Abstract
When human cells incur DNA damage, two fundamental responses can follow, cell cycle arrest or apoptosis. Human RAD9 (hRAD9) and p53 function in both processes, but the mechanistic relationship between their activities is unknown. p53 mediates checkpoint control at G1 by transcriptional regulation of p21. In this report, we show that hRAD9, like p53, can also regulate p21 at the transcriptional level. We demonstrate that overexpression of hRAD9 leads to increased p21 RNA and encoded protein levels. The promoter region of p21 fused to a luciferase reporter can be transactivated by either hRAD9 or p53, indicating that hRAD9 regulates the p21 promoter for transcriptional control of expression. Using an electrophoretic mobility-shift assay, we show that hRAD9 specifically binds to a p53-consensus DNA-binding sequence in the p21 promoter. Microarray screening coupled with Northern analysis reveals that hRAD9 regulates the abundance of other messages in addition to p21. Our data reveal a previously undescribed mechanism for regulation of p21 and demonstrate that hRAD9 can control gene transcription. We suggest that hRAD9 and p53 co-regulate p21 to direct cell cycle progression by similar molecular mechanisms. Furthermore, hRAD9 might regulate other cellular processes as well by modulating transcription of multiple down-stream target genes.
Human RAD9 was originally identified as a structural homologue of yeast Schizosaccharomyces pombe rad9 and is capable of partially complementing the radiosensitivity, hydroxyurea sensitivity, and associated checkpoint defects of rad9 null yeast (1). It has multiple cellular functions. Human RAD9 (hRAD9) is a nuclear protein (2, 3) that can be phosphorylated by ATM in response to DNA damage, an event important for the G1/S checkpoint (4). Additional phosphorylation events in the C-terminal region are also important for cell cycle control and radioresistance (5, 6). Furthermore, the protein demonstrates 3′ to 5′ exonuclease activity (7). hRAD9 also functions as a cell death mediator. The human and yeast genes are members of the BH3 domain-only proapoptotic family of Bcl-2 proteins (8, 9). Phosphorylation of Tyr-28 in the BH3 domain of hRAD9 by c-Abl is important for proapoptotic function (10). hRAD9 can bind the antiapoptotic proteins Bcl-2 and Bcl-XL with its N-terminal region and the checkpoint-related proteins hHUS1 and hRAD1 with its C-terminal region, suggesting that the protein possesses at least two functional domains, each involved in distinct cellular responses to DNA damage (11). Several groups demonstrated that hRAD9, hHUS1, and hRAD1 interact to form a heterotrimer with structural similarity to proliferating cell nuclear antigen (2, 11, 12). It is believed that a complex of hRAD17 and replication factor C aids in loading this structure onto DNA (13, 14). At least hRAD9 rapidly binds DNA containing double-strand breaks, likely facilitating subsequent binding of additional proteins that participate in checkpoint control or other pathways that respond to DNA damage (15).
Tumor suppressor gene p53 plays a fundamental role in controlling cell cycle checkpoints, apoptosis, and genetic stability (16). p53 acts as a sequence-specific DNA-binding protein and activates transcription through binding distinct DNA consensus sequences (17, 18). p53 is a negative regulator of cell cycle progression and controls transition from G1 to S phase of the cell cycle. p53 is required for the cell cycle response to DNA damage caused by exposure to radiation (19). Wild-type p53 controls these processes by regulating transcription of target genes through binding consensus DNA sites in promoter regions (18). Transcriptional targets of 53 have been identified and regulate cell growth, DNA repair, or cell death processes (19–26).
Perhaps one of the most important downstream target genes of p53 is p21 (27), also known as WAF1 and CIP1 (28, 29). p21 is directly induced by wild-type p53 through consensus p53-binding sequences in its promoter (28). p21 is a universal inhibitor of cyclin/cyclin-dependent kinases and functions as a brake for cell cycle progression through inhibition of cyclins and cyclin-dependent kinases, such as CDK4 and CDK6 (27). p21 is required for p53-mediated cell cycle arrest by radiation-induced damage (30–32). Because p21 is a key factor in cell cycle control, regulation of p21 is a critical step for the cell cycle response to DNA damage and other environmental stresses.
hRAD9 and p53 have important roles in cell cycle checkpoint control and apoptosis, suggesting that these two proteins might function coordinately. In this report, we demonstrate that, like p53, hRAD9 can transactivate p21 using p21 promoter sequences. Furthermore, we show that hRAD9 can physically bind a known p53-consensus sequence in the p21 promoter. These results provide evidence that hRAD9 and p53 coregulate p21 to control cell cycle progression from G1 to S. Using DNA microarray technology, we demonstrate that hRAD9 can regulate expression of other genes in addition to p21. Our results suggest that modulation of gene transcription might be a mechanism by which hRAD9 controls multiple cellular processes.
Materials and Methods
DNA Constructs. pGL2-p21-Luci, containing the p21 promoter-luciferase reporter fusion, was constructed by first PCR amplifying a 2,337-bp fragment of the human p21 promoter (base pairs 2258–4594) (32, 34), containing two p53-binding consensus sites, and adding HindIII and XhoI sites on the ends. This fragment was inserted into the corresponding sites within pGL2-basic (Promega) to make the final construct. pCMV-β-gal reporter plasmid was used as a transfection control. GAPDH cDNA was used as a probe for Northern analysis to verify that equal amounts of RNA were loaded per well. pZeoSV2(+)-RAD9 was made by inserting the hRAD9 cDNA ORF into pZeoSV2(+) (Invitrogen). hRAD9 insert was generated by PCR by using pHRAD9–1 cDNA (1) as template, and the following pair of primers: 5′-CTGACTCTCGAGGACTACAAAGACGATGACGA-3′ and 5′-CGCGCGACTAGTGACTACAAAGACGATGA-3′. The amplified fragment and pZeoSV2(+) were digested with HindIII and BamHI, then ligated by T4 DNA ligase following the manufacturer's instructions (New England Biolabs).
pcDNA3 was obtained commercially (Invitrogen). pC53-SN3 is described in ref. 33.
Cells, Culturing Conditions, and Transfection. H1299 cells were grown in Earle's MEM (GIBCO), with 10% FBS at 37°C. Lipofect AMINE Reagent (GIBCO) was used with 1–4 μg of each plasmid for transient transfection, according to the manufacturer's instructions.
Escherichia coli DH5α cells were grown in LB. Competent cells were purchased (Stratagene) and used for transfections as well as plasmid amplification by standard procedures (34).
Northern and Western Analyses. For Northern analysis, total RNA was prepared from H1299 cells (35) by using TRIzol (Life Technologies, Grand Island, NY), according to the manufacturer's instructions. A PCR-amplified human p21 cDNA fragment (450 bp) was used as probe. cDNA was labeled with [α-32P]dCTP (Amersham Pharmacia) by using Prime-It RmT (Stratagene) and purified by ProbeQuant G-50 micro columns (Pharmacia). Total RNA (20 μg) was fractionated by electrophoresis in a 1.2% (wt/vol) formaldehyde-agarose gel and transferred to Hybond-N membrane. Hybridization was carried out in QuikHyb hybridization solution (Stratagene) with 106 cpm/ml 32P-labeled p21 cDNA probe. The filter was used to expose x-ray film. The filter was reprobed with a 32P-labeled GAPDH DNA fragment as a loading control. Signal intensities on the autoradiogram were measured by densitometric scanning. p21 signals were normalized with respect to GAPDH bands in the same blot.
Northern blot analysis was also used to confirm some of the gene expression profiles indicated by microarray screening and was performed essentially as described above. To generate gene probes, oligo primer pairs for PCR were designed by using primerselect software (DNASTAR, Madison, WI), and those primers are listed in Table 2, which is published as supporting information on the PNAS web site. Five micrograms of total RNA was subjected to RT-PCR in a two-step protocol by using Moloney's murine leukemia virus reverse transcriptase and Taq polymerase (Invitrogen). PCR products were TA cloned into pCRII (Invitrogen), and the DNA sequence of inserts was verified. Plasmid DNA was cut with restriction endonucleases, and inserts were gel purified to prepare cDNA probes.
Western analysis was performed by routine procedures (11). Extracts were prepared from cells 48 h posttransfection with indicated plasmids. Equal amounts of each (40 μg per lane) were loaded onto and run through polyacrylamide gels, then probed with monoclonal antibodies against p21 (Oncogene Research Products, Nottingham, U.K.), or polyclonal antibodies against hRAD9, actin (Santa Cruz Biotechnology), or p53 (Oncogene Research Products).
Luciferase Activity. Transfected cells were cultured in a complete growth medium for 48 h and harvested for luciferase assays, performed according to available protocols (Promega). Luciferase activity was measured on a Berthold (Nashua, NH) Autolumat LB953 Rack Luminometer.
Electrophoretic Mobility-Shift Assay (EMSA). Complementary oligos containing the p53-binding site were synthesized commercially (Invitrogen) and annealed in buffer (100 mM NaCl/5 mM Tris·Cl, pH 7.5/10 mM MgCl2/0.02 mM EDTA/1 mM DTT). These probes were end-labeled with [γ-32P]ATP by using T4 polynucleotide kinase (New England Biolabs). U937 cell nuclear protein extract (2–5 μg) was incubated with 2 × 105 cpm of probe in binding buffer containing 20 mM Hepes (pH 7.9), 1 mM MgCl2, 4% Ficoll, 50 mM KCl, 0.5 mM DTT, 1 μg of BSA, and 2 μg of nonspecific competitor poly(dI-dC) (Sigma). Binding reaction mix was assembled on ice and incubated at room temperature for 30 min. DNA–protein complexes were resolved on a 4% native polyacrylamide gel, which was vacuum dried and used for autoradiography. For competitive binding reactions, 200 ng of cold probe was added during incubation. For supershift experiments, antibodies were preincubated with nuclear extract at 4°C for 20 min.
Microarray Analysis. H1299 cells, 60–80% confluent in 100-mm cell culture dishes, were transiently transfected with 1–4 μg of plasmid. Forty-eight hour later, cells were collected. Total RNA was extracted by TRIzol reagent (Invitrogen). mRNA was isolated with Oligotex (Qiagen, Chatsworth, CA). cDNA was synthesized from 1 μg of the mRNA by the MessageAmp aRNA kit (Ambion, Austin, TX). cRNA was biotin labeled by using the BioArray High Yield RNA Transcript labeling kit (Affymetrix). Labeled cRNA was fragmented, and 15 μg was sent to the Columbia DNA Microarray facility for hybridization to Human Genome U133A chips, washing, and scanning, following the instructions of the manufacturer (Affymetrix). The Affymetrix microarray 5.0 suite of software programs was used to analyze relative abundance of each message, including within cells receiving pZeoSV2(+)-RAD9, pC53-SN3, or control insertless vectors. Parameters used for analysis by the software were set to values recommended by Affymetrix. All chip data were normalized, then compared.
Results
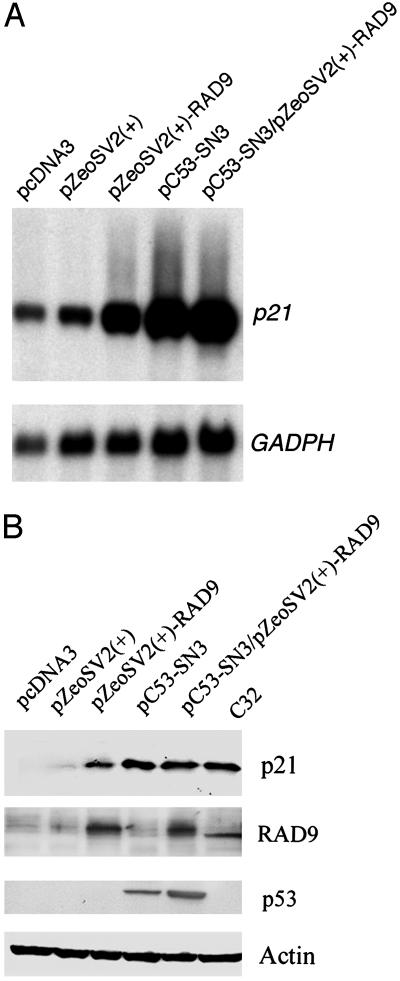
hRAD9 Can Regulate p21 RNA and Encoded Protein Levels. Human p53 protein regulates the transition of cells from G1 to S phases of the cell cycle by activating transcription of p21. We tested whether hRAD9 could also modulate expression of this gene, because both p53 and hRAD9 regulate the DNA damage-inducible G1 checkpoint. As shown in Fig. 1A, transient overexpression of p53 or hRAD9 in p53-null H1299 cells (35) caused a significant increase in p21 transcript levels (lanes 3 and 4), relative to the effects of insertless plasmid controls (lanes 1 and 2), as determined by Northern analyses (36). Furthermore, coexpression of p53 and hRAD9 increased the amount of p21 RNA in H1299 cells to a level comparable to that observed when either gene alone is tested (lane 5 relative to lanes 3 and 4). All p21 RNA levels were considered relative to the abundance of GAPDH message. This same profile was reflected in the inherent levels of p21 protein measured by Western analysis in the same cell-free extracts. As indicated in Fig. 1B, overexpression of hRAD9 (lane 3), p53 (lane 4), or the two genes together (lane 5) induced comparable levels of p21 protein, relative to vector controls (lanes 1 and 2), in equivalent amounts of the different cell-free extracts. The levels of hRAD9, p53, and actin (loading control) proteins are also presented. Levels of these proteins in C32 cell extracts are illustrated as controls. This suggests that p53 and hRAD9 regulate p21 by the same molecular mechanism because of the lack of additivity of increased p21 RNA or encoded protein levels during coexpression. These results indicate that, similar to p53, hRAD9 is a positive regulator of p21, likely at the level of transcription.
Fig. 1.
hRAD9 and p53 regulate p21 RNA and encoded protein levels. (A) Northern analysis of p21 expression. Exponentially growing H1299 cells were transfected with equal amounts (1 μg) of indicated plasmids and harvested for isolation of total RNA 48 h later. RNA (20 μg) was fractionated on a 1.2% formaldehyde agarose gel and transferred onto a nylon membrane for Northern blotting by using a [α-32P]dCTP-labeled human p21 probe following standard procedures. The blot was rehybridized with a human GAPDH cDNA probe as a loading control. Cells were transfected with: lane 1, pcDNA3; lane 2, pZeoSV2(+); lane 3, pZeoSV2(+)-RAD9; lane 4, pC53-SN3; lane 5, pZeoSV2(+)-RAD9 and pC53-SN3. (B) Western analysis of p21 protein. H1299 cells were transiently transfected, and cell-free protein extracts were prepared 48 h later. Equal amounts (40 μg) of each preparation were loaded onto a SDS-polyacrylamide gel for standard Western analysis using antibodies against p21, hRAD9, p53, or actin. H1299 cells were transfected with the following vectors: lane 1, pcDNA3; lane 2, pC53-SN3; lane 3, pZeoSV2(+)-RAD9; lane 4, pC53-SN3 and pZeoSV2(+)-RAD9; lane 5, pZeoSV2(+); lane 6, C32 whole cell lysate (Santa Cruz Biotechnology) control.
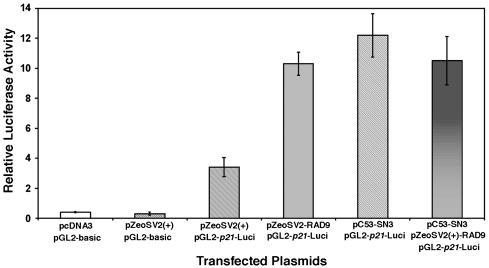
hRAD9 Can Transactivate a p21 Promoter-Luciferase Reporter Construct. A p21 promoter-luciferase reporter system was used to confirm results obtained by Northern and Western analyses and to verify that regulation of p21 by hRAD9 and p53 is at the level of transcription. The promoter region of p21, consisting of 2.3 kb of DNA, was fused to a luciferase reporter (in vector pGL2-basic) and cotransfected along with several plasmids into H1299 cells. Because the p21 promoter contains two p53-binding consensus sequences, as expected, overexpression of p53 induces luciferase activity (Fig. 2, column 5). Consistent with previous Northern and Western blot results, overexpression of hRAD9 alone also significantly induces luciferase activity (Fig. 2, column 4), suggesting that hRAD9, much like p53, acts as a transcription factor to regulate p21 expression. In fact, induction by hRAD9 is independent of p53, because the H1299 host cells for the assay are null for p53. Coexpression of p53 and hRAD9 induces high levels of luciferase activity (Fig. 2, column 6), but the level is comparable to that observed when either protein alone is overproduced. Background levels are low when insertless expression vectors are coupled with promoterless pGL2-basic (Fig. 2, columns 1 and 2). Some luciferase activity above background is detected when insertless expression vectors are present with pGL2-p21-Luci, and this may reflect activity of inherent hRAD9 protein in H1299 cells (Fig. 2, column 3; comparable data for pcDNA3 were obtained). These results essentially mirror those obtained when p21 RNA and encoded protein are monitored and strongly suggest that hRAD9 is a transcription factor capable of regulating p21 expression.
Fig. 2.
hRAD9 and p53 regulate p21 promoter activity in a transient expression assay. H1299 cells were transiently transfected with pcDNA3, pZeoSV2(+), pZeoSV2(+)-RAD9, or pC53-SN3, together with pGL2-p21-Luci, a luciferase reporter with the human p21 promoter upstream in pGL2. Extracts were assayed for luciferase activity on a Berthold Autolumat LB953 Rack Luminometer. All luciferase measurements are expressed as means ± SD of triplicate transfections and cultures. Transfection groups include the following (from left to right): insertless pcDNA3 vector and pGL2-basic; insertless pZeoSV2(+) vector and pGL2-basic; pZeoSV2(+) and pGL2-p21-Luci; pZeoSV2(+)-RAD9 and pGL2-p21-Luci; pC53-SN3 and pGL2-p21-Luci; (6) pZeoSV2(+)-RAD9 and pC53-SN3 and pGL2-p21-Luci.
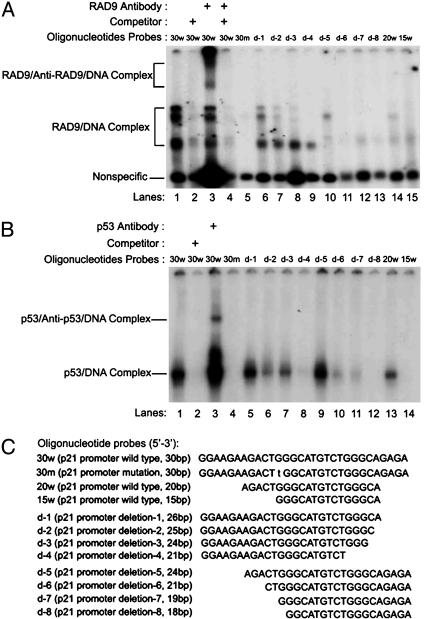
hRAD9 Can Bind p53 DNA-Binding Consensus Sequences in the p21 Promoter. EMSAs were performed to identify regions of the p21 promoter that could serve as a hRAD9 consensus DNA sequence-specific binding site, consistent with a role for this protein as a transcription transactivator. Two p53 consensus-binding sites in the p21 promoter were identified previously (28), and we found that hRAD9 can bind both. However, the interaction with the more downstream sequence was stronger based on results obtained by using EMSA (data not shown). Therefore, we focused on the downstream p53-binding region (i.e., 5′-GGAAGA AGACTGGGCATGTCTGGGCAGAGA-3′; underlined sequences indicate the binding consensus site and flanking nucleotides were included in the oligo for EMSA analysis) to identify and better define the hRAD9-binding sequence using nuclear extract from U937 cells as the source of hRAD9. As indicated in Fig. 3A, incubation of the nuclear extract with the 30-bp oligo containing the downstream p53 consensus sequence resulted in several shifted bands (lane 1). These bands were essentially eliminated by addition of excess cold 30-bp oligo as a competitor (lane 2), suggesting that these are specific protein–probe interactions. Because previous work demonstrated that hRAD9 is alternatively hyperphosphorylated and can exist in multiple forms in unperturbed cells (2, 11), the three EMSA-shifted bands may represent hRAD9–DNA complexes containing different versions of hRAD9 or the binding of different combinations of hRAD9 and other proteins in the cell extract. Nevertheless, at least two supershifted bands were detected by adding a specific antibody against hRAD9 in the reaction (lane 3), indicating that these are hRAD9/DNA complexes. The amount of the nonspecific band in lane 3 is relatively high compared to the levels in other lanes, suggesting that there was somewhat more total sample loaded. However, the hRAD9/DNA complex bands in lane 3 are fainter than in lane 1, but multiple prominent supershift bands are present. No such band shifts were detected when hRAD9 antibodies were added to the oligo in the absence of protein extract (data not shown), demonstrating that the findings are not simply due to antibody-DNA binding and are in fact the result of hRAD9–DNA interactions. Addition of cold oligo in the reaction also eliminated supershift bands (lane 4), but another 30-mer with an unrelated sequence was not an effective competitor (data not shown), confirming the specificity of this protein–DNA association as well. Excess nonspecific competitor poly(dI-dC) was included in every reaction, further indicating reaction specificity.
Fig. 3.
Identification of a common binding site for hRAD9 and p53 in the p21 promoter. (A) hRAD9 binds to oligos containing a p53 consensus site in the p21 promoter. 32P-labeled oligos were incubated with nuclear protein extract from growing U937 cells. Resulting protein–DNA complexes were resolved by 4% acrylamide gel electrophoresis and visualized by exposing x-ray film. Three gel-shift bands were formed in lane 1 as indicated. Cold competitor oligos were present in 100-fold excess to demonstrate specificity of the protein–DNA complexes (lanes 2 and 4). Anti-RAD9 antibodies were added to identify proteins in the complexes (lanes 3 and 4). Oligos with various lengths of p53 consensus sequence were used to confirm the minimum sequence necessary and identify critical sequences for hRAD9 binding. Lines indicate protein-antibody–DNA complex (Upper) and protein/DNA complex (Lower). (B) p53 binds to oligos containing the hRAD9 binding element. Human p53 protein (20) was incubated with 32P-labeled oligos and resolved by gel electrophoresis. Unlabeled competitor oligo was added in 100-fold excess for lane 2. Anti-p53 antibodies were used for the supershift in lane 3. Wild-type and modified probes were used as above (A). (C) Oligos used for EMSA. Wild-type (30-bp) and mutated (30-bp) oligo differ by 1 bp (lowercase letter). Deleted oligos were designed to omit base pairs at the ends of the starting 30-mer.
To define the binding region in more detail, we used oligos containing a mutation or a series of deletions as probes for EMSA (see Fig. 3C for details). We observed that maximum binding of all three bands occurred when the 30-mer remained intact. A single point mutation (G to T) at nucleotide 12 completely eliminated protein-DNA binding (Fig. 3A, lane 5). Two nested sets of deletions at the 3′ (Fig. 3C, d-1–d-4), the 5′ (Fig. 3C, d-5–d-8) or both 3′ and 5′ (20w, 15w) ends of the 30-mer were generated and tested for binding to cell-free extracts. As indicated in Fig. 3A, lanes 6–15, individual deletions differentially effected the formation of band shifts. For example, deletion d-1 reduced but did not eliminate all three bands (lane 6), but d-3 effected only two (lane 8). Together this series defines the DNA sequence patterns within the 30-mer critical for hRAD9 binding.
To compare binding characteristics of hRAD9 and p53, we incubated DNA probes used in Fig. 3A (and illustrated in Fig. 3C) with recombinant human p53 protein (20) for EMSA. As indicated in Fig. 3B, there is clearly a shifted band resulting from the interaction between p53 and the 30-bp oligo (lane 1). The specificity was confirmed by competition (lane 2). This is a p53–DNA complex because a supershifted band was formed in the presence of an anti-p53 monoclonal antibody (PAb 421; lane 3). Results of experiments designed to define boundaries and critical individual nucleotide of the DNA sequence interacting with p53 are indicated in Fig. 3B, lanes 4–14. Interestingly, the G-T alteration of the 30-bp sequence, which eliminates binding to hRAD9 (Fig. 3A, lane 5), also prevented formation of an EMSA band after incubation with p53 (Fig. 3B, lane 4). Some deletions effected band formation with p53 and hRAD9 comparably. For example, d-1 had little effect, whereas d-2 and d-3 reduced binding somewhat for both proteins. However, some deletions differentially effected band shift formation. Deletion d-5 had a greater adverse effect on the hRAD9-mediated shift, but 20w provided a better binding substrate for p53. In conclusion, these results demonstrate that the 30-mer can serve as a specific binding consensus site for p53 and hRAD9, but the precise DNA-binding sequence requirements for each are overlapping yet not identical.
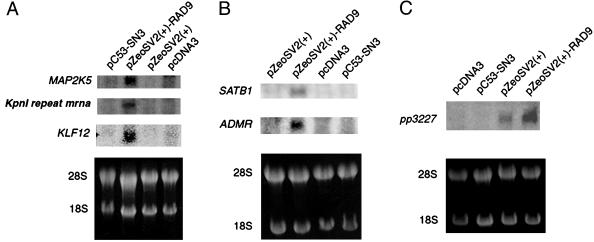
hRAD9 Can Regulate Abundance of Multiple RNAs in Addition to p21. Because hRAD9 can bind p53 consensus sequences in the p21 promoter region and transactivate p21, we tested whether hRAD9 can regulate transcription of other genes involved in cell cycle progression, apoptosis, DNA repair, and/or signal transduction. Microarray screening was used to address this question. We transfected p53-deficient H1299 cells with pC53-SN3, pZeoSV2(+)-RAD9, or corresponding empty vectors, isolated RNA 48 h later, then probed Affymetrix U133A chips to assess message abundance. This chip allows the analysis of expression for 15,106 independent known human genes. Using the increased abundance of p21 RNA in hRAD9 or p53 overexpressing cells (compared, respectively, with cells receiving corresponding empty vectors) as the minimum magnitude of induction considered significant, we identified other RNAs demonstrating the same or higher relatively increased levels under these conditions. Using the Affymetrix suite of programs, we found that RNA corresponding to 92 genes showed increased abundance when just hRAD9 was overexpressed, 54 when just p53 was tested, and 17 when either gene was overexpressed.
Northern analysis was used to begin to verify the microarray gene expression data. Sequences of several of the cDNAs representing genes potentially regulated by just hRAD9 were identified by a computer search of GenBank, and PCR was used to amplify a fragment of each from a human cDNA library. The products were used to probe Northern blots containing RNA from cells 48 h posttransfection with an hRAD9 or p53 expression vector or from those containing an insertless vector. As indicated in Fig. 4, we demonstrate that six genes in addition to p21 are up-regulated when hRAD9 is overexpressed. Furthermore, expression of these genes is low when cells are transfected with insertless vectors or a p53 expression vector (Fig. 4). Northern analysis was used to verify that the latter cells were expressing p53 (data not shown). Table 1 lists genes induced by hRAD9 overexpression, along with relevant information.
Fig. 4.
Northern blot analysis indicates that hRAD9 overexpression can increase the abundance of multiple RNA species. H1299 cells were transfected with pcDNA3, pC53-SN3, pZeoSV2(+), or pZeoSV2(+)-RAD9. Forty-eight hours later, cells were harvested, and RNA was purified, then subjected to Northern analysis with probes for the following genes: MAP2K5, KpnI repeat mrna, KLF12 (A); SATB1, ADMR (B); pp3227 (C). 28S and 18S RNAs in ethidium bromide-stained gels are illustrated (Lower) to demonstrate equivalent sample loading. When multiple RNA species are examined per gel, blots were stripped and reprobed.
Table 1. Genes induced by hRAD9.
| Affymetrix gene no. | Gene name* | Function/characteristics† |
|---|---|---|
| 202284 | p21 (WAF1/CIP1) | Regulates progression of cells from G1 to S |
| 214506 | ADMR (AMR, 7TMR, hrhAMR) | Adrenomedullin receptor; member of calcitonin gene-related peptide family |
| 206965 | KLFI2 (AP-2rep) | Kruppel-like factor 12; transcriptional repressor activity |
| 217137 | Kpnl repeat mrna | Unknown function |
| 210482 | MAP2K5 (MEK5, MAPKK5, PRKMK5) | Dual specificity protein kinase, member of mitogen-activated protein kinase kinase family; involved in signal cascade that mediates growth factor stimulated cell proliferation |
| 221200 | pp3227 | Inhibits cancer cell growth |
| 203408 | SATB1 | Special AT-rich sequence-binding protein 1; establishment and/or maintenance of chromatin architecture |
Alternate gene symbol in parentheses.
Information related to the function/characteristics of the genes is in links at www.affymetrix.com/index.affx, under “Analysis.”
Discussion
hRAD9 protein is a key player in multiple cellular responses to DNA damage, including G1 and G2 cell cycle checkpoints (1, 4, 37). However, the precise molecular mechanism used by hRAD9 to mediate these processes is not clear. In this report, we demonstrate that hRAD9 acts as a positive transcription regulator of p21, a gene whose product is important for transition of cells from G1 to the S phase of the cell cycle. p53 is also a critical regulator of the DNA damage-inducible G1 checkpoint, and its ability to transactivate p21 is thought to be a significant step in its competence to control cell cycle progression. It does so via its physical interaction with specific DNA consensus sequences within the promoter of p21 (28), and we demonstrate that hRAD9 can also interact with the same sequences. These results provide evidence that p53 and hRAD9 can regulate cell cycle progression from G1 to S via the control of p21 transcription. Furthermore, we demonstrate that hRAD9 can do so even in the absence of p53, suggesting that the two proteins can regulate this gene independently but through similar molecular mechanisms. This is an example of two different positive regulators of gene-specific transcription capable of interacting with the same DNA consensus sequence. In addition, this is evidence that hRAD9 plays a role in regulating transcription of a target gene.
p53 plays an important role in cell cycle checkpoint control and apoptosis via the ability to transactivate not just p21 but many different downstream target genes (16). hRAD9 functions in these two basic cellular processes, and the results presented suggest that hRAD9 might control G1 cell cycle progression by regulating transcription of p21. Furthermore, our results imply that, like p53, hRAD9 might also direct the more global response of cells to DNA damage by transactivating a set of downstream genes that at least partially overlap those controlled by p53. Microarray data presented herein support this possibility. Genes whose protein products can function as a transcription factor and are capable of controlling cell growth or participating in signal transduction have already been identified.
hRAD9 can physically interact with cell cycle checkpoint proteins hHUS1 and hRAD1 and form a proliferating cell nuclear antigen-like heterotrimer thought to bind DNA and function as a DNA damage sensor (2, 11, 12, 38). Subsequently, the protein–DNA interaction is believed to signal cell cycle machinery and ultimately influence cell cycle progression (11). However, it is not clear whether this protein complex is important for the ability of hRAD9 to transactivate p21. In fact, several lines of evidence suggest that the heterotrimer is not necessary for this activity. First, ectopic overexpression of just hRAD9, and not also hRAD1 and hHUS1, is sufficient to increase p21 message and encoded protein levels, as well as increase the activity of the p21 promoter fused to a luciferase reporter gene. Second, preliminary EMSA studies suggest that hRAD1 and hHUS1 in cell-free extracts cannot interact with the p53 DNA-binding consensus sequence within the p21 promoter under conditions that allow detection of binding by hRAD9 (unpublished data). These findings provide evidence that hRAD9 might mediate p21 transcription without the aid of hRAD1 or hHUS1. Nevertheless, hRAD9 might still be part of another complex of proteins when it directly, or perhaps indirectly through bridging proteins, binds the p21 promoter sequence because the EMSA studies presented made use of whole cell extracts.
Conclusion
We demonstrate that hRAD9 can bind p53 DNA-binding consensus sequences in the promoter region of p21 and enhance expression of the gene. These results suggest that hRAD9, like p53, can mediate several of its known activities by controlling a gene regulon, a group of coordinately regulated genes. Perhaps the ability of hRAD9 or p53 proteins to enhance p21 expression is simply a redundant activity. On the other hand, hRAD9 and p53 might activate p21 in response to different signals generated, for example, by different kinds of DNA damage. However, whatever the mechanism, because of the importance of hRAD9, p53, and p21 in cell cycle control and other fundamental cellular responses to DNA damage, as well as the established tumor-related activity of the latter two genes, understanding their relationships at the molecular level is clearly important.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants GM52493 (to H.B.L.), CA89816 (to H.B.L.), and CA73946 (to Y.Y.).
Abbreviations: EMSA, electrophoretic mobility-shift assay; hRAD9, human RAD9.
References
- 1.Lieberman, H. B., Hopkins, K. M., Nass, M., Demetrick, D. & Davey, S. (1996) Proc. Natl. Acad. Sci. USA 93, 13890–13895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.St. Onge, R. P., Udell, C. M., Casselman, R. & Davey, S. (1999) Mol. Biol. Cell 10, 1985–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burtelow, M. A., Kaufmann, S. H. & Karnitz, L. M. (2000) J. Biol. Chem. 275, 26343–26348. [DOI] [PubMed] [Google Scholar]
- 4.Chen, M. J., Lin, Y. T., Lieberman, H. B., Chen, G. & Lee, E. Y. (2001) J. Biol. Chem. 276, 16580–16586. [DOI] [PubMed] [Google Scholar]
- 5.Roos-Mattjus, P., Hopkins, K. M., Oestreich, A. J., Vroman, B. T., Johnson, K. L., Naylor, S., Lieberman, H. B. & Karnitz, L. M. (2003) J. Biol. Chem. 278, 24428–24437. [DOI] [PubMed] [Google Scholar]
- 6.St. Onge, R. P., Besley, B. D., Pelley, J. L. & Davey, S. (2003) J. Biol. Chem. 278, 26620–26628. [DOI] [PubMed] [Google Scholar]
- 7.Bessho, T. & Sancar, A. (2000) J. Biol. Chem. 275, 7451–7454. [DOI] [PubMed] [Google Scholar]
- 8.Komatsu, K., Miyashita, T., Hang, H., Hopkins, K. M., Zheng, W., Cuddeback, S., Yamada, M., Lieberman, H. B. & Wang, H. G. (2000) Nat. Cell Biol. 2, 1–6. [DOI] [PubMed] [Google Scholar]
- 9.Komatsu, K., Hopkins, K., Lieberman, H. & Wang, H. (2000) FEBS Lett. 481, 122–126. [DOI] [PubMed] [Google Scholar]
- 10.Yoshida, K., Komatsu, K., Wang, H. G. & Kufe, D. (2002) Mol. Cell. Biol. 22, 3292–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hang, H. & Lieberman, H. B. (2000) Genomics 65, 24–33. [DOI] [PubMed] [Google Scholar]
- 12.Volkmer, E. & Karnitz, L. M. (1999) J. Biol. Chem. 274, 567–570. [DOI] [PubMed] [Google Scholar]
- 13.Zou, L., Cortez, D. & Elledge, S. J. (2002) Genes Dev. 16, 198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bermudez, V. P., Lindsey-Boltz, L. A., Cesare, A. J., Maniwa, Y., Griffith, J. D., Hurwitz, J. & Sancar, A. (2003) Proc. Natl. Acad. Sci. USA 100, 1633–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greer, D. A., Besley, B. D. A., Kennedy, K. B. & Davey, S. (2003) Cancer Res. 63, 4829–4835. [PubMed] [Google Scholar]
- 16.Levine, A. J. (1997) Cell 88, 323–331. [DOI] [PubMed] [Google Scholar]
- 17.Kern, S. E., Kinzler, K. W., Bruskin, A., Jarosz, D., Friedman, P., Prives, C. & Vogelstein, B. (1991) Science 252, 1708–1711. [DOI] [PubMed] [Google Scholar]
- 18.El-Deiry, W. S., Kern, S. E., Pietenpol, J. A., Kinzler, K. W. & Vogelstein, B. (1992) Nat. Genet. 1, 45–49. [DOI] [PubMed] [Google Scholar]
- 19.Kastan, M. B., Zhan, Q., el Deiry, W. S., Carrier, F., Jacks, T., Walsh, W. V., Plunkett, B. S., Vogelstein, B. & Fornace, A. J., Jr. (1992) Cell 71, 587–597. [DOI] [PubMed] [Google Scholar]
- 20.Barak, Y., Juven, T., Haffner, R. & Oren, M. (1993) EMBO J. 12, 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Momand, J., Zambetti, G. P., Olson, D. C., George, D. & Levine, A. J. (1992) Cell 69, 1237–1245. [DOI] [PubMed] [Google Scholar]
- 22.Oliner, J. D., Pietenpol, J. A., Thiagalingam, S., Gyuris, J., Kinzler, K. W. & Vogelstein, B. (1993) Nature 362, 857–860. [DOI] [PubMed] [Google Scholar]
- 23.Barak, Y., Gottlieb, E., Juven Gershon, T. & Oren, M. (1994) Genes Dev. 8, 1739–1749. [DOI] [PubMed] [Google Scholar]
- 24.Oliner, J. D., Kinzler, K. W., Meltzer, P. S., George, D. L. & Vogelstein, B. (1992) Nature 358, 80–83. [DOI] [PubMed] [Google Scholar]
- 25.Okamoto, K. & Beach, D. (1994) EMBO J. 13, 4816–4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korsmeyer, S. J. (1995) Trends Genet. 11, 101–105. [DOI] [PubMed] [Google Scholar]
- 27.Xiong, Y., Hannon, G. J., Zhang, H., Casso, D., Kobayashi, R. & Beach, D. (1993) Nature 366, 701–704. [DOI] [PubMed] [Google Scholar]
- 28.El-Deiry, W. S., Tokino, T., Velculescu, V. E., Levy, D. B., Parsons, R., Trent, J. M., Lin, D., Mercer, W. E., Kinzler, K. W. & Vogelstein, B. (1993) Cell 75, 817–825. [DOI] [PubMed] [Google Scholar]
- 29.Harper, J. W., Adami, G. R., Wei, N., Keyomarsi, K. & Elledge, S. J. (1993) Cell 75, 805–816. [DOI] [PubMed] [Google Scholar]
- 30.Brugarolas, J., Chandrasekaran, C., Gordon, J. I., Beach, D., Jacks, T. & Hannon, G. J. (1995) Nature 377, 552–557. [DOI] [PubMed] [Google Scholar]
- 31.Deng, C., Zhang, P., Harper, J. W., Elledge, S. J. & Leder, P. (1995) Cell 82, 675–684. [DOI] [PubMed] [Google Scholar]
- 32.El-Deiry, W. S., Tokino, T., Waldman, T., Oliner, J. D., Velculescu, V. E., Burrell, M., Hill, D. E., Healy, E., Rees, J. L., Hamilton, S. R., et al. (1995) Cancer Res. 55, 2910–2919. [PubMed] [Google Scholar]
- 33.Baker, S. J., Markowitz, S., Fearon, E. R., Willson, J. K. & Vogelstein, B. (1990) Science 249, 912–915. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed.
- 35.Yin, Y., Terauchi, T., Solomon, G. G., Aizawa, S., Rangarajan, P. N., Yazaki, Y., Kadowaki, T. & Barrett, J. C. (1998) Nature 391, 707–710. [DOI] [PubMed] [Google Scholar]
- 36.Yin, Y., Liu, Y. X., Jin, Y. J., Hall, E. J. & Barrett, J. C. (2003) Nature 422, 527–531. [DOI] [PubMed] [Google Scholar]
- 37.Hirai, I & Wang, H. G. (2002) J. Biol. Chem. 277, 25722–25727. [DOI] [PubMed] [Google Scholar]
- 38.Griffith, J. D., Lindsey-Boltz, L. A. & Sancar, A. (2002) J. Biol. Chem. 277, 15233–15236. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.