Two articles, one by Welcker et al. (1) in this issue of PNAS and one in the EMBO Journal (2), report that the F-box protein Fbw7, a component of an SCF-class ubiquitin ligase (E3) complex, mediates recognition and ubiquitination of the c-Myc transcription factor when the latter is phosphorylated on Thr 58. This regulatory connection is conserved throughout evolution, because the Fbw7 homologue Archipelago fulfills the same function for the Drosophila Myc protein, dMyc (3). Here, these findings are discussed alongside recent progress in our understanding of the posttranslational modifications regulating Myc turnover in vivo (4) and previous work connecting another F-box protein, Skp2, to Myc regulation (5, 6).
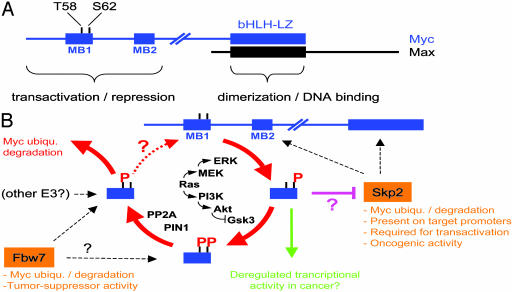
c-Myc (Myc, Fig. 1A) is a transcription factor of the basic helix–loop–helix leucine zipper (bHLH-LZ) family. Through dimerization with Max, Myc binds the DNA element CACGTG and contributes to transcriptional activation. Myc also represses transcription at alternative DNA sites through association with other transcriptional activators such as Miz-1. The first 143 residues contain sequences required for both transcriptional activation and repression (7–10) and include two highly conserved “Myc boxes” (MB). MB1 includes threonine-58 (T58) and serine-62 (S62), that are phosphorylated in an interdependent manner: phosphorylation of S62, believed to be catalyzed by ERK kinases (11), is a prerequisite for phosphorylation of T58 by GSK3 (10, 12).
Fig. 1.
Emergence of a “Myc modification cycle” regulating protein turnover and activity. (A) Structure of Myc and Max. (B) A series of posttranslational modifications in MB1 regulates Myc's interactions with ubiquitin ligases and may impact its transcriptional activities. Question marks indicate unresolved or hypothetical connections.
The importance of T58 in the regulation of Myc function was suggested by observations in human lymphomas. Oncogenic activation of c-myc (e.g., by gene translocation or amplification) commonly results in deregulated expression of c-myc, and there is no strict requirement for alterations in primary sequence. However, a subset of Burkitt's lymphomas show secondary missense mutations within the translocated c-myc allele, suggesting that such mutations endow tumor cells with a selective advantage in vivo. One of the hot spots for mutations is T58. A series of studies suggested that T58 mutation results in decreased ubiquitination and proteasome-mediated turnover of Myc, as well as in enhanced oncogenic activity (10, 12, 13).
These findings prompted a search for the ubiquitin ligase(s) responsible for degradation of Myc. One year ago, two groups reported that the F-box protein Skp2, the substrate-binding subunit of the SCFSkp2 E3–ligase complex, associates with Myc and can induce its ubiquitination and degradation (5, 6, 14). Skp2, however, did not fulfill the prerequisites for a T58 phosphorylation-dependent ligase, because it recognized the MB2 and bHLH-LZ regions of Myc but did not require MB1 (Fig. 1B). Now, the groups of Clurman and Eisenman (1) and Nakayama (2) report that another F-box protein, Fbw7, fulfills this function.
The new data demonstrate that Fbw7 binds Myc in vivo and induces Myc degradation when overexpressed in transfected cells. Mutation of T58 and/or S62 in Myc renders the protein resistant to Fbw7-induced degradation. Overexpression of a dominant-negative form of Fbw7, Fbw7 knock-down, or gene deletion, invariably leads to decreased Myc turnover. Inhibition of GSK3 has a similar effect and prevents interaction of Myc and Fbw7. In vitro, Fbw7 interacts specifically with MB1 peptides, and SCFFbw7 catalyzes ubiquitination of Myc; both of these activities depend on T58 phosphorylation. In a parallel study, the Hariharan group (3) demonstrates that Drosophila Fbw7 (Archipelago or Ago) similarly regulates degradation of dMyc and antagonizes dMyc function during development (3).
Collectively, these findings establish that Fbw7/Ago is a conserved SCF subunit that controls Myc turnover, emphasizing the importance of understanding how this interaction is regulated. The simplest scenario would be that sequential phosphorylation of Myc on S62 by ERK and T58 by GSK3 leads to Fbw7 binding. Although this scenario is certainly part of the answer, the situation seems actually more complex.
Sears, Nevins, and collaborators have shown that Ras signaling can stabilize Myc in vivo (15), most likely through concomitant activation of ERK (inducing S62 phosphorylation) and inhibition of GSK3 (suppressing T58 phosphorylation) (11) (Fig. 1B). These authors now report that the connection between T58 and S62 extends further: phosphorylated T58 constitutes a recognition site for the prolyl isomerase Pin1, which is proposed to act on P59 of Myc (4). The action of Pin1 is a prerequisite for the recruitment of protein phosphatase 2A (PP2A), which selectively dephosphorylates S62. Whether T58 is the target of a distinct phosphatase remains unknown, but, in any instance, Myc phosphorylated on T58 alone seems to be rapidly ubiquitinated and degraded. Thus, T58 and S62 appear to be tightly inter-connected through a phosphorylation–dephosphorylation cycle that leads to the ultimate disposal of Myc (Fig. 1B). It is tempting to draw a direct parallel between this mechanism and the action of Fbw7, although some discrepancies remain to be resolved.
In the experiments of Welcker et al. (1), mutating either T58 or S62 prevented the degradation of Myc induced by Fbw7 overexpression. This finding is consistent with the model in which S62 and T58 are phosphorylated in succession to generate the recognition site for Fbw7. In contrast, when Myc was overexpressed in mitogen-deprived cells, the protein was stabilized by mutation of T58 but not of S62 (11). These opposite effects of S62 mutation suggest that the two experimental conditions reveal different modes of Myc degradation.
S62 mutation prevents phosphorylation of T58 altogether. Thus, from the instability of the S62 mutant in quiescent cells (11), it can be inferred that we are looking at degradation of the “nonphosphorylated MB1” form of Myc, which must be recognized by an E3 ligase other than Fbw7. Skp2 is the best, although not the exclusive, candidate at this time (Fig. 1B). T58 mutation generated a stable Myc protein that was phosphorylated on S62 (11). Thus, it must be further inferred that, when S62 alone is phosphorylated, Myc is not ubiquitinated and remains stable. We speculate that S62 phosphorylation negatively regulates association with the E3 ligase (Fig. 1B). Although Skp2 requires neither MB1 nor T58/S62 for binding to Myc (5, 6), a negative effect of phospho-S62 on this interaction remains to be tested. Upon mitogenic stimulation of normal cells, phosphorylation of S62 is likely to be followed rapidly by that of T58. Thus, the initial event that stabilizes Myc against Skp2 (or another E3) simultaneously sets the stage for its disposal by Fbw7 (Fig. 1B).
Yeh et al. (4) also suggest that Pin1/PP2A-mediated dephosphorylation of S62 must occur before ubiquitination of Myc and, hence, one would presume, before Fbw7 binding. This possibility conveys another apparent contradiction with the data on Fbw7, because this ligase binds doubly T58/S62-phosphorylated MB1 peptides in vitro as efficiently as those phosphorylated on T58 alone (Fig. 1B) (1). The fine-tuning of this interaction, however, remains to be addressed on full-length Myc; it remains possible that Fbw7 binds MB1 in vivo only when it is phosphorylated on T58 and in the Pin1-isomerized form. In other words, Fbw7 may bind only the “final” form of Myc that requires obligatory passage, through the cycle shown in Fig. 1B (4). Alternatively, we may not be through with the E3 ligases that regulate Myc's fate.
Skp2 and Fbw7 not only recognize Myc in different manners but also have opposite functional and biological effects. Besides Myc, Fbw/Ago also ubiquitinates cyclin E and Notch (see references in ref. 2) and appears to function as a tumor suppressor gene (15). Skp2 ubiquitinates negative growth regulators, like p27, p21, and p57 (see ref. 2), and possesses growth-promoting and oncogenic activities (17, 18). Most remarkably, Skp2 appears to play a positive role in Myc-induced transcription (5, 6). Skp2, ubiquitin, and the 19S proteasome subunit Sug1 associate with at least one Myc-regulated promoter in vivo (5), extending previous observations connecting ubiquitin and 19S to transcriptional activation (19). Consistent with a role for Skp2 as a cofactor of Myc, this ligase also induces S-phase entry (20) and does so in a Myc-dependent manner (5).
Based on these observations, it is tempting to speculate that, during mitogenic stimulation, the phosphorylation–ubiquitination cycle shown in Fig. 1B occurs at least partly on chromatin and is coordinated with other activities of Myc, such as histone acetylation (21). If so, blocking the completion of this cycle in cancer cells (e.g., with oncogenic Ras or T58 mutation) may have profound consequences not only on Myc homeostasis but also on its transcriptional and biological activities (Fig. 1B, green arrow). Although experiments based on transient transfections pointed to a role of T58/S62 in transcription (e.g., refs. 10 and 22), the precise contribution of posttranslational modifications on Myc to the regulation of its target genes remains to be explored.
The same remark holds true for cellular transformation by Myc. Although earlier experiments did suggest reduced apoptotic and enhanced transforming activities of T58 mutants (10, 23), the correlation was not systematic, and the available data were not wholly conclusive. Yeh et al. (4) now provide stronger support for the notion that the activity of Myc is indeed constrained by the T58/S62 phosphorylation–ubiquitination cycle (Fig. 1B). The authors have used a transformation assay in human cells, based on the coexpression of Ras, hTERT, and SV40 large T and small T (24). In this assay, small T can be functionally substituted by a T58 mutant of Myc but not by WT Myc. In this context, small T acts as an inhibitor of PP2A, reducing dephosphorylation of S62 and stabilizing endogenous Myc (4).
These considerations take us back where we started: have we understood why T58 mutations are selected in Burkitt's lymphomas? We certainly have witnessed a big step forward with the chain of events described in Fig. 1B (4) and with the current demonstration that phospho-T58 is a recognition site for Fbw7 (1, 2). It should be stressed, however, that no existing experiment demonstrates that augmented Myc levels are the basis for positive selection of T58 mutations in patients. Instead, cooperating genetic lesions in mouse models clearly demonstrate that the main selective advantage for a tumor cell in vivo stems from dampening Myc-induced apoptosis (e.g., refs. 25 and 26). Thus, further augmenting Myc levels may even be a counterselected event.
In conclusion, the effects of T58 and other Burkitt's-associated mutations will need to be carefully studied on cellular chromatin and in mouse tumor models. The jury is still out about whether Myc activation is about quantity alone, as textbooks would say, or also about qualitative changes in molecular function.
See companion article on page 9085.
References
- 1.Welcker, M., Orian, A., Jin, J., Grim, J. A., Harper, J. W., Eisenman, R. N. & Clurman, B. E. (2004) Proc. Natl. Acad. Sci. USA 101, 9085–9090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yada, M., Hatakeyama, S., Kamura, T., Nishiyama, M., Tsunematsu, R., Imaki, H., Ishida, N., Okumura, F., Nakayama, K. & Nakayama, K. I. (2004) EMBO J., in press. [DOI] [PMC free article] [PubMed]
- 3.Moberg, K. H., Mukherjee, A., Veraksa, A., Artavanis-Tsakonas, S. & Hariharan, I. K. (2004) Curr. Biol., in press.. [DOI] [PubMed]
- 4.Yeh, E., Cunningham, M., Arnold, H., Chasse, D., Monteith, T., Ivaldi, G., Hahn, W. C., Stukenberg, P. T., Shenolikar, S., Uchida, T., et al. (2004) Nat. Cell Biol. 6, 308–318. [DOI] [PubMed] [Google Scholar]
- 5.Kim, S. Y., Herbst, A., Tworkowski, K. A., Salghetti, S. E. & Tansey, W. P. (2003) Mol. Cell 11, 1177–1188. [DOI] [PubMed] [Google Scholar]
- 6.von der Lehr, N., Johansson, S., Wu, S., Bahram, F., Castell, A., Cetinkaya, C., Hydbring, P., Weidung, I., Nakayama, K., Nakayama, K. I., et al. (2003) Mol. Cell 11, 1189–1200. [DOI] [PubMed] [Google Scholar]
- 7.Amati, B., Frank, S. R., Donjerkovic, D. & Taubert, S. (2001) Biochim. Biophys. Acta 1471, M135–M145. [DOI] [PubMed] [Google Scholar]
- 8.Eisenman, R. N. (2001) Genes Dev. 15, 2023–2030. [DOI] [PubMed] [Google Scholar]
- 9.Wanzel, M., Herold, S. & Eilers, M. (2003) Trends Cell Biol. 13, 146–150. [DOI] [PubMed] [Google Scholar]
- 10.Oster, S. K., Ho, C. S., Soucie, E. L. & Penn, L. Z. (2002) Adv. Cancer Res. 84, 81–154. [DOI] [PubMed] [Google Scholar]
- 11.Sears, R., Nuckolls, F., Haura, E., Taya, Y., Tamai, K. & Nevins, J. R. (2000) Genes Dev. 14, 2501–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gregory, M. A., Qi, Y. & Hann, S. R. (2003) J. Biol. Chem. 278, 51606–51612. [DOI] [PubMed] [Google Scholar]
- 13.Bahram, F., von der Lehr, N., Cetinkaya, C. & Larsson, L. G. (2000) Blood 95, 2104–2110. [PubMed] [Google Scholar]
- 14.von der Lehr, N., Johansson, S. & Larsson, L. G. (2003) Cell Cycle 2, 403–407. [PubMed] [Google Scholar]
- 15.Sears, R., DeGregori, J., Leone, G. & Nevins, J. R. (1999) Mol. Cell 3, 169–179. [DOI] [PubMed] [Google Scholar]
- 16.Spruck, C. H., Strohmaier, H., Sangfelt, O., Muller, H. M., Hubalek, M., Muller-Holzner, E., Marth, C., Widschwendter, M. & Reed, S. I. (2002) Cancer Res. 62, 4535–4539. [PubMed] [Google Scholar]
- 17.Gstaiger, M., Jordan, R., Lim, M., Catzavelos, C., Mestan, J., Slingerland, J. & Krek, W. (2001) Proc. Natl. Acad. Sci. USA 98, 5043–5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Latres, E., Chiarle, R., Schulman, B. A., Pavletich, N. P., Pellicer, A., Inghirami, G. & Pagano, M. (2001) Proc. Natl. Acad. Sci. USA 98, 2515–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muratani, M. & Tansey, W. P. (2003) Nat Rev. Mol. Cell Biol. 4, 192–201. [DOI] [PubMed] [Google Scholar]
- 20.Sutterlüty, H., Chatelain, E., Marti, A., Wirbelauer, C., Senften, M., Müller, U. & Krek, W. (1999) Nat. Cell Biol. 1, 207–214. [DOI] [PubMed] [Google Scholar]
- 21.Frank, S. R., Schroeder, M., Fernandez, P., Taubert, S. & Amati, B. (2001) Genes Dev. 15, 2069–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta, S. & Davis, R. J. (1994) FEBS Lett. 353, 281–285. [DOI] [PubMed] [Google Scholar]
- 23.Chang, D. W., Claassen, G. F., Hann, S. R. & Cole, M. D. (2000) Mol. Cell. Biol. 20, 4309–4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hahn, W. C., Dessain, S. K., Brooks, M. W., King, J. E., Elenbaas, B., Sabatini, D. M., DeCaprio, J. A. & Weinberg, R. A. (2002) Mol. Cell. Biol. 22, 2111–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmitt, C. A., Fridman, J. S., Yang, M., Baranov, E., Hoffman, R. M. & Lowe, S. W. (2002) Cancer Cell 1, 289–298. [DOI] [PubMed] [Google Scholar]
- 26.Pelengaris, S., Khan, M. & Evan, G. I. (2002) Cell 109, 321–334. [DOI] [PubMed] [Google Scholar]