T mutation in targeted ES clones. (A) The c.451 C>T (R151X) mutations in targeted ES clones were confirmed by direct DNA sequencing. The arrow shows the mutation site (C>T). (B) Schematic of the PCR-based genotype analysis. The sequences of primers were listed in the Materials and Methods.
Figure S2. Biochemical analysis of Catalase and NeuN levels. (A) The Catalase-mRNA levels in the brain tissues of Ppt1-KI as well as those of their WT littermates. The Catalase-mRNA levels in Ppt1−/− mice were used as a control. The results are presented as the mean (n = 12) +SD. (B) Western blot analysis of brain tissues from Ppt1-KI mice and those of their WT littermates. Brain tissues from Ppt1−/− mice were used for comparison. β-actin was used as a loading control. (C) The NeuN-mRNA levels in the brain tissues of Ppt1-KI as well as in those of their WT littermates. The NeuN-mRNA levels in Ppt1−/− mice were used as control. The results are presented as the mean (n = 12) +SD. (D) Western blot analysis of brain tissues from Ppt1-KI mice and those of their WT littermates. Brain tissues from Ppt1−/− mice were used as a control. β-actin was used as a loading control. *P < 0.05, **P < 0.01, ***P < 0.001.