Abstract
The activity of transcription factors is tightly modulated by posttranslational modifications affecting stability, localization, and protein–protein interactions. Conjugation to SUMO is a reversible posttranslational modification that has been shown to regulate important transcription factors involved in cell proliferation, differentiation, and tumor suppression. Here, we demonstrate that the erythroid transcription factor GATA-1 is sumoylated in vitro and in vivo and map the single lysine residue involved in SUMO-1 attachment. We show that the nuclear RING finger protein PIASy promotes sumoylation of GATA-1 and dramatically represses its transcriptional activity. We present evidence that a nonsumoylatable GATA-1 mutant is indistinguishable from the WT protein in its ability to transactivate a reporter gene in mammalian cells and in its ability to trigger endogenous globin expression in Xenopus explants. These observations open interesting questions about the biological role of this posttranslational modification of GATA-1.
GATA proteins constitute a family of zinc finger transcription factors that bind the core consensus DNA sequence (T/A)GATA(A/G) and play essential roles in diverse developmental processes (1). Several GATA proteins have been identified in vertebrates (GATA-1–GATA-6) as well as in yeast, fungi, Drosophila melanogaster, Caenorhabditis elegans, and Arabidopsis thaliana (2). Of these, GATA-1 is abundantly expressed in erythroid, megakaryocytic, and mast-cell lineages, as well as in Sertoli cells of the testis (3, 4). GATA-binding sites are found in the promoters of virtually all erythroidand megakaryocyte-specific genes studied, including GATA-1 (3). Gene-targeting and loss-of-function studies have proved that GATA-1 plays an essential role in erythroand megakaryopoiesis. GATA-1 knockout mice die at day 10.5 of gestation because of severe anemia with arrest of erythroid maturation (5, 6). Accordingly, embryonic stem cell mutants at the GATA-1 locus fail to contribute to the erythroid lineage in chimeric mice (7); formation of other hematopoietic lineages is not affected, but GATA-1–/–megakaryocytes hyperproliferate and fail to complete maturation (8).
The function of GATA-1 is tightly modulated by interaction with transcriptional cofactors such as the FOG proteins (9) and PU.1 (10), as well as by an array of posttranslational modifications (11). GATA-1 is phosphorylated in vivo within the N terminus (12), and inhibition of phosphatases increases the binding of GATA-1 to target sequences in the human erythroid cell line K562 (13). GATA-1 is also acetylated on sequences surrounding the C-terminal finger, and this modification stimulates its transcriptional activity in vivo (14). Finally, it has been shown that in erythroid cells GATA-1 localizes to specific subnuclear compartments that might favor protein–protein interactions and further posttranslational modifications (15).
SUMO-1 is a small ubiquitin-related protein that, similarly to ubiquitin, can be covalently linked to protein substrates (16). The pathways for conjugation of the two peptides are distinct but share several similarities, and SUMO-specific E3 ligases have been recently identified (16). Among the ligases is the family of PIAS [protein inhibitors of activated STATs (signal transducers and activators of transcription)] nuclear proteins that function as SUMO ligases for STATs and a number of other proteins (17–21). In contrast to ubiquitination, sumoylation does not target a protein for degradation but may affect its localization, stability, and activity with crucial implications for many cellular processes (16, 22). Notably, the activity of several transcription factors such as p53, c-Jun, androgen receptor, and Lef1/Tcf is modulated by conjugation to SUMO (23–25).
Here we show that GATA-1 is conjugated to SUMO-1 both in vitro and in vivo, and we map the single lysine residue involved in the modification. We also show that PIASy physically interacts with GATA-1 and enhances its sumoylation. The interaction between PIASy and GATA-1 is not affected by mutation of the sumoylatable lysine and results in a dramatic inhibition of GATA-1-dependent transcription. We finally show that a non-sumoylatable GATA-1 mutant is indistinguishable from the WT protein in a number of experiments, suggesting that sumoylation may provide a fine modulation of GATA-1 activity that escapes detection in transient overexpression assays.
Methods
Cell Lines. Cells were cultured at 37°C in DMEM or RPMI medium 1640 supplemented with 10% FCS and antibiotics. U2OS and MG63 are human osteosarcomas. MEL is a murine erythroleukemia, and HEL and K-562 are human erythroleukemias. 293T is a derivative of a human embryonic kidney cell line.
Plasmids. The cDNA-encoding murine GATA-1 was cloned in pcDNA3 (Invitrogen). Mutant K137R was generated by PCR-directed mutagenesis. Both WT and mutant GATA-1 were transferred in pCS2+ vectors for in vitro synthesis of capped mRNA for microinjection. The luciferase reporter plasmid used in transactivation assays contains three repeats of the GATA consensus cloned upstream of a minimal metallothionein promoter in the pGL3-basic vector (Promega). The plasmid pCMV-T7-PIASy is described in ref. 20, and the myc-LUC reporter is described in ref. 26.
In Vitro SUMO Conjugation. GATA-1 was translated in vitro by using the TNT rabbit reticulocyte lysate system (Promega) and [35S]methionine. Murine Ubc9 and GST-SUMO-1 were expressed in Escherichia coli and purified as described in ref. 27, and as a source of SUMO-activating enzyme (E1), protein extracts were prepared from NIH 3T3 fibroblasts and fractionated by anion exchange chromatography (27). In vitro sumoylation assays were performed as described in ref. 28.
Immunoprecipitation and Western Blotting. For immunoprecipitations, cells were harvested 36 h after transfection in 1 ml of ice-cold radioimmunoprecipitation assay buffer containing 10 mM N-ethylmaleimide, 1 mM PMSF, and protease inhibitors. Lysis was performed at 4°C for 20 min. Lysates were incubated for 4 h at 4°C with primary antibodies prebound to 20 μl of protein A-sepharose (Amersham Biosciences). Beads were washed three times with 1 ml of ice-cold lysis buffer before elution with Lemmli sample buffer.
For coimmunoprecipitation experiments, cells were lysed in a buffer containing 50 mM Hepes at pH 7.5, 150 mM NaCl, 10% glycerol, 0.1% Tween 20, 10 mM N-ethylmaleimide, 1 mM PMSF, and protease inhibitors. The following primary antibodies were used: 21C7 monoclonal anti-SUMO (Zymed), polyclonal anti-GFP (Invitrogen), N6 monoclonal anti-GATA-1 (Santa Cruz Biotechnology), and monoclonal anti-T7 epitope (Novagen).
RT-PCR. Blood induction in animal caps was performed as described in ref. 29. Xenopus laevis embryos were obtained by in vitro fertilization, dejellied in 2% cysteine, and grown in 0.1× Marc's modified Ringer solution. Capped mRNAs were transcribed by using the mMESSAGE mMACHINE SP6 Transcription Kit (Ambion, Austin, TX) and injected at a volume of 4 nl per blastomere. Animal caps were dissected at stages 8–9 and incubated in 0.5× MMR containing 50 ng/ml recombinant human basic fibroblast growth factor (Roche Diagnostics) until sibling embryos reached stages 30–35. Total RNA was extracted by using the RNeasy procedure (Qiagen, Valencia, CA). Radioactive semiquantitative RT-PCR was performed on random primed cDNA by using primers described in ref. 29.
Transfections and Luciferase Assays. Transfections were performed by using the calcium phosphate precipitate method or by lipofection with FuGENE (Roche Diagnostics). For luciferase assays, U2OS cells in 3-cm Petri dishes were lipofected with 400 ng of the reporter, 250 ng of GATA-1-expression plasmids, and 200 or 400 ng of pCMV-T7-PIASy. In all samples, 40 ng of the plasmid pRL-CMV (Promega) encoding Renilla luciferase were included for normalization of transfection efficiency. After 36 h, cells were lysed and assayed by using the Dual Luciferase kit (Promega). Relative luciferase activity is the ratio of firefly to Renilla luciferase activity, normalized to the activity of the reporter alone. Expression levels of transfected proteins were verified by immunoblotting of the lysates normalized for transfection efficiency. MEL, HEL, and K-562 erythroid cells were lipofected by using Tfx-50 (Promega) as described in ref. 30.
Results
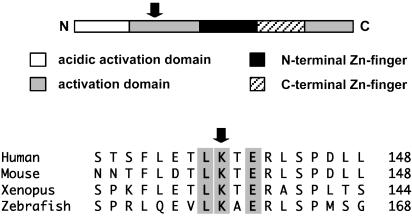
Identification of a Functional Sumoylation Site in GATA-1. Inspection of the sequence of murine GATA-1 revealed the presence of the tetrapeptide LKTE centered on lysine 137 within the N-terminal transactivation domain. This sequence conforms to the consensus found in most sumoylated proteins (16) and is conserved among mammals, amphibians, and fish (Fig. 1). To test whether GATA-1 might be a previously unrecognized substrate for SUMO modification, we used an in vitro sumoylation assay in which radioactively labeled GATA-1 is incubated in the presence of recombinant SUMO conjugating enzymes and bacterially expressed GST-SUMO-1 (27). As shown in Fig. 2A, in vitro translated GATA-1 migrates as two bands, the shorter isoform (GATA-1s) being generated by using an internal initiation codon (31). When E1, mUbc9, and GST-SUMO-1 were included in the reaction mixture, slower migrating forms of the two GATA-1 proteins were detected. These forms were only produced when all components of the sumoylation pathway were present and were not detected when we used a SUMO-1 deletion lacking the C-terminal glycine required for attachment to substrates (GST-SUMOΔ6). These observations suggest that the slower migrating bands are sumoylated GATA-1 and GATA-1s. Based on the apparent molecular weights, a single molecule of GST-SUMO-1 is attached to GATA-1 under these conditions.
Fig. 1.
Schematic representation of GATA-1 and alignment of the region surrounding the site of SUMO attachment. Amino acids conforming to the consensus motif for sumoylation are highlighted. The arrow points to lysine 137 in murine GATA-1.
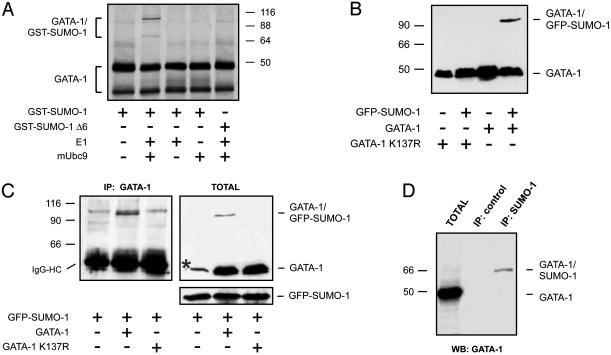
Fig. 2.
GATA-1 is sumoylated in vitro and in vivo.(A) In vitro sumoylation of GATA-1. Radioactively labeled in vitro translated GATA-1 was incubated in the presence of recombinant components of the SUMO conjugation pathway as indicated. Two isoforms of GATA-1 are visible (see text). GST-SUMOΔ6 is a C-terminal truncation lacking the glycine residues required for attachment to substrates. Reactions were separated by SDS/PAGE and visualized by autoradiography. (B) Sumoylation of GATA-1 in 293T cells. WT GATA-1 or the K137R mutant were transfected in 293T cells with or without a vector expressing GFP-SUMO-1. Lysates were separated by SDS/PAGE, and GATA-1 was detected by immunoblotting. (C) Sumoylation of GATA-1 in osteosarcoma cells. WT GATA-1 or the K137R mutant were transfected in MG63 cells together with GFP-SUMO-1. Lysates were immunoprecipitated with a monoclonal antibody to GATA-1 and immunoblotted with an antibody to SUMO-1 (Left). IgG heavy chains are indicated. GATA-1 and GFP-SUMO-1 were also detected in total lysates (Right). The asterisk indicates a nonspecific band recognized by the GATA-1 antibody in these cells. (D) Sumoylation of GATA-1 in mouse erythroleukemia cells. Lysates from murine erythroleukemia cells were immunoprecipitated with an antibody to SUMO-1 and immunoblotted with an antibody to GATA-1. Immunoprecipitation with an unrelated monoclonal antibody served as a negative control.
GATA-1 Is Sumoylated in Mammalian Cells. To verify that sumoylation of GATA-1 occurs in vivo, we cotransfected 293T cells with GATA-1 and GFP-SUMO-1 expression plasmids and visualized GATA-1 by immunoblotting. We used a monoclonal antibody (N6) that recognizes an N-terminal epitope not present in the short isoform GATA-1s (31). As shown in Fig. 2 B and C, a slower-migrating band is clearly detected in cells ectopically expressing GATA-1 and GFP-SUMO-1. We demonstrated that the higher-molecular-weight band corresponds to sumoylated GATA-1 by immunoprecipitating GATA-1 and blotting the immune complexes with an antibody to SUMO-1 (Fig. 2C). To test the requirement of lysine 137 for conjugation, we replaced it with arginine by site-directed mutagenesis. As shown in Fig. 2 B and C, the resulting protein (GATA-1 K137R) was no longer modified, demonstrating that this conserved residue is the major SUMO-1 attachment site.
Because the above experiments were performed in cells that do not express GATA-1, we asked whether endogenous GATA-1 might be sumoylated in erythroid cells. To this aim we immunoprecipitated sumoylated proteins from murine erythroleukemia cells by using a monoclonal antibody to SUMO-1 and blotted the immune complexes with an antibody to GATA-1. As shown in Fig. 2D, a band of ≈70 kDa was specifically detected in the anti-SUMO-1 immunoprecipitate, confirming that GATA-1 is sumoylated in murine erythroleukemia cells. The electrophoretic mobility is consistent with addition of a single SUMO-1 chain. We conclude that in these erythroid cells, under normal growth conditions, a fraction of endogenous GATA-1 is monosumoylated.
Sumoylation Is Not Required for GATA-1 Transcriptional Activity in a Number of Assays. We set out to analyze whether sumoylation might affect GATA-1 transcriptional activity. Because lysine 137 resides within the GATA-1 transactivation domain, we decided to focus on the intrinsic transactivation activity of GATA-1, using a system that would not be affected by the complexity of GATA-1 interactions with transcriptional cofactors. We chose a luciferase reporter construct in which three copies of the GATA sequence are cloned upstream of a minimal promoter; this construct has a moderate basal activity in nonerythroid cells and responds efficiently to GATA-1 overexpression. We transfected this construct, together with GATA-1 and increasing amounts of GFP-SUMO-1 in human MG63, SaOS-2, and U2OS cells, and assayed for luciferase activity. Although coexpression of GFP-SUMO increases the fraction of sumoylated GATA-1 (Fig. 2), we found no reproducible differences in the transcriptional activity of GATA-1 in the presence or in the absence of GFP-SUMO-1 (data not shown). Importantly, we obtained identical results with the nonsumoylatable GATA-1 K137R mutant (data not shown). Taken together, these observations suggest that sumoylation does not affect the activity of GATA-1 on this specific promoter under these experimental conditions.
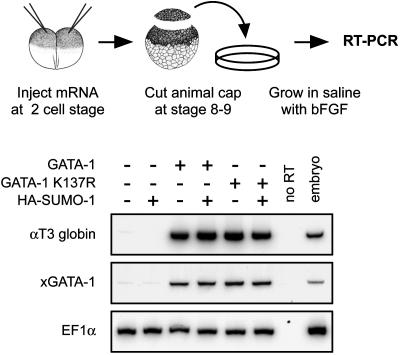
We next tested whether sumoylation might modulate GATA-1 activity in a well established experimental system where embryonic hematopoiesis is recapitulated in animal pole explants from X. laevis blastulae (32). Treatment with basic fibroblast growth factor can induce some blood differentiation in animal caps but produces only few erythrocytes (32); under these conditions GATA-1 overexpression is sufficient to greatly increase erythroid differentiation, which can be measured by expression of larval globin (33). We injected capped mRNA for WT GATA-1 or GATA-1 K137R in the animal pole of Xenopus embryos at the twoto four-cell stage. Animal cap explants were excised at blastula stage and cultured in the presence of basic fibroblast growth factor until sibling embryos reached tailbud stage. At this point, expression of αT3 globin was analyzed by RT-PCR. Because GATA-1 transactivates its own promoter in a positive regulatory feedback (34, 35) we used Xenopus-specific primers to analyze expression of endogenous xGATA-1 in the same samples. As shown in Fig. 3, injection of GATA-1 mRNA efficiently induced expression of larval globin and xGATA-1; in the same experiments, coinjection of HA-SUMO-1 mRNA had no reproducible effects. Importantly, αT3 globin and xGATA-1 were efficiently induced by injection of the mRNA encoding the nonsumoylatable GATA-1 K137R mutant, indicating that mutation of lysine 137 does not impair GATA-1's ability to transactivate endogenous target genes in Xenopus animal cap explants.
Fig. 3.
Blood induction in animal caps, and RT-PCR analysis of erythroid markers. Eighty picograms of capped mRNA encoding WT GATA-1 or the K137R mutant were injected in the animal pole of Xenopus embryos, with or without 2 ng of mRNA-encoding HA-SUMO-1. Animal caps were excised at stages 8–9 and incubated in the presence of 50 ng/ml basic fibroblast growth factor. The erythroid markers αT3 globin and xGATA-1 were analyzed by radioactive RT-PCR when sibling embryos reached stages 30–35. Endogenous xGATA-1 was amplified by using primers not recognizing the injected mouse GATA-1 mRNA. EF1α was amplified as a control for RNA levels.
PIASy Is a SUMO Ligase for GATA-1 and Represses GATA-1 Transcriptional Activity. As shown in Fig. 2, coexpression of SUMO-1 increases the fraction of sumoylated GATA-1, but this fraction is very small compared with the levels of nonconjugated protein. Thus, to uncover the effects of sumoylation it might be necessary to find experimental conditions that would increase the fraction of sumoylated GATA-1. Because members of the PIAS family of proteins can function as SUMO ligases (18, 20), we asked whether coexpression of PIASy would increase GATA-1 sumoylation and whether such an increase might result in appreciable effects on GATA-1 transcriptional activity. We cotransfected 293T cells with expression vectors encoding GATA-1, GFP-SUMO-1, and T7-PIASy and visualized GATA-1 by immunoblotting. The same experiment was performed with the GATA-1 K137R mutant. As shown in Fig. 4A, a slower-migrating band corresponding to monosumoylated GATA-1 is readily detected upon transfection of GFP-SUMO-1, but the intensity of the band is significantly increased by coexpression of PIASy, indicating that PIASy behaves as an E3 ligase for GATA-1. There is no evidence for sumoylation of the K137R mutant, even in the presence of high levels of PIASy. This observation excludes the existence of latent low-affinity sumoylation sites and further confirms that lysine 137 is the only requirement for SUMO-1 attachment to GATA-1.
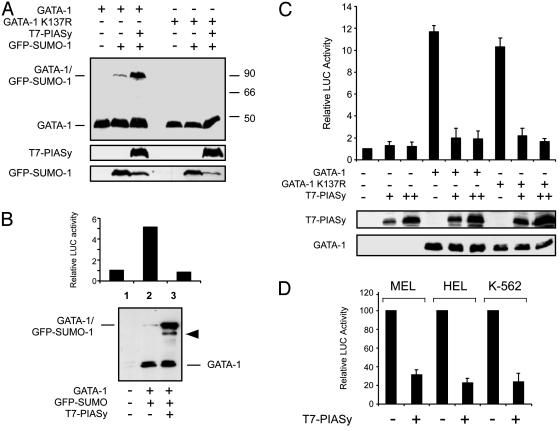
Fig. 4.
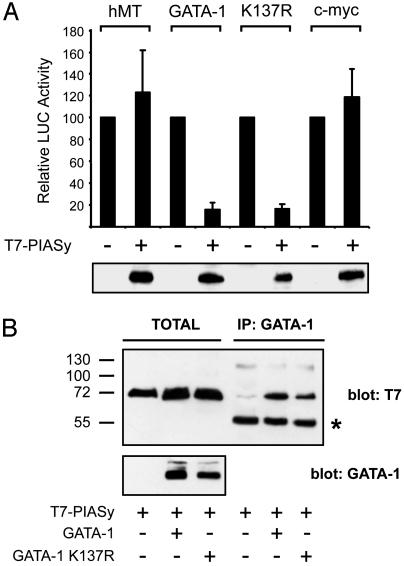
PIASy enhances GATA-1 sumoylation and represses GATA-1 transcriptional activity. (A) PIASy increases GATA-1 sumoylation. WT GATA-1 or the K137R mutant were transfected in 293T cells together with GFP-SUMO-1 and a vector expressing T7-PIASy as indicated. Lysates were separated by SDS/PAGE, and GATA-1 was detected by immunoblotting. (B) PIASy inhibits GATA-1-dependent transactivation. A GATA-1-responsive luciferase reporter plasmid was transfected in U2OS cells together with vectors expressing GATA-1, GFP-SUMO-1, and T7-PIASy as indicated. A plasmid constitutively expressing Renilla luciferase (pRL-CMV) was included as a control for transfection efficiency. GATA-1 transcriptional activity was measured by a dual luciferase assay (Upper), and GATA-1 protein was analyzed by immunoblotting (Lower). The band indicated by an arrowhead in lane 3 likely corresponds to GATA-1 conjugated to endogenous SUMO. (C) Sumoylation is not required for PIASy inhibition of GATA-1 transcriptional activity. Reporter experiments were performed in U2OS cells by using either WT GATA-1 or the nonsumoylatable K137R mutant as described above. Dual luciferase assays were done 36 h after transfection. Lower shows the expression levels of GATA-1 and T7-PIASy in one representative experiment as detected by immunoblotting. (D) PIASy represses GATA-1 transcriptional activity in erythroid cells. Reporter experiments were performed in the indicated cell lines transfecting WT GATA-1 with or without T7-PIASy as indicated. Dual luciferase assays were done 36 h after transfection. Relative luciferase activity is expressed as percent of the activity of the GATA reporter in the absence of PIASy.
We next analyzed whether the transcriptional activity of GATA-1 might be affected by PIASy-induced sumoylation with the luciferase reporter described above. U2OS human cells were transfected in duplicate with the same plasmid mixtures: one set of plates was used for luciferase assays, the other was used for immunoblotting. As shown in Fig. 4B, luciferase assays revealed that coexpression of PIASy dramatically inhibited GATA-1 transcriptional activity. In parallel, immunoblotting established that similar levels of GATA-1 were expressed in the samples and that PIASy had increased the fraction of sumoylated GATA-1. Upon cotransfection of PIASy a fraction of GATA-1 conjugated to endogenous SUMO-1 could also be detected.
To test whether the strong transcriptional repression induced by PIASy might be the consequence of increased GATA-1 sumoylation, we repeated the experiments without addition of GFP-SUMO-1 and with the nonsumoylatable GATA-1 mutant. As summarized in Fig. 4C, under these conditions GATA-1 K137R transactivated the reporter as efficiently as WT GATA-1, and PIASy repressed the transcriptional activity of the K137R mutant as efficiently as that of the WT protein. Identical results were obtained in the presence of cotransfected GFP-SUMO-1 (data not shown). In these assays PIASy had no significant effect on the reporter alone, and GATA-1 proteins were expressed at comparable levels in all samples. Thus, although PIASy greatly enhances the fraction of sumoylated GATA-1, its transcriptional repression activity does not require GATA-1 sumoylation.
Because the above experiments were performed in nonerythroid cells, which lack specific cofactors such as FOG, we asked whether PIASy would also repress GATA-1 transcriptional activity in erythroid cells. As summarized in Fig. 4D, luciferase reporter assays in three different erythroleukemia cell lines gave results very similar to those obtained in nonerythroid cells.
PIASy Repression of GATA-1 Transcriptional Activity Is Specific. To assess the specificity of PIASy repression of GATA-1 transcriptional activity, we tested the effects of PIASy overexpression on a construct where luciferase is under the control of the human c-myc promoter (26). As summarized in Fig. 5A, activity of the c-myc promoter was not affected. Also, the activity of the human metallothionein minimal promoter was not inhibited by PIASy, and we never observed significant changes in the activity of the human cytomegalovirus promoter driving Renilla luciferase, which was included in all transfections. Taken together, these observations support the notion that PIASy inhibition of GATA-1 transcriptional activity is specific.
Fig. 5.
PIASy interacts with GATA-1 and specifically represses its transcriptional activity. (A) PIASy does not inhibit a GATA-1-independent promoter. Constructs expressing luciferase under control of human metallothionein minimal promoter (hMT), human c-myc promoter, or the GATA-responsive promoter was transfected in U2OS cells with or without T7-PIASy as described in Fig. 4. Experiments with the GATA reporter also contained expression vectors for the indicated GATA-1 proteins. Relative luciferase activity is expressed as percent of the activity of the promoters in the absence of PIASy. Lower shows the expression levels of PIASy in one representative experiment. (B) Interaction between PIASy and GATA-1. WT GATA-1 or the K137R mutant was transfected in 293T cells together with T7-PIASy as indicated. After 36 h, lysates were immunoprecipitated with a monoclonal antibody to GATA-1 and immunoblotted with an antibody to the T7 epitope (Right). Expression of GATA-1 and T7-PIASy was also analyzed in total lysates (Left). IgG heavy chains are indicated by an asterisk.
We next asked whether PIASy might physically interact with GATA-1. To this aim, we cotransfected T7-PIASy with either WT GATA-1 or the K137R mutant. GATA-1 proteins were immunoprecipitated with a monoclonal antibody, and immune complexes were analyzed with an antibody to the T7 epitope. As shown in Fig. 5B, under these conditions PIASy coimmunoprecipitated with both the WT and K137R GATA-1 proteins. We conclude that PIASy binds to GATA-1 and specifically represses its transcriptional activity through a mechanism not requiring GATA-1 sumoylation.
Discussion
Sumoylation is a posttranslational modification that can modulate the activity of many nuclear proteins, although the molecular basis of such effects are still poorly understood (23–25). In the present work we demonstrate that GATA-1, a key transcriptional regulator of erythroid and megakaryocytic differentiation, is sumoylated both in vitro and in vivo. We found that a fraction of endogenous GATA-1 is sumoylated in mouse erythroleukemia cells under normal culture conditions, and it will be interesting to analyze whether the proportion of sumoylated GATA-1 changes during erythtroid differentiation in vivo.
Proteins may have multiple sites for sumoylation, and occasionally SUMO may be attached to lysines not conforming to the canonical consensus (16, 36). However, when we mapped the site of sumoylation in GATA-1 we found that lysine 137, perfectly matching the consensus sequence, is the only residue involved.
In an attempt to increase the fraction of sumoylated GATA-1 for functional studies, we found that PIASy is a potent SUMO ligase for GATA-1. PIASy is a member of the PIAS family of RING finger nuclear proteins, which interact with activated STATs and inhibit transcription of STAT-regulated genes after IFN stimulation (18, 37). Different PIAS proteins inhibit different STATs via diverse mechanisms (38), in some cases correlating with PIAS-mediated STAT sumoylation (17, 19). We found that PIASy binds to GATA-1 and that this interaction is not dependent on the presence of the sumoylatable lysine 137. Finally, we found that PIASy is a powerful inhibitor of GATA-1 transcriptional activity. This inhibition is also observed with the GATA-1 K137R mutant, so the molecular mechanism whereby PIASy blocks GATA-1 activity does not involve GATA-1 sumoylation. This result is not totally surprising because it was reported for other proteins. For example, serum response factor, Lef1/Tcf, and Smad3 are all sumoylated by PIASy, and their transcriptional activity is inhibited by PIASy coexpression, but in none of these cases does transcriptional repression require sumoylation (20, 39, 40). In the case of Lef1/Tcf, interaction with PIASy results in accumulation within subnuclear compartments (20). In the case of Smad3, PIASy appears to recruit histone deacetylases to Smad3/Smad4/PIASy complexes on the promoters of type β transforming growth factor target genes (41). We did not observe GATA-1 relocation to nuclear bodies upon PIASy overexpression, and treatment with the histone deacetylase inhibitor trichostatin A did not relieve PIASy repression of GATA-1 transcriptional activity in reporter assays (data not shown), suggesting that the mechanism by which PIASy blocks GATA-1 transcriptional activity might be different from that proposed for Lef1/Tcf or Smad3.
Recently, it has been reported that PIASy enhances SUMO conjugation to GATA-2, a member of the GATA family that is expressed in primitive hematopoietic cells and is critical for survival and growth of multipotential progenitors (42, 43). GATA-2 is also present in adult endothelia, and overexpression of PIASy inhibits GATA-2-dependent transcription in endothelial cell lines. Even in the case of GATA-2, PIASy-mediated repression does not appear to require sumoylation (43), suggesting that PIASy's inhibitory effect on both GATA proteins might involve a common mechanism, which awaits investigation. PIASy is detected in many tissues and cell lines (43, 44), so it is tempting to hypothesize some functional interplay between PIASy and GATA proteins in regulating the fate of hematopoietic cells.
In the present study we focused on GATA-1 intrinsic transcriptional activity, performing transient overexpression experiments in mammalian cell lines. We also assayed GATA-1 capacity to trigger globin expression in a simple hematopoietic differentiation system by using explants from Xenopus embryos. In our experiments, we never observed a significant difference in the activity of the nonsumoylatable GATA-1 K137R mutant with respect to the WT. Nonetheless, it is conceivable that sumoylation might affect the biochemical activity of GATA-1, perhaps impinging on fine regulatory mechanisms that cannot be easily detected in transient overexpression experiments. One possibility is that sumoylation modulates interaction of GATA-1 with one or more of its transcriptional partners (11). Although GATA-1 interaction with most cofactors appears to be through the zinc fingers (9, 10), it is conceivable that SUMO attachment to the N-terminal region might modulate such binding. Sumoylation might in turn regulate the affinity of GATA-1 for different promoters, resulting in activation or repression of selected target genes.
Another possibility is that sumoylation modulates GATA-1 interaction with kinases responsible for its phosphorylation or with enzymes involved in its acetylation. Finally, we should also consider the possibility that under some conditions sumoylation might affect the turnover of GATA-1, resulting in accumulation or degradation of the protein.
In conclusion, our data support the notion that SUMO-1 attachment to GATA-1 provides a fine-tuning mechanism affecting one or more of these regulatory interactions, rather than a major switch triggering specific transcriptional responses. Additional studies are required to understand the biological role of this posttranslational modification of GATA-1, possibly employing more sensitive assays, within more complex experimental systems.
Acknowledgments
We thank Federico Mauri for enthusiastic help with some of the experiments, members of Laboratorio Nazionale Consorzio Interuniversitario per le Biotecnologie (Italy) for discussion, and Stefano Piccolo and all members of his lab for their kind hospitality during Xenopus microinjection experiments. This work was supported by research grants from Associazione Italiana per la Ricerca sul Cancro (Italy) (to G.D.S.) and Telethon (to C.S. and G.D.S.).
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Orkin, S. H. (1992) Blood 80, 575–581. [PubMed] [Google Scholar]
- 2.Patient, R. K. & McGhee, J. D. (2002) Curr. Opin. Genet. Dev. 12, 416–422. [DOI] [PubMed] [Google Scholar]
- 3.Weiss, M. J. & Orkin, S. H. (1995) Exp. Hematol. (Charlottesville, VA) 23, 99–107. [PubMed] [Google Scholar]
- 4.Yamamoto, M., Ko, L. J., Leonard, M. W., Beug, H., Orkin, S. H. & Engel, J. D. (1990) Genes Dev. 4, 1650–1662. [DOI] [PubMed] [Google Scholar]
- 5.Weiss, M. J., Keller, G. & Orkin, S. H. (1994) Genes Dev. 8, 1184–1197. [DOI] [PubMed] [Google Scholar]
- 6.Fujiwara, Y., Browne, C. P., Cunniff, K., Goff, S. C. & Orkin, S. H. (1996) Proc. Natl. Acad. Sci. USA 93, 12355–12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pevny, L., Simon, M. C., Robertson, E., Klein, W. H., Tsai, S. F., D'Agati, V., Orkin, S. H. & Costantini, F. (1991) Nature 349, 257–260. [DOI] [PubMed] [Google Scholar]
- 8.Pevny, L., Lin, C. S., D'Agati, V., Simon, M. C., Orkin, S. H. & Costantini, F. (1995) Development (Cambridge, U.K.) 121, 163–172. [DOI] [PubMed] [Google Scholar]
- 9.Kowalski, K., Liew, C. K., Matthews, J. M., Gell, D. A., Crossley, M. & Mackay, J. P. (2002) J. Biol. Chem. 277, 35720–35729. [DOI] [PubMed] [Google Scholar]
- 10.Rekhtman, N., Radparvar, F., Evans, T. & Skoultchi, A. I. (1999) Genes Dev. 13, 1398–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cantor, A. B. & Orkin, S. H. (2002) Oncogene 21, 3368–3376. [DOI] [PubMed] [Google Scholar]
- 12.Crossley, M. & Orkin, S. H. (1994) J. Biol. Chem. 269, 16589–16596. [PubMed] [Google Scholar]
- 13.Partington, G. A. & Patient, R. K. (1999) Nucleic Acids Res. 27, 1168–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyes, J., Byfield, P., Nakatani, Y. & Ogryzko, V. (1998) Nature 396, 594–598. [DOI] [PubMed] [Google Scholar]
- 15.Elefanty, A. G., Antoniou, M., Custodio, N., Carmo-Fonseca, M. & Grosveld, F. G. (1996) EMBO J. 15, 319–333. [PMC free article] [PubMed] [Google Scholar]
- 16.Muller, S., Hoege, C., Pyrowolakis, G. & Jentsch, S. (2001) Nat. Rev. Mol. Cell Biol. 2, 202–210. [DOI] [PubMed] [Google Scholar]
- 17.Ungureanu, D., Vanhatupa, S., Kotaja, N., Yang, J., Aittomaki, S., Janne, O. A., Palvimo, J. J. & Silvennoinen, O. (2003) Blood 102, 3311–3313. [DOI] [PubMed] [Google Scholar]
- 18.Jackson, P. K. (2001) Genes Dev. 15, 3053–3058. [DOI] [PubMed] [Google Scholar]
- 19.Rogers, R. S., Horvath, C. M. & Matunis, M. J. (2003) J. Biol. Chem. 278, 30091–30097. [DOI] [PubMed] [Google Scholar]
- 20.Sachdev, S., Bruhn, L., Sieber, H., Pichler, A., Melchior, F. & Grosschedl, R. (2001) Genes Dev. 15, 3088–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sapetschnig, A., Rischitor, G., Braun, H., Doll, A., Schergaut, M., Melchior, F. & Suske, G. (2002) EMBO J. 21, 5206–5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alarcon-Vargas, D. & Ronai, Z. (2002) Cancer Biol. Ther. 1, 237–242. [DOI] [PubMed] [Google Scholar]
- 23.Seeler, J. S. & Dejean, A. (2003) Nat. Rev. Mol. Cell Biol. 4, 690–699. [DOI] [PubMed] [Google Scholar]
- 24.Verger, A., Perdomo, J. & Crossley, M. (2003) EMBO Rep. 4, 137–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gill, G. (2003) Curr. Opin. Genet. Dev. 13, 108–113. [DOI] [PubMed] [Google Scholar]
- 26.Frazier, M. W., He, X., Wang, J., Gu, Z., Cleveland, J. L. & Zambetti, G. P. (1998) Mol. Cell. Biol. 18, 3735–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwarz, S. E., Matuschewski, K., Liakopoulos, D., Scheffner, M. & Jentsch, S. (1998) Proc. Natl. Acad. Sci. USA 95, 560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gostissa, M., Hengstermann, A., Fogal, V., Sandy, P., Schwarz, S. E., Scheffner, M. & Del Sal, G. (1999) EMBO J. 18, 6462–6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collavin, L. & Kirschner, M. W. (2003) Development (Cambridge, U.K.) 130, 805–816. [DOI] [PubMed] [Google Scholar]
- 30.Elnitski, L. & Hardison, R. (1999) Blood Cells Mol. Dis. 25, 299–304. [DOI] [PubMed] [Google Scholar]
- 31.Calligaris, R., Bottardi, S., Cogoi, S., Apezteguia, I. & Santoro, C. (1995) Proc. Natl. Acad. Sci. USA 92, 11598–11602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyanaga, Y., Shiurba, R. & Asashima, M. (1999) Dev. Genes Evol. 209, 69–76. [DOI] [PubMed] [Google Scholar]
- 33.Huber, T. L., Perkins, A. C., Deconinck, A. E., Chan, F. Y., Mead, P. E. & Zon, L. I. (2001) Curr. Biol. 11, 1456–1461. [DOI] [PubMed] [Google Scholar]
- 34.Ohneda, K. & Yamamoto, M. (2002) Acta Haematol. 108, 237–245. [DOI] [PubMed] [Google Scholar]
- 35.Nishikawa, K., Kobayashi, M., Masumi, A., Lyons, S. E., Weinstein, B. M., Liu, P. P. & Yamamoto, M. (2003) Mol. Cell. Biol. 23, 8295–8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, P. S., Chang, C., Liu, D. & Derynck, R. (2003) J. Biol. Chem. 278, 27853–27863. [DOI] [PubMed] [Google Scholar]
- 37.Starr, R. & Hilton, D. J. (1999) BioEssays 21, 47–52. [DOI] [PubMed] [Google Scholar]
- 38.Liu, B., Liao, J., Rao, X., Kushner, S. A., Chung, C. D., Chang, D. D. & Shuai, K. (1998) Proc. Natl. Acad. Sci. USA 95, 10626–10631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsuzaki, K., Minami, T., Tojo, M., Honda, Y., Uchimura, Y., Saitoh, H., Yasuda, H., Nagahiro, S., Saya, H. & Nakao, M. (2003) Biochem. Biophys. Res. Commun. 306, 32–38. [DOI] [PubMed] [Google Scholar]
- 40.Imoto, S., Sugiyama, K., Muromoto, R., Sato, N., Yamamoto, T. & Matsuda, T. (2003) J. Biol. Chem. 278, 34253–34258. [DOI] [PubMed] [Google Scholar]
- 41.Long, J., Matsuura, I., He, D., Wang, G., Shuai, K. & Liu, F. (2003) Proc. Natl. Acad. Sci. USA 100, 9791–9796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsai, F. Y., Keller, G., Kuo, F. C., Weiss, M., Chen, J., Rosenblatt, M., Alt, F. W. & Orkin, S. H. (1994) Nature 371, 221–226. [DOI] [PubMed] [Google Scholar]
- 43.Chun, T. H., Itoh, H., Subramanian, L., Iniguez-Lluhi, J. A. & Nakao, K. (2003) Circ. Res. 92, 1201–1208. [DOI] [PubMed] [Google Scholar]
- 44.Liu, B., Gross, M., ten Hoeve, J. & Shuai, K. (2001) Proc. Natl. Acad. Sci. USA 98, 3203–3207. [DOI] [PMC free article] [PubMed] [Google Scholar]