SUMMARY
Bacterial culture was the first method used to describe the human microbiota, but this method is considered outdated by many researchers. Metagenomics studies have since been applied to clinical microbiology; however, a “dark matter” of prokaryotes, which corresponds to a hole in our knowledge and includes minority bacterial populations, is not elucidated by these studies. By replicating the natural environment, environmental microbiologists were the first to reduce the “great plate count anomaly,” which corresponds to the difference between microscopic and culture counts. The revolution in bacterial identification also allowed rapid progress. 16S rRNA bacterial identification allowed the accurate identification of new species. Mass spectrometry allowed the high-throughput identification of rare species and the detection of new species. By using these methods and by increasing the number of culture conditions, culturomics allowed the extension of the known human gut repertoire to levels equivalent to those of pyrosequencing. Finally, taxonogenomics strategies became an emerging method for describing new species, associating the genome sequence of the bacteria systematically. We provide a comprehensive review on these topics, demonstrating that both empirical and hypothesis-driven approaches will enable a rapid increase in the identification of the human prokaryote repertoire.
INTRODUCTION
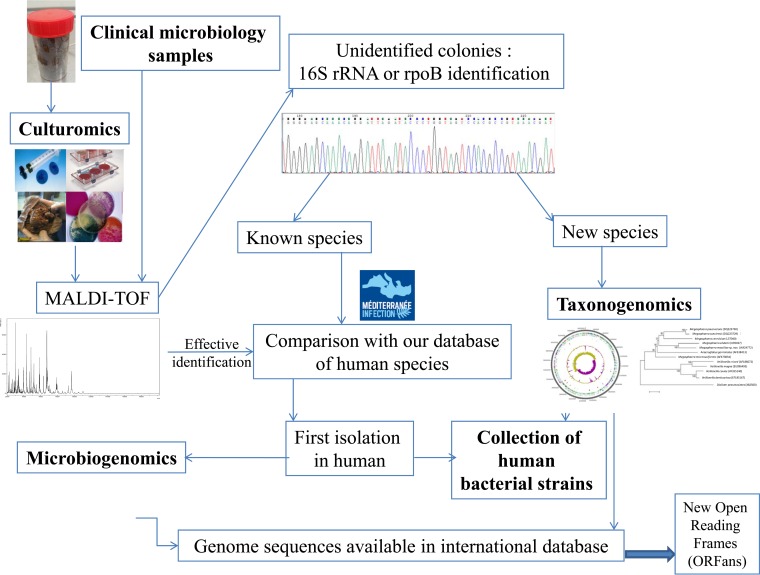
Metagenomic studies (the study of metagenomes by using high-throughput sequencing directly from a complex ecosystem) appear to be able to replace bacterial culture (1). Indeed, the first metagenomic studies, which were specifically performed to study environmental samples, highlighted that 80% of the bacteria identified by metagenomics or by pyrosequencing targeting the 16S rRNA gene had not yet been cultured (1, 2). Because the term was a misnomer, where “uncultured” was transformed into “uncultivable,” 80% of bacteria from the environment or from the human gut microbiota were accepted as being uncultivable. Of course, this conclusion is not realistic because all microorganisms are susceptible to being cultured; the best route and proper tools for culturing must be found. One of the major biases of metagenomic studies is the depth bias (3, 4). Indeed, in complex ecosystems such as the gut microbiota, which consists of ∼1012 bacteria per gram of stool, current metagenomic studies are unable to detect bacteria at concentrations of <105 bacteria per gram (4). For example, Salmonella enterica serovar Typhi, which is a formidable pathogen, is undetectable by current metagenomic analysis methods (4). Metagenomics and culturomics (diversification of culture conditions together with identification by matrix-assisted laser desorption ionization–time of flight mass spectrometry [MALDI-TOF MS], to increase the bacterial repertoire) can detect comparable numbers of species. The complementarity between culture-dependent and culture-independent studies has been demonstrated, because only 15% of species detected were detected concomitantly by the 2 techniques (4–7). Environmental microbiologists have used culture conditions to mimic the natural environment of bacteria to culture microorganisms previously considered uncultivable (8). Indeed, culture efforts and easy identification through 16S rRNA amplification and sequencing and then the recent use of MALDI-TOF in routine bacteriology have caused a dramatic increase in the number of identified species. In fact, only 1,791 official bacterial species had been recognized in 1980, whereas currently, >12,000 species have been recorded (9). Recently, culturomics performed in our laboratory greatly participated in this effort (Table 1). In addition to the accompanying large review summarizing the past and current techniques used in clinical microbiology (10), we propose here a focus on the bacterial identification progresses that allow culturomics, drawing on environmental microbiologists' techniques and a rapid method of bacterial identification by MALDI-TOF MS to dramatically extend the human gut microbiota repertoire. Finally, we developed taxonogenomics, a method proposed to include genome sequencing in addition to classic criteria in order to describe a new bacterial species.
TABLE 1.
Bacterial species first cultured or first isolated from human samples at URMITE, Marseille, France
| Detection method | No. of bacterial species detecteda |
Source and/or references | ||
|---|---|---|---|---|
| 1st human case | New species | Total | ||
| Isolation of fastidious bacteria | 4 | 23 | http://www.mediterranee-infection.com/article.php?laref=258&titer=new-microbes | |
| Isolation of anaerobic species | NA | 6 | 96, 97 | |
| Isolation of new bacterial species using 16S rRNA sequencing | 16 | 29 | 47, 53 | |
| Other | 2 | 16 | http://www.mediterranee-infection.com/article.php?laref=258&titer=new-microbes | |
| MALDI-TOF mass spectrometry in clinical microbiology laboratory | 31 | 0 | 42, 80 | |
| Culturomics | 168 | 91 | 4–7, 121; http://www.mediterranee-infection.com/article.php?laref=258&titer=new-microbes | |
| Total | 221 | 165 | 386 | |
NA, not applicable.
BACTERIAL IDENTIFICATION STRATEGIES
Detection of Growth
Staining.
Direct observation of microorganisms has frequently been the first step in identification. Dark-field microscopy has been used to visualize spirochetes observed against a black background (11). Gram staining is the most useful and cost-effective direct-observation method used in clinical microbiology. The most significant bacteria that have not been Gram stained include Mycoplasma, which does have not a cell wall, and Rickettsia and Chlamydia, which do not stain well (11). In addition, Gram staining is not constantly a robust method because the genera Bacillus, Gemella, and Listeria (12, 13) and certain Gram-positive anaerobes (14) can display aberrant Gram staining and appear Gram variable or Gram negative (15). In fact, in a recent study of the human gut microbiota, Hugon et al. revealed that electron microscopy, which is considered a reference method for studying the bacterial cell wall, allowed the identification of an average of 2-fold more Gram-positive prokaryotes than did Gram staining (16). Ziehl-Neelsen staining detects acid-alcohol-resistant bacteria, such as Mycobacterium spp. or Nocardia spp. (11). Intracellular bacteria might not be stained by Gram staining; however, detection of bacterial growth within cells can be identified after Gimenez or Giemsa staining for Rickettsia spp., Coxiella burnetii, and Tropheryma whipplei, or Giemsa staining can be used for Ehrlichia spp., Anaplasma spp., and Chlamydia spp. (17, 18). Acridine orange can also be used for the detection of bacteria.
4′,6′-Diamidino-2-phenylindole (DAPI) is a fluorescent molecule that strongly binds to DNA, specifically with adenine and thymine bases (19). The design of specific probes has allowed the direct visualization of diverse bacterial species such as C. burnetii (20), Staphylococcus spp. (21), and Salmonella spp. (22) directly from clinical samples.
Electron microscopy.
Electron microscopy improved knowledge of viruses (23) and bacteria (24), with the assistance of greatly improved resolution. Nevertheless, electron microscopy has been used more in bacterial research to observe microorganisms for the first time, to study the structure and function of cells (25), or by environmental microbiologists (26–28). Bacterial morphologies (29) and cell wall structures can be easily observed, and Gram-negative and Gram-positive types of bacteria can be distinguished (16). Over the past decade, cryo-electron microscopy techniques, which can be divided into cryo-electron tomography (to visualize cell structures at the protein level) (29), single-particle cryo-electron microscopy (the most commonly used) (30), and electron crystallography, are techniques based on cryofixation that emerged from sample preparation steps, which can affect specimens (30). These methods have permitted the exploration of macromolecules and the study of cell architecture as well as observations of viruses and protein molecules at the molecular level (30).
Microcolony detection by autofluorescence.
Naked-eye discrimination of bacterial growth is most likely associated with a relative delay. Both nondestructive fluorescence-based staining procedures and microcolony visualization using a scanner can be used to detect bacterial colonies more rapidly and easily than with the naked eye (31), notably in environmental microbiology and in the agroalimentary industry, in which such technologies were first used to detect food or water contamination (31–33). The expansion of the use of this technique will permit cost reduction. For example, Asano et al. identified slowly growing lactic acid bacteria by the microcolony method, using carboxyfluorescein diacetate (CFDA) staining (31). The strains were detected within 3 days of incubation, compared with 3 to 6 days using traditional culture methods. This method was highly discriminative, with results equal to those with fluorescence in situ hybridization (FISH). Baumstummler et al. demonstrated the same results by using a cocktail of 5 fluorescent dyes and a FACSCalibur flow cytometer, with detection of bacterial contamination of filterable products that was 3 to 5 times faster than with conventional methods (34).
In clinical microbiology, microscopically observed drug susceptibility (MODS) testing has been compared with traditional culture on solid media or automated liquid culture (35, 36). den Hertog et al. recently proposed an alternative method, initially using a porous aluminum oxide (PAO) support before transfer to a selective medium, with detection by using an automated microscope system (FluXXscan) and then a lipid fluorescent probe (Nile Red) to determine growth at early time points (37). Microcolonies were first detected after 4 to 5 days, demonstrating that the potential of this method was at least equivalent to that of MODS or thin-layer agar assays (37).
Liquid and agar culture media.
The turbidity of liquid culture media can be observed and represents the basis of growth detection in blood culture bottles for detection of bacteremia in clinical microbiology. Since 1990, the optimization of commercially available automated systems has permitted a reduction of the initially large proportion of false-positive blood culture bottles (38, 39).
The naked eye permits observation of the morphology of colonies on solid agar cultures, which can help an experienced microbiologist to determine the presumptive species group of the bacterial isolate. The use of blood-based agar facilitates the observation of the hemolytic ability of the bacterial species, which is frequently the first step in identification, particularly for streptococci (11). Nevertheless, the usual rule that the macroscopic aspect of the colonies frequently represents the first step in bacterial identification in routine bacteriology should be reconsidered. Because MALDI-TOF mass spectrometry permits the rapid testing of a large number of colonies, Lagier et al. demonstrated that colonies that are indistinguishable in appearance can be different bacterial species, particularly for the genus Enterococcus, and this finding should motivate the comprehensive testing of colonies (4).
Identification
Classic phenotypic identification.
Schematically, different system reactivities are available and combined, such as pH-based reactions; the enzyme profile; carbon source utilization (tetrazolium-based indicators); acid detection; as well as analysis of carbohydrate utilization, preformed enzymes, organic products, and cellular fatty acids (40, 41). The primary tests for the mode of energy metabolism are oxidase and catalase tests, which can be easily obtained in a few minutes. Carbohydrate metabolism is analyzed primarily by examining acid production (pH-based reactions). The tested end products of carbohydrate metabolism included primarily CO2, acetate, and lactate. Later, diverse tests for enzymes such as glucuronidase, glucosidase, galactosidase, and fucosidase were introduced (40). The tests for protein and amino acid metabolism include the production of indole and H2S; subsequently, tests for gelatin and casein digestion were introduced. Nitrogen metabolism is highlighted by tests for lysine, ornithine, arginine decarboxylase, arginine dihydrolase, phenylalanine deaminase, and urease (40). A breakthrough in the characterization of asaccharolytic bacteria emerged with tests for specific arylamidase activities. Regarding lipid metabolism, the primary tests are those for lipase and lecithinase on egg yolk agar and the digestion of Tween. Tests based on cell wall receptors, including optochin, lysozyme susceptibility, and bile solubility tests, are frequently used for the differentiation of Gram-positive cocci (40).
Many of these phenotypic characteristics can be simultaneously tested by using commercial kits and/or automated phenotypic systems, with the time to obtain an identification varying from 4 to 48 h (42). The relatively high cost is related to the reagents' costs and can amount to €4.6 to €12.65 for each identified bacterial colony (42). Until the MALDI-TOF revolution, these systems were the “gold standard” used in clinical laboratories worldwide for bacterial species identification (40).
Molecular tools.
(i) 16S rRNA.
The development of both molecular tools and sequence databases has constituted one of the most important advances of the late 20th century in clinical microbiology (43). The ability to detect and identify bacterial nucleic acids has permitted the rapid identification of both cultured and not-yet-cultivable bacteria (44, 45). In addition, this ability was also an unquestionable advance in the identification of extremely fastidious microorganisms, for which conventional biochemical methods were sometimes difficult (43). The development of the largest databank, known as GenBank, facilitated this progress. Janda and Abbot reported that 1,791 valid bacterial species names were recorded in 1980, whereas 8,168 species were recognized in 2007 and 12,391 are recognized today (http://www.bacterio.net/-number.html [accessed 1 August 2013]). This rapid increase is directly linked to the performance of 16S rRNA sequencing (9). In parallel, for a few years, the increasing number of genome sequences available in databases such as the Genomes OnLine Database (GOLD) has offered an effective in silico approach for choosing DNA targets for identification (46).
Before the advent of molecular tools, routine reference bacteriology laboratories estimated that 0.5 to 1% of the cultured bacteria were unidentified by classic phenotypic identification methods (47). A universal bacterial detection and identification system, using a gene present exclusively in bacteria and based on 16S rRNA gene amplification and sequencing, has been proposed. This gene was not discriminant for several bacterial species, such as Rickettsia species (48), Brucella species (49), Streptococcus spp. (50), Corynebacterium spp. (51), and Bacillus spp. (52).
16S rRNA amplification and sequencing have offered a large opportunity to describe new bacterial species and cultured bacteria (47, 53–57) and have increased the efficiency of bacterial identification. Drancourt et al. described phenotypic misidentification of 58.7% of 138 bacterial species from environmental and clinical samples identified at the species level by 16S rRNA amplification and sequencing (53). In a 5-year study, those same authors described 27 bacterial species newly determined to be associated with humans, including 11 entirely new bacterial species (47). Tortoli et al. estimated a prevalence of unidentified mycobacteria of >1% (55), and Bosshard et al. described 27 aerobic Gram-positive rods in an 18-month evaluation (58). In addition, for identification to the genus level, broad-spectrum PCR targeting the 23S rRNA gene was used (59). The 16S-23S rRNA gene intergenic transcribed spacer (ITS) has great variability in many bacterial species, such as Gram-positive anaerobic cocci, Streptococcus spp., Bartonella spp., and T. whipplei (60–63).
(ii) rpoB and other genes.
For identification to the species level, the RNA polymerase beta subunit-encoding gene, rpoB, was proposed early as an efficient tool for microbiologists (64). The use of the rpoB gene has been superior to the 16S rRNA gene for the identification of Enterobacteriaceae (65), Staphylococcus spp. (66), Streptococcus spp. and other aerobic Gram-positive species (50), Mycobacterium spp. (67), Leptospira spp. (68), and Corynebacterium spp. (51) as well as for the identification of spirochetes (69). The superoxide dismutase (SOD) gene also permits reliable identification of Streptococcus spp. (70, 71) and C. burnetii (72) to the species level; the sodA gene was particularly efficient at distinguishing different species among nonenterococcal group D streptococci (73). The Tuf gene has better discrimination for coagulase-negative Staphylococcus (74), and the gap gene could be an alternative for increasing the taxonomical distinction among Staphylococcus species (75). Molecular identification of intracellular bacteria could be based on the amplification of the specific gene used, usually for the detection of these bacteria by real-time PCR. The targets most often used for real-time PCR are the IS1111 intergenic sequence for C. burnetii, the 16S-23S rRNA intergenic region for Bartonella, a repeated sequence for T. whipplei, a hypothetical protein (RC0338 gene) for spotted fever group Rickettsia, and a hypothetical protein (RP278 gene) for typhus group Rickettsia. Sequencing of the 16S RNA gene might be required in some cases (18, 76, 77), if real-time PCR is noncontributive to bacterial identification. Other genes could be used for more precise and discriminant identification of intracellular bacteria, for example, rpoB for Bartonella species (78) or citrate synthase (gltA), ompA, or ompB for Rickettsia spp. (79).
The time required for bacterial identification using 16S rRNA amplification and sequencing is, at a minimum, 24 h, and this method is ∼100 times more expensive than MALDI-TOF MS identification (80). Molecular identification must remain the gold standard for bacterial identification, particularly when no spectra are available in the MALDI-TOF database. Sequencing will allow corroboration of the MALDI spectra and permit reliable additions to protein databases (80).
MALDI-TOF MS, a rapid and low-cost identification method.
For 10 to 20 years, automated phenotypic methods were rapidly developed in clinical microbiology laboratories (81). Nevertheless, MALDI-TOF MS, although developed for 3 decades, was originally used only anecdotally for bacterial identification (82–84). In 2009, Seng et al. were the first researchers to anticipate the ongoing revolution in routine clinical microbiology, correctly identifying 95.4% of 1,660 isolates tested, including 81.4% to the species level (42). van Veen et al. reported similar results, with efficient identification ranging from 84.8 to 97.7% at the species level (85). Bizzini et al. obtained 93.2% correct identifications at the species level for 1,278 species tested but with a protein extraction step for 25.6% of these isolates (86). Importantly, this revolution in bacterial identification has been the keystone in facilitating the design of broad culture studies, including microbial culturomics studies.
(i) Methods.
Three systems, the Andromas database (Andromas SAS, Paris, France), the Vitek-MS platform (bioMérieux, Marcy l'Etoile, France), and the Bruker Biotyper (Bruker Daltonics, Heidelberg, Germany, in collaboration with Becton Dickinson, Franklin Lakes, NJ, USA), are available for MALDI identification of microorganisms (81). The MALDI-TOF principle corresponds to a soft ionization mechanism, which is obtained by using a matrix, added to the bacterial colonies on metal plates. Ionization was performed by using a UV laser beam. The measurement of the time of flight into the tube to reach a detector generated spectra. Spectral comparison with data from the defined database available was automatically performed and permitted identification.
(ii) Effective bacterial identification by MALDI-TOF MS in clinical microbiology.
Coagulase-negative staphylococci and Staphylococcus aureus are generally well identified by MALDI-TOF MS (42). Dupont et al. (using sodA amplification and sequencing as a reference) and Carpaij et al. (using tuf amplification and sequencing as a reference), who tested 234 and 62 coagulase-negative Staphylococcus strains, respectively, reported that 93.2% to 100% of the strains were correctly identified compared with the reference molecular technique used (87, 88). All 132 of the tested Enterococcus species strains were correctly identified by Fang et al. by MALDI-TOF MS (89). Cherkaoui et al. demonstrated the superiority of MALDI-TOF MS compared with the Vitek-2 system coupled with agglutination tests in identifying 386 beta-hemolytic Streptococcus clinical strains. All the strains were accurately identified by MALDI-TOF MS, along with 39% to 92% of the isolates (90). Friedrichs et al. observed 100% concordant results in identifying viridians group streptococci by both MALDI-TOF and conventional methods (91). Nevertheless, discordant results were observed for the identification of nonenterococcal group D streptococci (92, 93). Neisseria meningitidis and Neisseria gonorrhoeae were accurately identified by MALDI-TOF MS, as reported by Ilina et al. (94). Overall, Enterobacteriaceae were accurately identified by MALDI-TOF MS (85). Degand et al. reported that 549 nonfermenting Gram-negative bacteria from clinical samples were well identified by MALDI-TOF MS, whereas 9 strains of the Burkholderia cepacia complex were not identified to the species level (95). The increasing identification allowed by this method was demonstrated by 2 studies performed with anaerobes in that same laboratory. In 2011, La Scola et al. identified 61% of 544 isolates by MALDI-TOF (96). In 2013, the same group, using the same cutoff, tested 1,325 anaerobes species, with accurate identification to the species level for 92.5% of the strains (97). Similar results were observed by Nagy et al., who correctly identified 77% of the 283 strains tested, and Fedorko et al., who correctly identified 82% of the 152 isolates, both using a cutoff of >2, the usually referred-to cutoff, whereas La Scola et al. used a cutoff of >1.9 (96–99).
(iii) Discordant results or remaining challenges in clinical microbiology.
Two primary difficulties with bacterial identification by MALDI-TOF MS have appeared.
(a) Still-limited databases.
For rare species, the lack of spectra decreases the safety of identification; however, this limitation can be easily surmounted. In fact, Verroken et al. (100) demonstrated that interest in an expanded database increased the accurate identification of Nocardia spp. In the first step, those authors identified 110 species of Nocardia. In the second step, after the implementation of the database with these 110 previously generated spectra, those authors identified 43 other strains, with the identification rate increasing at the genus and species levels from 44 to 88% and from 23 to 79%, respectively, using this optimized database. The feasibility of Mycobacterium species identification by MALDI-TOF MS was well established; nevertheless, incrementation of the database will be required to increase the correct identification rate (81). Using an original protocol, El Khechine et al. reported the accurate identification of >100 different strains of heat-inactivated Mycobacterium tuberculosis and M. avium (101). Couturier et al. showed that identification of Haemophilus, Actinobacillus, Cardiobacterium, Eikenella, and Kingella (HACEK) species was accurate at the genus level for 93 to 100% of isolates with a customized database including clinical spectra but that identification at the species level was efficient for only 66% of the isolates (102). In 2013, with an optimized database, Powell et al. identified a larger number of isolates to the species level with MALDI-TOF MS than with conventional phenotypic methods (86% and 60 to 77%, respectively) (103).
(b) Remaining challenges.
Despite the large databases available for certain genera, there are still discrepancies in the identification of some species sometimes frequently observed in clinical microbiology. One of the major challenges will be to differentiate Streptococcus pneumoniae from Streptococcus mitis by MALDI-TOF MS, which currently remains inaccurate (104). As another example, routinely used protocols do not allow the distinction of Bacillus anthracis and Bacillus cereus, although combination with artificial neural networks for spectral analysis increased this discrimination (105). Salmonella spp. were well identified to the genus level; however, progress should be observed to precisely and safely determine the subspecies and the serovar levels (106, 107). Different authors have shown that MALDI-TOF MS was unable to distinguish Escherichia coli from Shigella spp. (108, 109). Finally, Pavlovic et al. demonstrated that MALDI-TOF MS did not allow differentiation between different tested species of Enterobacter (110).
(iv) Rare and fastidious bacteria and archaea.
MALDI-TOF MS is an identification tool that has permitted the extension of biodiversity knowledge. In our laboratory, Seng et al. compared a period using conventional phenotypic tools, annually identifying a mean of 44 different species, and a period using MALDI-TOF MS, annually identifying a mean of 112 different species (80). Importantly, 77% of the bacterial species rarely reported to be human pathogens and phenotypically identified were found by MALDI-TOF MS (80). In addition, among the 128 rarely isolated pathogenic bacteria during this period, nearly 30% were already identified by MALDI-TOF MS, and the proportion of these isolates identified by molecular tools decreased significantly in the last few years (80) with permanent incrementation of MALDI-TOF databases. MALDI-TOF MS was applied for archaeal species identification. Dridi et al. reported that Methanobrevibacter smithii, Methanobrevibacter oralis, Methanosphaera stadtmanae, and the recently described Methanomassiliicoccus luminyensis were correctly identified (111). Other rare bacterial species were correctly identified, including Brucella spp. at the genus level (112) and Pasteurella spp. (113), Bartonella spp. (114), Francisella spp., Leptospira spp. (115, 116), and Legionella spp. (117, 118) at the species level.
(v) Time-effective and cost-effective method.
Mass spectrometry is a method requiring a low level of training and can identify bacterial strains in a few minutes (i.e., 6 min to 8 min 30 s with an AutoFlex II system [Bruker Daltonics] and only 1 min 46 s with the next-generation MicroFlex LT mass spectrometer [Bruker Daltonics, Heidelberg, Germany]) (119). One individual can easily test at least 1,000 different colonies per week, as in culturomics studies (4); this technology has revolutionized approaches in clinical microbiology, completely removing the time requirements of classic phenotypic identification (i.e., 5 to 8 h for the Vitek-2 system and 18 to 48 h for API system identification) (42, 80). Concerns over potential contamination of the culture or polymicrobial infection can be easily allayed, permitting both safe bacterial identification and antibiotic susceptibility testing (42, 119).
MALDI-TOF MS has permitted a dramatic reduction in identification costs. In fact, the identification cost of 1 bacterial colony was €1.35 to €1.43 per strain, whereas the costs of classic phenotypic identification and 16S rRNA sequencing were €4.6 to €12.65 per strain and €137.7 per strain, respectively, in a same laboratory (42, 80). Martiny et al. used a MALDI-TOF MS network, most likely showing the future of shared technical platforms (120). Over a 1-month period, 1,055 isolates were analyzed in a laboratory located 7.5 km away. The median time to identification was 5 h 11 min, which was faster than the identification performed in parallel using conventional techniques (120). In addition to sharing the costs to acquire a mass spectrometer, these networks permit the sharing of competence regarding updated software and reducing maintenance costs.
(vi) From clinical to culturomic studies.
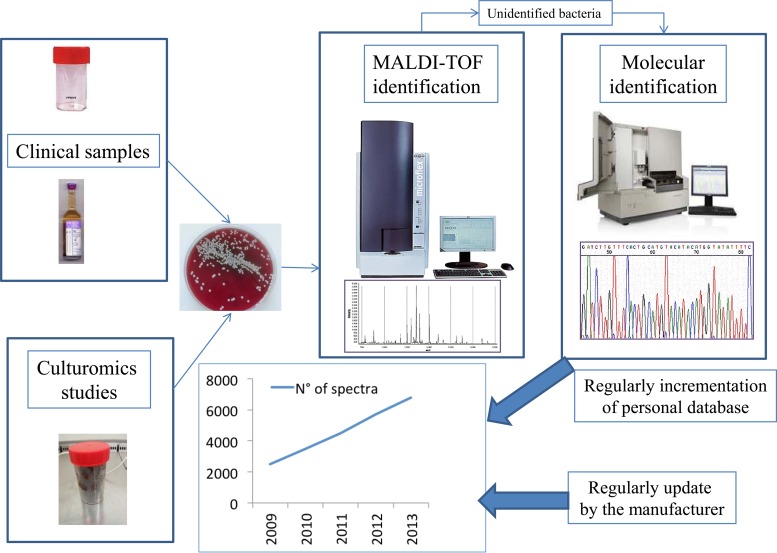
MALDI-TOF MS, when used for culturomics studies, highlighted that the incrementation of the spectral database also significantly decreased the use of molecular biology (Fig. 1) (80). Indeed, between current and previous stool samples tested in culturomics studies, we observed a decrease in the number of colonies required to perform 16S rRNA sequencing to obtain a correct identification (80). We tested 4.5 to 5% of colonies from stool samples of Senegalese, obese French, and anorexia nervosa patients by 16S rRNA sequencing (4, 6), whereas only a mean of 1.4% of the other stool samples recently tested required molecular identification (121).
FIG 1.
A MALDI-TOF incrementation database in a 4,000-bed university hospital in Marseille, France. The number of spectra has tripled in 3 years because we added the spectra of all the bacterial species identified by molecular tools that were cultured from routine laboratory samples and in culturomics studies.
Antibiotic Susceptibility Testing
Bacterial culture allowed antibiotic susceptibility testing that might have potential clinical impact. As an example, Whipple's disease was empirically treated by trimethoprim-sulfamethoxazole for a long time (122). The first culture of T. whipplei, which is the causative bacterium, revealed that this bacterium was naturally resistant to trimethoprim and that the combination of doxycycline and hydroxychloroquine was the sole bactericidal treatment (123). Clinical studies demonstrated a better outcome predicted by these in vitro results (124). In addition, reducing the culture time for M. tuberculosis is a hot topic with considerable consequences for clinical outcomes, as a worldwide outbreak of multidrug-resistant tuberculosis is ongoing (125). As another example, the now possible culture of anaerobic species, including most methanogenic bacteria, aerobically due to the addition of antioxidants, suggests a great potential to more easily test antibiotic susceptibility in all bacteriology laboratories (126). Finally, the rapid emergence of outbreaks of extended-spectrum beta-lactamases (ESBLs) and carbapenemases in Gram-negative bacteria will be a challenge for their rapid detection in routine laboratories to manage such spread. Recently, we demonstrated as a proof of concept that real-time video imaging coupled with bacterial culture allowed the detection of such resistance in 3 to 4 h (J. M. Rolain and D. Raoult, unpublished data).
ENVIRONMENTAL MICROBIOLOGY AS A SOURCE OF MEDIA FOR CLINICAL MICROBIOLOGY
Empirical Approach
In the environment, it is believed that only 1% of bacterial species are cultivable by using current techniques (8). Among these species, more than half of the known phyla have not yet been isolated in culture (127, 128). Environmental microbiologists have been pioneers in resolving the great plate count anomaly, explaining why only a small fraction of this diverse population has been grown in artificial media (129, 130). The bacterial cultures obtained by successive dilutions of the original population, to near extinction of the ability to grow, are named dilution cultures. These techniques were originally used by environmental microbiologists to explore minority populations (131, 132). These scientists were also pioneers in revitalizing techniques, facilitating the mimicking of the natural environment to increase the proportions of cultured bacterial species (129, 133). The most famous example of empirical culture was recently observed. SAR11, which is an alphaproteobacterial clade representing, in some regions, 26 to 50% of all rRNA genes in the surface water of the oceans, remained uncultivated until recently (134). Because of an empirical approach using sterile Oregon seawater with a mixture of defined organic and inorganic components, one of the most common bacteria was cultured and named “Candidatus Pelagibacter ubique” (135). The authors of that study suggested that this bacterium plays a central role in the recycling of dissolved organic carbon to CO2 in the ocean (136).
Diffusion Chamber and ichip
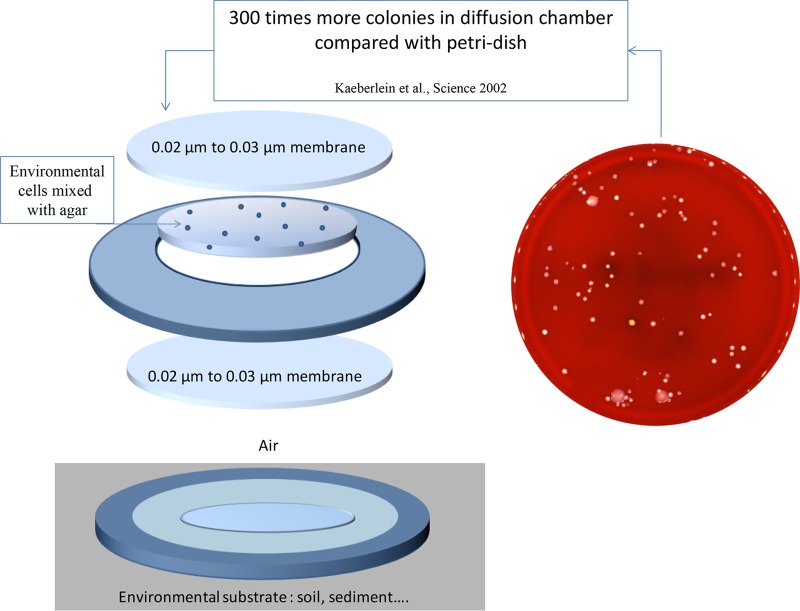
Chemical compounds enabling increases in the numbers of bacterial species that can be cultured remain difficult to characterize; however, several authors have applied an empirical approach by using a “diffusion chamber” (137). Kaeberlein et al. introduced a method of in situ cultivation that bypassed the difficulties of replicating the natural environment caused by usual petri dish-based approaches (8). The authors placed bacteria in a diffusion chamber that was then introduced back into the environment from which the sample originated. The diffusion chamber was composed of a mix of agar and a diluted environmental sample, which was sandwiched between two membranes glued onto a washer (Fig. 2). This diffusion chamber permitted the cultivation of up to 40% of bacterial cells from a marine sediment environment, compared to 0.05% that grew on a petri dish. A parallel can be drawn to bacteria that are able to grow only in satellitism with another bacterial species. Among the most famous examples of microcolonies growing in satellitism around another bacterial species include Haemophilus influenzae growing around S. aureus secreting V factor (NAD) (138) or Helcococcus ovis (139).
FIG 2.
Contribution of environmental microbiologists to improving bacterial culture due to diffusion chambers, allowing the number of colonies cultured to increase 300 times by mimicking the natural environment (8).
For culturing of 3 different environment samples, Aoi et al. compared the performance of standard cultures with that of a system using a hollow-fiber porous membrane chamber, permitting the rapid exchange of chemical compounds from the natural environment. A total of 10 to 48% of the isolates were potentially new bacterial species identified by using the diffusion chamber, compared to 0 to 4% by using standard culture (140). Gavrish et al. used a trap formed by two semipermeable membranes (0.2- to 0.6-μm pore size) glue dried and then placed onto moist soil. Most of the bacteria isolated by the trap were Actinomycetes, including members of rare groups such as Dactylosporangium, Catellatospora, Catenulispora, Lentzea, and Streptacidiphilus (141). Ferrari et al. proposed a protocol using a polycarbonate membrane as support for growth and a soil extract as a growing promoter coupled with FISH for the rapid visualization of microcolonies (142).
Nichols et al. designed an isolation chip (ichip) for high-throughput bacterial cultivation, consisting of several hundred miniature diffusion chambers, each inoculated with a single environmental cell. This technique allowed these authors to demonstrate that the microbial repertoire of the ichip exceeded the repertoire afforded by standard cultivation by manyfold, particularly for Deltaproteobacteria. In addition, the species isolated by using the ichip method had significant phylogenetic novelty (143).
Signaling Compounds
Quorum sensing is important for interspecies competition in complex ecosystems (144). Chandler et al. demonstrated this fact recently in a coculture model using the soil saprophytes Burkholderia thailandensis and Chromobacterium violaceum (145). Bruns et al. demonstrated that signaling compounds were able to trigger microbial growth (146). The use of cyclic AMP (cAMP), N-butyryl homoserine lactone, or N-oxohexanoyl-dl-homoserine lactone at a low concentration of 10 μM permitted an increase in the microbial recovery of heterotrophic bacteria from the central Baltic Sea (146). The concept of mutualism between bacterial species emerged and was significantly highlighted by the role of siderophores (147–149). D'Onofrio et al. identified 5 new acyl-desferrioxamine siderophores isolated from Micrococcus luteus KLE1011, permitting the culture of a substantial number of marine bacterial species previously considered to be uncultivable (147).
Evidence that small signaling molecules, such as short peptides, might be essential factors in initiating the growth of nongrowing cells has been demonstrated (150, 151). For culture of Psychrobacter sp. strain MSC33, one 5-amino-acid peptide, LQPEV, induced the otherwise “uncultivable” strain to grow in standard media (150). This finding illustrates that deficiencies in nutrient composition and concentrations in standard media provide remarkable opportunities for access to culture for some of these species through identification of the signaling compounds required for growth and their addition to standard medium formulations (8, 150).
Recently, a model of the microbial life cycle was proposed by Epstein (152). Dormancy is used by bacteria, for example, spore-forming bacteria, to survive (153). Epstein hypothesized that dormant bacteria (named scout) would awaken stochastically, without implications of external and environmental signals (152). If these hypotheses provided partially microbiological explanations, including the existence of viable but nonculturable cells (153), then extremely long-term incubations (>3 months) would not increase the proportion of new species isolation (154). The results of these experiments strongly suggest that the success in culturing of new species depends on the amount of cultivation effort rather than on the length of incubation. Those authors concluded that the single-cell approach allows the recovery of many more colonies from the same inoculum, as does cultivation by dilution to extinction (154).
From Environmental to Clinical Microbiology
ichip in clinical microbiology.
Recently, Sizova et al. extended the cultured bacterial repertoire of the human oral cavity specifically as a result of the growth of anaerobic species (155). Those authors concomitantly developed in vivo culture methods to specifically enrich for species belonging to the oral microbiota (minitrap method), single-cell long-term cultivation to decrease the effects of fast-growing microorganisms, and modifications of usual enrichment techniques, notably the use of culture conditions without sugar. Minitrap enrichment was the best method. The complementarity among the different methods was highlighted because none of the species was isolated concomitantly by the three methods (155). In addition, 10 different bacterial strains that were previously detected only by molecular tools, including three new microbial genera, were cultured (155).
The example of Akkermansia muciniphila.
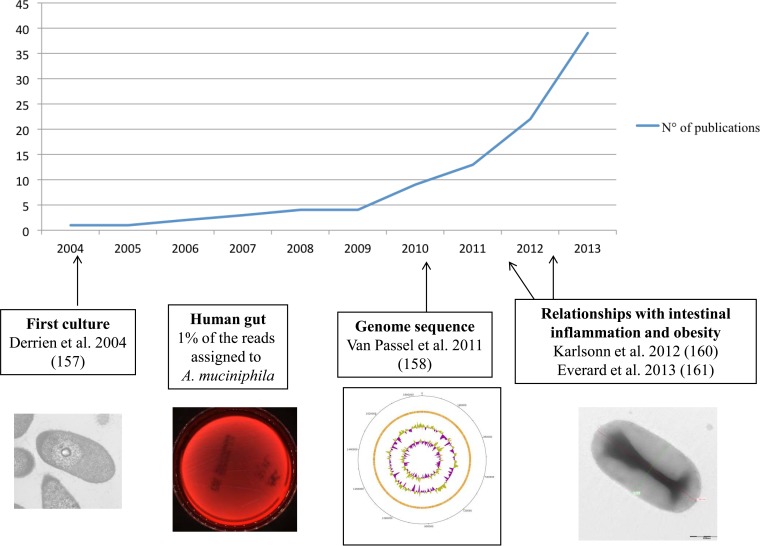
The phylum Verrucomicrobia includes primarily environmental bacterial species (156). In 2004, Derrien et al. cultured Akkermansia muciniphila gen. nov., sp. nov., which is a mucin-degrading bacterium in the human gut, by using dilution of stool samples to extinction in an anaerobic medium containing gastric mucin as the sole carbon and nitrogen source (157) and then sequenced its genome (158). Using a specific 16S rRNA-targeted probe, that same team revealed that this species represented an average of 1% of the bacteria in the human gut, and using pyrosequencing, Dubourg et al. recently showed that this species may represent 44 to 85% of the sequences in patients under heavy antibiotic treatment (159). Finally, this species was detected in lower concentrations in obese subjects than in controls (160), and an experimental study in mice recently showed that the use of A. muciniphila could be protective against obesity and its associated metabolic disorders (161) (Fig. 3). This bacterial species represents a recent example of the rapid extension of knowledge following pure culture of microorganisms such as T. whipplei, where a comprehensive understanding of T. whipplei infection was acquired only after the first culture (122, 162).
FIG 3.
Brief history of Akkermansia muciniphila (2004 to 2014) from its first culture to its relationships with humans, including its suggested role in obesity (157, 158, 160, 161).
Future challenges.
(i) Halophilic bacteria.
Halophilic microorganisms are encountered in two domains of life: Bacteria and Archaea (163, 164). The former domain contains primarily low-halophilic bacteria (marine) or moderately and some extremely halophilic bacteria, which are limited to a few species of strictly fermentative and photosynthetic purple bacteria (165). Originally, halophilic organisms held little interest except for their development in salty foods (166). Today, these organisms are used for the expansion of fundamental knowledge regarding life in extreme environments, biodiversity, and their particular adaptive physiology as well as knowledge applied in the search for enzymes with special properties (167). In the environment, their development is limited to hypersaline environments, with the consequences of significant evaporation of salt water in regions where drought conditions and the accumulation of salts are optimal (165). Only halophilic Archaea have been detected in the human digestive tract. Indeed, DNA sequences from halophilic Archaea belonging to the Halobacteriales were recently detected in intestinal biopsy specimens from patients who ingested a salt solution before a colonoscopic examination (166). Currently, human halophilic bacteria remain poorly studied, and their DNAs have never been detected in the human digestive tract to the best of our knowledge. This lack of detection could be due to the detection systems and to culture conditions that target only halophilic Archaea and that discriminate against halophilic bacteria. A future challenge would be to grow such bacteria from the human gut.
(ii) Planctomycetes.
The first axenic culture of a planctomycete species was achieved in 1973, and the species was renamed Pirellula staleyi in 1987 (168). In 2001, the phylum Planctomycetes was proposed, and in 2006, the superphyla Planctomycetes, Verrucomicrobia, and Chlamydiae were proposed (169, 170). These bacterial species represent examples of a small number of microorganisms isolated in pure culture (2%) compared with the clone sequences detected by molecular techniques. Schlesner demonstrated that Planctomycetes species required low-carbon and -nitrogen sources compared with the fast overgrowth of other bacterial species of complex ecosystems using N-acetylglucosamine (171). Because of the absence of the peptidoglycan responsible for β-lactam resistance in Planctomycetes (172), the use of these antibiotics also facilitates the fight against fast-growing bacterial species, thus assisting in pure Planctomycetes culture (169, 173). Davis et al. found that extending the incubation time improved the culture of Planctomycetes, including previously uncultured species within the WPS-1 lineage (174). Using a complex culture medium including aqueous extracts of spring sediments and rumen fluid, Elshahed et al. facilitated culturing of the Pirellula-Rhodopirellula-Blastopirellula clade (175). Nevertheless, to the best of our knowledge, no Planctomycetes pure cultures from human samples have been achieved to date. However, Planctomycetes species have been detected in the human gut by both pyrosequencing and specific PCR (176, 177). This culturing will be a challenge for future culturomic studies, and recent studies regarding possible identification by MALDI-TOF MS should offer broad perspectives (178).
In conclusion, based on these examples, significant progress in clinical microbiology has been made by designing specific devices that permit pure culture of Planctomycetes or extremophile bacterial species, such as halophilic bacteria, to be obtained from human samples. Based on the model used by Sizova et al. to study oral microbiota (155), diffusion chambers could be used to study human stool samples or, more interestingly, duodenal samples (179). Broad use of gastric mucin will also permit a comprehensive exploration of the diversity of Verrucomicrobia from human samples.
CULTUROMICS: THE EXAMPLE OF HUMAN GUT MICROBIOTA
Historical
Culture was the first tool for the exploration of the digestive bacterial ecosystem (180–182). Stool samples consist of between 1011 and 1012 bacteria/g of feces by traditional methods (181), corresponding to 60% of the dry mass of feces (183). The gastrointestinal microbiome consists primarily of bacteria belonging to 4 phyla (Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria) (184). Most of the species isolated during the 1970s by culture belonged to the Enterobacteriaceae and Veillonellaceae families, and before molecular methods were developed, the number of bacterial species of the gastrointestinal microbiota was estimated to be ∼400 (185).
Interestingly, relationships among gut composition, diet, and geographic provenance have been observed for a long time (181, 186–188). Differences in fecal flora were observed among individuals living in Uganda and British individuals eating a Western diet (189). Based on these culture studies, Hill and Drasar proposed a normal colonic flora composition overwhelmingly predominated by non-spore-forming, anaerobic, rod-like organisms (190).
Since the advent of metagenomics, molecular tools have supplanted culture techniques, which were both time-consuming and difficult (3). Nevertheless, some studies suggested a potentially imperfect overlap between culture-dependent and culture-independent studies. Hayashi et al., comparing the gastrointestinal microbiota by cloning and sequencing versus anaerobic culture (191), obtained between 48 and 65 phylotypes for each individual by cloning and 48 species for 3 individuals, including 3 potentially new species, by culture. In a previous study, of the 48 species, ∼50% were detected only by cloning, 20% were identified by both techniques, and 30% were identified only by culture (192). A gap between molecular studies including pyrosequencing of the 16S rRNA gene and direct observation (16) was highlighted by Hugon et al., who emphasized that pyrosequencing neglected nearly 15% of apparently Gram-negative prokaryotes (16). The team of Jeffrey Gordon also demonstrated that culturing of a large repertoire of an individual's gut microbiota by using straightforward anaerobic culturing conditions was possible (193). These findings have motivated the design of large culture studies to complete the identification of the bacterial human gut repertoire.
Microbial Culturomics
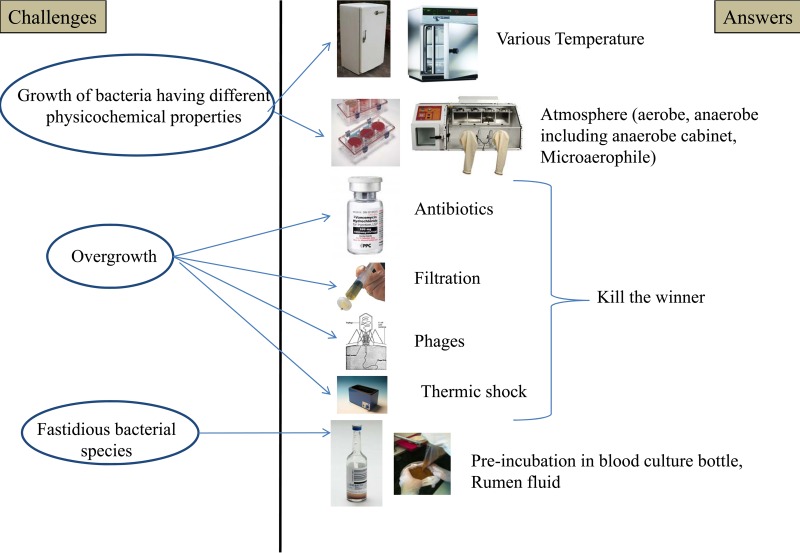
For 3 years, our group used culturing techniques to demonstrate that culture was not less effective than pyrosequencing for studying the gut microbiota repertoire (4, 6, 7). In this section, we summarize this approach and our primary findings. The different challenges that have been identified and their answers are summarized in Fig. 4. Lagier et al. applied culturomics to stool samples from 2 rural Senegalese subjects and from 1 obese French subject under different culture conditions (4). Pfleiderer et al. designed supplementary culture conditions and applied the 70 best culture conditions to a stool sample from an anorexia nervosa patient (6), and Dubourg et al. applied these conditions to samples from patients treated with antibiotics (7). Colony identification was performed by mass spectrometry (MALDI-TOF), permitting rapid and effective identification (119) (see above). The colonies not identified by MALDI-TOF MS were tested by 16S rRNA or rpoB amplification and sequencing (4–7).
FIG 4.
Challenges for culturomics and specific answers, including the techniques used to limit the overgrowth of common bacteria and to increase the growth of fastidious bacteria.
Nonselective media.
Based on the model proposed by environmental microbiologists, the effects of various growth media, atmospheres, and times of incubation were first tested (174). First, nonselective commercial culture media, such as 5% sheep blood agar, were used (4). Each gram of stool was diluted in 9 ml of Dulbecco's phosphate-buffered saline (DPBS) and was inoculated into different culture media under various conditions in a dilution series ranging from 1/10 to 1/1010. Different atmospheric conditions (aerobic conditions, aerobic conditions with 2.5% CO2 or 5% CO2, microaerophilic conditions, and anaerobic conditions) and temperatures ranging from 4°C to 55°C were tested (10). The incubation time ranged from 24 h to 2 months to identify slow-growing bacterial species by using wet compresses in a jar to fight against agar dryness. After 24 and 48 h and then once per week after the initial seeding, each agar plate was observed, and new colonies were identified (4). By applying only these routine culture conditions, 4 new bacterial species were cultured in the first work (4). Nevertheless, the high concentration of bacteria in the human gut limited the contribution of nonselective media. Indeed, the plates were frequently overloaded despite serial dilutions, and supplementary strategies had to be designed.
Selective culture conditions.
Next, the aim was to select and identify specifically the minority bacterial populations that were not detected by pyrosequencing due to low threshold concentrations (≤105 bacteria/g) (3). Indeed, diverse antibiotics or nonantibiotic inhibitors were used to identify the species that were present in smaller quantities in a strategy that we named “kill the winner,” which was previously used in a model for population dynamics of phage bacteria.
(i) Antibiotics and other inhibitors.
Gram-negative bacterial growth was inhibited by the addition of colimycin (50 to 100 μg/ml) or kanamycin (10 μg/ml), and Gram-positive bacterial growth was limited by the addition of vancomycin (10 to 50 μg/ml) (4). Squalamine (5 to 15 μg/ml), which is a natural aminosterol isolated from the dogfish shark and which presents a large spectrum of antimicrobial activities (194), was tested. Diverse inhibitors usually used in clinical microbiology to facilitate the identification of enterobacteria were tested, such as bile extract, eosin, methylene blue, sodium citrate, and sodium thiosulfate (19).
(ii) Active and passive filtration.
To decrease the bacterial load and to detect bacteria present in low concentrations, syringe filters with successive pore sizes from 5 to 0.2 μm were used. After each successive pore size, the filtrate was inoculated in 5% sheep blood agar and brain heart infusion agar at 37°C in aerobic and anaerobic atmospheres, respectively (4). Lagier et al. selected cells based on their motility. The initial aim was to detect spirochetes from human stool samples, as previously reported (195). A similar technique was also recently used to isolate Campylobacter fetus from bovine preputial samples (196). Passive filtration to select motile species using companion plate and cell culture inserts with 0.4-μm membranes was also used. The diluted stool with broth was placed on one side of the membrane, and sterile broth (Leptospira broth or modified Barbour-Stonner-Kelly medium [BSK-H]) was placed on the other side. Every day, the supernatant was observed by using a dark-field microscope to detect the eventual presence of spirochetes. In cases of contamination, the supernatant used was inoculated in 5% sheep blood agar using different growth atmospheres. Among the motile species detected by this method, 2 new bacterial species (Herbaspirillum massiliense and Cellulomonas massiliensis) were cultured (4, 197, 198).
(iii) Heat shock.
Metagenomic studies of the human gut bacterial repertoire revealed that many species were strictly anaerobes and belonged to the Clostridiaceae family (6). Most of these species sporulated. To select these bacteria specifically, heat shock was applied to the studied stool samples, as previously reported for Clostridium spp. and Bacillus spp. (199, 200). In the first culturomics work, 2 different protocols (65°C for 20 min and 80°C for 20 min) were tested. Recently, new strategies were developed, adding heat shock for a longer time (1 h), followed immediately by inoculation of the diluted stool sample in a blood culture bottle (121).
(iv) Phages.
Bacteriophages are bacterium-specific viruses that infect and, in the case of lytic phages, destroy their host bacteria. Phage therapy was previously used therapeutically in humans (201) and to eliminate the contamination of food by food-borne bacterial pathogens (202). Bacteriophages were previously used by Subramanyam et al. to decontaminate sputum samples instead of using antiseptics and/or antibiotics before performance of Mycobacterium species culture (203, 204). The use of T1, T4, and phiX174-like E. coli phages was initially undertaken to decrease the number of E. coli colonies adhering to the urothelium and causing persistent urinary tract infections (205).
As previously reported for environmental cultures with fast-growing species, for one of the first studies of stool specimens by culturomics, petri dishes were overloaded with E. coli, particularly for enterobacterium-selective media (4). To decrease the growth of E. coli from these stool samples, T1 and T4 lytic phages were used as previously reported (205). A 50% decrease in bacterial growth was observed in the petri dishes, permitting the identification of one new species (Enterobacter massiliensis) (4, 206).
Enrichment conditions.
(i) Inoculation of stool samples in blood culture bottles.
The components included in blood culture bottles have increased the growth of certain bacteria, such as Kingella kingae, which is the first arthritis-causing bacterium identified in young children (207, 208). In the past, this preincubation also permitted culturing of a new strictly anaerobic bacterial genus (Phocaeicola abscessus), which was isolated from a human brain abscess sample (209). To increase the growth of bacterial species from the human gut, particularly anaerobic bacteria, Lagier et al. inoculated stool specimens in both aerobic and anaerobic blood culture bottles for 1, 5, 10, 14, 21, 26, and 30 days before inoculating the broth onto different agar media. The dramatic efficiency of this blood culture bottle preincubation was highlighted by 50 of 91 new bacterial species being isolated by this technique (4, 6, 7, 121).
(ii) Addition of stool extract.
Fresh stool extracts were previously used extensively as growth promoters for culturing Archaea including Methanobacterium ruminantium (210), anaerobic species from the human gut (211) or from adult swine feces (212), and E. coli (213). Goodman et al. also used fresh stool extracts to improve techniques for high-throughput anaerobic culturing of samples from the human gut (193).
In culturomics studies, a mixture was prepared from several different volunteer donor stool samples. First, this mixture was lyophilized and then ground by using a mortar and pestle. The resulting powder was resuspended in sterile water (2-g pellet in 100 ml of water) and then centrifuged at low speed (5,000 × g for 12 min) to remove cell debris. The recovered supernatant underwent several treatments: a first sonication (50 μm/4 min and then 70 μm/2 min) was performed by using the Qsonica sonicator apparatus, followed by treatment in a French press and, finally, a second sonication (50 μm/3 min and then 70 μm/1 min). The obtained stool solution was filtered to 0.2 μm. The final culture media were prepared with 20% and 50% stool filtrate, agar, and antibiotics (50 μg/ml vancomycin and 50 μg/ml colimycin) (121).
(iii) Addition of rumen fluid.
To apply the model developed by environmental microbiologists, consisting of mimicking the natural environment of the bacteria to facilitate their growth (8), culturomics studies were performed by using the nutrient properties of rumen fluid (193). Originally described as a growth promoter for spirochetes (214), including Treponema hyodysenteriae and Treponema innocens (215), Elshahed et al. used rumen fluid in a complex medium, permitting the culturing of 2 new bacterial species belonging to the Pirellula-Rhodopirellula-Blastopirellula clade within the Planctomycetes phylum (175). Nottingham and Hungate also used rumen fluid for the growth of archaeal species (210). Rumen fluid was also used to isolate cellulolytic Bacteroides species (216) or to extend the bacterial repertoire in the human gut by the team of J. I. Gordon (217).
Sixteen different digestives contents from sheep paunches were used to prepare the rumen fluid. After a first filtration in a funnel, the suspension was centrifuged (10,000 rpm for 90 min). The supernatant was then collected, and after 3 successive filtrations using filters with 0.8-μm, 0.45-μm, and 0.2-μm pore sizes, respectively, rumen fluid was obtained (4). Next, the rumen fluid was stored at −80°C and primarily used by adding 5 ml to a blood culture bottle. In parallel, the inoculation of only rumen fluid in a blood culture bottle served as a negative control to verify the sterility of the nutrient (4). This substrate permitted the culturing of 17 different bacterial strains not detected by classic culture techniques in the first culturomics work. Overall, among the 91 new bacterial species identified by culturomics, 30 were identified by using rumen fluid as a specific nutrient.
(iv) Lipid addition.
Some bacteria, such as Mycoplasma spp., require the addition of fatty acids for their growth (218). Stool samples were diluted 1:10 in phosphate-buffered saline (PBS) and then inoculated with 6 ml of 20% Medialipid (B-Braun Medical SA, Boulogne Billancourt, France), which is a compound rich in medium-chain triglycerides and soy oil, in anaerobic blood culture bottles for 2 to 60 days. The broth was then inoculated in 5% sheep blood agar under anaerobic conditions at 37°C. Using this nutrient, Bacillus okuhidensis was first isolated in humans.
(v) Ascorbic acid addition.
Ascorbic acid has been used as a growth factor for Spirochaeta gallinarum, Sarcina flava, Staphylococcus aureus, and several anaerobic species (219). To analyze these properties, one stool sample was inoculated in a blood culture bottle with rumen fluid and 500 μg/ml of ascorbic acid (B. La Scola and D. Raoult, patent pending). In addition, the same concentration of ascorbic acid was added to 5% sheep blood agar. This strategy permitted the culturing of Enterococcus canintestini, which was first isolated in the human gut (121).
Evolution of culturomics.
(i) From 212 to 70 culture conditions.
Lagier et al. analyzed 212 different culture conditions (4) and identified 340 new bacterial species, including 174 bacterial species first detected in the human gut. Nevertheless, a refined analysis of the results showed that the 20 more effective culture conditions permitted the identification of 73% of the 340 bacteria cultured in their study, in at least 1 of the 3 analyzed stool samples (4). In addition, all the bacteria identified in the first study were cultured at least once using one of the 70 culture conditions (4) and were used for the following studies with various stool samples. Thus far, between 3,000 and >34,000 different colonies were analyzed in each stool sample (5–7, 121). With 14 stool samples already reported and completely analyzed by culturomics (4, 5, 7, 121), with a large study using a few conditions applied to 347 stool samples (220), and with the analysis of >170,000 colonies by MALDI-TOF MS, 559 bacterial species were cultured, including 281 species from the Firmicutes phylum, 135 from the Actinobacteria phylum, 82 from the Proteobacteria phylum, 52 from the Bacteroidetes phylum, 6 from the Fusobacterium phylum, 2 from the Synergistetes phylum, and 1 from the Deinococcus-Thermus phylum (4–6, 121). Among these species, 304 bacterial species were first described in the human gut, including 59 new bacterial species (4-7, 121). In addition to the ongoing projects, culturomics has permitted the identification of 717 different bacterial species, including 91 new bacteria, 168 species first isolated in humans, and 155 species already described in humans but first isolated from the gut.
(ii) Reducing the number of culture conditions.
Reducing the workload will be the first objective to standardize culturomics. Currently, culturomics requires a large workforce and involves a considerable workload for the operators (4–7). Therefore, to standardize culturomics, the number of culture conditions used must be reduced. Preliminary results selected 18 different culture conditions to standardize culturomics (Table 2). These conditions were used to monitor bacterial cultures in a liquid medium over a period of 30 to 40 days, with subcultures on solid medium every 3 days (S. Khelaifia and D. Raoult, unpublished data). Our objective is to analyze 12,000 colonies for each stool sample because the number of supplementary bacterial species isolated for each sample decreased significantly when this number of colonies was analyzed (121).
TABLE 2.
Description of the 18 different culture conditions used for culturomics standardization
| Culture conditions for culturomics standardization |
|---|
| Preincubation in aerobic blood culture bottle with rumen fluid and then 5% sheep blood agar under aerobic conditions at 37°C |
| Preincubation in anaerobic blood culture bottle with rumen fluid and then 5% sheep blood agar under anaerobic conditions at 37°C |
| Preincubation in anaerobic blood culture bottle and then 5% sheep blood agar under anaerobic conditions at 37°C |
| Preincubation under aerobic conditions in Trypticase soy broth and then 5% sheep blood agar under aerobic conditions at 37°C |
| Preincubation under anaerobic conditions in 5% sheep blood broth and then 5% sheep blood agar under anaerobic conditions at 28°C |
| Preincubation under aerobic conditions in 5% sheep blood broth and then 5% sheep blood agar under aerobic conditions at 28°C |
| Preincubation under anaerobic conditions in 5% sheep blood broth and then 5% sheep blood agar under anaerobic conditions at 37°C |
| Preincubation under aerobic conditions in 5% sheep blood broth and then 5% sheep blood agar under aerobic conditions at 37°C |
| Preincubation in anaerobic blood culture bottle with stool filtered at 5 μm and then 5% sheep blood agar under anaerobic conditions at 37°C |
| Preincubation in aerobic blood culture bottle with stool filtered at 5 μm and then 5% sheep blood agar under aerobic conditions at 37°C |
| Preincubation in aerobic blood culture bottle with 5 ml sheep blood and then 5% sheep blood agar under aerobic conditions at 37°C |
| Preincubation in anaerobic blood culture bottle with 5 ml sheep blood and then 5% sheep blood agar under anaerobic conditions at 37°C |
| Preincubation in anaerobic blood culture bottle after thermic shock at 80°C during 20 min and then 5% sheep blood agar under anaerobic conditions at 37°C |
| Preincubation in anaerobic blood culture bottle with 5 ml rumen fluid and sheep blood and then 5% sheep blood agar under anaerobic conditions at 37°C |
| Preincubation in aerobic blood culture bottle with 5 ml rumen fluid and sheep blood and then 5% sheep blood agar under aerobic conditions at 37°C |
| Preincubation under aerobic conditions in brain heart infusion broth with 5% sheep blood and then 5% sheep blood agar under aerobic conditions at 37°C |
| Preincubation under anaerobic conditions in marine broth and then 5% sheep blood agar under anaerobic conditions at 37°C |
| Preincubation in aerobic marine broth and then 5% sheep blood agar under aerobic conditions at 37°C |
(iii) Use of culturomics on many samples.
Recently, Samb-Ba et al. performed culturomics (under selection of the more effective culture conditions) with identification by MALDI-TOF MS for 347 different stool samples from Senegalese subjects with or without diarrhea (220). Patients with diarrhea had a relative decrease in diversity, particularly for commensal bacterial species, including E. coli, several Enterococcus spp., and anaerobes. This fundamental study is the first example of the application of culturomics to a large number of samples.
New bacterial species.
Overall, 91 new bacterial species were cultured, including 58 from the Firmicutes phylum, 21 from Actinobacteria, 8 from the Bacteroidetes phylum, and 4 from the Proteobacteria phylum (Table 3) (197, 198, 206, 221–247). In an effort to evaluate the efficiency of culturomics in identifying new bacterial species from the human gut, we compared our results with the number of bacterial species identified in the human gut and validated by the International Journal of Systematic and Evolutionary Microbiology (IJSEM) (http://www.bacterio.cict.fr/). Consequently, in 3 years, culturomics has identified 91 of the 121 new bacterial species (75%) cultured in the gut by the rest of the world.
TABLE 3.
New bacterial species and genera isolated from human gut by culturomics and culture conditions used, 2011 to 2014
| Phylum | New bacterial species | Stool sample typea | Culture methodc | Atmosphere | Temp (°C) | Reference(s) |
|---|---|---|---|---|---|---|
| Firmicutes | Oceanobacillus massiliensis | A | Active filtration (0.45-μm filter) and then inoculation in brain heart infusion agar with 5% sheep blood | Aerobic | 37 | 4, 244 |
| Bacillus timonensis | A | Brain heart infusion agar with 5% sheep blood | Aerobic | 37 | 4, 223 | |
| Kurthia massiliensis | B | CNA agar | Aerobic, 2.5% CO2 | 37 | 4, 232 | |
| Kurthia senegalensis | B | Active filtration (1.2-μm filter) and then inoculation in 5% sheep blood agar | Aerobic | 37 | 4 | |
| Kurthia timonensis | B | Haemophilus test medium | Aerobic, 2.5% CO2 | 37 | 4 | |
| Anaerococcus senegalensis | B | Brucella agar | Anaerobic | 37 | 4, 226 | |
| Paenibacillus senegalensis | B | Schaedler agar + kanamycin and vancomycin | Aerobic | 37 | 4, 230 | |
| Bacillus massiliosenegalensis | B | 5% sheep blood agar | Aerobic | 28 | 4, 239 | |
| Clostridium senegalense | B | Inoculation for 5 days in blood culture bottle with sheep blood and then 5% sheep blood agar | Anaerobic | 37 | 4, 228 | |
| Peptoniphilus senegalensis | B | Inoculation for 10 days in blood culture bottle with sheep blood and then 5% sheep blood agar | Anaerobic | 37 | 4, 221 | |
| Peptoniphilus timonensis | B | Inoculation for 14 days in blood culture bottle with rumen fluid and sheep blood and then 5% sheep blood agar | Anaerobic | 37 | 4, 229 | |
| Ruminococcus massiliensis | B | Inoculation for 14 days in blood culture bottle with rumen fluid and sheep blood and then 5% sheep blood agar | Anaerobic | 37 | 4b | |
| Dielma fastidiosa | B | Inoculation for 10 days in blood culture bottle and then brain heart infusion agar | Anaerobic | 37 | 4, 237 | |
| Anaerococcus obesiensis | C | Inoculation for 5 days in blood culture bottle with thioglycolate and then 5% sheep blood agar | Anaerobic | 37 | 4 | |
| Brevibacillus massiliensis | C | M17 agar | Aerobic | 37 | 4, 233 | |
| Peptoniphilus grossensis | C | Inoculation for 26 days in blood culture bottle with rumen fluid and sheep blood and then 5% sheep blood agar | Anaerobic | 37 | 4, 247 | |
| Peptoniphilus obesi | C | Inoculation for 26 days in blood culture bottle with rumen fluid and sheep blood and then 5% sheep blood agar | Anaerobic | 37 | 4, 222 | |
| Kallipyga massiliensis | C | Inoculation for 26 days in blood culture bottle with rumen fluid and sheep blood and then 5% sheep blood agar | Anaerobic | 37 | 4, 241 | |
| Paenibacillus antibioticophila | D | Inoculation for 21 days in blood culture bottle with cow rumen and sheep blood and then 5% sheep blood agar | Aerobic | 37 | 7 | |
| Pytheasella massiliensis | F | 5% sheep blood agar during 14 days | Aerobic | 28 | 7 | |
| Paenibacillus reamassiliensis | H | Inoculation for 14 days in blood culture bottle with rumen fluid and sheep blood and then 5% sheep blood agar | Aerobic | 37 | 7 | |
| Soleaferrea massiliensis | E | Inoculation in blood culture bottle and then 5% sheep blood agar | Anaerobic | 37 | 6 | |
| Stoquefichus massiliensis | E | 5% sheep blood agar | Anaerobic | 28 | 6 | |
| Dorea massiliensis | E | Inoculation in blood culture bottle with rumen fluid and then 5% sheep blood agar | Anaerobic | 37 | 6 | |
| Holdemania massiliensis | E | Inoculation in blood culture bottle with thioglycolate and then 5% sheep blood agar | Anaerobic | 37 | 6, 246 | |
| Clostridium ihumii | E | Inoculation in blood culture bottle with sheep blood and then 5% sheep blood agar | Anaerobic | 37 | 6 | |
| Bacillus massilioanorexius | E | Inoculation in blood culture bottle for 1 mo with rumen fluid and then 5% sheep blood agar | Anaerobic | 37 | 6, 240 | |
| Clostridium polynesiense | I | Active filtration (0.45-μm filter) and then inoculation in 5% sheep blood agar | Anaerobic | 37 | Our unpublished data | |
| Bacillus saudii | K | Inoculation in blood culture bottle with sheep blood and then 5% sheep blood agar | Aerobic | 37 | Our unpublished data | |
| Polynesia massiliensis | I | Inoculation for 5 days in blood culture bottle and then inoculation in 5% sheep blood agar | Anaerobic | 37 | Our unpublished data | |
| Bacillus jeddahense | K | Inoculation in blood culture bottle with rumen fluid and sheep blood and then 5% sheep blood agar | Aerobic | 37 | Our unpublished data | |
| Clostridium saudii | K | 5% sheep blood agar | Anaerobic cabinet | 37 | Our unpublished data | |
| Clostridium jeddahense | K | Inoculation in blood culture bottle with rumen fluid and then 5% sheep blood agar | Anaerobic | 37 | 286 | |
| Clostridium massilioamazoniensis | J | Inoculation for 5 days in blood culture bottle and then Schaedler agar with neomycin + vancomycin | Anaerobic | 37 | Our unpublished data | |
| Anaerosalibacter massiliensis | J | Thermic shock (20 min, 65°C) and then inoculation for 3 days in blood culture bottle and then 5% sheep blood agar | Anaerobic | 37 | Our unpublished data | |
| Nosocomiicoccus massiliensis | L | Inoculation for 14 days in blood culture bottle with rumen fluid and then 5% sheep blood agar | Aerobic | 37 | 243 | |
| Megasphaera massiliensis | L | Inoculation for 7 days in blood culture bottle with rumen fluid and sheep blood and then 5% sheep blood agar | Anaerobic | 37 | 242 | |
| Bacillus casamancensis | M | 5% sheep blood agar | Anaerobic | 37 | Our unpublished data | |
| Clostridium dakarense | M | 5% sheep blood agar | Anaerobic | 37 | 245 | |
| Bacillus numidis | O | Inoculation for 21 days in liquid brain heart infusion broth with 5% sheep blood then 5% sheep blood agar | Aerobic | 37 | Our unpublished data | |
| Numidum massiliensis | O | Inoculation for 21 days in liquid brain heart infusion broth with 5% sheep blood and then 5% sheep blood agar | Aerobic | 37 | Our unpublished data | |
| Bacillus rubiinfantis | P | Inoculation for 21 days in blood culture bottle with sheep blood and then 5% sheep blood agar | Aerobic | 37 | Our unpublished data | |
| Amazonia massiliensis | Q | Inoculation in Columbia broth during 30 days and then 5% sheep blood agar | Anaerobic | 37 | Our unpublished data | |
| Clostridium culturomicsense | S | Inoculation for 7 days in blood culture bottle with rumen fluid and then 5% sheep blood agar | Anaerobic | 37 | Our unpublished data | |
| Clostridium jeddahtimonense | S | Inoculation for 7 days in blood culture bottle with rumen fluid and then 5% sheep blood agar | Anaerobic | 37 | Our unpublished data | |
| Oceanobacillus jeddahense | S | Inoculation for 15 days in Columbia broth medium with 100 g/liter NaCl and then 5% sheep blood agar | Aerobic | 37 | Our unpublished data | |
| Bacillus jeddahtimonense | T | Inoculation for 15 days in Columbia broth medium with 100 g/liter NaCl and then 5% sheep blood agar | Aerobic | 37 | Our unpublished data | |
| Paraliobacillus massiliensis | U | Inoculation for 21 days in Columbia broth medium with 100 g/liter NaCl and then 5% sheep blood agar | Aerobic | 37 | Our unpublished data | |
| Thalassobacillus massiliensis | U | Inoculation for 21 days in Columbia broth medium with 100 g/liter NaCl and then 5% sheep blood agar | Aerobic | 37 | Our unpublished data | |
| Peptoniphilus ihumii | V | Inoculation for 21 days in blood culture bottle with rumen fluid and sheep blood and then 5% sheep blood agar | Anaerobic | 37 | Our unpublished data | |
| Bacillus andreraoultii | Y | Inoculation for 10 days in blood culture bottle with sheep blood and then 5% sheep blood agar | Anaerobic | 37 | Our unpublished data | |
| Rubiinfantum massiliensis | X | Inoculation in marine liquid medium during 3 days and then 5% sheep blood agar | Aerobic | 37 | Our unpublished data | |
| Bacillus massilioamazoniensis | Y | Inoculation for 10 days in blood culture bottle with sheep blood and then 5% sheep blood agar | Aerobic | 37 | Our unpublished data | |
| Clostridium amazonitimonense | Y | Inoculation for 15 days in blood culture bottle with rumen fluid and sheep blood and then 5% sheep blood agar | Anaerobic | 37 | Our unpublished data | |
| Guyana massiliensis | Y | Inoculation for 15 days in blood culture bottle with rumen fluid and sheep blood and then 5% sheep blood agar | Anaerobic | 37 | Our unpublished data | |
| Virgibacillus senegalensis | Z | Inoculation for 3 days in Columbia broth medium with 100 g/liter NaCl and then 5% sheep blood agar | Aerobic | 37 | Our unpublished data | |
| Virgibacillus massiliensis | Y2 | Inoculation for 21 days in Columbia broth medium with 100 g/liter NaCl and then 5% sheep blood agar | Aerobic | 37 | Our unpublished data | |
| Planomicrobium massiliensis | Z2 | Inoculation for 2 days in Columbia broth medium with 100 g/liter NaCl and then 5% sheep blood agar | Aerobic | 37 | Our unpublished data | |
| Bacteroidetes | Alistipes senegalensis | B | Schaedler agar with kanamycin + vancomycin | Anaerobic | 37 | 4, 227 |
| Alistipes timonensis | B | Inoculation for 5 days in blood culture bottle and then Schaedler agar with kanamycin + vancomycin | Anaerobic | 37 | 4, 228 | |
| Alistipes obesi | C | Inoculation for 11 days in blood culture bottle with rumen fluid and then 5% sheep blood agar | Anaerobic | 37 | 4, 235 | |
| Bacteroides timonensis | E | 5% sheep blood agar for 1 mo | Anaerobic | 37 | 6, 291 | |
| Bacteroides neonati | N | Inoculation in blood culture bottle and then 5% sheep blood agar | Anaerobic | 37 | 283 | |
| Alistipes ihumii | E | Inoculation in blood culture bottle with thioglycolate and then 5% sheep blood agar | Anaerobic | 37 | 6 | |
| Butyricimonas massiliensis | G | Inoculation for 14 days in blood culture bottle with rumen fluid and sheep blood and then 5% sheep blood agar | Anaerobic | 37 | 7b | |
| Alistipes jeddahensis | S | Incubation 3 days in PolyViteX chocolate agar medium | Anaerobic | 37 | Our unpublished data | |
| Actinobacteria | Timonella senegalensis | B | Inoculation for 14 days in blood culture bottle with rumen fluid and sheep blood and then 5% sheep blood agar | Anaerobic | 37 | 4, 238 |
| Senegalemassilia anaerobia | B | Inoculation for 5 days in blood culture bottle and then 5% sheep blood agar | Anaerobic | 37 | 4, 236 | |
| Cellulomonas massiliensis | B | Passive filtration (0.4-μm filter) using Leptospira broth and then inoculation in 5% sheep blood agar | Aerobic | 37 | 4, 197 | |
| Aeromicrobium massiliense | B | 5% sheep blood agar | Aerobic | 37 | 4, 231 | |
| Brevibacterium senegalense | B | Brucella agar | Aerobic | 37 | 4, 224 | |
| Actinomyces grossensis | C | Inoculation for 4 days in blood culture bottle with thioglycolate and then 5% sheep blood agar | Anaerobic | 37 | 4 | |
| Streptomyces massiliensis | E | Active filtration (0.45-μm filter) and then inoculation in brain heart infusion agar | Aerobic | 37 | 6 | |
| Blastococcus massiliensis | E | Brucella agar | Aerobic | 37 | 6 | |
| Collinsella massiliensis | G | Inoculation for 14 days in blood culture bottle with rumen fluid and sheep blood and then 5% sheep blood agar | Anaerobic | 37 | 7 | |
| Enorma massiliensis | C | Inoculation for 4 days in blood culture bottle with thioglycolate and then 5% sheep blood agar | Anaerobic | 37 | 4, 234 | |
| Enorma timonensis | G | Inoculation for 14 days in blood culture bottle with rumen fluid and sheep blood and then 5% sheep blood agar | Anaerobic | 37 | 284 | |
| Corynebacterium ihumii | H | PolyViteX chocolate agar during 21 days | Aerobic with 5% CO2 | 37 | 287 | |
| Nocardioides massiliensis | H | PolyViteX chocolate agar during 14 days | Aerobic | 28 | Our unpublished data | |
| Actinomyces polynesiense | I | Inoculation in blood culture bottle with coconut milk and then 5% sheep blood agar | Anaerobic | 37 | Our unpublished data | |
| Nesterenkonia massiliensis | L | Inoculation for 14 days in blood culture bottle with rumen fluid and then 5% sheep blood agar | Aerobic | 37 | 277 | |
| Corynebacterium jeddahense | K | Inoculation for 14 days in blood culture bottle with rumen fluid and sheep blood and then 5% sheep blood agar | Aerobic | 37 | 285 | |
| Collinsella massilioamazoniensis | Q | Inoculation in marine liquid medium during 30 days and then 5% sheep blood agar | Anaerobic | 37 | Our unpublished data | |
| Jeddahella massiliensis | S | Inoculation for 3 days in blood culture bottle with TSA medium and then 5% sheep blood agar | Aerobic | 37 | Our unpublished data | |
| Nigerium massiliensis | W | Inoculation for 21 days in blood culture bottle with liquid brain heart infusion medium, yeast extract, and proteose peptone and then 5% sheep blood agar | Anaerobic | 28 | Our unpublished data | |
| Flaviflexus massiliensis | X | Inoculation in marine liquid medium during 3 days and then 5% sheep blood agar | Aerobic | 37 | Our unpublished data | |
| Tessaracoccus massiliensis | X | Inoculation in marine liquid medium during 10 days and then 5% sheep blood agar | Aerobic | 37 | Our unpublished data | |
| Proteobacteria | Enterobacter massiliensis | B | Use of both T1 and T4 phage and then inoculation in 5% sheep blood agar | Aerobic | 37 | 4, 206 |
| Herbaspirillum massiliense | B | Passive filtration (0.4-μm filter) using Leptospira broth and then inoculation in 5% sheep blood agar | Aerobic | 37 | 4, 198 | |
| Microvirga massiliensis | B | MOD 2 | Aerobic | 37 | 10 | |
| Dakarella massiliensis | R | Inoculation for 1 day in blood culture bottle with rumen fluid and sheep blood and then 5% sheep blood agar | Anaerobic | 37 | Our unpublished datab |
A, N’Diop, Senegal (healthy individual A); B, Dielmo, Senegal, healthy individual; C, French obese patient; D, XDR tuberculosis patient; E, French anorexia nervosa patient; F, Q fever patient; G, intensive care unit (patient A); H, intensive care unit (patient B); I, Raiatea Island (French Polynesia) healthy individual; J, Amazonian (healthy individual A); K, obese Saudi (patient A); L, AIDS (patient A); M, Senegalese subjects (684 samples); N, preterm infant with necrotizing enterocolitis; O, Touareg healthy individual; P, Kwashiorkor malnutrition (patient A); Q, Amazonian (healthy individual B); R, fresh sample from French healthy individual; S, obese Saudi (patient B); T, lean Saudi; U, Amazonian (healthy individual C); V, AIDS (patient B); W, Kwashiorkor malnutrition (patient B); X, Kwashiorkor malnutrition (patient C); Y, Amazonian (healthy individual D); Z, N’Diop, Senegal (healthy individual B); Y2, Amazonian (healthy individual E); Z2, N’Diop, Senegal (healthy individual C).
Bacterial species were not characterized because subculturing was impossible.
TSA, Trypticase soy agar.
Future perspectives.
(i) Culture of bacterial species considered unculturable.
Diverse recent pyrosequencing studies of the human gut have revealed that most of the sequences to date were assigned to unculturable species belonging to the Firmicutes and Proteobacteria phyla, within the Clostridiales order, and within the Bacteroidaceae, Rikenellaceae, Ruminococcaceae, Lachnospiraceae, and Erysipelotrichaceae families (6, 248, 249). Most of these bacterial species are strictly anaerobic species, revealing a future challenge for culturomics. Strategies including a reduction of the sample transport time before inoculation and the use of antioxidant agents such as ascorbic acid and glutathione will dramatically improve the culture of anaerobic bacteria (126).
(ii) Comparison of diversity among individuals.
A relative decrease in biodiversity was observed for HIV-infected patients and patients treated with heavy antibiotic regimens for long durations (5, 121). In addition, the decrease of both biodiversity and bacterial load of the gut microbiota of a patient treated for extensively drug-resistant (XDR) tuberculosis may have been a reflection of the general desertification of mucosal microbiota and could have been one of the substrates of deadly pneumococcemia developed by this patient (5). The next step for culturomics will be to apply the 18 more effective culture conditions to more stool samples to compare biodiversity among individuals. After a comprehensive approach in culturomics (4–7, 121), it will be exciting to perform the same studies to compare the repertoires among the different samples analyzed. It would be an exciting challenge to apply culturomics to samples from patients suffering from diseases associated with modification of microbiota as metabolic diseases, such as obesity or malnutrition, including kwashiorkor or marasmus (250, 251).
From Culturomics to Taxonogenomics
The current classification of prokaryotes, generally called polyphasic taxonomy, relies on a combination of phenotypic and genotypic characteristics (252, 253). However, the validity of several of the criteria used has been debated (254). Currently, the primary genotypic criteria used in bacterial taxonomy include DNA-DNA hybridization, G+C content, and 16S rRNA sequence similarity (252, 255). Although empirically designed, DNA-DNA hybridization is considered the gold-standard criterion for estimating the genetic relatedness between prokaryotes. A DNA-DNA hybridization value of ≤70% is considered the cutoff, permitting the definition of new species (256, 257). However, various limitations have been reported, including its elevated cost; the lack of interlaboratory and interassay reproducibility, preventing the creation of a reference database (254); and the cutoff value not being applicable to all prokaryotic genera, as is the case for members of the genus Rickettsia (48). Similarly, the16S rRNA sequence identity discriminatory power varies greatly from one genus to another (252, 258). Originally, the cutoffs used to classify bacterial isolates within new species and/or genera were 97% and 95%, respectively (259); however, the cutoff value at the species level was reevaluated to be 98.7% in 2006 (255). Stackebrandt et al. proposed that 16S rRNA identity values could replace DNA-DNA hybridization (255, 259). More recently, Meier-Kolthoff et al. proposed that a threshold of between 98.2 and 99.0% 16S rRNA sequence identity could be used as a safe alternative to DNA-DNA hybridization (257). However, 16S rRNA sequencing also exhibits several biases, notably a high degree of conservation in some genera (49). Genomic sequences from microorganisms constitute a major source of genetic information that could provide unequaled data for the development and identification of genotyping tools as well as for the design of culture media (260–263). We believe that microbiogenomics, which consists of sequencing of the genomes of all microorganisms cultured for which no such sequences are yet available, will be a major advance in clinical microbiology (Fig. 5).
FIG 5.
Overall vision of the strategy from clinical sample to genomic applications. All the bacteria identified by MALDI-TOF MS and by molecular tools were compared with our database of species from humans. We determined the genomes of all the bacteria first isolated in humans (microbiogenomics), and all the new bacterial species were described by using taxonogenomics. All these bacteria were deposited in our collection of bacterial strains (CSUR). GenBank accession numbers are shown in parentheses.
Although whole-genome sequencing constituted a revolution by providing access to the complete genetic information for a bacterial strain, this method has not yet been officially accepted as a source of taxonomic information. However, although this technique was previously time- and money-consuming, because of high-throughput next-generation sequencing (NGS) methods, sequencing of a complete bacterial genome currently requires only a few days and limited cost (260). As of January 2014, >37,000 bacterial genome sequencing projects have been completed or are ongoing (http://www.genomesonline.org/). These sequences are then made available in public databases, facilitating whole-genome sequence comparisons of new bacterial strains (264, 265).
In 1999, Fitz-Gibbon and House were the first to propose that the presence or absence of genes within genomes could be used to describe taxonomic relations among prokaryotes (266). Phylogenomic studies, which are based on comparisons of orthologous genes, have demonstrated good similarity with studies based on comparisons of 16S rRNA sequences (267). In 2005, it was suggested that the average nucleotide identity (ANI) between two bacterial species could constitute an effective alternative to DNA-DNA hybridization (268, 269). An ANI value of ≥95% between genomes was proposed by those authors of to be equivalent to a DNA-DNA hybridization value of ≥70% (269). In 2009, Richter and Rossello-Mora (270) and, in 2010, Tindall et al. (252) proposed that ANI could be used to replace DNA-DNA hybridization in characterizing prokaryotic species, and thus far, this method has been used to describe several new taxa (271–273). However, several drawbacks of using genomic sequences for taxonomic purposes have been highlighted. Criticism emerged regarding the overrepresented sequences from bacterial species of medical or technological interest in comparison to other species (267), the lack of sequenced genomes from many strain types (252), the questionable quality of some genome sequences available in public databases (274), and the high proportion (63%) of presumably less informative draft genomes compared with complete genomes (275).
However, the rapidly increasing number of available genome sequences has solved some of these drawbacks (276). Similarly to other authors, we believe that genome sequences should be systematically included in the description of new taxa. Thus far, we have proposed the creation of 45 new bacterial species based on this strategy (67, 197, 198, 206, 221–247, 277–291), which we call taxonogenomics (Fig. 5) (292). Taxonogenomics consists of a polyphasic approach, incorporating both phenotypic and genotypic data (292), to describe a new bacterial species. First, we consider bacterial strains that exhibit a 16S rRNA sequence identity of <98.7% compared with the phylogenetically most closely related species with standing in nomenclature, as previously recommended (255), as belonging to a putative new species, provided that this identity value is in the range of identities observed among validly published species within the same genus. Next, we study the primary phenotypic characteristics of the bacterial isolate, such as habitat, Gram staining, electron microscopy, primary culture, and metabolic characteristics; proteic spectra obtained by MALDI-TOF MS; and genomic characteristics (genome size; G+C content; percentage of coding sequences; gene content; gene distribution in COG categories [293]; numbers and types of RNA genes; the presence or absence of mobile genetic elements, signal peptides, and transmembrane helices; and average genomic identity of orthologous gene sequences [AGIOS]), compared with the characteristics of the most closely related species with standing in nomenclature (292). Type strains are also deposited into two international culture collections, including our collection (Collection de Souche de l'Unité des Rickettsies [CSUR], WDCM 875). All new species isolated by microbial culturomics (4–6) have been, or will be, described based on this model.
REMAINING CHALLENGES
Despite the spectacular advances in recent years thanks to culturomics, there are yet unsatisfactory results remaining. Many anaerobic bacteria detected in feces by metagenomic methods remain uncultured. Development of new methods for collection, transport, and culture, including the use of antioxidant agents to clarify the still unknown part of the gut microbiota repertoire, will be required (294). The pioneering works of microbial culturomics that permitted significant extension of the microbial repertoire (4–6) have as a central fault that these works were not easily transferred to other laboratories. One of the major future objectives of our laboratory will be to standardize culturomics for 18 culture conditions to define comparisons between laboratories precisely.
Due to its low cost for each test performed and its rapid results, the generalization of the use of MALDI-TOF MS in most clinical microbiology laboratories will clearly change identification strategies and will facilitate a dramatic rebirth regarding the increase in the human microbial repertoire (42, 80). Indeed, the ever-increasing number and sizes of databases allow every laboratory to identify all the bacteria that have been found in the history of humanity and to discover new bacterial species. In total, 107 different bacterial species are assumed to exist on the surface of the earth, and we have currently identified slightly more than 104 (295). The conditions that will allow us to significantly increase the knowledge regarding microbes will continue to require the development of new cultivation techniques.
CONCLUSION
The technologically driven approach to identification methods in clinical microbiology has dramatically extended the bacterial repertoire (296). The systematic use of 16S rRNA or rpoB gene amplification and sequencing has allowed us to identify 29 new bacterial species and 16 bacterial species first isolated from humans (47, 53). The second revolution consists of demonstrating the efficient use of MALDI-TOF MS in clinical microbiology laboratories (42). The seminal publication has been cited >400 times since 2010 (http://apps.webofknowledge.com/). This publication allowed us to identify 30 bacterial species for the first time in humans (80). Currently, culturomics has identified 717 different bacterial species, including 91 new bacterial species and 168 bacteria first isolated from humans.
In conclusion, by applying an empirical approach driven by technology, our laboratory has cultured 386 bacterial species (with ∼18% of these species being identified in humans) for the first time from human samples, including 165 new bacterial species (Table 1), in the diverse fields of clinical microbiology (10, 96, 97, 296). We believe that culture is essential for discoveries and could increase our knowledge in the future (294).
ACKNOWLEDGMENTS
We thank Gregory Dubourg for his technical assistance with Fig. 2, all the technicians working in our laboratory, and all the Ph.D. and M.Sc. students working on the culturomics team.
Bernard La Scola and Didier Raoult are coinventors of a pending patent on the culture of anaerobes, which is owned by Aix Marseille University.
Biographies

Jean-Christophe Lagier, M.D., Ph.D., who specializes in infectious diseases, is an associate professor at the Faculty of Medicine of Marseille, Aix Marseille University, since 2013. His clinical medical activity occurs at the University Hospital Institute Mediterranée Infection of Marseille. His research interests include Tropheryma whipplei infections and gut microbiota diversity, which he has specifically explored by culture with the D. Raoult team since 2007. He coordinates with S. Khelaifia the culturomics team directed by Prof. D. Raoult. By October 2014, Dr. Lagier had coauthored more than 85 publications in the international literature.

Perrine Hugon, Pharm.D., studied pharmacy at the Pharmacy Faculty in Paris 5 and subsequently started a Ph.D. in human pathologies specializing in transmissible diseases at the Faculty of Medicine of Marseille in 2011, supervised by Professor Didier Raoult. Her research domain focuses on bacteria inhabiting the human gut and on several techniques to study these bacteria, such as bacterial culture, electron microscopy, and high-throughput sequencing.

Saber Khelaifia received his Ph.D. from the Faculty of Medicine, Aix Marseille University, France, investigating the detection and culture of Archaea associated with the intestinal and oral human mucosa. He succeeded in growing and isolating new methanogenic Archaea from the human gut. Currently, he is a young research investigator within the Research Unit in Infectious and Tropical Emergent Diseases directed by Professor Didier Raoult. With J.-C. Lagier, he coordinates the culturomics team directed by Prof. D. Raoult, working on the exploration of human intestinal microbiota by culture. He is also working on developing a new culture process for isolating and cultivating fastidious bacteria.

Pierre-Edouard Fournier, M.D., Ph.D., is a clinical microbiologist and professor of microbiology at the Faculty of Medicine of Marseille, Aix Marseille University, France. Within the Research Unit in Infectious and Tropical Emergent Diseases at this facility, he is the Director of the French reference center for the diagnosis of rickettsioses, bartonelloses, and Q fever. He specializes in diagnosing infectious diseases, specifically infections caused by intracellular bacteria. Dr. Fournier has authored or coauthored more than 250 indexed articles.

Bernard La Scola, M.D., Ph.D., is a professor of microbiology at the Faculty of Medicine of Marseille, Aix Marseille University, France, where he is in charge of the isolation of fastidious microorganisms, including all those microorganisms isolated in the NSB3 laboratory. His research interests include emerging bacteria and viruses primarily associated with protozoa. Recently, his research has primarily focused on giant viruses. By October 2014, Prof. La Scola had coauthored more than 220 publications in the international literature. He is the director of the Centre National de Reference of Francisella and a worldwide-recognized expert on amoeba-associated bacteria and viruses. At the university, he is in charge of the program for M.Sc. research in infectious diseases.

Didier Raoult, M.D., Ph.D., who specializes in infectious diseases, is a professor of microbiology at the Faculty of Medicine of Marseille, Aix Marseille University. In 1984, he created ex nihilo his research laboratory, the Rickettsia Unit. This unit has now become the Research Unit in Infectious and Tropical Emergent Diseases (URMITE), collaborating with the CNRS (National Center for Scientific Research), the IRD (Institute of Research for Development), and INSERM (National Institute of Health and Medical Research). In 2011, he became the director of the University Hospital Institute Mediterranée Infection, which is a 600-person medical institute focused on infectious diseases. This facility includes the largest diagnostic and research microbiology laboratory in France. As of 2014, Prof. Raoult has published more than 1,900 indexed publications. In the last 30 years, he has cultured approximately 16% of the bacteria isolated for the first time in humans, including T. whipplei, in his laboratory and developed a culturomics team in 2011.
REFERENCES
- 1.Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers YH, Smith HO. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 2.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lagier JC, Million M, Hugon P, Armougom F, Raoult D. 2012. Human gut microbiota: repertoire and variations. Front Cell Infect Microbiol 2:136. doi: 10.3389/fcimb.2012.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lagier JC, Armougom F, Million M, Hugon P, Pagnier I, Robert C, Bittar F, Fournous G, Gimenez G, Maraninchi M, Trape JF, Koonin EV, La Scola B, Raoult D. 2012. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect 18:1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- 5.Dubourg G, Lagier JC, Armougom F, Robert C, Hamad I, Brouqui P, Raoult D. 2013. The gut microbiota of a patient with resistant tuberculosis is more comprehensively studied by culturomics than by metagenomics. Eur J Clin Microbiol Infect Dis 32:637–645. doi: 10.1007/s10096-012-1787-3. [DOI] [PubMed] [Google Scholar]
- 6.Pfleiderer A, Lagier JC, Armougom F, Robert C, Vialettes B, Raoult D. 2013. Culturomics identified 11 new bacterial species from a single anorexia nervosa stool sample. Eur J Clin Microbiol Infect Dis 32:1471–1481. doi: 10.1007/s10096-013-1900-2. [DOI] [PubMed] [Google Scholar]
- 7.Dubourg G, Lagier JC, Robert C, Armougom F, Hugon P, Metidji S, Dione N, Makaya Dangui NP, Pfleiderer A, Abrahao J, Musso D, Papazian L, Brouqui P, Bibi F, Yasir M, Vialettes B, Raoult D. 2014. Culturomics and pyrosequencing evidence of the reduction in gut microbiota diversity in patients with broad-spectrum antibiotics. Int J Antimicrob Agents 44:117–124. doi: 10.1016/j.ijantimicag.2014.04.020. [DOI] [PubMed] [Google Scholar]
- 8.Kaeberlein T, Lewis K, Epstein SS. 2002. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science 296:1127–1129. doi: 10.1126/science.1070633. [DOI] [PubMed] [Google Scholar]
- 9.Janda JM, Abbott SL. 2007. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J Clin Microbiol 45:2761–2764. doi: 10.1128/JCM.01228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lagier J-C, Edouard S, Pagnier I, Mediannikov O, Drancourt M, Raoult D. 2015. Current and past strategies for bacterial culture in clinical microbiology. Clin Microbiol Rev 28:208–236. doi: 10.1128/CMR.00110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chapin K. 2007. Clinical microscopy, p 33–51. In Murray PR, Baron EJ, Jorgensen JH, Landry ML, Pfaller MA (ed), Manual of clinical microbiology, 9th ed, vol 1 ASM Press, Washington, DC. [Google Scholar]
- 12.Carlone GM, Valadez MJ, Pickett MJ. 1982. Methods for distinguishing gram-positive from gram-negative bacteria. J Clin Microbiol 16:1157–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fenollar F, Raoult D. 2000. Comparison of a commercial disk test with vancomycin and colimycin susceptibility testing for identification of bacteria with abnormal gram staining reactions. Eur J Clin Microbiol Infect Dis 19:33–38. doi: 10.1007/s100960050006. [DOI] [PubMed] [Google Scholar]
- 14.Johnson MJ, Thatcher E, Cox ME. 1995. Techniques for controlling variability in gram staining of obligate anaerobes. J Clin Microbiol 33:755–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beveridge TJ. 1990. Mechanism of gram variability in select bacteria. J Bacteriol 172:1609–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hugon P, Lagier JC, Robert C, Lepolard C, Papazian L, Musso D, Vialettes B, Raoult D. 2013. Molecular studies neglect apparently Gram-negative populations in the human gut microbiota. J Clin Microbiol 51:3286–3293. doi: 10.1128/JCM.00473-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gimenez DF. 1964. Staining rickettsiae in yolk-sac cultures. Stain Technol 39:135–140. [DOI] [PubMed] [Google Scholar]
- 18.Gouriet F, Fenollar F, Patrice JY, Drancourt M, Raoult D. 2005. Use of shell-vial cell culture assay for isolation of bacteria from clinical specimens: 13 years of experience. J Clin Microbiol 43:4993–5002. doi: 10.1128/JCM.43.10.4993-5002.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapin KC. 2007. Principles of stains and media, p 182–191. In Murray PR, Baron EJ, Jorgensen JH, Landry ML, Pfaller MA (ed), Manual of clinical microbiology, 9th ed ASM Press, Washington, DC. [Google Scholar]
- 20.Jensen TK, Montgomery DL, Jaeger PT, Lindhardt T, Agerholm JS, Bille-Hansen V, Boye M. 2007. Application of fluorescent in situ hybridisation for demonstration of Coxiella burnetii in placentas from ruminant abortions. APMIS 115:347–353. doi: 10.1111/j.1600-0463.2007.apm_591.x. [DOI] [PubMed] [Google Scholar]
- 21.Laub RR, Knudsen JD. 2014. Clinical consequences of using PNA-FISH in staphylococcal bacteraemia. Eur J Clin Microbiol Infect Dis 33:599–601. doi: 10.1007/s10096-013-1990-x. [DOI] [PubMed] [Google Scholar]
- 22.Almeida C, Azevedo NF, Fernandes RM, Keevil CW, Vieira MJ. 2010. Fluorescence in situ hybridization method using a peptide nucleic acid probe for identification of Salmonella spp. in a broad spectrum of samples. Appl Environ Microbiol 76:4476–4485. doi: 10.1128/AEM.01678-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horne RW, Wildy P. 1963. Virus structure revealed by negative staining. Adv Virus Res 10:101–170. [DOI] [PubMed] [Google Scholar]
- 24.Isalska BJ, Curry A, Stanbridge TN, Tweddle D, Caul EO. 1996. Electron microscopy and serological features of a patient with Q fever prosthetic valve endocarditis. J Clin Pathol 49:679–681. doi: 10.1136/jcp.49.8.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Curry A, Appleton H, Dowsett B. 2006. Application of transmission electron microscopy to the clinical study of viral and bacterial infections: present and future. Micron 37:91–106. doi: 10.1016/j.micron.2005.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amako K, Takade A, Taniai H, Yoshida S. 2008. Electron microscopic examination of uncultured soil-dwelling bacteria. Microbiol Immunol 52:265–269. doi: 10.1111/j.1348-0421.2008.00037.x. [DOI] [PubMed] [Google Scholar]
- 27.Bae HC, Cota-Robles EH, Casida LE. 1972. Microflora of soil as viewed by transmission electron microscopy. Appl Microbiol 23:637–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eltsov M, Zuber B. 2006. Transmission electron microscopy of the bacterial nucleoid. J Struct Biol 156:246–254. doi: 10.1016/j.jsb.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 29.Boekema EJ, Scheffers DJ, van Bezouwen LS, Bolhuis H, Folea IM. 2013. Focus on membrane differentiation and membrane domains in the prokaryotic cell. J Mol Microbiol Biotechnol 23:345–356. doi: 10.1159/000351361. [DOI] [PubMed] [Google Scholar]
- 30.Milne JL, Borgnia MJ, Bartesaghi A, Tran EE, Earl LA, Schauder DM, Lengyel J, Pierson J, Patwardhan A, Subramaniam S. 2013. Cryo-electron microscopy—a primer for the non-microscopist. FEBS J 280:28–45. doi: 10.1111/febs.12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asano S, Iijima K, Suzuki K, Motoyama Y, Ogata T, Kitagawa Y. 2009. Rapid detection and identification of beer-spoilage lactic acid bacteria by microcolony method. J Biosci Bioeng 108:124–129. doi: 10.1016/j.jbiosc.2009.02.016. [DOI] [PubMed] [Google Scholar]
- 32.Baba T, Inoue N, Yamaguchi N, Nasu M. 2012. Rapid enumeration of active Legionella pneumophila in freshwater environments by the microcolony method combined with direct fluorescent antibody staining. Microbes Environ 27:324–326. doi: 10.1264/jsme2.ME11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meder H, Baumstummler A, Chollet R, Barrier S, Kukuczka M, Olivieri F, Welterlin E, Beguin V, Ribault S. 2012. Fluorescence-based rapid detection of microbiological contaminants in water samples. ScientificWorldJournal 2012:234858. doi: 10.1100/2012/234858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baumstummler A, Chollet R, Meder H, Olivieri F, Rouillon S, Waiche G, Ribault S. 2011. Development of a nondestructive fluorescence-based enzymatic staining of microcolonies for enumerating bacterial contamination in filterable products. J Appl Microbiol 110:69–79. doi: 10.1111/j.1365-2672.2010.04859.x. [DOI] [PubMed] [Google Scholar]
- 35.Caviedes L, Lee TS, Gilman RH, Sheen P, Spellman E, Lee EH, Berg DE, Montenegro-James S. 2000. Rapid, efficient detection and drug susceptibility testing of Mycobacterium tuberculosis in sputum by microscopic observation of broth cultures. The Tuberculosis Working Group in Peru. J Clin Microbiol 38:1203–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leung E, Minion J, Benedetti A, Pai M, Menzies D. 2012. Microcolony culture techniques for tuberculosis diagnosis: a systematic review. Int J Tuberc Lung Dis 16:16–23. doi: 10.5588/ijtld.10.0065. [DOI] [PubMed] [Google Scholar]
- 37.den Hertog AL, Visser DW, Ingham CJ, Fey FH, Klatser PR, Anthony RM. 2010. Simplified automated image analysis for detection and phenotyping of Mycobacterium tuberculosis on porous supports by monitoring growing microcolonies. PLoS One 5:e11008. doi: 10.1371/journal.pone.0011008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caslow M, Ellner PD, Kiehn TE. 1974. Comparison of the BACTEC system with blind subculture for the detection of bacteremia. Appl Microbiol 28:435–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thiemke WA, Wicher K. 1975. Laboratory experience with a radiometric method for detecting bacteremia. J Clin Microbiol 1:302–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller JM, O'Hara CM. 2007. Substrate utilization systems for the identification of bacteria and yeasts, p 103–109. In Murray PR, Baron EJ, Jorgensen JH, Landry ML, Pfaller MA (ed), Manual of clinical microbiology, 9th ed ASM Press, Washington, DC. [Google Scholar]
- 41.Magee JT. 2001. Applied systematics, p 9–32. In Cemolai N. (ed), Laboratory diagnosis of bacterial infections. Marcel Dekker, New York, NY. [Google Scholar]
- 42.Seng P, Drancourt M, Gouriet F, La Scola B, Fournier PE, Rolain JM, Raoult D. 2009. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis 49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 43.Fenollar F, Raoult D. 2004. Molecular genetic methods for the diagnosis of fastidious microorganisms. APMIS 112:785–807. doi: 10.1111/j.1600-0463.2004.apm11211-1206.x. [DOI] [PubMed] [Google Scholar]
- 44.Relman DA, Loutit JS, Schmidt TM, Falkow S, Tompkins LS. 1990. The agent of bacillary angiomatosis. An approach to the identification of uncultured pathogens. N Engl J Med 323:1573–1580. doi: 10.1056/NEJM199012063232301. [DOI] [PubMed] [Google Scholar]
- 45.Relman DA, Schmidt TM, MacDermott RP, Falkow S. 1992. Identification of the uncultured bacillus of Whipple's disease. N Engl J Med 327:293–301. doi: 10.1056/NEJM199207303270501. [DOI] [PubMed] [Google Scholar]
- 46.Fenollar F, Fournier PE, Robert C, Raoult D. 2004. Use of genome selected repeated sequences increases the sensitivity of PCR detection of Tropheryma whipplei. J Clin Microbiol 42:401–403. doi: 10.1128/JCM.42.1.401-403.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Drancourt M, Berger P, Raoult D. 2004. Systematic 16S rRNA gene sequencing of atypical clinical isolates identified 27 new bacterial species associated with humans. J Clin Microbiol 42:2197–2202. doi: 10.1128/JCM.42.5.2197-2202.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fournier PE, Raoult D. 2009. Current knowledge on phylogeny and taxonomy of Rickettsia spp. Ann N Y Acad Sci 1166:1–11. doi: 10.1111/j.1749-6632.2009.04528.x. [DOI] [PubMed] [Google Scholar]
- 49.Gandara B, Merino AL, Rogel MA, Martinez-Romero E. 2001. Limited genetic diversity of Brucella spp. J Clin Microbiol 39:235–240. doi: 10.1128/JCM.39.1.235-240.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Drancourt M, Roux V, Fournier PE, Raoult D. 2004. rpoB gene sequence-based identification of aerobic Gram-positive cocci of the genera Streptococcus, Enterococcus, Gemella, Abiotrophia, and Granulicatella. J Clin Microbiol 42:497–504. doi: 10.1128/JCM.42.2.497-504.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khamis A, Raoult D, La Scola B. 2005. Comparison between rpoB and 16S rRNA gene sequencing for molecular identification of 168 clinical isolates of Corynebacterium. J Clin Microbiol 43:1934–1936. doi: 10.1128/JCM.43.4.1934-1936.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ki JS, Zhang W, Qian PY. 2009. Discovery of marine Bacillus species by 16S rRNA and rpoB comparisons and their usefulness for species identification. J Microbiol Methods 77:48–57. doi: 10.1016/j.mimet.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 53.Drancourt M, Bollet C, Carlioz A, Martelin R, Gayral JP, Raoult D. 2000. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol 38:3623–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ferroni A, Sermet-Gaudelus I, Abachin E, Quesne G, Lenoir G, Berche P, Gaillard JL. 2002. Use of 16S rRNA gene sequencing for identification of nonfermenting gram-negative bacilli recovered from patients attending a single cystic fibrosis center. J Clin Microbiol 40:3793–3797. doi: 10.1128/JCM.40.10.3793-3797.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tortoli E, Bartoloni A, Bottger EC, Emler S, Garzelli C, Magliano E, Mantella A, Rastogi N, Rindi L, Scarparo C, Urbano P. 2001. Burden of unidentifiable mycobacteria in a reference laboratory. J Clin Microbiol 39:4058–4065. doi: 10.1128/JCM.39.11.4058-4065.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tang YW, Ellis NM, Hopkins MK, Smith DH, Dodge DE, Persing DH. 1998. Comparison of phenotypic and genotypic techniques for identification of unusual aerobic pathogenic gram-negative bacilli. J Clin Microbiol 36:3674–3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roux V, Drancourt M, Stein A, Riegel P, Raoult D, La Scola B. 2004. Corynebacterium species isolated from bone and joint infections identified by 16S rRNA gene sequence analysis. J Clin Microbiol 42:2231–2233. doi: 10.1128/JCM.42.5.2231-2233.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bosshard PP, Abels S, Altwegg M, Bottger EC, Zbinden R. 2004. Comparison of conventional and molecular methods for identification of aerobic catalase-negative gram-positive cocci in the clinical laboratory. J Clin Microbiol 42:2065–2073. doi: 10.1128/JCM.42.5.2065-2073.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van Camp G, Chapelle S, De Wachter R. 1993. Amplification and sequencing of variable regions in bacterial 23S ribosomal RNA genes with conserved primer sequences. Curr Microbiol 27:147–151. doi: 10.1007/BF01576012. [DOI] [PubMed] [Google Scholar]
- 60.Roux V, Raoult D. 1995. Inter- and intraspecies identification of Bartonella (Rochalimaea) species. J Clin Microbiol 33:1573–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hill KE, Davies CE, Wilson MJ, Stephens P, Lewis MA, Hall V, Brazier J, Thomas DW. 2002. Heterogeneity within the gram-positive anaerobic cocci demonstrated by analysis of 16S-23S intergenic ribosomal RNA polymorphisms. J Med Microbiol 51:949–957. [DOI] [PubMed] [Google Scholar]
- 62.Hassan AA, Khan IU, Abdulmawjood A, Lammler C. 2003. Inter- and intraspecies variations of the 16S-23S rDNA intergenic spacer region of various streptococcal species. Syst Appl Microbiol 26:97–103. doi: 10.1078/072320203322337371. [DOI] [PubMed] [Google Scholar]
- 63.Maiwald M, Lepp PW, Relman DA. 2003. Analysis of conserved non-rRNA genes of Tropheryma whipplei. Syst Appl Microbiol 26:3–12. doi: 10.1078/072320203322337254. [DOI] [PubMed] [Google Scholar]
- 64.Adekambi T, Drancourt M, Raoult D. 2009. The rpoB gene as a tool for clinical microbiologists. Trends Microbiol 17:37–45. doi: 10.1016/j.tim.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 65.Mollet C, Drancourt M, Raoult D. 1997. rpoB sequence analysis as a novel basis for bacterial identification. Mol Microbiol 26:1005–1011. doi: 10.1046/j.1365-2958.1997.6382009.x. [DOI] [PubMed] [Google Scholar]
- 66.Drancourt M, Raoult D. 2002. rpoB gene sequence-based identification of Staphylococcus species. J Clin Microbiol 40:1333–1338. doi: 10.1128/JCM.40.4.1333-1338.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ben Sallah I, Adekambi T, Raoult D, Drancourt M. 2008. rpoB sequence-based identification of Mycobacterium avium complex species. Microbiology 154:3715–3723. doi: 10.1099/mic.0.2008/020164-0. [DOI] [PubMed] [Google Scholar]
- 68.La Scola B, Bui LT, Baranton G, Khamis A, Raoult D. 2006. Partial rpoB gene sequencing for identification of Leptospira species. FEMS Microbiol Lett 263:142–147. doi: 10.1111/j.1574-6968.2006.00377.x. [DOI] [PubMed] [Google Scholar]
- 69.Renesto P, Lorvellec-Guillon K, Drancourt M, Raoult D. 2000. rpoB gene analysis as a novel strategy for identification of spirochetes from the genera Borrelia, Treponema, and Leptospira. J Clin Microbiol 38:2200–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Poyart C, Berche P, Trieu-Cuot P. 1995. Characterization of superoxide dismutase genes from gram-positive bacteria by polymerase chain reaction using degenerate primers. FEMS Microbiol Lett 131:41–45. doi: 10.1111/j.1574-6968.1995.tb07751.x. [DOI] [PubMed] [Google Scholar]
- 71.Poyart C, Quesne G, Coulon S, Berche P, Trieu-Cuot P. 1998. Identification of streptococci to species level by sequencing the gene encoding the manganese-dependent superoxide dismutase. J Clin Microbiol 36:41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stein A, Raoult D. 1992. Detection of Coxiella burnetti by DNA amplification using polymerase chain reaction. J Clin Microbiol 30:2462–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Romero B, Morosini MI, Loza E, Rodriguez-Banos M, Navas E, Canton R, Campo RD. 2011. Reidentification of Streptococcus bovis isolates causing bacteremia according to the new taxonomy criteria: still an issue? J Clin Microbiol 49:3228–3233. doi: 10.1128/JCM.00524-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hwang SM, Kim MS, Park KU, Song J, Kim EC. 2011. Tuf gene sequence analysis has greater discriminatory power than 16S rRNA sequence analysis in identification of clinical isolates of coagulase-negative staphylococci. J Clin Microbiol 49:4142–4149. doi: 10.1128/JCM.05213-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ghebremedhin B, Layer F, Konig W, Konig B. 2008. Genetic classification and distinguishing of Staphylococcus species based on different partial gap, 16S rRNA, hsp60, rpoB, sodA, and tuf gene sequences. J Clin Microbiol 46:1019–1025. doi: 10.1128/JCM.02058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fournier PE, Thuny F, Richet H, Lepidi H, Casalta JP, Arzouni JP, Maurin M, Celard M, Mainardi JL, Caus T, Collart F, Habib G, Raoult D. 2010. Comprehensive diagnostic strategy for blood culture-negative endocarditis: a prospective study of 819 new cases. Clin Infect Dis 51:131–140. doi: 10.1086/653675. [DOI] [PubMed] [Google Scholar]
- 77.Renvoise A, Rolain JM, Socolovschi C, Raoult D. 2012. Widespread use of real-time PCR for rickettsial diagnosis. FEMS Immunol Med Microbiol 64:126–129. doi: 10.1111/j.1574-695X.2011.00899.x. [DOI] [PubMed] [Google Scholar]
- 78.Renesto P, Gouvernet J, Drancourt M, Roux V, Raoult D. 2001. Use of rpoB gene analysis for detection and identification of Bartonella species. J Clin Microbiol 39:430–437. doi: 10.1128/JCM.39.2.430-437.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.La Scola B, Raoult D. 1997. Laboratory diagnosis of rickettsioses: current approaches to diagnosis of old and new rickettsial diseases. J Clin Microbiol 35:2715–2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Seng P, Abat C, Rolain JM, Colson P, Lagier JC, Gouriet F, Fournier PE, Drancourt M, La Scola B, Raoult D. 2013. Identification of rare pathogenic bacteria in a clinical microbiology laboratory: impact of matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 51:2182–2194. doi: 10.1128/JCM.00492-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Clark AE, Kaleta EJ, Arora A, Wolk DM. 2013. Matrix-assisted laser desorption ionization–time of flight mass spectrometry: a fundamental shift in the routine practice of clinical microbiology. Clin Microbiol Rev 26:547–603. doi: 10.1128/CMR.00072-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Anhalt JP, Fenselau C. 1975. Identification of bacteria using mass spectrometry. Anal Chem 47:219–225. doi: 10.1021/ac60352a007. [DOI] [Google Scholar]
- 83.Claydon MA, Davey SN, Edwards-Jones V, Gordon DB. 1996. The rapid identification of intact microorganisms using mass spectrometry. Nat Biotechnol 14:1584–1586. doi: 10.1038/nbt1196-1584. [DOI] [PubMed] [Google Scholar]
- 84.Krishnamurthy T, Ross PL. 1996. Rapid identification of bacteria by direct matrix-assisted laser desorption/ionization mass spectrometric analysis of whole cells. Rapid Commun Mass Spectrom 10:1992–1996. doi:. [DOI] [PubMed] [Google Scholar]
- 85.van Veen SQ, Claas EC, Kuijper EJ. 2010. High-throughput identification of bacteria and yeast by matrix-assisted laser desorption ionization–time of flight mass spectrometry in conventional medical microbiology laboratories. J Clin Microbiol 48:900–907. doi: 10.1128/JCM.02071-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bizzini A, Durussel C, Bille J, Greub G, Prod'hom G. 2010. Performance of matrix-assisted laser desorption ionization–time of flight mass spectrometry for identification of bacterial strains routinely isolated in a clinical microbiology laboratory. J Clin Microbiol 48:1549–1554. doi: 10.1128/JCM.01794-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dupont C, Sivadon-Tardy V, Bille E, Dauphin B, Beretti JL, Alvarez AS, Degand N, Ferroni A, Rottman M, Herrmann JL, Nassif X, Ronco E, Carbonnelle E. 2010. Identification of clinical coagulase-negative staphylococci, isolated in microbiology laboratories, by matrix-assisted laser desorption/ionization-time of flight mass spectrometry and two automated systems. Clin Microbiol Infect 16:998–1004. doi: 10.1111/j.1469-0691.2009.03036.x. [DOI] [PubMed] [Google Scholar]
- 88.Carpaij N, Willems RJ, Bonten MJ, Fluit AC. 2011. Comparison of the identification of coagulase-negative staphylococci by matrix-assisted laser desorption ionization time-of-flight mass spectrometry and tuf sequencing. Eur J Clin Microbiol Infect Dis 30:1169–1172. doi: 10.1007/s10096-011-1204-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fang H, Ohlsson AK, Ullberg M, Ozenci V. 2012. Evaluation of species-specific PCR, Bruker MS, VITEK MS and the VITEK 2 system for the identification of clinical Enterococcus isolates. Eur J Clin Microbiol Infect Dis 31:3073–3077. doi: 10.1007/s10096-012-1667-x. [DOI] [PubMed] [Google Scholar]
- 90.Cherkaoui A, Emonet S, Fernandez J, Schorderet D, Schrenzel J. 2011. Evaluation of matrix-assisted laser desorption ionization–time of flight mass spectrometry for rapid identification of beta-hemolytic streptococci. J Clin Microbiol 49:3004–3005. doi: 10.1128/JCM.00240-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Friedrichs C, Rodloff AC, Chhatwal GS, Schellenberger W, Eschrich K. 2007. Rapid identification of viridans streptococci by mass spectrometric discrimination. J Clin Microbiol 45:2392–2397. doi: 10.1128/JCM.00556-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hinse D, Vollmer T, Erhard M, Welker M, Moore ER, Kleesiek K, Dreier J. 2011. Differentiation of species of the Streptococcus bovis/equinus-complex by MALDI-TOF mass spectrometry in comparison to sodA sequence analyses. Syst Appl Microbiol 34:52–57. doi: 10.1016/j.syapm.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 93.Schulthess B, Brodner K, Bloemberg GV, Zbinden R, Bottger EC, Hombach M. 2013. Identification of Gram-positive cocci by use of matrix-assisted laser desorption ionization–time of flight mass spectrometry: comparison of different preparation methods and implementation of a practical algorithm for routine diagnostics. J Clin Microbiol 51:1834–1840. doi: 10.1128/JCM.02654-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ilina EN, Borovskaya AD, Malakhova MM, Vereshchagin VA, Kubanova AA, Kruglov AN, Svistunova TS, Gazarian AO, Maier T, Kostrzewa M, Govorun VM. 2009. Direct bacterial profiling by matrix-assisted laser desorption-ionization time-of-flight mass spectrometry for identification of pathogenic Neisseria. J Mol Diagn 11:75–86. doi: 10.2353/jmoldx.2009.080079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Degand N, Carbonnelle E, Dauphin B, Beretti JL, Le Bourgeois M, Sermet-Gaudelus I, Segonds C, Berche P, Nassif X, Ferroni A. 2008. Matrix-assisted laser desorption ionization–time of flight mass spectrometry for identification of nonfermenting gram-negative bacilli isolated from cystic fibrosis patients. J Clin Microbiol 46:3361–3367. doi: 10.1128/JCM.00569-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.La Scola B, Fournier PE, Raoult D. 2011. Burden of emerging anaerobes in the MALDI-TOF and 16S rRNA gene sequencing era. Anaerobe 17:106–112. doi: 10.1016/j.anaerobe.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 97.Barreau M, Pagnier I, La Scola B. 2013. Improving the identification of anaerobes in the clinical microbiology laboratory through MALDI-TOF mass spectrometry. Anaerobe 22:123–125. doi: 10.1016/j.anaerobe.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 98.Fedorko DP, Drake SK, Stock F, Murray PR. 2012. Identification of clinical isolates of anaerobic bacteria using matrix-assisted laser desorption ionization-time of flight mass spectrometry. Eur J Clin Microbiol Infect Dis 31:2257–2262. doi: 10.1007/s10096-012-1563-4. [DOI] [PubMed] [Google Scholar]
- 99.Nagy E, Becker S, Kostrzewa M, Barta N, Urban E. 2012. The value of MALDI-TOF MS for the identification of clinically relevant anaerobic bacteria in routine laboratories. J Med Microbiol 61:1393–1400. doi: 10.1099/jmm.0.043927-0. [DOI] [PubMed] [Google Scholar]
- 100.Verroken A, Janssens M, Berhin C, Bogaerts P, Huang TD, Wauters G, Glupczynski Y. 2010. Evaluation of matrix-assisted laser desorption ionization–time of flight mass spectrometry for identification of Nocardia species. J Clin Microbiol 48:4015–4021. doi: 10.1128/JCM.01234-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.El Khechine A, Couderc C, Flaudrops C, Raoult D, Drancourt M. 2011. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry identification of mycobacteria in routine clinical practice. PLoS One 6:e24720. doi: 10.1371/journal.pone.0024720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Couturier MR, Mehinovic E, Croft AC, Fisher MA. 2011. Identification of HACEK clinical isolates by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 49:1104–1106. doi: 10.1128/JCM.01777-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Powell EA, Blecker-Shelly D, Montgomery S, Mortensen JE. 2013. Application of matrix-assisted laser desorption ionization–time of flight mass spectrometry for identification of the fastidious pediatric pathogens Aggregatibacter, Eikenella, Haemophilus, and Kingella. J Clin Microbiol 51:3862–3864. doi: 10.1128/JCM.02233-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Werno AM, Christner M, Anderson TP, Murdoch DR. 2012. Differentiation of Streptococcus pneumoniae from nonpneumococcal streptococci of the Streptococcus mitis group by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 50:2863–2867. doi: 10.1128/JCM.00508-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lasch P, Beyer W, Nattermann H, Stammler M, Siegbrecht E, Grunow R, Naumann D. 2009. Identification of Bacillus anthracis by using matrix-assisted laser desorption ionization–time of flight mass spectrometry and artificial neural networks. Appl Environ Microbiol 75:7229–7242. doi: 10.1128/AEM.00857-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dieckmann R, Helmuth R, Erhard M, Malorny B. 2008. Rapid classification and identification of salmonellae at the species and subspecies levels by whole-cell matrix-assisted laser desorption ionization–time of flight mass spectrometry. Appl Environ Microbiol 74:7767–7778. doi: 10.1128/AEM.01402-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kuhns M, Zautner AE, Rabsch W, Zimmermann O, Weig M, Bader O, Gross U. 2012. Rapid discrimination of Salmonella enterica serovar Typhi from other serovars by MALDI-TOF mass spectrometry. PLoS One 7:e40004. doi: 10.1371/journal.pone.0040004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.He Y, Li H, Lu X, Stratton CW, Tang YW. 2010. Mass spectrometry biotyper system identifies enteric bacterial pathogens directly from colonies grown on selective stool culture media. J Clin Microbiol 48:3888–3892. doi: 10.1128/JCM.01290-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tan KE, Ellis BC, Lee R, Stamper PD, Zhang SX, Carroll KC. 2012. Prospective evaluation of a matrix-assisted laser desorption ionization–time of flight mass spectrometry system in a hospital clinical microbiology laboratory for identification of bacteria and yeasts: a bench-by-bench study for assessing the impact on time to identification and cost-effectiveness. J Clin Microbiol 50:3301–3308. doi: 10.1128/JCM.01405-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pavlovic M, Konrad R, Iwobi AN, Sing A, Busch U, Huber I. 2012. A dual approach employing MALDI-TOF MS and real-time PCR for fast species identification within the Enterobacter cloacae complex. FEMS Microbiol Lett 328:46–53. doi: 10.1111/j.1574-6968.2011.02479.x. [DOI] [PubMed] [Google Scholar]
- 111.Dridi B, Raoult D, Drancourt M. 2012. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry identification of Archaea: towards the universal identification of living organisms. APMIS 120:85–91. doi: 10.1111/j.1600-0463.2011.02833.x. [DOI] [PubMed] [Google Scholar]
- 112.Lista F, Reubsaet FA, De Santis R, Parchen RR, de Jong AL, Kieboom J, van der Laaken AL, Voskamp-Visser IA, Fillo S, Jansen HJ, Van der Plas J, Paauw A. 2011. Reliable identification at the species level of Brucella isolates with MALDI-TOF-MS. BMC Microbiol 11:267. doi: 10.1186/1471-2180-11-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zangenah S, Guleryuz G, Borang S, Ullberg M, Bergman P, Ozenci V. 2013. Identification of clinical Pasteurella isolates by MALDI-TOF—a comparison with VITEK 2 and conventional microbiological methods. Diagn Microbiol Infect Dis 77:96–98. doi: 10.1016/j.diagmicrobio.2013.06.024. [DOI] [PubMed] [Google Scholar]
- 114.Fournier PE, Couderc C, Buffet S, Flaudrops C, Raoult D. 2009. Rapid and cost-effective identification of Bartonella species using mass spectrometry. J Med Microbiol 58:1154–1159. doi: 10.1099/jmm.0.009647-0. [DOI] [PubMed] [Google Scholar]
- 115.Rettinger A, Krupka I, Grunwald K, Dyachenko V, Fingerle V, Konrad R, Raschel H, Busch U, Sing A, Straubinger RK, Huber I. 2012. Leptospira spp. strain identification by MALDI TOF MS is an equivalent tool to 16S rRNA gene sequencing and multi locus sequence typing (MLST). BMC Microbiol 12:185. doi: 10.1186/1471-2180-12-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Djelouadji Z, Roux V, Raoult D, Kodjo A, Drancourt M. 2012. Rapid MALDI-TOF mass spectrometry identification of Leptospira organisms. Vet Microbiol 158:142–146. doi: 10.1016/j.vetmic.2012.01.028. [DOI] [PubMed] [Google Scholar]
- 117.Gaia V, Casati S, Tonolla M. 2011. Rapid identification of Legionella spp. by MALDI-TOF MS based protein mass fingerprinting. Syst Appl Microbiol 34:40–44. doi: 10.1016/j.syapm.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 118.Moliner C, Ginevra C, Jarraud S, Flaudrops C, Bedotto M, Couderc C, Etienne J, Fournier PE. 2010. Rapid identification of Legionella species by mass spectrometry. J Med Microbiol 59:273–284. doi: 10.1099/jmm.0.014100-0. [DOI] [PubMed] [Google Scholar]
- 119.Seng P, Rolain JM, Fournier PE, La Scola B, Drancourt M, Raoult D. 2010. MALDI-TOF-mass spectrometry applications in clinical microbiology. Future Microbiol 5:1733–1754. doi: 10.2217/fmb.10.127. [DOI] [PubMed] [Google Scholar]
- 120.Martiny D, Cremagnani P, Gaillard A, Miendje Deyi VY, Mascart G, Ebraert A, Attalibi S, Dediste A, Vandenberg O. 2014. Feasibility of matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-TOF MS) networking in university hospitals in Brussels. Eur J Clin Microbiol Infect Dis 33:745–754. doi: 10.1007/s10096-013-2006-6. [DOI] [PubMed] [Google Scholar]
- 121.Lagier JC. 2013. Human gut microbiota diversity studied by culturomics and pyrosequencing. Ph.D. thesis. Aix Marseille Université, Marseille, France. http://www.mediterranee-infection.com/arkotheque/client/ihumed/_depot_arko/articles/269/these-lagier_doc.pdf.
- 122.Fenollar F, Puechal X, Raoult D. 2007. Whipple's disease. N Engl J Med 356:55–66. doi: 10.1056/NEJMra062477. [DOI] [PubMed] [Google Scholar]
- 123.Boulos A, Rolain JM, Raoult D. 2004. Antibiotic susceptibility of Tropheryma whipplei in MRC5 cells. Antimicrob Agents Chemother 48:747–752. doi: 10.1128/AAC.48.3.747-752.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lagier JC, Fenollar F, Lepidi H, Giorgi R, Million M, Raoult D. 2014. Treatment of classic Whipple's disease: from in vitro results to clinical outcome. J Antimicrob Chemother 69:219–227. doi: 10.1093/jac/dkt310. [DOI] [PubMed] [Google Scholar]
- 125.Ghodbane R, Raoult D, Drancourt M. 2014. Dramatic reduction of culture time of Mycobacterium tuberculosis. Sci Rep 4:4236. doi: 10.1038/srep04236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.La Scola B, Khelaifia S, Lagier JC, Raoult D. 2014. Aerobic culture of anaerobic bacteria using antioxidants: a preliminary report. Eur J Clin Microbiol Infect Dis 33:1781–1783. doi: 10.1007/s10096-014-2137-4. [DOI] [PubMed] [Google Scholar]
- 127.Rappe MS, Giovannoni SJ. 2003. The uncultured microbial majority. Annu Rev Microbiol 57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- 128.Zengler K, Toledo G, Rappe M, Elkins J, Mathur EJ, Short JM, Keller M. 2002. Cultivating the uncultured. Proc Natl Acad Sci U S A 99:15681–15686. doi: 10.1073/pnas.252630999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Epstein S. 2013. The phenomenon of microbial uncultivability. Curr Opin Microbiol 16:636–642. doi: 10.1016/j.mib.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 130.Staley JT, Konopka A. 1985. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu Rev Microbiol 39:321–346. doi: 10.1146/annurev.mi.39.100185.001541. [DOI] [PubMed] [Google Scholar]
- 131.Baik KS, Park SC, Kim EM, Bae KS, Ahn JH, Ka JO, Chun J, Seong CN. 2008. Diversity of bacterial community in freshwater of Woopo wetland. J Microbiol 46:647–655. doi: 10.1007/s12275-008-0135-x. [DOI] [PubMed] [Google Scholar]
- 132.Button DK, Schut F, Quang P, Martin R, Robertson BR. 1993. Viability and isolation of marine bacteria by dilution culture: theory, procedures, and initial results. Appl Environ Microbiol 59:881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Puspita ID, Kamagata Y, Tanaka M, Asano K, Nakatsu CH. 2012. Are uncultivated bacteria really uncultivable? Microbes Environ 27:356–366. doi: 10.1264/jsme2.ME12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Field KG, Gordon D, Wright T, Rappe M, Urback E, Vergin K, Giovannoni SJ. 1997. Diversity and depth-specific distribution of SAR11 cluster rRNA genes from marine planktonic bacteria. Appl Environ Microbiol 63:63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rappe MS, Connon SA, Vergin KL, Giovannoni SJ. 2002. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418:630–633. doi: 10.1038/nature00917. [DOI] [PubMed] [Google Scholar]
- 136.Sun J, Steindler L, Thrash JC, Halsey KH, Smith DP, Carter AE, Landry ZC, Giovannoni SJ. 2011. One carbon metabolism in SAR11 pelagic marine bacteria. PLoS One 6:e23973. doi: 10.1371/journal.pone.0023973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bollmann A, Lewis K, Epstein SS. 2007. Incubation of environmental samples in a diffusion chamber increases the diversity of recovered isolates. Appl Environ Microbiol 73:6386–6390. doi: 10.1128/AEM.01309-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Evans NM, Bell SM, Smith DD. 1975. New satellitism test for isolation and identification of Haemophilus influenzae and Haemophilus parainfluenzae in sputum. J Clin Microbiol 1:89–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kutzer P, Schulze C, Engelhardt A, Wieler LH, Nordhoff M. 2008. Helcococcus ovis, an emerging pathogen in bovine valvular endocarditis. J Clin Microbiol 46:3291–3295. doi: 10.1128/JCM.00867-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Aoi Y, Kinoshita T, Hata T, Ohta H, Obokata H, Tsuneda S. 2009. Hollow-fiber membrane chamber as a device for in situ environmental cultivation. Appl Environ Microbiol 75:3826–3833. doi: 10.1128/AEM.02542-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gavrish E, Bollmann A, Epstein S, Lewis K. 2008. A trap for in situ cultivation of filamentous actinobacteria. J Microbiol Methods 72:257–262. doi: 10.1016/j.mimet.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ferrari BC, Winsley T, Gillings M, Binnerup S. 2008. Cultivating previously uncultured soil bacteria using a soil substrate membrane system. Nat Protoc 3:1261–1269. doi: 10.1038/nprot.2008.102. [DOI] [PubMed] [Google Scholar]
- 143.Nichols D, Cahoon N, Trakhtenberg EM, Pham L, Mehta A, Belanger A, Kanigan T, Lewis K, Epstein SS. 2010. Use of ichip for high-throughput in situ cultivation of “uncultivable” microbial species. Appl Environ Microbiol 76:2445–2450. doi: 10.1128/AEM.01754-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Dandekar AA, Chugani S, Greenberg EP. 2012. Bacterial quorum sensing and metabolic incentives to cooperate. Science 338:264–266. doi: 10.1126/science.1227289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chandler JR, Heilmann S, Mittler JE, Greenberg EP. 2012. Acyl-homoserine lactone-dependent eavesdropping promotes competition in a laboratory co-culture model. ISME J 6:2219–2228. doi: 10.1038/ismej.2012.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bruns A, Cypionka H, Overmann J. 2002. Cyclic AMP and acyl homoserine lactones increase the cultivation efficiency of heterotrophic bacteria from the central Baltic Sea. Appl Environ Microbiol 68:3978–3987. doi: 10.1128/AEM.68.8.3978-3987.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.D'Onofrio A, Crawford JM, Stewart EJ, Witt K, Gavrish E, Epstein S, Clardy J, Lewis K. 2010. Siderophores from neighboring organisms promote the growth of uncultured bacteria. Chem Biol 17:254–264. doi: 10.1016/j.chembiol.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Radzki W, Gutierrez Manero FJ, Algar E, Lucas Garcia JA, Garcia-Villaraco A, Ramos SB. 2013. Bacterial siderophores efficiently provide iron to iron-starved tomato plants in hydroponics culture. Antonie Van Leeuwenhoek 104:321–330. doi: 10.1007/s10482-013-9954-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Eto D, Watanabe K, Saeki H, Oinuma K, Otani K, Nobukuni M, Shiratori-Takano H, Takano H, Beppu T, Ueda K. 2013. Divergent effects of desferrioxamine on bacterial growth and characteristics. J Antibiot (Tokyo) 66:199–203. doi: 10.1038/ja.2012.111. [DOI] [PubMed] [Google Scholar]
- 150.Nichols D, Lewis K, Orjala J, Mo S, Ortenberg R, O'Connor P, Zhao C, Vouros P, Kaeberlein T, Epstein SS. 2008. Short peptide induces an “uncultivable” microorganism to grow in vitro. Appl Environ Microbiol 74:4889–4897. doi: 10.1128/AEM.00393-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lewis K, Epstein S, D'Onofrio A, Ling LL. 2010. Uncultured microorganisms as a source of secondary metabolites. J Antibiot (Tokyo) 63:468–476. doi: 10.1038/ja.2010.87. [DOI] [PubMed] [Google Scholar]
- 152.Epstein SS. 2009. Microbial awakenings. Nature 457:1083. doi: 10.1038/4571083a. [DOI] [PubMed] [Google Scholar]
- 153.Buerger S, Spoering A, Gavrish E, Leslin C, Ling L, Epstein SS. 2012. Microbial scout hypothesis, stochastic exit from dormancy, and the nature of slow growers. Appl Environ Microbiol 78:3221–3228. doi: 10.1128/AEM.07307-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Buerger S, Spoering A, Gavrish E, Leslin C, Ling L, Epstein SS. 2012. Microbial scout hypothesis and microbial discovery. Appl Environ Microbiol 78:3229–3233. doi: 10.1128/AEM.07308-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sizova MV, Hohmann T, Hazen A, Paster BJ, Halem SR, Murphy CM, Panikov NS, Epstein SS. 2012. New approaches for isolation of previously uncultivated oral bacteria. Appl Environ Microbiol 78:194–203. doi: 10.1128/AEM.06813-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Janssen PH, Yates PS, Grinton BE, Taylor PM, Sait M. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl Environ Microbiol 68:2391–2396. doi: 10.1128/AEM.68.5.2391-2396.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Derrien M, Vaughan EE, Plugge CM, de Vos WM. 2004. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol 54:1469–1476. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- 158.van Passel MW, Kant R, Zoetendal EG, Plugge CM, Derrien M, Malfatti SA, Chain PS, Woyke T, Palva A, de Vos WM, Smidt H. 2011. The genome of Akkermansia muciniphila, a dedicated intestinal mucin degrader, and its use in exploring intestinal metagenomes. PLoS One 6:e16876. doi: 10.1371/journal.pone.0016876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Dubourg G, Lagier JC, Armougom F, Robert C, Audoly G, Papazian L, Raoult D. 2013. High-level colonisation of the human gut by Verrucomicrobia following broad-spectrum antibiotic treatment. Int J Antimicrob Agents 41:149–155. doi: 10.1016/j.ijantimicag.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 160.Karlsson CL, Onnerfalt J, Xu J, Molin G, Ahrne S, Thorngren-Jerneck K. 2012. The microbiota of the gut in preschool children with normal and excessive body weight. Obesity (Silver Spring) 20:2257–2261. doi: 10.1038/oby.2012.110. [DOI] [PubMed] [Google Scholar]
- 161.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, de Vos WM, Cani PD. 2013. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A 110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Belzer C, de Vos WM. 2012. Microbes inside—from diversity to function: the case of Akkermansia. ISME J 6:1449–1458. doi: 10.1038/ismej.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Woese CR, Gupta R. 1981. Are archaebacteria merely derived ‘prokaryotes’? Nature 289:95–96. doi: 10.1038/289095a0. [DOI] [PubMed] [Google Scholar]
- 164.Ventosa A, Nieto JJ, Oren A. 1998. Biology of moderately halophilic aerobic bacteria. Microbiol Mol Biol Rev 62:504–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Oren A. 2002. Diversity of halophilic microorganisms: environments, phylogeny, physiology, and applications. J Ind Microbiol Biotechnol 28:56–63. doi: 10.1038/sj/jim/7000176. [DOI] [PubMed] [Google Scholar]
- 166.Oxley AP, Lanfranconi MP, Wurdemann D, Ott S, Schreiber S, McGenity TJ, Timmis KN, Nogales B. 2010. Halophilic archaea in the human intestinal mucosa. Environ Microbiol 12:2398–2410. doi: 10.1111/j.1462-2920.2010.02212.x. [DOI] [PubMed] [Google Scholar]
- 167.Margesin R, Schinner F. 2001. Potential of halotolerant and halophilic microorganisms for biotechnology. Extremophiles 5:73–83. doi: 10.1007/s007920100184. [DOI] [PubMed] [Google Scholar]
- 168.Schlesner H, Hirsch P. 1987. Rejection of the genus name Pirella for pear-shaped budding bacteria and proposal to create the genus Pirellula gen. nov. Int J Syst Evol Microbiol 37:441. [Google Scholar]
- 169.Lage OM, Bondoso J. 2012. Bringing Planctomycetes into pure culture. Front Microbiol 3:405. doi: 10.3389/fmicb.2012.00405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wagner M, Horn M. 2006. The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr Opin Biotechnol 17:241–249. doi: 10.1016/j.copbio.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 171.Schlesner H. 1994. The development of media suitable for the microorganisms morphologically resembling Planctomyces spp., Pirellula spp., and other Planctomycetes from various aquatic habitats using dilute media. Syst Appl Microbiol 17:135–145. [Google Scholar]
- 172.Cayrou C, Raoult D, Drancourt M. 2010. Broad-spectrum antibiotic resistance of Planctomycetes organisms determined by Etest. J Antimicrob Chemother 65:2119–2122. doi: 10.1093/jac/dkq290. [DOI] [PubMed] [Google Scholar]
- 173.Fuerst JA, Sagulenko E. 2011. Beyond the bacterium: planctomycetes challenge our concepts of microbial structure and function. Nat Rev Microbiol 9:403–413. doi: 10.1038/nrmicro2578. [DOI] [PubMed] [Google Scholar]
- 174.Davis KE, Joseph SJ, Janssen PH. 2005. Effects of growth medium, inoculum size, and incubation time on culturability and isolation of soil bacteria. Appl Environ Microbiol 71:826–834. doi: 10.1128/AEM.71.2.826-834.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Elshahed MS, Youssef NH, Luo Q, Najar FZ, Roe BA, Sisk TM, Buhring SI, Hinrichs KU, Krumholz LR. 2007. Phylogenetic and metabolic diversity of Planctomycetes from anaerobic, sulfide- and sulfur-rich Zodletone Spring, Oklahoma. Appl Environ Microbiol 73:4707–4716. doi: 10.1128/AEM.00591-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. 2010. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A 107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Cayrou C, Sambe B, Armougom F, Raoult D, Drancourt M. 2013. Molecular diversity of the Planctomycetes in the human gut microbiota in France and Senegal. APMIS 121:1082–1090. doi: 10.1111/apm.12087. [DOI] [PubMed] [Google Scholar]
- 178.Cayrou C, Raoult D, Drancourt M. 2010. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry for the identification of environmental organisms: the Planctomycetes paradigm. Environ Microbiol Rep 2:752–760. doi: 10.1111/j.1758-2229.2010.00176.x. [DOI] [PubMed] [Google Scholar]
- 179.Raoult D, Henrissat B. 2013. Are stool samples suitable for studying the link between gut microbiota and obesity? Eur J Epidemiol 29:307–309. doi: 10.1007/s10654-014-9905-4. [DOI] [PubMed] [Google Scholar]
- 180.Moore WE, Holdeman LV. 1974. Human fecal flora: the normal flora of 20 Japanese-Hawaiians. Appl Microbiol 27:961–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Savage DC. 1977. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol 31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 182.Finegold SM, Attebery HR, Sutter VL. 1974. Effect of diet on human fecal flora: comparison of Japanese and American diets. Am J Clin Nutr 27:1456–1469. [DOI] [PubMed] [Google Scholar]
- 183.Stephen AM, Cummings JH. 1980. The microbial contribution to human faecal mass. J Med Microbiol 13:45–56. doi: 10.1099/00222615-13-1-45. [DOI] [PubMed] [Google Scholar]
- 184.Dethlefsen L, Huse S, Sogin ML, Relman DA. 2008. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Rajilic-Stojanovic M, Smidt H, de Vos WM. 2007. Diversity of the human gastrointestinal tract microbiota revisited. Environ Microbiol 9:2125–2136. doi: 10.1111/j.1462-2920.2007.01369.x. [DOI] [PubMed] [Google Scholar]
- 186.Simon GL, Gorbach SL. 1984. Intestinal flora in health and disease. Gastroenterology 86:174–193. [PubMed] [Google Scholar]
- 187.Finegold SM, Sutter VL, Sugihara PT, Elder HA, Lehmann SM, Phillips RL. 1977. Fecal microbial flora in Seventh Day Adventist populations and control subjects. Am J Clin Nutr 30:1781–1792. [DOI] [PubMed] [Google Scholar]
- 188.Attebery HR, Sutter VL, Finegold SM. 1972. Effect of a partially chemically defined diet on normal human fecal flora. Am J Clin Nutr 25:1391–1398. [DOI] [PubMed] [Google Scholar]
- 189.Aries V, Crowther JS, Drasar BS, Hill MJ, Williams RE. 1969. Bacteria and the aetiology of cancer of the large bowel. Gut 10:334–335. doi: 10.1136/gut.10.5.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Hill MJ, Drasar BS. 1975. The normal colonic bacterial flora. Gut 16:318–323. doi: 10.1136/gut.16.4.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Hayashi H, Sakamoto M, Benno Y. 2002. Phylogenetic analysis of the human gut microbiota using 16S rDNA clone libraries and strictly anaerobic culture-based methods. Microbiol Immunol 46:535–548. doi: 10.1111/j.1348-0421.2002.tb02731.x. [DOI] [PubMed] [Google Scholar]
- 192.Wilson KH, Blitchington RB. 1996. Human colonic biota studied by ribosomal DNA sequence analysis. Appl Environ Microbiol 62:2273–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, Dantas G, Gordon JI. 2011. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci U S A 108:6252–6257. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Alhanout K, Malesinki S, Vidal N, Peyrot V, Rolain JM, Brunel JM. 2010. New insights into the antibacterial mechanism of action of squalamine. J Antimicrob Chemother 65:1688–1693. doi: 10.1093/jac/dkq213. [DOI] [PubMed] [Google Scholar]
- 195.Chandler FW Jr, Clark JW Jr. 1970. Membrane filtration of the Reiter treponeme. Appl Microbiol 20:786–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Chaban B, Guerra AG, Hendrick SH, Waldner CL, Hill JE. 2013. Isolation rates of Campylobacter fetus subsp venerealis from bovine preputial samples via passive filtration on nonselective medium versus selective medium, with and without transport medium. Am J Vet Res 74:1066–1069. doi: 10.2460/ajvr.74.8.1066. [DOI] [PubMed] [Google Scholar]
- 197.Lagier JC, Ramasamy D, Rivet R, Raoult D, Fournier PE. 2012. Non contiguous-finished genome sequence and description of Cellulomonas massiliensis sp. nov. Stand Genomic Sci 7:258–270. doi: 10.4056/sigs.3316719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Lagier JC, Gimenez G, Robert C, Raoult D, Fournier PE. 2012. Non-contiguous finished genome sequence and description of Herbaspirillum massiliense sp. nov. Stand Genomic Sci 7:200–209. doi: 10.4056/sigs.3086474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Marler LM, Siders JA, Wolters LC, Pettigrew Y, Skitt BL, Allen SD. 1992. Comparison of five cultural procedures for isolation of Clostridium difficile from stools. J Clin Microbiol 30:514–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Hecker M, Schumann W, Volker U. 1996. Heat-shock and general stress response in Bacillus subtilis. Mol Microbiol 19:417–428. [DOI] [PubMed] [Google Scholar]
- 201.Lu TK, Koeris MS. 2011. The next generation of bacteriophage therapy. Curr Opin Microbiol 14:524–531. doi: 10.1016/j.mib.2011.07.028. [DOI] [PubMed] [Google Scholar]
- 202.Sulakvelidze A. 2013. Using lytic bacteriophages to eliminate or significantly reduce contamination of food by foodborne bacterial pathogens. J Sci Food Agric 93:3137–3146. doi: 10.1002/jsfa.6222. [DOI] [PubMed] [Google Scholar]
- 203.Subramanyam B, Sivaramakrishnan GN, Dusthackeer A, Nagamiah S, Kumar V. 2012. Phage lysin as a substitute for antibiotics to detect Mycobacterium tuberculosis from sputum samples with the BACTEC MGIT 960 system. Clin Microbiol Infect 18:497–501. doi: 10.1111/j.1469-0691.2011.03601.x. [DOI] [PubMed] [Google Scholar]
- 204.Subramanyam B, Sivaramakrishnan G, Dusthackeer A, Kumar V. 2013. Phage lysin to control the overgrowth of normal flora in processed sputum samples for the rapid and sensitive detection of Mycobacterium tuberculosis by luciferase reporter phage assay. BMC Infect Dis 13:44. doi: 10.1186/1471-2334-13-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Sillankorva S, Oliveira D, Moura A, Henriques M, Faustino A, Nicolau A, Azeredo J. 2011. Efficacy of a broad host range lytic bacteriophage against E. coli adhered to urothelium. Curr Microbiol 62:1128–1132. doi: 10.1007/s00284-010-9834-8. [DOI] [PubMed] [Google Scholar]
- 206.Lagier JC, El Karkouri K, Mishra AK, Robert C, Raoult D, Fournier PE. 2013. Non contiguous-finished genome sequence and description of Enterobacter massiliensis sp. nov. Stand Genomic Sci 7:399–412. doi: 10.4056/sigs.3396830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Yagupsky P. 1999. Use of blood culture systems for isolation of Kingella kingae from synovial fluid. J Clin Microbiol 37:3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Yagupsky P, Dagan R, Howard CW, Einhorn M, Kassis I, Simu A. 1992. High prevalence of Kingella kingae in joint fluid from children with septic arthritis revealed by the BACTEC blood culture system. J Clin Microbiol 30:1278–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Al Masalma M, Raoult D, Roux V. 2009. Phocaeicola abscessus gen. nov., sp. nov., an anaerobic bacterium isolated from a human brain abscess sample. Int J Syst Evol Microbiol 59:2232–2237. doi: 10.1099/ijs.0.007823-0. [DOI] [PubMed] [Google Scholar]
- 210.Nottingham PM, Hungate RE. 1968. Isolation of methanogenic bacteria from feces of man. J Bacteriol 96:2178–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Eller C, Crabill MR, Bryant MP. 1971. Anaerobic roll tube media for nonselective enumeration and isolation of bacteria in human feces. Appl Microbiol 22:522–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Salanitro JP, Blake IG, Muirhead PA. 1977. Isolation and identification of fecal bacteria from adult swine. Appl Environ Microbiol 33:79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Ahn Y, Sung K, Rafii F, Cerniglia CE. 2012. Effect of sterilized human fecal extract on the sensitivity of Escherichia coli ATCC 25922 to enrofloxacin. J Antibiot (Tokyo) 65:179–184. doi: 10.1038/ja.2012.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Binek M, Szynkiewicz Z. 1985. Multiplication in liquid medium of Treponema sp. isolated from intestinal contents of swine. Acta Microbiol Pol 34:167–175. [PubMed] [Google Scholar]
- 215.Szynkiewicz ZM, Binek M. 1986. Evaluation of selective media for primary isolation of Treponema hyodysenteriae and Treponema innocens. Comp Immunol Microbiol Infect Dis 9:71–77. [DOI] [PubMed] [Google Scholar]
- 216.Betian HG, Linehan BA, Bryant MP, Holdeman LV. 1977. Isolation of a cellulotytic Bacteroides sp. from human feces. Appl Environ Microbiol 33:1009–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Wozny MA, Bryant MP, Holdeman LV, Moore WE. 1977. Urease assay and urease-producing species of anaerobes in the bovine rumen and human feces. Appl Environ Microbiol 33:1097–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Edward DG, Fitzgerald WA. 1951. Cholesterol in the growth of organisms of the pleuropneumonia group. J Gen Microbiol 5:576–586. [DOI] [PubMed] [Google Scholar]
- 219.Eddy BP, Ingram M. 1953. Interactions between ascorbic acid and bacteria. Bacteriol Rev 17:93–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Samb-Ba B, Mazenot C, Gassama-Sow A, Dubourg G, Richet H, Hugon P, Lagier JC, Raoult D, Fenollar F. 2014. MALDI-TOF identification of the human gut microbiome in people with and without diarrhea in Senegal. PLoS One 9:e87419. doi: 10.1371/journal.pone.0087419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Mishra AK, Lagier JC, Nguyen TT, Raoult D, Fournier PE. 2013. Non contiguous-finished genome sequence and description of Peptoniphilus senegalensis sp. nov. Stand Genomic Sci 7:370–381. doi: 10.4056/sigs.3366764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Mishra AK, Hugon P, Lagier JC, Nguyen TT, Robert C, Couderc C, Raoult D, Fournier PE. 2013. Non contiguous-finished genome sequence and description of Peptoniphilus obesi sp. nov. Stand Genomic Sci 7:357–369. doi: 10.4056/sigs.32766871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Kokcha S, Mishra AK, Lagier JC, Million M, Leroy Q, Raoult D, Fournier PE. 2012. Non contiguous-finished genome sequence and description of Bacillus timonensis sp. nov. Stand Genomic Sci 6:346–355. doi: 10.4056/sigs.2776064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Kokcha S, Ramasamy D, Lagier JC, Robert C, Raoult D, Fournier PE. 2012. Non-contiguous finished genome sequence and description of Brevibacterium senegalense sp. nov. Stand Genomic Sci 7:233–245. doi: 10.4056/sigs.3256677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Lagier JC, Armougom F, Mishra AK, Nguyen TT, Raoult D, Fournier PE. 2012. Non-contiguous finished genome sequence and description of Alistipes timonensis sp. nov. Stand Genomic Sci 6:315–324. doi: 10.4056/sigs.2685917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Lagier JC, El Karkouri K, Nguyen TT, Armougom F, Raoult D, Fournier PE. 2012. Non-contiguous finished genome sequence and description of Anaerococcus senegalensis sp. nov. Stand Genomic Sci 6:116–125. doi: 10.4056/sigs.2415480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Mishra AK, Gimenez G, Lagier JC, Robert C, Raoult D, Fournier PE. 2012. Genome sequence and description of Alistipes senegalensis sp. nov. Stand Genomic Sci 6:1–16. doi: 10.4056/sigs.2625821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Mishra AK, Lagier JC, Robert C, Raoult D, Fournier PE. 2012. Non-contiguous finished genome sequence and description of Clostridium senegalense sp. nov. Stand Genomic Sci 6:386–395. doi: 10.4056/sigs.2766062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Mishra AK, Lagier JC, Robert C, Raoult D, Fournier PE. 2012. Non contiguous-finished genome sequence and description of Peptoniphilus timonensis sp. nov. Stand Genomic Sci 7:1–11. doi: 10.4056/sigs.2956294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Mishra AK, Lagier JC, Rivet R, Raoult D, Fournier PE. 2012. Non-contiguous finished genome sequence and description of Paenibacillus senegalensis sp. nov. Stand Genomic Sci 7:70–81. doi: 10.4056/sigs.3056450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Ramasamy D, Kokcha S, Lagier JC, Nguyen TT, Raoult D, Fournier PE. 2012. Genome sequence and description of Aeromicrobium massiliense sp. nov. Stand Genomic Sci 7:246–257. doi: 10.4056/sigs.3306717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Roux V, El Karkouri K, Lagier JC, Robert C, Raoult D. 2012. Non-contiguous finished genome sequence and description of Kurthia massiliensis sp. nov. Stand Genomic Sci 7:221–232. doi: 10.4056/sigs.3206554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Hugon P, Mishra AK, Lagier JC, Nguyen TT, Couderc C, Raoult D, Fournier PE. 2013. Non-contiguous finished genome sequence and description of Brevibacillus massiliensis sp. nov. Stand Genomic Sci 8:1–14. doi: 10.4056/sigs.3466975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Mishra AK, Hugon P, Lagier JC, Nguyen TT, Couderc C, Raoult D, Fournier PE. 2013. Non contiguous-finished genome sequence and description of Enorma massiliensis gen. nov., sp. nov., a new member of the family Coriobacteriaceae. Stand Genomic Sci 8:290–305. doi: 10.4056/sigs.3426906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Hugon P, Ramasamy D, Lagier JC, Rivet R, Couderc C, Raoult D, Fournier PE. 2013. Non contiguous-finished genome sequence and description of Alistipes obesi sp. nov. Stand Genomic Sci 7:427–439. doi: 10.4056/sigs.3336746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Lagier JC, El Karkouri K, Rivet R, Couderc C, Raoult D, Fournier PE. 2013. Non contiguous-finished genome sequence and description of Senegalemassilia anaerobia gen. nov., sp. nov. Stand Genomic Sci 7:343–356. doi: 10.4056/sigs.3246665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Ramasamy D, Lagier JC, Nguyen TT, Raoult D, Fournier PE. 2013. Non contiguous-finished genome sequence and description of Dielma fastidiosa gen. nov., sp. nov., a new member of the family Erysipelotrichaceae. Stand Genomic Sci 8:336–351. doi: 10.4056/sigs.3567059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Mishra AK, Lagier JC, Robert C, Raoult D, Fournier PE. 2013. Genome sequence and description of Timonella senegalensis gen. nov., sp. nov., a new member of the suborder Micrococcinae. Stand Genomic Sci 8:318–335. doi: 10.4056/sigs.3476977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Ramasamy D, Lagier JC, Gorlas A, Raoult D, Fournier PE. 2013. Non contiguous-finished genome sequence and description of Bacillus massiliosenegalensis sp. nov. Stand Genomic Sci 8:264–278. doi: 10.4056/sigs.3496989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Mishra AK, Pfleiderer A, Lagier JC, Robert C, Raoult D, Fournier PE. 2013. Non contiguous-finished genome sequence and description of Bacillus massilioanorexius sp. nov. Stand Genomic Sci 8:465–479. doi: 10.4056/sigs.4087826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Hugon P, Ramasamy D, Robert C, Couderc C, Raoult D, Fournier PE. 2013. Non-contiguous finished genome sequence and description of Kallipyga massiliensis gen. nov., sp. nov., a new member of the family Clostridiales Sedis XI. Stand Genomic Sci 8:500–515. doi: 10.4056/sigs.4047997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Padhmanabhan R, Lagier JC, Makaya Dangui NP, Blanc-Tailleur C, Couderc C, Raoult D. 2013. Non-contiguous finished genome sequence and description of Megasphaera massiliensis. Stand Genomic Sci 8:525–538. doi: 10.4056/sigs.4077819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Mishra AK, Edouard S, Makaya Dangui NP, Lagier JC, Caputo A, Blanc-Tailleur C, Ravaux I, Raoult D, Fournier PE. 2013. Non-contiguous finished genome sequence and description of Nosocomiicoccus massiliensis sp. nov. Stand Genomic Sci 9:205–219. doi: 10.4056/sigs.4378121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Roux V, Million M, Robert C, Magne A, Raoult D. 2013. Non-contiguous finished genome sequence and description of Oceanobacillus massiliensis. Stand Genomic Sci 9:370–384. doi: 10.4056/sigs.4267953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Lo CI, Mishra AK, Padhmanabhan R, Samb-Ba B, Gassama Sow A, Robert C, Couderc C, Faye N, Raoult D, Fournier PE. 2013. Non-contiguous finished genome sequence and description of Clostridium dakarense sp. nov. Stand Genomic Sci 9:14–27. doi: 10.4056/sigs.4097825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Mishra AK, Lagier JC, Pfleiderer A, Nguyen TT, Caputo A, Raoult D, Fournier PE. 2013. Non-contiguous finished genome sequence and description of Holdemania massiliensis sp. nov. Stand Genomic Sci 9:395–409. doi: 10.4056/sigs.4628316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Mishra AK, Hugon P, Robert C, Raoult D, Fournier PE. 2012. Non contiguous-finished genome sequence and description of Peptoniphilus grossensis sp. nov. Stand Genomic Sci 7:320–330. doi: 10.4056/sigs.3076460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Li K, Bihan M, Methe BA. 2013. Analyses of the stability and core taxonomic memberships of the human microbiome. PLoS One 8:e63139. doi: 10.1371/journal.pone.0063139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, Rosenbaum M, Gordon JI. 2013. The long-term stability of the human gut microbiota. Science 341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Trehan I, Goldbach HS, LaGrone LN, Meuli GJ, Wang RJ, Maleta KM, Manary MJ. 2013. Antibiotics as part of the management of severe acute malnutrition. N Engl J Med 368:425–435. doi: 10.1056/NEJMoa1202851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Million M, Lagier JC, Yahav D, Paul M. 2013. Gut bacterial microbiota and obesity. Clin Microbiol Infect 19:305–313. doi: 10.1111/1469-0691.12172. [DOI] [PubMed] [Google Scholar]
- 252.Tindall BJ, Rossello-Mora R, Busse HJ, Ludwig W, Kampfer P. 2010. Notes on the characterization of prokaryote strains for taxonomic purposes. Int J Syst Evol Microbiol 60:249–266. doi: 10.1099/ijs.0.016949-0. [DOI] [PubMed] [Google Scholar]
- 253.Kampfer P, Glaeser SP. 2012. Prokaryotic taxonomy in the sequencing era—the polyphasic approach revisited. Environ Microbiol 14:291–317. doi: 10.1111/j.1462-2920.2011.02615.x. [DOI] [PubMed] [Google Scholar]
- 254.Sentausa E, Fournier PE. 2013. Advantages and limitations of genomics in prokaryotic taxonomy. Clin Microbiol Infect 19:790–795. doi: 10.1111/1469-0691.12181. [DOI] [PubMed] [Google Scholar]
- 255.Stackebrandt E, Ebers J. 2006. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today 33:152–155. [Google Scholar]
- 256.Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WE, Murray RGE, Stackebrandt E, Starr MP, Truper HG. 1987. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464. doi: 10.1099/00207713-37-4-463. [DOI] [Google Scholar]
- 257.Meier-Kolthoff JP, Goker M, Sproer C, Klenk HP. 2013. When should a DDH experiment be mandatory in microbial taxonomy? Arch Microbiol 195:413–418. doi: 10.1007/s00203-013-0888-4. [DOI] [PubMed] [Google Scholar]
- 258.Vandamme P, Pot B, Gillis M, de Vos P, Kersters K, Swings J. 1996. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol Rev 60:407–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Stackebrandt E, Frederiksen W, Garrity GM, Grimont PA, Kampfer P, Maiden MC, Nesme X, Rossello-Mora R, Swings J, Truper HG, Vauterin L, Ward AC, Whitman WB. 2002. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int J Syst Evol Microbiol 52:1043–1047. doi: 10.1099/ijs.0.02360-0. [DOI] [PubMed] [Google Scholar]
- 260.Fournier PE, Drancourt M, Raoult D. 2007. Bacterial genome sequencing and its use in infectious diseases. Lancet Infect Dis 7:711–723. doi: 10.1016/S1473-3099(07)70260-8. [DOI] [PubMed] [Google Scholar]
- 261.Renesto P, Crapoulet N, Ogata H, La Scola B, Vestris G, Claverie JM, Raoult D. 2003. Genome-based design of a cell-free culture medium for Tropheryma whipplei. Lancet 362:447–449. doi: 10.1016/S0140-6736(03)14071-8. [DOI] [PubMed] [Google Scholar]
- 262.Omsland A, Sager J, Nair V, Sturdevant DE, Hackstadt T. 2012. Developmental stage-specific metabolic and transcriptional activity of Chlamydia trachomatis in an axenic medium. Proc Natl Acad Sci U S A 109:19781–19785. doi: 10.1073/pnas.1212831109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Fournier PE, Raoult D. 2011. Prospects for the future using genomics and proteomics in clinical microbiology. Annu Rev Microbiol 65:169–188. doi: 10.1146/annurev-micro-090110-102922. [DOI] [PubMed] [Google Scholar]
- 264.Wolf YI, Rogozin IB, Grishin NV, Tatusov RL, Koonin EV. 2001. Genome trees constructed using five different approaches suggest new major bacterial clades. BMC Evol Biol 1:8. doi: 10.1186/1471-2210-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Mirkin BG, Fenner TI, Galperin MY, Koonin EV. 2003. Algorithms for computing parsimonious evolutionary scenarios for genome evolution, the last universal common ancestor and dominance of horizontal gene transfer in the evolution of prokaryotes. BMC Evol Biol 3:2. doi: 10.1186/1471-2210-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Fitz-Gibbon ST, House CH. 1999. Whole genome-based phylogenetic analysis of free-living microorganisms. Nucleic Acids Res 27:4218–4222. doi: 10.1093/nar/27.21.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Zhi XY, Zhao W, Li WJ, Zhao GP. 2012. Prokaryotic systematics in the genomics era. Antonie Van Leeuwenhoek 101:21–34. doi: 10.1007/s10482-011-9667-x. [DOI] [PubMed] [Google Scholar]
- 268.Konstantinidis KT, Tiedje JM. 2005. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci U S A 102:2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. 2007. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 270.Richter M, Rossello-Mora R. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A 106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Ritalahti KM, Justicia-Leon SD, Cusick KD, Ramos-Hernandez N, Rubin M, Dornbush J, Loffler FE. 2012. Sphaerochaeta globosa gen. nov., sp. nov. and Sphaerochaeta pleomorpha sp. nov., free-living, spherical spirochaetes. Int J Syst Evol Microbiol 62:210–216. doi: 10.1099/ijs.0.023986-0. [DOI] [PubMed] [Google Scholar]
- 272.Hoffmann M, Monday SR, Allard MW, Strain EA, Whittaker P, Naum M, McCarthy PJ, Lopez JV, Fischer M, Brown EW. 2012. Vibrio caribbeanicus sp. nov., isolated from the marine sponge Scleritoderma cyanea. Int J Syst Evol Microbiol 62:1736–1743. doi: 10.1099/ijs.0.032375-0. [DOI] [PubMed] [Google Scholar]
- 273.Loffler FE, Yan J, Ritalahti KM, Adrian L, Edwards EA, Konstantinidis KT, Muller JA, Fullerton H, Zinder SH, Spormann AM. 2013. Dehalococcoides mccartyi gen. nov., sp. nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov. and family Dehalococcoidaceae fam. nov., within the phylum Chloroflexi. Int J Syst Evol Microbiol 63:625–635. doi: 10.1099/ijs.0.034926-0. [DOI] [PubMed] [Google Scholar]
- 274.Ricker N, Qian H, Fulthorpe RR. 2012. The limitations of draft assemblies for understanding prokaryotic adaptation and evolution. Genomics 100:167–175. doi: 10.1016/j.ygeno.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 275.Klassen JL, Currie CR. 2012. Gene fragmentation in bacterial draft genomes: extent, consequences and mitigation. BMC Genomics 13:14. doi: 10.1186/1471-2164-13-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Klenk HP, Goker M. 2010. En route to a genome-based classification of Archaea and Bacteria? Syst Appl Microbiol 33:175–182. doi: 10.1016/j.syapm.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 277.Edouard S, Sankar S, Makaya Dangui NP, Lagier JC, Michelle C, Raoult D, Fournier PE. 2014. Genome sequence and description of Nesterenkonia massiliensis sp. nov. strain NP1T. Stand Genomic Sci 9:866–882. doi: 10.4056/sigs.5631022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Keita MB, Diene SM, Robert C, Raoult D, Fournier PE. 2013. Non-contiguous finished genome sequence and description of Bacillus massiliogorillae sp. nov. Stand Genomic Sci 9:93–105. doi: 10.4056/sigs.4388124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Hassani II, Robert C, Michelle C, Raoult D, Hacene H, Desnues C. 2013. Non-contiguous finished genome sequence and description of Halopiger djelfamassiliensis sp. nov. Stand Genomic Sci 9:160–174. doi: 10.4056/sigs.4578289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Pagnier I, Croce O, Robert C, Raoult D, La Scola B. 2013. Non-contiguous finished genome sequence and description of Anaerococcus pacaensis sp. nov. Stand Genomic Sci 8:548–560. doi: 10.4056/sigs.4177252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Mediannikov O, El Karkouri K, Robert C, Fournier PE, Raoult D. 2013. Non-contiguous finished genome sequence and description of Bartonella florenciae sp. nov. Stand Genomic Sci 9:185–196. doi: 10.4056/sigs.4358060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Mediannikov O, El Karkouri K, Diatta G, Robert C, Fournier PE, Raoult D. 2013. Non-contiguous finished genome sequence and description of Bartonella senegalensis sp. nov. Stand Genomic Sci 8:279–289. doi: 10.4056/sigs.3807472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Cassir N, Croce O, Pagnier I, Benamar S, Couderc C, Robert C, Raoult D, La Scola B. 2014. Non-contiguous finished genome sequence and description of Bacteroides neonati sp. nov., a new species of anaerobic bacterium. Stand Genomic Sci 9:794–806. doi: 10.4056/sigs.5159098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Ramasamy D, Dubourg G, Robert C, Caputo A, Papazian L, Raoult D, Fournier PE. 2014. Non contiguous-finished genome sequence and description of Enorma timonensis sp. nov. Stand Genomic Sci 9:970–986. doi: 10.4056/sigs.4878632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Edouard S, Bibi F, Ramasamy D, Lagier JC, Azhar EI, Robert C, Yasir M, Jiman-Fatani AA, Alawi M, Fournier PE, Raoult D. 2014. Non-contiguous finished genome sequence and description of Corynebacterium jeddahense sp. nov. Stand Genomic Sci 9:987–1002. doi: 10.4056/sigs.5561028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Lagier JC, Bibi F, Ramasamy D, Azhar EI, Robert C, Yasir M, Jiman-Fatani AA, Alshali KZ, Fournier PE, Raoult D. 2014. Non contiguous-finished genome sequence and description of Clostridium jeddahense sp. nov. Stand Genomic Sci 9:1003–1019. doi: 10.4056/sigs.5571026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Padhmanabhan R, Dubourg G, Lagier JC, Couderc C, Michelle C, Raoult D, Fournier PE. 2014. Genome sequence and description of Corynebacterium ihumii sp. nov. Stand Genomic Sci 9:1128–1143. doi: 10.4056/sigs.5149006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Pfleiderer A, Mishra AK, Lagier JC, Robert C, Caputo A, Raoult D, Fournier PE. 2014. Non-contiguous finished genome sequence and description of Alistipes ihumii sp. nov. Stand Genomic Sci 9:1221–1235. doi: 10.4056/sigs.4698398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Padhmanabhan R, Dubourg G, Lagier JC, Nguyen TT, Couderc C, Rossi-Tamisier M, Caputo A, Raoult D, Fournier PE. 2014. Non-contiguous finished genome sequence and description of Collinsella massiliensis sp. nov. Stand Genomic Sci 9:1144–1158. doi: 10.4056/sigs.5399696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Roux V, Lagier JC, Gorlas A, Robert C, Raoult D. 2014. Non contiguous finished genome sequence and description of Kurthia senegalensis sp. nov. Stand Genomic Sci 9:1319–1330. doi: 10.4056/sigs.5078947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Ramasamy D, Lagier JC, Rossi-Tamisier M, Pfleiderer A, Michelle C, Couderc C, Raoult D, Fournier PE. 2014. Genome sequence and description of Bacteroides timonensis sp. nov. Stand Genomic Sci 9:1181–1197. doi: 10.4056/sigs.5389564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Ramasamy D, Mishra AK, Lagier JC, Padhmanabhan R, Rossi-Tamisier M, Sentausa E, Raoult D, Fournier PE. 2014. A polyphasic strategy incorporating genomic data for the taxonomic description of new bacterial species. Int J Syst Evol Microbiol 64:384–391. doi: 10.1099/ijs.0.057091-0. [DOI] [PubMed] [Google Scholar]
- 293.Tatusov RL, Natale DA, Garkavtsev IV, Tatusova TA, Shankavaram UT, Rao BS, Kiryutin B, Galperin MY, Fedorova ND, Koonin EV. 2001. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res 29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, Hedlund BP, Tsiamis G, Sievert SM, Liu WT, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM, Hugenholtz P, Woyke T. 2013. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 295.Youle M, Haynes M, Rohwer F. 2012. Scratching the surface of biology’s dark matter, p 61–83. In Witzany G. (ed), Viruses: essential agents of life. Springer, New York, NY. [Google Scholar]
- 296.Raoult D. 2010. Technology-driven research will dominate hypothesis-driven research: the future of microbiology. Future Microbiol 5:135–137. doi: 10.2217/fmb.09.119. [DOI] [PubMed] [Google Scholar]