Abstract
Inhibition of myosin phosphatase is critical for agonist-induced contractility of vascular smooth muscle. The protein CPI-17 is a phosphorylation-dependent inhibitor of myosin phosphatase and, in response to agonists, Thr-38 is phosphorylated by protein kinase C, producing a >1,000-fold increase in inhibitory potency. Here, we addressed how CPI-17 could selectively inhibit myosin phosphatase among other protein phosphatase-1 (PP1) holoenzymes. PP1 in cell lysates was separated by sequential affinity chromatography into at least two fractions, one bound specifically to thiophospho-CPI-17, and another bound specifically to inhibitor-2. The MYPT1 regulatory subunit of myosin phosphatase was concentrated only in the fraction bound to thiophospho-CPI-17. This binding was eliminated by addition of excess microcystin-LR to the lysate, showing that binding at the active site of PP1 is required. Phospho-CPI-17 failed to inhibit glycogen-bound PP1 from skeletal muscle, composed primarily of PP1 with the striated muscle glycogen-targeting subunit (GM) regulatory subunit. Phospho-CPI-17 was dephosphorylated during assay of glycogen-bound PP1, not MYPT1-associated PP1, even though these two holoenzymes have the same PP1 catalytic subunit. Phosphorylation of CPI-17 in rabbit arteries was enhanced by calyculin A but not okadaic acid or fostriecin, consistent with PP1-mediated dephosphorylation. We propose that CPI-17 binds at the PP1 active site where it is dephosphorylated, but association of MYPT1 with PP1C allosterically retards this hydrolysis, resulting in formation of a complex of MYPT1·PP1C·P-CPI-17, leading to an increase in smooth muscle contraction.
Protein phosphatase-1 (PP1) is a dominant Ser/Thr phosphatase, controlling a plethora of events in cells, from yeast to mammals. Cellular PP1 holoenzymes consist of a catalytic subunit (PP1C) and various regulatory (targeting) subunits. PP1C in mammalian cells exists as four isoforms that contain a conserved catalytic domain plus a variable region in the C-terminal tails (1). On the other hand, >50 PP1 regulatory subunits have been discovered in mammalian cells (reviewed in refs. 2 and 3). These subunits have a common PP1-binding sequence motif, VxF, that associates with PP1C on the backside opposite the active site, tethering PP1C to compartmentalize PP1 activity (4). Additionally, the interaction with regulatory subunits results in allosteric modulation of PP1C substrate specificity (5). Thus, PP1 subunits modify properties of PP1C to generate diversity of function in cells (2, 3).
In addition to regulatory subunits, several PP1-specific inhibitor proteins are present in mammalian cells. These inhibitor proteins potently suppress the activity of purified PP1C at nanomolar concentrations (6). Most PP1 inhibitor proteins are phosphoproteins, suggesting that cellular PP1 activities are modulated in response to kinase signaling via phosphorylation of PP1 inhibitor proteins. Originally, PP1 inhibitor proteins such as inhibitor-1 (I-1) and inhibitor-2 (Inh2) were believed to inhibit only the free PP1C released from regulatory subunits but not the PP1 holoenzymes themselves. This concept was based on results showing that neither I-1 nor Inh2 blocked activity of the glycogen-bound PP1 holoenzyme (7) or myosin phosphatase holoenzyme (8). More recently, several lines of evidence show regulation of particular PP1 holoenzymes by the inhibitor proteins CPI-17 (9), I-1 (10), and Inh2 (11–13). These new data bring into question how inhibitor proteins recognize different PP1 holoenzymes with common catalytic subunits.
CPI-17 was purified as a myosin phosphatase inhibitor protein from pig aorta (9, 14). The inhibitory potency of CPI-17 is increased >1,000-fold by phosphorylation at Thr-38 (9). Several kinases purified from smooth muscles, such as PKCα/δ, ZIP-kinase, and integrin-linked kinase, activate CPI-17 by phosphorylation at Thr-38 (15–17). Phosphorylation of CPI-17 at Thr-38 in smooth muscle cells occurs in response to various agonists, such as histamine, endothelin-1, and angiotensin II, in parallel with induction of myosin phosphorylation and contraction (18, 19). On the other hand, phosphorylation of CPI-17 is reversed during vasodilation induced by nitric oxide production (20). Thus, phosphorylation of CPI-17 suppresses myosin phosphatase activity, resulting in phosphorylation of myosin and contraction of smooth muscle. In addition, specific depletion of endogenous CPI-17 by small interfering RNA or antibody microinjection eliminated the cerebellar long-term synaptic depression of Purkinje cells mediated by PKC, demonstrating involvement of CPI-17 in neuronal signaling (21). Although phospho-CPI-17 inhibits monomeric PP1C in addition to myosin phosphatase, myosin phosphatase was proposed as a preferred target of phospho-CPI-17 in smooth muscle (22), fibroblasts (23), and cerebellar Purkinje cells (21). Here we investigated how phospho-CPI-17 discriminates myosin phosphatase from among other cellular PP1 holoenzymes, to mediate specific signaling.
Experimental Procedures
Materials. Recombinant His-6, S-tag (H6S)-CPI-17, and (H6S)-Inh2 were prepared as described (6). Thiophosphorylation and phosphorylation were performed by using ATPγS (Roche Applied Science, Indianapolis) and ATP (Sigma), respectively. Antibodies for pan-PP1C and MYPT1 were purchased from Transduction Laboratories (Lexington, KY) and Babco (Richmond, CA), respectively. Anti-myc epitope (9E10) antibody was obtained from the Lymphocyte Culture Center at the University of Virginia. Antibodies for catalytic subunit of PP2A (PP2Ac), CPI-17, and P-CPI-17(T38) were prepared as described (18, 22, 24). S-protein agarose and glutathione-agarose were purchased from Novagen and Sigma, respectively. Microcystin-LR (MC-LR) was obtained from Calbiochem and coupled with Affigel 10 (Bio-Rad, Hercules, CA) according to the manufacturer's protocol. Preparation of the expression vector for myc epitope-tagged human MYPT1 (myc-MYPT1) was described previously (25). An Ala-mutant of myc-MYPT1 at Phe-38, F38A, was prepared by the QuikChange protocol of Stratagene. A PCR fragment encoding MYPT1 (1–300) was ligated into pGEX4T-2 at BamH1/EcoRI sites. The fusion protein, GST-MYPT1 (1–300), was expressed in Escherichia coli, by induction with isopropyl thio-β-galactoside and purified by using glutathione-agarose chromatography. Myosin phosphatase was isolated from pig aorta (9). Glycogen particles from a rabbit skeletal muscle extract were prepared by the protocol of Gruppuso and Brautigan (26). This preparation contains major glycogen-associating proteins, such as glycogen phosphorylase and glycogen synthase (26), and has been used for starting material of glycogen-bound PP1 preparation (7). Cell culture materials were obtained from Invitrogen. COS7 and NIH 3T3 cells were obtained from American Type Culture Collection and maintained with 10% newborn calf serum in DMEM. Rat aorta smooth muscle cells were a generous gift from G. Owen's lab at the University of Virginia and were maintained with 10% FBS in DMEM/F12 (1:1) medium.
Binding Assay (“Pull-Down Assay”). Cells in a 10-cm dish were lysed with 1 ml of lysis buffer (50 mM 4-morpholinepropanesulfonic acid–NaOH, pH 7.0, including 0.1 M NaCl/1 mM EGTA/0.1% Triton X-100/5% glycerol/0.2% 2-mercaptoethanol/0.4 mM Pefabloc). After centrifugation at 20,000 ×g for 10 min, an aliquot (0.25 ml) of the lysate was mixed for 30 min at 4°C with 5 μg of either H6S-CPI-17 or H6S-Inh2 immobilized on a slurry (10 μl) of S-protein agarose beads (Novagen). The beads were recovered, washed three times by centrifugation with 0.15 ml of lysis buffer, and mixed with 20 μl of 2× Laemmli buffer. Eluted proteins were immunoblotted with anti-MYPT1 (1:5,000 dilution), anti-PP1C (1:1,000) or anti-PP2Ac (1:5,000). Using these conditions, ≈70% of total MYPT1 is recovered in the soluble fraction, as described (27).
Sequential Pull-Down Assay. Rat aorta smooth muscle cells in a 15-cm dish were lysed with 1 ml of lysis buffer. The extract clarified by centrifugation at 20,000 × g for 20 min was mixed for 3 h at 4°C with 5 μg of either unphosphorylated H6S-CPI-17 thiophosphorylated H6S-CPI-17, or H6S-Inh2 plus 10 μl of S-protein agarose slurry. These conditions were determined to be limiting for the amount of extracts so the inhibitors would fully deplete the sample of their binding partners. The suspension was centrifuged as before, and the supernatant was transferred into another tube. The beads collected were washed by centrifugation three times with 0.15 ml of lysis buffer, and bound proteins were analyzed by anti-PP1C immunoblotting. An aliquot (0.3 ml) of the supernatant was mixed for 1.5 h at 4°C with 5 μg of either H6S-CPI-17 or H6S-Inh2 plus 10 μl of S-protein agarose slurry. The beads were collected, washed, and proteins eluted and analyzed as described above.
Immunoprecipitation Assay. COS7 cells in a 10-cm dish were transiently transfected for 24 h with myc-MYPT1 vector (5 μg) by using 15 μl of FuGENE6 transfection reagent (Roche Applied Sciences), as described (23). Cells expressing myc-MYPT1 were lysed with 1 ml of lysis buffer. The extract was clarified by centrifugation for 10 min at 20,000 × g and split into two portions. Half was incubated overnight with anti-myc antibody (4 μg) on ice. The anti-myc immunocomplex was captured for 30 min with 10 μl of Protein G-agarose slurry (Sigma) at 4°C. The other half of the extract was subjected to pull-down assay by using thiophospho-CPI-17 (TP-CPI-17) beads. After washing three times with 0.15 ml of lysis buffer, proteins on beads were solubilized by addition of Laemmli buffer, and the samples were subjected to immunoblotting analysis.
Phosphatase Assay. Phosphatase activity was measured by using 32P-labeled substrate, 1 μM 32P-LC20 for myosin phosphatase, or 0.3 mg/ml 32P-phosphorylase a, for glycogen-bound phosphatase, as described (28). Conditions used were 25 mM 4-morpholinepropanesulfonic acid–NaOH, pH 7.0, including 50 mM NaCl/0.1 mM EGTA/1 nM okadaic acid/1 mM DTT/0.02% Brij-35/0.4 mM Pefabloc. Caffeine (5 mM) was added to phosphorylase-a phosphatase assay. Reaction was initiated by addition of myosin phosphatase (3.9 microunits) or glycogen-bound phosphatase (56 microunits). Phosphorylation of Thr-38 in CPI-17 in the mixture was measured by immunoblotting by using anti-P-CPI-17(T38) (1:1,000). The blot was reprobed with anti-CPI-17 (1:5,000) for a loading control.
Smooth Muscle Tissue Preparation and Measurement of CPI-17 Phosphorylation. All animal procedures were approved by the Animal Care and Use Committee of the Boston Biomedical Research Institute (Boston). Femoral artery strips from adult male New Zealand White rabbits were equipped on a transducer throughout experiments and were subjected to permeabilization with Staphylococcus aureus α-toxin (20 μg/ml; List, Campbell, CA), as described (29). Permeabilized strips were first treated with the ATP-free Ca2+-free solution for 30 min and then incubated for 20 min in the presence of phosphatase inhibitors with 30 μM GTPγS. Phosphorylation was initiated by addition of 4.5 mM MgATP and 10 mM creatine phosphate at 20°C. After 4 min incubation, strips were quickly frozen and subjected to the immunoassay for phosphorylation of CPI-17. Phosphorylation of CPI-17 was measured by anti-P-CPI-17(T38) immunoblotting, as described (18).
Others. SDS/PAGE, immunoblotting, densitometry, and protein determination were carried out as described (6).
Results
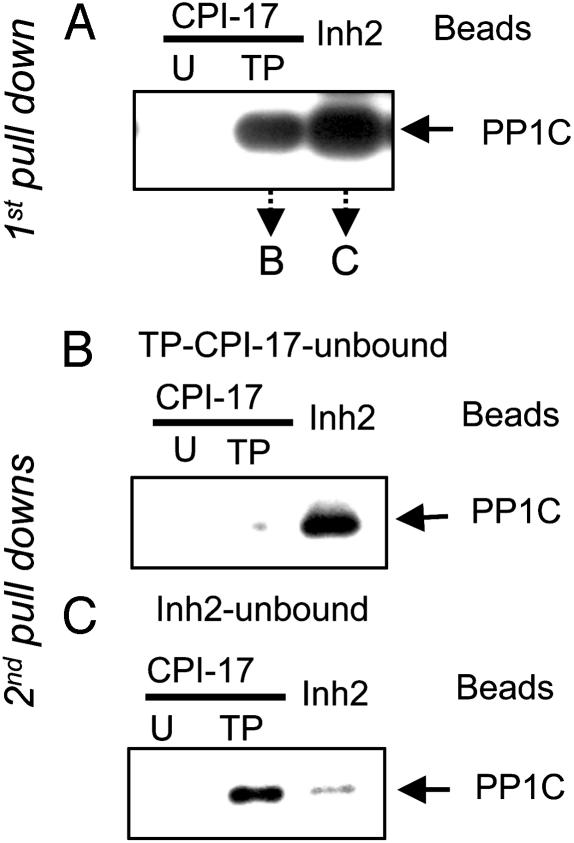
Selective Binding of Inhibitor Proteins to PP1 Holoenzymes. Selective binding of PP1 holoenzymes to TP-CPI-17 and Inh2 was analyzed by a two-stage “sequential pull-down assay” (Fig. 1). In the first stage, a cell lysate was mixed with TP-CPI-17 or Inh2 coupled to beads. In the second stage, the unbound fractions from each pull down were split in half and subjected to a second pull-down assay with fresh batches of TP-CPI-17 and Inh2 beads. The amount of PP1 holoenzyme bound was estimated by immunoblotting, using anti-PP1C antibody. In the first stage, substantial amounts of PP1C were bound to either TP-CPI-17 or Inh2, compared to control, where no PP1C was bound to unphosphorylated CPI-17 (Fig. 1 A). Using the unbound fraction from TP-CPI-17 beads yielded little PP1C binding in the second stage with TP-CPI-17 beads, indicating this pool of PP1 holoenzymes was depleted in the first stage. However, this same unbound fraction had substantial amounts of PP1 holoenzymes that bound to Inh2 beads in the second stage (Fig. 1B). Conversely, PP1 holoenzymes that did not bind Inh2 beads in the first stage were captured on TP-CPI-17 beads in the second stage, but not on Inh2 beads (Fig. 1C). The data show that CPI-17 and Inh2 exclusively recognized and bound to different subsets of PP1 holoenzymes.
Fig. 1.
CPI-17 and Inh2 bind particular subsets of PP1 holoenzymes (sequential pull-down assay). (A) First pull-down. Smooth muscle cell extracts were split into parallel samples and mixed with agarose beads conjugated with recombinant unphosphorylated (U)-, thiophosphorylated (TP)-CPI-17, or Inh2. After centrifugation, the supernatant (“unbound fraction”) was used in a second pull-down assay, the beads in the pellet were eluted, and proteins were subjected to immunoblotting by using anti-PP1C. (B and C) Second pull down. The unbound fractions from A were split into three portions and mixed with fresh beads as in A. The bound PP1C was eluted and detected by immunoblotting with anti-PP1C. The results were reproduced in two independent experiments.
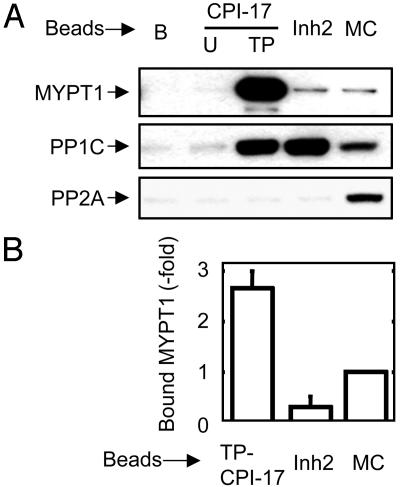
The binding of myosin phosphatase to TP-CPI-17 and Inh2 was analyzed further by immunoblotting (Fig. 2). MYPT1 was recovered only from TP-CPI-17 (Fig. 2A Top), even though about equal amounts of PP1C holoenzymes bound to TP-CPI-17 and Inh2 (Fig. 2A Middle). As a control, lysates were adsorbed to MC-LR, an active site inhibitor, to bind the PP1C and PP2A phosphatases in the lysate (Fig. 2A). The amount of bound MYPT1·PP1C (myosin phosphatase) relative to the amount of PP1C was compared for each inhibitor (Fig. 2B). From this analysis, MYPT1·PP1C binding to TP-CPI-17 beads was enriched by 9.0-fold compared to Inh2 beads and 2.6-fold compared to MC-LR beads. The results indicate that TP-CPI-17 preferentially captures the MYPT1·PP1C complex from among various other PP1 holoenzymes in cells. Similar results were obtained by using lysates of other cultured cells or extracts of tissues, such as brain and smooth muscles.
Fig. 2.
CPI-17 selectively associates with myosin phosphatase among PP1 holoenzymes in cell lysate. (A) Pull-down assay was performed as described for Fig. 1 using beads conjugated with inhibitor proteins or MC-LR. The lane marked B indicates blank beads. After incubation with a NIH 3T3 cell lysate, beads were washed, and the bound proteins were eluted and analyzed by immunoblotting using anti-MYPT1 (130 kDa; Top), anti-PP1C (37 kDa; Middle), and anti-PP2Ac (36 kDa; Bottom). (B) Bar graph shows relative recovery of MYPT1 and PP1C. Staining intensity of MYPT1 was measured by densitometer, compared to intensity of PP1C stain. Mean value and SEM from three independent experiments are shown.
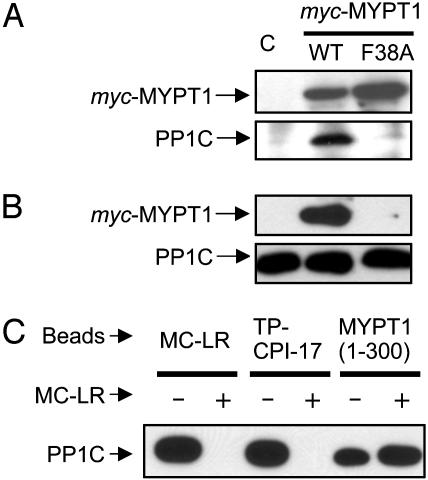
We determined that binding of TP-CPI-17 to myosin phosphatase occurs at the active site of PP1C (Fig. 3). COS7 cells were transiently transfected with WT myc-tagged MYPT1 or MYPT1 mutated (F38A) in the putative PP1C-binding site. The cell lysate was split, and one-half was subjected to immunoprecipitation with anti-myc antibody (Fig. 3A). Both myc-MYPT1(WT) and myc-MYPT1(F38A) were expressed and precipitated effectively, but PP1C was detected only with myc-MYPT1(WT), showing that the F38A mutation eliminated PP1C binding to MYPT1. The other half of the lysate was used in a binding assay with TP-CPI-17 beads (Fig. 3B). The myc-MYPT1(WT)·PP1C holoenzyme bound to TP-CPI-17 beads, whereas the F38A mutant of MYPT1 was not recovered, even though endogenous PP1C bound to these beads. We concluded that MYPT1 did not bind directly to TP-CPI-17 in these assays, but the interaction is indirect and bridged via PP1C. Sites on PP1C for interaction of MYPT1 and TP-CPI-17 are functionally separate. We tested whether the active site inhibitor MC-LR would compete with TP-CPI-17 binding to myosin phosphatase (Fig. 3C). Binding assays were performed by using MC-LR, TP-CPI-17 or GST-MYPT1(1–300) as ligands, in the presence or absence of excess MC-LR in cell lysates. In the control, addition of MC-LR fully eliminated PP1C binding to MC-LR beads. Likewise, addition of MC-LR prevented PP1C binding to TP-CPI-17 beads. However, MC-LR added to the lysate did not reduce binding of PP1C to GST-MYPT1 (1–300). Binding to TP-CPI-17 requires the unoccupied active site of PP1C, but MYPT1 can bind PP1C with MC-LR at the active site.
Fig. 3.
CPI-17 docks at active site of myosin phosphatase. (A and B) Immunoprecipitation compared to CPI-17 binding of myc-MYPT1·PP1C. A myc-tagged MYPT1 WT or F38A mutant (F38A) was transiently expressed in COS7 cells. The control experiment used empty vector, and the sample is shown in lane C. Half of the cell lysate was subjected to immunoprecipitation by using anti-myc conjugated beads (A). The other half of the lysate was mixed with TP-CPI-17 beads, and bound proteins were eluted and analyzed by immunoblotting (B). The results were reproduced in two independent experiments. (C) MC-LR competes with TP-CPI-17 for binding to PP1C. Smooth muscle cell lysate was mixed with beads conjugated with MC-LR, TP-CPI-17, or MYPT1 (1–300) fragment, in the absence or presence of 1 μM MC-LR. The bound PP1C was detected by anti-PP1C immunoblotting. The result was reproduced in two independent experiments.
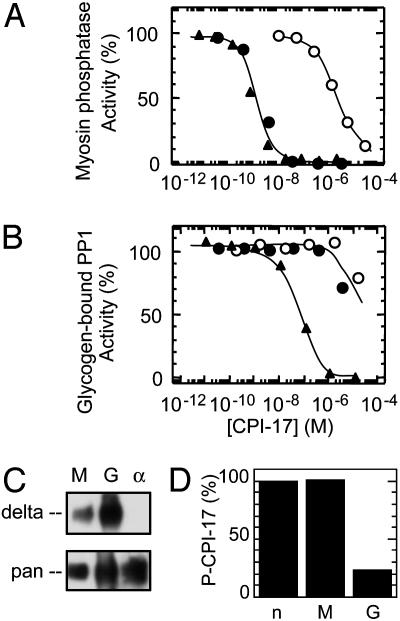
Specific Inhibition of Myosin Phosphatase by Differential Dephosphorylation of CPI-17. Consistent with its preferential binding, CPI-17 is a highly specific inhibitor of myosin phosphatase (Fig. 4). Myosin phosphatase prepared from porcine aorta smooth muscle consisted of the PP1C δ isoform and a fragment of MYPT1 (9). Both TP-CPI-17 and P-CPI-17 inhibited myosin phosphatase with the same potency (IC50 = 3 nM), as described (9, 22, 30). In contrast, Inh2 up to micromolar concentrations did not inhibit myosin phosphatase (23). For comparison, the PP1 holoenzyme associated with glycogen particles was prepared from rabbit skeletal muscle. This PP1 holoenzyme is composed of the striated muscle glycogen-targeting subunit (GM) plus PP1C, and it was inhibited by TP-CPI-17 with an IC50 of 100 nM, >30-fold higher compared with myosin phosphatase (Fig. 4 A and B). The glycogen-bound PP1 mainly consists of PP1 δ isoform (Fig. 4C), consistent with the report by Colbran et al. (31). Surprisingly, P-CPI-17 was a relatively poor inhibitor of this glycogen-bound PP1, with a potency the same as the unphosphorylated inactive CPI-17 (Fig. 4B). This raised the possibility that CPI-17 was dephosphorylated in the assay. Indeed, Thr-38 of P-CPI-17 was dephosphorylated during the assay with the glycogen-bound PP1 (Fig. 4D). In contrast, P-CPI-17 remained phosphorylated in the parallel assay with myosin phosphatase (Fig. 4D). Because these assays were carried out in the presence of 1 nM okadaic acid and 0.1 mM EDTA, any traces of PP2A, PP2B, or PP2C activities were unlikely to be responsible for the dephosphorylation of the P-CPI-17. The glycogen-bound phosphatase was exclusively PP1, because both TP-CPI-17 and phospho-PHI-1, an analog of CPI-17 (28), could completely inhibit the activity. These results show glycogen-bound PP1 dephosphorylates P-CPI-17 to alleviate inhibition, whereas myosin phosphatase binds P-CPI-17 but hydrolyzes it so slowly that an inactive complex accumulates.
Fig. 4.
P-CPI-17 specifically inhibits myosin phosphatase. Phosphatase assays were carried out by using purified myosin phosphatase (A) or glycogen-bound PP1 (B), in the presence of unphosphorylated CPI-17 (open circle)-, phosphorylated CPI-17 (filled circle)-, or thiophosphorylated (filled triangle)-CPI-17 at the indicated concentrations. Phosphatase activity without added CPI-17 is set as 100%. Assay was initiated by addition of phosphatase preparation. Okadaic acid (1 nM) and EDTA (0.1 mM) were added in the reaction mixture to inhibit any PP2A and PP2B activities that possibly contaminated the glycogen-bound PP1 preparation. (C) Myosin phosphatase (M), glycogen-bound PP1 (G), and recombinant PP1C α isoform (α) were subjected to immunoblotting by using PP1C δ isoform specific antibody (Upper) and pan-PP1C antibody (Lower). (D) Aliquots of the inhibition assay mixture including phospho-CPI-17 (100 nM) were subjected to immunoblotting by using anti-P-CPI-17(T38) and anti-CPI-17 antibodies. Mean values from duplicate assays are shown. n indicates mixture without added phosphatases.
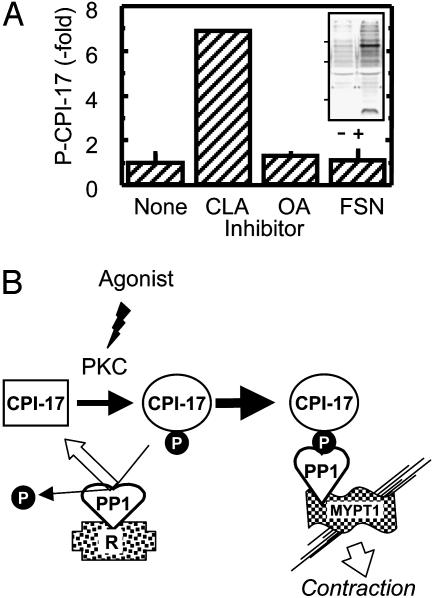
Is PP1 responsible for dephosphorylation of CPI-17 in smooth muscle? α-Toxin-permeabilized rabbit femoral artery was treated with various phosphatase inhibitor compounds, and phosphorylation of Thr-38 in endogenous CPI-17 was assayed (Fig. 5A). Addition of calyculin A (1 μM) increased CPI-17 phosphorylation 7-fold compared with basal levels. On the other hand, okadaic acid (1 μM) or fostriecin (3 μM), both inhibitors more selective for PP2A relative to PP1, did not increase phosphorylation of CPI-17. Inhibition of PP2A by fostriecin was verified by the appearance of increased P-Thr in 90- and 20-kDa proteins by phospho-specific immunoblotting (Fig. 5A Inset). The results indicate that Thr-38 in P-CPI-17 is dephosphorylated by PP1 in smooth muscles.
Fig. 5.
PP1 dephosphorylation of P-CPI-17 in smooth muscle. (A) Phosphorylation of CPI-17 in rabbit femoral artery. After permeabilization with α-toxin, rabbit femoral artery was exposed for 30 min to calyculin A (CLA, 1 μM), okadaic acid (OA, 1 μM), or fostriecin (FSN, 3 μM), in the presence of EGTA and 30 μM GTPγS, at 20°C. After 4-min incubation with ATP and creatine phosphate, the muscle strip was quickly frozen and phosphorylation of endogenous CPI-17 was measured by anti-P-CPI-17(T38) immunoblotting. Phosphorylation of CPI-17(T38) was normalized with CPI-17 in the control (none). Relative CPI-17(T38) phosphorylation is shown as the mean value ± SEM from five independent assays. Aliquots of the sample from control and FSN-treated strips were subjected to immunoblotting by using anti-phospho-Thr antibody (Cell Signaling Technology, Beverly, MA; 1:1,000) as a control for the action of the inhibitor (Inset). Bars at left indicate position of molecular mass markers of 100, 50, and 25 kDa from top. (B) Proposed mechanism for selective inhibition of myosin phosphatase by CPI-17 in smooth muscle. CPI-17 is phosphorylated by active PKC in response to agonist stimuli, resulting in conformational change to the active form (35). Phospho-CPI-17 preferentially associates with PP1C bound to MYPT1 and forms a stable-inactive complex with myosin phosphatase that causes an increase in myosin phosphorylation and contraction. On the other hand, phospho-CPI-17 binds to PP1C associated with other regulatory subunits (R), such as GM, but undergoes dephosphorylation and dissociates from PP1. By this model, only myosin phosphatase is effectively regulated by PKC/CPI-17 signaling.
Discussion
In this study, we provide evidence that phosphorylated CPI-17 binds and inhibits myosin phosphatase and is selective for myosin phosphatase among various PP1 holoenzymes in cells. Immunoblotting using an isoform-specific antibody of PP1C showed that both myosin phosphatase and glycogen-bound phosphatase contain the same δ isoform of PP1 (Fig. 4C) (31), so the selectivity of P-CPI-17 cannot be ascribed to sequence differences in the catalytic subunit. Instead, we propose that the inhibitor phosphoprotein detects distinctive conformations of PP1C produced by interactions with different regulatory subunits. Regulatory subunit association with PP1C is known to exert allosteric control on substrate specificity (5), and here we show binding to different PP1 inhibitor proteins is affected by PP1 regulatory subunits. Affinity chromatography (22) and the sequential pull-down assay done here separate PP1 holoenzymes in cell lysates into at least two groups, those that bind or do not bind to CPI-17. Specific binding of CPI-17 to myosin phosphatase was eliminated by addition of MC-LR, indicating CPI-17 docks primarily at the active site of PP1C associated with MYPT1. Alternatively, interaction of GM with PP1C affects the conformation at the active site to reduce the relative affinity for CPI-17 and also allow PP1C to dephosphorylate CPI-17. Thus, glycogen-bound PP1 evades inhibition by CPI-17 by inactivating the inhibitor, by dephosphorylation. The PP1 holoenzymes that dephosphorylate CPI-17 appear insensitive to inhibition, whereas PP1 holoenzymes (i.e., myosin phosphatase) that poorly dephosphorylate CPI-17 are thereby sensitive to inhibition. Previous studies showed that the dephosphorylation rate of P-CPI-17 by myosin phosphatase is <0.1 sec–1, which is 50-fold slower compared with P-LC20 substrate (30). Furthermore, P-CPI-17 and TP-CPI-17 were equally potent toward myosin phosphatase, consistent with no dephosphorylation during assay (Fig. 4 A and D). Therefore, P-CPI-17 binds myosin phosphatase and forms a transiently inactive complex. We propose that the inhibitory specificity of CPI-17 is determined by relative rates of dephosphorylation by different PP1 holoenzymes (Fig. 5B).
This model has the advantage that it does not require other phosphatases to dephosphorylate and inactivate CPI-17. Thiophosphorylated CPI-17 captured essentially no PP2A from cell lysates, indicative of relatively weak binding. Even though there are reports that PP2A can dephosphorylate purified CPI-17 in biochemical assays, we think it unlikely that PP2A dephosphorylates CPI-17 in vivo. Supporting this view, inhibition of PP1, but not PP2A, -2B, or -2C, enhanced phosphorylation of CPI-17 in permeabilized artery strips (Fig. 5A). Another consideration is the high expression level of CPI-17 in arteries, where the concentration is estimated at 7 μM (32). Upon activation (phosphorylation of Thr-38) of CPI-17, one would predict that all PP1 in the cell would be inhibited, because, as an example, CPI-17 has an IC50 of 100 nM for glycogen-bound PP1. A disaster is averted by efficient dephosphorylation of CPI-17 by different PP1 holoenzymes. This concept is critical to account for restricted inhibition of a select pool of PP1 holoenzymes. Myosin phosphatase containing the MYPT1 subunit is one such pool of CPI-17-sensitive phosphatase. However, this does not imply that MYPT1-containing phosphatase is the only CPI-17-sensitive holoenzyme. Related regulatory subunits such as p85 (33), or other subunits, may also produce the conformation of PP1C that binds phospho-CPI-17 with high affinity and restricted catalytic activity. These would be CPI-17-sensitive by the same proposed mechanism.
P-CPI-17 inhibits native PP1C monomer purified from rabbit muscle with IC50 of 10 nM (9). The IC50 value is 5-fold lower against myosin phosphatase and 10-fold higher against glycogen-bound PP1. This implies that binding of MYPT1 alters the conformation of PP1C to increase sensitivity to CPI-17, whereas interaction with other targeting subunits, such as GM, reduces affinity for CPI-17 and facilitates hydrolysis of P-CPI-17. The interaction of PP1 and CPI-17 is sensitive to the inhibitor conformation as well, because the Y41A mutant of CPI-17 is dephosphorylated by myosin phosphatase as efficiently as the preferred substrate phospho-myosin light chain (30). The NMR solution structure of CPI-17 reveals an N-terminal loop consisting of the phosphorylation site, called the P-loop, followed by a four-helix bundle (34, 35). The Asp mutation at Thr-38, which mimics phosphorylation, exposes the P-loop on the molecular surface (35). In the T38D mutant and phosphoT38 structure, the side chain of Tyr-41 tethers P-loop to B-helix of CPI-17, suggesting that Tyr-41 could stabilize P-loop structure to prevent hydrolysis of P-CPI-17 at the active site of myosin phosphatase.
Purified myosin phosphatase is also inhibited by phosphorylated forms of PHI-1 and KEPI, members of a family of inhibitors related to CPI-17 (28, 36, 37). Amino acid sequences are highly conserved within the domain required for potent inhibition of PP1 holoenzymes (called the phosphatase holoenzyme inhibitory domain) that includes the Tyr-41 residue in the P-loop. Therefore PHI-1 and KEPI are also likely to interact with the PP1C active site. We propose that the mechanism described here might apply in parallel to these other PP1 phosphoinhibitor proteins. Interestingly, unlike CPI-17, P-PHI-1 inhibits glycogen-bound PP1 with an IC50 of 30 nM (9). Thus, differences in the conformation of PHI-1 relative to CPI-17 may protect it from dephosphorylation by glycogen-bound PP1. Conformations of PP1C induced by regulatory subunit association would allow either (i) binding and dephosphorylation or (ii) binding and inhibition.
Because both phospho-I-1 and -Inh2 were unable to inhibit glycogen-bound PP1, since that time it has been accepted that the action of these inhibitors required dissociation of PP1 holoenzymes to release the monomeric catalytic subunit (38). The KIQF sequence of I-1 and IKGI sequence of Inh2 are thought to function as the canonical VXF motif for PP1C binding. This concept makes interaction of PP1C with inhibitor proteins and targeting subunits mutually exclusive (4, 39). Nevertheless, there are now several lines of evidence that indicate direct interaction of I-1 and Inh2 with PP1 holoenzymes, without dissociation (11–13). These results lead us to propose a model for specific regulation of PP1 holoenzymes by inhibitor proteins, where the inhibitor has narrow specificity for a pool of PP1 holoenzymes. All of the inhibitor proteins can inhibit PP1C monomer. However, interaction with regulatory subunits occludes some sites and allosterically modifies the available active site to restrict PP1C binding to particular inhibitor proteins. Indeed, I-1 and Inh2, as well as CPI-17, each exhibits activity against a select pool of PP1 holoenzymes. It remains unclear how, or even whether, the motifs that resemble VXF in I-1, Inh2, PHI-1, and KEPI interact with PP1 in holoenzymes. In fact, most structure/function analysis of inhibitor proteins to date has been done using isolated PP1C or Mn2+-activated recombinant PP1C, leaving regulation of PP1 holoenzymes by inhibitor proteins mostly an unexplored issue. Over 50 PP1 targeting subunits are reported, and there are less than a dozen PP1 inhibitor proteins to date. To complete the circuitry that gives specific signaling to individual pools of PP1, we might expect more inhibitor proteins or means of restricted intracellular distribution. In addition, there is regulation via direct phosphorylation of PP1C and regulatory subunits (2, 3). Last, discrimination of PP1C conformations within the context of different holoenzymes holds out hope that small molecule inhibitors of individual PP1 holoenzymes remain a possibility for therapeutic applications.
Acknowledgments
This work was supported in part by a Scientist Development Grant from the American Heart Association National Center (to M.E.) and by grants from the U.S. Public Health Service and the National Institutes of Health [Grants GM56362 and CA40042 (to D.L.B.) and HL51824 and HL70881 (to T.K.)].
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PP1, protein phosphatase-1; MC-LR, microcystin-LR; TP-CPI-17, thiophospho-CPI-17; I-1, inhibitor-1; Inh2, inhibitor-2.
References
- 1.Shima, H., Hatano, Y., Chun, Y. S., Sugimura, T., Zhang, Z., Lee, E. Y. & Nagao, M. (1993) Biochem. Biophys. Res. Commun. 192, 1289–1296. [DOI] [PubMed] [Google Scholar]
- 2.Ceulemans, H. & Bollen, M. (2004) Physiol. Rev. 84, 1–39. [DOI] [PubMed] [Google Scholar]
- 3.Cohen, P. T. (2002) J. Cell Sci. 115, 241–256. [DOI] [PubMed] [Google Scholar]
- 4.Egloff, M. P., Johnson, D. F., Moorhead, G., Cohen, P. T., Cohen, P. & Barford, D. (1997) EMBO J. 16, 1876–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanaka, J., Ito, M., Feng, J., Ichikawa, K., Hamaguchi, T., Nakamura, M., Hartshorne, D. J. & Nakano, T. (1998) Biochemistry 37, 16697–16703. [DOI] [PubMed] [Google Scholar]
- 6.Eto, M., Leach, C., Tountas, N. A. & Brautigan, D. L. (2003) in Methods in Enzymology, eds. Klumpp, S. & Krieglstein, J. (Academic, San Diego), Vol. 366, pp. 243–260. [DOI] [PubMed] [Google Scholar]
- 7.Stralfors, P., Hiraga, A. & Cohen, P. (1985) Eur. J. Biochem. 149, 295–303. [DOI] [PubMed] [Google Scholar]
- 8.Tulloch, A. G. & Pato, M. D. (1991) J. Biol. Chem. 266, 20168–20174. [PubMed] [Google Scholar]
- 9.Eto, M., Ohmori, T., Suzuki, M., Furuya, K. & Morita, F. (1995) J. Biochem. 118, 1104–1107. [DOI] [PubMed] [Google Scholar]
- 10.Connor, J. H., Weiser, D. C., Li, S., Hallenbeck, J. M. & Shenolikar, S. (2001) Mol. Cell. Biol. 21, 6841–6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eto, M., Elliott, E., Prickett, T. D. & Brautigan, D. L. (2002) J. Biol. Chem. 277, 44013–44020. [DOI] [PubMed] [Google Scholar]
- 12.Terry-Lorenzo, R. T., Elliot, E., Weiser, D. C., Prickett, T. D., Brautigan, D. L. & Shenolikar, S. (2002) J. Biol. Chem. 277, 46535–46543. [DOI] [PubMed] [Google Scholar]
- 13.Wang, H. & Brautigan, D. L. (2002) J. Biol. Chem. 277, 49605–49612. [DOI] [PubMed] [Google Scholar]
- 14.Eto, M., Senba, S., Morita, F. & Yazawa, M. (1997) FEBS Lett. 410, 356–360. [DOI] [PubMed] [Google Scholar]
- 15.Eto, M., Kitazawa, T., Yazawa, M., Mukai, H., Ono, Y. & Brautigan, D. L. (2001) J. Biol. Chem. 276, 29072–29078. [DOI] [PubMed] [Google Scholar]
- 16.MacDonald, J. A., Eto, M., Borman, M. A., Brautigan, D. L. & Haystead, T. A. (2001) FEBS Lett. 493, 91–94. [DOI] [PubMed] [Google Scholar]
- 17.Deng, J. T., Sutherland, C., Brautigan, D. L., Eto, M. & Walsh, M. P. (2002) Biochem. J. 367, 517–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitazawa, T., Eto, M., Woodsome, T. P. & Brautigan, D. L. (2000) J. Biol. Chem. 275, 9897–9900. [DOI] [PubMed] [Google Scholar]
- 19.Kitazawa, T., Eto, M., Woodsome, T. P. & Khalequzzaman, M. (2003) J. Physiol. 546, 879–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Etter, E. F., Eto, M., Wardle, R. L., Brautigan, D. L. & Murphy, R. A. (2001) J. Biol. Chem. 276, 34681–34685. [DOI] [PubMed] [Google Scholar]
- 21.Eto, M., Bock, R., Brautigan, D. & Linden, D. J. (2002) Neuron 36, 1145–1158. [DOI] [PubMed] [Google Scholar]
- 22.Senba, S., Eto, M. & Yazawa, M. (1999) J. Biochem. 125, 354–362. [DOI] [PubMed] [Google Scholar]
- 23.Eto, M., Wong, L., Yazawa, M. & Brautigan, D. L. (2000) Cell Motil. Cytoskeleton 46, 222–234. [DOI] [PubMed] [Google Scholar]
- 24.Murata, K., Wu, J. & Brautigan, D. L. (1997) Proc. Natl. Acad. Sci. USA 94, 10624–10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Broustas, C. G., Grammatikakis, N., Eto, M., Dent, P., Brautigan, D. L. & Kasid, U. (2002) J. Biol. Chem. 277, 3053–3059. [DOI] [PubMed] [Google Scholar]
- 26.Gruppuso, P. A. & Brautigan, L. D. (1988) Biochem. Int. 16, 1027–1032. [PubMed] [Google Scholar]
- 27.Murata, K., Hirano, K., Villa-Moruzzi, E., Hartshorne, D. J. & Brautigan, D. L. (1997) Mol. Biol. Cell 8, 663–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eto, M., Karginov, A. & Brautigan, D. L. (1999) Biochemistry 38, 16952–16957. [DOI] [PubMed] [Google Scholar]
- 29.Masuo, M., Readon, S., Ikebe, M. & Kitazawa, T. (1994) J. Gen. Physiol. 104, 265–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hayashi, Y., Senba, S., Yazawa, M., Brautigan, D. L. & Eto, M. (2001) J. Biol. Chem. 276, 39858–39863. [DOI] [PubMed] [Google Scholar]
- 31.Colbran, R. J., Carmody, L. C., Bauman, P. A., Wadzinski, B. E. & Bass, M. A. (2003) in Methods in Enzymology, eds. Klumpp, S. & Krieglstein, J. (Academic, San Diego), Vol. 366, pp. 156–175. [DOI] [PubMed] [Google Scholar]
- 32.Woodsome, T. P., Eto, M., Everett, A., Brautigan, D. L. & Kitazawa, T. (2001) J. Physiol. 535, 553–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan, I., Ng, C. H., Lim, L. & Leung, T. (2001) J. Biol. Chem. 276, 21209–21216. [DOI] [PubMed] [Google Scholar]
- 34.Ohki, S., Eto, M., Kariya, E., Hayano, T., Hayashi, Y., Yazawa, M., Brautigan, D. & Kainosho, M. (2001) J. Mol. Biol. 314, 839–849. [DOI] [PubMed] [Google Scholar]
- 35.Ohki, S., Eto, M., Shimizu, M., Takada, R., Brautigan, D. L. & Kainosho, M. (2003) J. Mol. Biol. 326, 1539–1547. [DOI] [PubMed] [Google Scholar]
- 36.Liu, Q. R., Zhang, P. W., Zhen, Q., Walther, D., Wang, X. B. & Uhl, G. R. (2002) J. Biol. Chem. 277, 13312–13320. [DOI] [PubMed] [Google Scholar]
- 37.Erdodi, F., Kiss, E., Walsh, M. P., Stefansson, B., Deng, J. T., Eto, M., Brautigan, D. L. & Hartshorne, D. J. (2003) Biochem. Biophys. Res. Commun. 306, 382–387. [DOI] [PubMed] [Google Scholar]
- 38.Liu, J. & Brautigan, D. L. (2000) J. Biol. Chem. 275, 15940–15947. [DOI] [PubMed] [Google Scholar]
- 39.Huang, H. B., Horiuchi, A., Watanabe, T., Shih, S. R., Tsay, H. J., Li, H. C., Greengard, P. & Nairn, A. C. (1999) J. Biol. Chem. 274, 7870–7878. [DOI] [PubMed] [Google Scholar]