Abstract
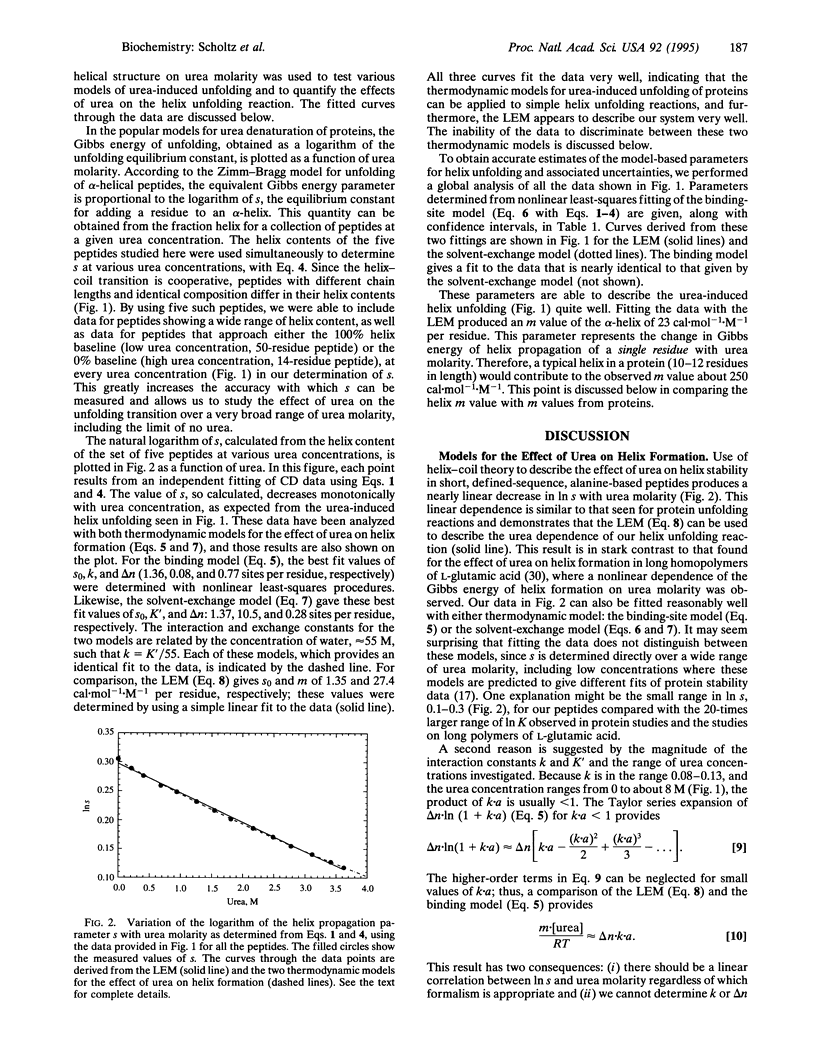
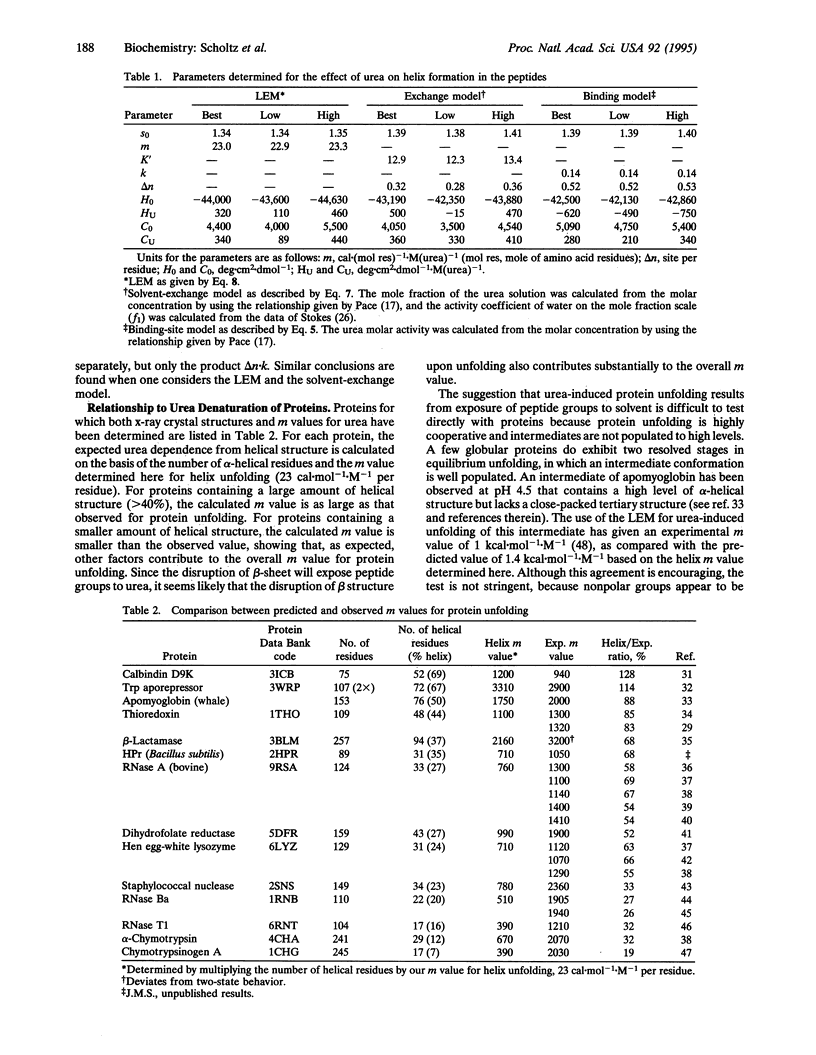
To provide a model system for understanding how the unfolding of protein alpha-helices by urea contributes to protein denaturation, urea unfolding was measured for a homologous series of helical peptides with the repeating sequence Ala-Glu-Ala-Ala-Lys-Ala and chain lengths varying from 14 to 50 residues. The dependence of the helix propagation parameter of the Zimm-Bragg model for helix-coil transition theory (s) on urea molarity ([urea]) was determined at 0 degree C with data for the entire set of peptides, and a linear dependence of In s on [urea] was found. The results were fitted by the binding-site model and by the solvent-exchange model for the interaction of urea with the peptides. Each of these thermodynamic models is able to describe the data quite well and we are not able to discern any difference between the ability of each model to fit the data. Thus a linear relation, ln s = ln s0 - (m/RT).[urea], fits the data for alpha-helix unfolding, just as others have found for protein unfolding. When the m value determined here for alpha-helix unfolding is multiplied by the number of helical residues in partly helical protein molecules, the resulting values agree within a factor of 2 with observed m values for these proteins. This result indicates that the interaction between urea and peptide groups accounts for a major part of the denaturing action of urea on proteins, as predicted earlier by some model studies with small molecules.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahmad F., Bigelow C. C. Estimation of the free energy of stabilization of ribonuclease A, lysozyme, alpha-lactalbumin, and myoglobin. J Biol Chem. 1982 Nov 10;257(21):12935–12938. [PubMed] [Google Scholar]
- Ahmad F. Free energy changes in ribonuclease A denaturation. Effect of urea, guanidine hydrochloride, and lithium salts. J Biol Chem. 1983 Sep 25;258(18):11143–11146. [PubMed] [Google Scholar]
- Aune K. C., Tanford C. Thermodynamics of the denaturation of lysozyme by guanidine hydrochloride. II. Dependence on denaturant concentration at 25 degrees. Biochemistry. 1969 Nov;8(11):4586–4590. doi: 10.1021/bi00839a053. [DOI] [PubMed] [Google Scholar]
- Barrick D., Baldwin R. L. Three-state analysis of sperm whale apomyoglobin folding. Biochemistry. 1993 Apr 13;32(14):3790–3796. doi: 10.1021/bi00065a035. [DOI] [PubMed] [Google Scholar]
- Brandts J. F., Kaplan L. J. Derivative sspectroscopy applied to tyrosyl chromophores. Studies on ribonuclease, lima bean inhibitors, insulin, and pancreatic trypsin inhibitor. Biochemistry. 1973 May 8;12(10):2011–2024. doi: 10.1021/bi00734a027. [DOI] [PubMed] [Google Scholar]
- Chen Y. H., Yang J. T., Chau K. H. Determination of the helix and beta form of proteins in aqueous solution by circular dichroism. Biochemistry. 1974 Jul 30;13(16):3350–3359. doi: 10.1021/bi00713a027. [DOI] [PubMed] [Google Scholar]
- Gittelman M. S., Matthews C. R. Folding and stability of trp aporepressor from Escherichia coli. Biochemistry. 1990 Jul 31;29(30):7011–7020. doi: 10.1021/bi00482a009. [DOI] [PubMed] [Google Scholar]
- Greene R. F., Jr, Pace C. N. Urea and guanidine hydrochloride denaturation of ribonuclease, lysozyme, alpha-chymotrypsin, and beta-lactoglobulin. J Biol Chem. 1974 Sep 10;249(17):5388–5393. [PubMed] [Google Scholar]
- Hughson F. M., Barrick D., Baldwin R. L. Probing the stability of a partly folded apomyoglobin intermediate by site-directed mutagenesis. Biochemistry. 1991 Apr 30;30(17):4113–4118. doi: 10.1021/bi00231a001. [DOI] [PubMed] [Google Scholar]
- Johnson M. L. Evaluation and propagation of confidence intervals in nonlinear, asymmetrical variance spaces. Analysis of ligand-binding data. Biophys J. 1983 Oct;44(1):101–106. doi: 10.1016/S0006-3495(83)84281-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley R. F., Shalongo W., Jagannadham M. V., Stellwagen E. Equilibrium and kinetic measurements of the conformational transition of reduced thioredoxin. Biochemistry. 1987 Mar 10;26(5):1406–1411. doi: 10.1021/bi00379a029. [DOI] [PubMed] [Google Scholar]
- Makhatadze G. I., Privalov P. L. Protein interactions with urea and guanidinium chloride. A calorimetric study. J Mol Biol. 1992 Jul 20;226(2):491–505. doi: 10.1016/0022-2836(92)90963-k. [DOI] [PubMed] [Google Scholar]
- Marqusee S., Baldwin R. L. Helix stabilization by Glu-...Lys+ salt bridges in short peptides of de novo design. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8898–8902. doi: 10.1073/pnas.84.24.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchinson C., Pain R. H. Effects of sulphate and urea on the stability and reversible unfolding of beta-lactamase from Staphylococcus aureus. Implications for the folding pathway of beta-lactamase. J Mol Biol. 1985 Jul 20;184(2):331–342. doi: 10.1016/0022-2836(85)90384-5. [DOI] [PubMed] [Google Scholar]
- NOZAKI Y., TANFORD C. THE SOLUBILITY OF AMINO ACIDS AND RELATED COMPOUNDS IN AQUEOUS UREA SOLUTIONS. J Biol Chem. 1963 Dec;238:4074–4081. [PubMed] [Google Scholar]
- Nandi P. K., Robinson D. R. Effects of urea and guanidine hydrochloride on peptide and nonpolar groups. Biochemistry. 1984 Dec 18;23(26):6661–6668. doi: 10.1021/bi00321a058. [DOI] [PubMed] [Google Scholar]
- Nozaki Y., Tanford C. The solubility of amino acids and two glycine peptides in aqueous ethanol and dioxane solutions. Establishment of a hydrophobicity scale. J Biol Chem. 1971 Apr 10;246(7):2211–2217. [PubMed] [Google Scholar]
- Pace C. N. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 1986;131:266–280. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- Pace C. N., Laurents D. V., Erickson R. E. Urea denaturation of barnase: pH dependence and characterization of the unfolded state. Biochemistry. 1992 Mar 17;31(10):2728–2734. doi: 10.1021/bi00125a013. [DOI] [PubMed] [Google Scholar]
- Pace C. N., Laurents D. V., Thomson J. A. pH dependence of the urea and guanidine hydrochloride denaturation of ribonuclease A and ribonuclease T1. Biochemistry. 1990 Mar 13;29(10):2564–2572. doi: 10.1021/bi00462a019. [DOI] [PubMed] [Google Scholar]
- Pace C. N. The stability of globular proteins. CRC Crit Rev Biochem. 1975 May;3(1):1–43. doi: 10.3109/10409237509102551. [DOI] [PubMed] [Google Scholar]
- Pace C. N., Vanderburg K. E. Determining globular protein stability: guanidine hydrochloride denaturation of myoglobin. Biochemistry. 1979 Jan 23;18(2):288–292. doi: 10.1021/bi00569a008. [DOI] [PubMed] [Google Scholar]
- Perry K. M., Onuffer J. J., Touchette N. A., Herndon C. S., Gittelman M. S., Matthews C. R., Chen J. T., Mayer R. J., Taira K., Benkovic S. J. Effect of single amino acid replacements on the folding and stability of dihydrofolate reductase from Escherichia coli. Biochemistry. 1987 May 19;26(10):2674–2682. doi: 10.1021/bi00384a004. [DOI] [PubMed] [Google Scholar]
- Prakash V., Loucheux C., Scheufele S., Gorbunoff M. J., Timasheff S. N. Interactions of proteins with solvent components in 8 M urea. Arch Biochem Biophys. 1981 Sep;210(2):455–464. doi: 10.1016/0003-9861(81)90209-5. [DOI] [PubMed] [Google Scholar]
- ROBINSON D. R., JENCKS W. P. THE EFFECT OF COMPOUNDS OF THE UREA-GUANIDINIUM CLASS ON THE ACTIVITY COEFFICIENT OF ACETYLTETRAGLYCINE ETHYL ESTER AND RELATED COMPOUNDS. J Am Chem Soc. 1965 Jun 5;87:2462–2470. doi: 10.1021/ja01089a028. [DOI] [PubMed] [Google Scholar]
- Rohl C. A., Scholtz J. M., York E. J., Stewart J. M., Baldwin R. L. Kinetics of amide proton exchange in helical peptides of varying chain lengths. Interpretation by the Lifson-Roig equation. Biochemistry. 1992 Feb 11;31(5):1263–1269. doi: 10.1021/bi00120a001. [DOI] [PubMed] [Google Scholar]
- SCHELLMAN J. A. The stability of hydrogen-bonded peptide structures in aqueous solution. C R Trav Lab Carlsberg Chim. 1955;29(14-15):230–259. [PubMed] [Google Scholar]
- SCHELLMAN J. A. The thermodynamics of urea solutions and the heat of formation of the peptide hydrogen bond. C R Trav Lab Carlsberg Chim. 1955;29(14-15):223–229. [PubMed] [Google Scholar]
- Santoro M. M., Bolen D. W. A test of the linear extrapolation of unfolding free energy changes over an extended denaturant concentration range. Biochemistry. 1992 May 26;31(20):4901–4907. doi: 10.1021/bi00135a022. [DOI] [PubMed] [Google Scholar]
- Santoro M. M., Bolen D. W. Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl alpha-chymotrypsin using different denaturants. Biochemistry. 1988 Oct 18;27(21):8063–8068. doi: 10.1021/bi00421a014. [DOI] [PubMed] [Google Scholar]
- Schellman J. A. A simple model for solvation in mixed solvents. Applications to the stabilization and destabilization of macromolecular structures. Biophys Chem. 1990 Aug 31;37(1-3):121–140. doi: 10.1016/0301-4622(90)88013-i. [DOI] [PubMed] [Google Scholar]
- Schellman J. A. Selective binding and solvent denaturation. Biopolymers. 1987 Apr;26(4):549–559. doi: 10.1002/bip.360260408. [DOI] [PubMed] [Google Scholar]
- Schellman J. A. The thermodynamics of solvent exchange. Biopolymers. 1994 Aug;34(8):1015–1026. doi: 10.1002/bip.360340805. [DOI] [PubMed] [Google Scholar]
- Scholtz J. M., Baldwin R. L. The mechanism of alpha-helix formation by peptides. Annu Rev Biophys Biomol Struct. 1992;21:95–118. doi: 10.1146/annurev.bb.21.060192.000523. [DOI] [PubMed] [Google Scholar]
- Scholtz J. M., Marqusee S., Baldwin R. L., York E. J., Stewart J. M., Santoro M., Bolen D. W. Calorimetric determination of the enthalpy change for the alpha-helix to coil transition of an alanine peptide in water. Proc Natl Acad Sci U S A. 1991 Apr 1;88(7):2854–2858. doi: 10.1073/pnas.88.7.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholtz J. M., Qian H., York E. J., Stewart J. M., Baldwin R. L. Parameters of helix-coil transition theory for alanine-based peptides of varying chain lengths in water. Biopolymers. 1991 Nov;31(13):1463–1470. doi: 10.1002/bip.360311304. [DOI] [PubMed] [Google Scholar]
- Serrano L., Kellis J. T., Jr, Cann P., Matouschek A., Fersht A. R. The folding of an enzyme. II. Substructure of barnase and the contribution of different interactions to protein stability. J Mol Biol. 1992 Apr 5;224(3):783–804. doi: 10.1016/0022-2836(92)90562-x. [DOI] [PubMed] [Google Scholar]
- Shortle D., Meeker A. K. Mutant forms of staphylococcal nuclease with altered patterns of guanidine hydrochloride and urea denaturation. Proteins. 1986 Sep;1(1):81–89. doi: 10.1002/prot.340010113. [DOI] [PubMed] [Google Scholar]
- Sijpkes A. H., van de Kleut G. J., Gill S. C. Urea-diketopiperazine interactions: a model for urea induced denaturation of proteins. Biophys Chem. 1993 Apr;46(2):171–177. doi: 10.1016/0301-4622(93)85024-c. [DOI] [PubMed] [Google Scholar]
- Thomson J. A., Shirley B. A., Grimsley G. R., Pace C. N. Conformational stability and mechanism of folding of ribonuclease T1. J Biol Chem. 1989 Jul 15;264(20):11614–11620. [PubMed] [Google Scholar]
- Wendt B., Hofmann T., Martin S. R., Bayley P., Brodin P., Grundström T., Thulin E., Linse S., Forsén S. Effect of amino acid substitutions and deletions on the thermal stability, the pH stability and unfolding by urea of bovine calbindin D9k. Eur J Biochem. 1988 Aug 15;175(3):439–445. doi: 10.1111/j.1432-1033.1988.tb14214.x. [DOI] [PubMed] [Google Scholar]


