Abstract
P transposable elements in Drosophila are mobilized via a cut-and-paste mechanism. The broken DNA ends generated during transposition can be repaired via the homology-directed synthesis-dependent strand annealing or by nonhomologous end joining (NHEJ). Genetic studies have demonstrated an interaction between the gene (mus309, for mutagen-sensitive) encoding the Drosophila Bloom's syndrome helicase homolog (DmBLM) and the Ku70 gene, which is involved in NHEJ. We have used RNA interference (RNAi) to knock down expression of DmBLM and one or both of the Drosophila Ku subunits, DmKu70 or DmKu80. Our results show that upon reduction of DmKu, an increase in small deletions (1–49 bp) and large deletions (≥50 bp) flanking the site of P element-induced breaks is observed, and a reduction in large deletions at these sites is found upon reduction of DmBLM. Moreover, double RNAi of DmKu and DmBLM results in an increase in small deletions characteristic of the DmKu RNAi and also partially suppresses the reduction in repair efficiency observed with DmKu RNAi. These results suggest that there are DNA double-strand break recognition and/or processing events involving DmKu and DmBLM that, when eliminated by RNAi, lead to deletions. Finally, these results raise the possibility that, unlike the situation in mammals, where BLM appears to function exclusively in the homologous repair pathway, in Drosophila, DmBLM may be directly involved in, or at least influence the double-strand break recognition that leads to the NHEJ repair pathway.
Transposable elements are mobile segments of DNA found in many prokaryotic and eukaryotic organisms (for review, see ref. 1) and are now known to make up half of the human genome (2, 3). The P element family of transposable elements in the fruit fly, Drosophila melanogaster, is one of the most studied eukaryotic transposons (4). These elements transpose through a DNA intermediate and create an 8-bp target site duplication upon insertion. The P element termini are required for transposition and include the 31-bp terminal inverted repeats, the 11-bp internal inverted repeats, and unique DNA sequences encompassing ≈150 bp at each end (4). Complete full-length P elements encode an 87-kDa sequence-specific DNA-binding transposase protein that recognizes internal sites at each end (4). Biochemical studies and genetic experiments have shown that P elements transpose via a cut-and-paste mechanism similar to the bacterial transposons Tn10, Tn5, and Tn7 (1, 4–8).
The mode by which P element transposition occurs generates a double-strand break (DSB) at the donor site after an excision event (5, 9). This DSB must be repaired to prevent chromosomal loss. Cells may repair DNA DSBs via either a homology-dependent pathway or by nonhomologous end joining (NHEJ) (10, 11). In the case of P element-induced DSBs, when a homologous chromosome or sister chromatid is present, repair may occur by a gene-conversion type of mechanism similar to double-strand gap repair, termed synthesis-dependent strand annealing (SDSA) (12–14). Additionally, repair of the donor site also can occur via NHEJ (15). Indeed, there seems to be a preferential use of NHEJ in somatic cells, whereas template-directed repair is more widespread in the germline (9, 16). Thus, repair of P element-induced DNA breaks, like those generated by ionizing radiation or x-ray mimetic chemicals, can be repaired via both homologous recombination (HR)-dependent or homology-independent (NHEJ) pathways.
Biochemical and genetic studies in mammals and yeast have provided general outlines of the components and enzymatic activities required for both the HR and NHEJ DNA repair pathways (10, 17). Further studies with mutant x-ray-sensitive mammalian cells that are defective for DSB repair and Ig variable (diversity) joining recombination (18, 19) have led to a general understanding of the NHEJ pathway. In Drosophila, genetic screens have identified many mutations that affect distinct repair pathways. These mutants, termed mus (for mutagen-sensitive), have been useful for investigating DNA repair after P element excision. For instance, mutations in two genes, mei41 (a PI3-like kinase family member) and mus101, can affect the recovery of chromosomes that have undergone P element excision (20, 21). More recently, mus309, the gene corresponding to the homolog of the human Bloom's syndrome DNA helicase, was shown to be defective for the repair of P element-induced DNA breaks, by both the NHEJ and HR-SDSA repair pathways (22, 23). In mammals, Bloom's helicases appear to function in the HR repair pathway, and Bloom's syndrome patients exhibit dwarfism, infertility, frequent infections, and a predispostion to cancer of all types diagnosed at a mean age of 24 years, usually leading to death (24, 25). In Drosophila, mus309 mutants are defective for repair of P element-induced chromosomal breaks, resulting in chromosome loss or defects at the donor site after P element excision (23, 26). Interestingly, a single transgenic genomic copy of the Drosophila DmKu70 gene can partially rescue the defects associated with the mus309 DmBLM mutation (23, 26). These genetic results suggest a direct or indirect functional association between DmBLM and the NHEJ factor Ku70.
To investigate more directly the functional associations of DmBLM and DmKu, we used an extrachromosomal assay to detect P element excision and donor site repair in Drosophila L2 tissue culture cells. RNA interference (RNAi) was used to specifically reduce the levels of DmBLM, DmKu70, and DmKu80 and to test the effects of reduction of these repair factors on NHEJ repair after P element excision. Our results indicate that reduction of the two DmKu subunits results in decreased repair efficiency and leads to increases in both small (1–49 bp) and large (≥50 bp) deletions flanking a P element-induced DNA break. A more dramatic deletion phenotype was observed when RNAi was targeted to DmKu70 compared with DmKu80. Depletion of DmBLM, although only slightly reducing repair efficiency, leads to a reduction in the class of large deletions flanking the break site. Moreover, reduction of both DmBLM and DmKu leads to an increase in repair efficiency compared to loss of DmKu alone and an increase in smaller deletions characteristic of the DmBLM-alone RNAi. Thus, loss of DmBLM suppresses both the reduction in repair efficiency caused by DmKu RNAi and the generation of large deletions. These data suggest interplay of DmBLM and DmKu in NHEJ repair of P element-induced DNA breaks, and unlike the situation in mammals, the possible direct involvement of DmBLM in DSB recognition or in NHEJ repair. These results may be rationalized in light of the biochemical activities of DmBLM (helicase) and the DmKu proteins (binding DNA ends).
Materials and Methods
Generation of Anti-DmBLM Antibodies. The following primer sequences were used to amplify a portion of the Drosophila Bloom's syndrome helicase (mus309) for generating and purifying antibodies: (i) GGGGGAATTCCAG CTAAAAGCGGAACAG base pair with nucleotides 1,312–1,329 of the Dm-BLM ORF, (ii) GGGGTCGACAACGCAATGGGCCTC, and (iii) GGGGAATTCAACGCAATGGGCCTC are antisense oligonucleotides that base pair with nucleotides 2,596–2,610 of the DmBLM ORF. DNA fragments generated by PCR with primers 1 + 2or1 + 3 were cut with EcoRI and SalIor EcoRI and NheI, respectively, and cloned into the expression vectors pVCH6 and pMH6, respectively (obtained from D. Schatz, Yale University, New Haven, CT). The DmBLM-pseudomonous exotoxin fusion protein was expressed in BL21 (DE3) cells that were transformed with the pVCH6 construct. An ≈140-kDa protein was induced by treatment with isopropyl β-d-thiogalactoside and bacterial extracts (20 mM sodium phsosphate, pH 8.0/500 mM NaCl/6 M urea/20 mM imidazole) were bound to Ni2+-NTA agarose (Qiagen, Valencia, CA) and eluted with the above urea buffer plus 500 mM imidazole. A single New Zealand White rabbit was injected with 100 μg of fusion protein in Ribi adjuvant every 3 weeks for a total of six injections. Polyclonal antisera was purified from crude serum by using a maltose binding protein (MBP)-mus309 fusion protein expressed in DH5α cells transformed with the pMH6 construct (see above). The MBP-DmBLM438–870 fusion protein was purified from bacterial extracts (20 mM Tris·HCl, pH 7.5/500 mM NaCl/0.1% Nonidet P-40/1 mM EDTA) with amylose resin (New England Biolabs). The ≈100-kDa purified fusion protein was coupled to CNBr-activated Sepharose 4B (Pharmacia), and the resultant affinity resin was used to purify polyclonal anti-mus309 antibody from rabbit serum (27, 28). DmKu70, DmKu80, and P element transposase antibodies are described in refs. 27 and 29. DmKu80 antibodies were prepared from expression in bacteria as described for DmBLM. Antibodies to Drosophila U2AF50 are described in ref. 30.
Double-Stranded RNA Synthesis. RNA of ≈400 bp, complementary to cDNA of interest, was synthesized in vitro by using T7 RNA polymerase (31–33). DNA oligonucleotides were synthesized with nonspecific spacer and T7 RNA polymerase promoter sequences 5′ of the PCR primer sequence (21 bp) specific to the targeted gene cDNA. The sequence of the primer 5′–3′ is: CGG CCA GTG AAT TGT TTA ATA CGA CTC ACT ATA GGG N21 or 20. The PCR products of ≈400 bp were amplified by using AmpliTaq (Applied Biosystems) from plasmid templates carrying the specific cDNA. RNA was transcribed from the purified and quantified PCR products by using T7 RNA polymerase (31–33). The RNeasy mini kit (Qiagen) was used to clean up the RNA after DNase RQ1 digestion, and RNA was quantified by using UV260.
N sequences in the oligonucleotides were as follows: pBSK1, forward 5′-GTT AAA ATT CGC GTT AAA TTT and reverse 5′-GTG TGG TGG TTA CGC GCA GCG; DmBlm, forward 5′-CCT CTC CAT CAC CAG CGG CAC and reverse 5′-TCC GCA AAG GTC GTG TAT AA; DmKu70, forward 5′-GCT TCA AGC ATC GAT CCT CTC and reverse 5′-TGG ATC GTT GAT GAG ATT CG; DmKu80, forward 5′-GCC GCC GTG AAG CTG GAC GCT and reverse 5′-TGG CAC TCG CCA GAA TAC AT.
RNAi and P Element Excision and Repair Assay. By using 5% FBS-M3 medium, 4 × 106 L2 cells were plated in 60-mm tissue culture dishes and allowed to adhere overnight. The cells were washed with 2 ml of serum-free M3 medium and overlaid with 1 ml of serum-free M3 medium. Ten micrograms of dsRNA in ddH2O was added. After a 1- to 2-h incubation, 1 ml of 10% FBS-M3 medium was added to make final 5%FBS-M3. The cells then were kept in an incubator for 48 h before the tissue culture transfection assay was carried out as described in ref. 29. The number of ampicillin-resistant colonies per ml of bacteria in SOC medium gives the estimate of total plasmids collected, and the number of kanamycin- and ampicillin-resistant colonies per ml of bacteria in SOC medium provides an estimate of reporter plasmids that are excised and repaired. The ratio of repaired plasmids to total plasmids gives the excision and repair activity.
Immunoblot Analysis. Ten microliters of protein sample from ≈2 × 106 L2 cells collected in the excision assay was run on 8% or 15% SDS/PAGE gels and transferred to nitrocellulose. The membranes were blocked with 5% milk in PBS/0.2% Tween 20, and polyclonal rabbit antibodies DmBLM, DmKu70, DmKu80, U2AF, or P element transposase were added in PBS-0.2% Tween 20. Goat anti-rabbit IgG-horseradish peroxidase-conjugated secondary antibody (Bio-Rad) was added at a 1:10,000 dilution, and enhanced chemiluminescence reagent (Amersham Biosciences) was used to detect the proteins.
DNA Sequencing. The plasmid DNA samples were primed 150 bp upstream of the P element insertion site to sequence repair products. Fluorescent thermocycle DNA sequencing was performed, as described in ref. 34, by using the following primer: pISP2/KM forward 150-bp 5′ P element, 5′-TTA GGC ACC CCA GGC.
Results
An Assay to Determine the Effects of DmBLM and DmKu on DNA NHEJ Repair After P Element Excision. In previous studies we have used an extrachromosomal plasmid-based assay to detect P element-induced excision and donor site repair. In this assay, two plasmids are cotransfected into Drosophila L2 tissue culture cells (Fig. 1). One plasmid encodes WT P element transposase, and the second plasmid contains a 0.6-kb P element inserted into a kanamycin-resistance gene, so that when P element excision occurs, NHEJ repair leads to restoration of the ORF of the kanamycin-resistance gene, which can be scored after recovery of DNA from the Drosophila cells, transfomation of E. coli, and plating on the appropriate antibiotic plates (26). In the current experiments, we have modified this procedure to incorporate a treatment of the cell culture with 400-bp duplex RNA for 24–48 h before transfection with the plasmid DNA (Fig. 1). It has been shown previously that these conditions will lead to RNAi in these cells and to dramatically reduced expression of the corresponding mRNA and protein (35). Here, we tested RNA duplexes corresponding to DmBLM, DmKu70, DmKu80, and a control sequence from the bacterial plasmid pBSKS(+) as a negative control. Immunoblot analysis showed that DmBLM, DmKu70, and DmKu80 could be dramatically reduced by using RNAi (Fig. 2B).
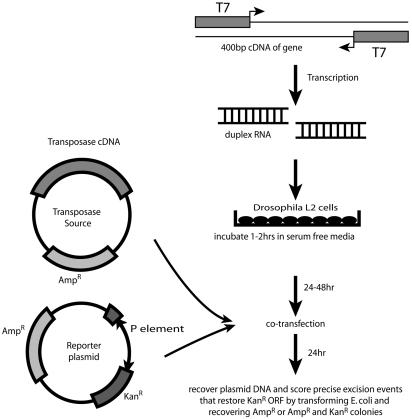
Fig. 1.
RNAi and P element excision and repair assay. The in vivo P element excision assay estimates the fraction of transposition by using two plasmids. The helper plasmid has an ampicillin resistance marker and the transposase cDNA under the Drosophila actin promoter. In addition to the AmpR gene, the excision indicator plasmid has a P element insert after the start codon of the N-terminal β-galactosidase fusion to kanamycin-resistance gene. E. coli is transformed by using collected plasmid DNA and plated on selective agar. The number of AmpR colonies gives an estimate of total plasmids collected. The number of KanR and AmpR colonies indicates reporter plasmids that have excised the P element and repaired the break to restore the kanamycin ORF. The percent transposase excision activity and repair was calculated by dividing the number of KanRAmpR colonies per ml of E. coli by AmpR colonies per ml of E. coli. Because the frequency of efficient repair varies from assay to assay, we normalized the numbers within each assay by expressing it in terms of the control RNAi.
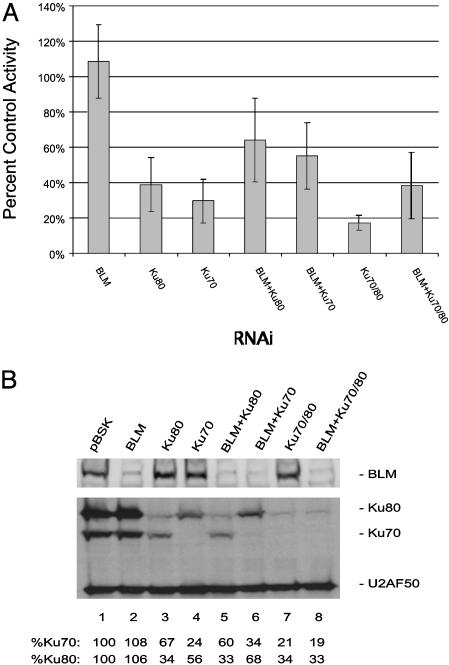
Fig. 2.
RNAi depletion of DmBLM and DmKu affects P element excision and repair activity. (A) P element excision and repair activity. The percent of the control excision and repair activity was calculated within each assay, and activities from at least three different assays were averaged with the standard deviations indicated. (B) Immunoblot of DmBLM and DmKu RNAi-treated Drosophila L2 cells. Immunoblot analysis of RNAi-treated samples after transfection shows specific depletions in the protein level of DmBLM, DmKu70, and DmKu80. DmBLM RNAi decreases the DmBLM protein level (lanes 2, 5, 6, and 8). Depletion of one DmKu subunit results in the decrease in the other as shown in lanes 3 and 4. U2AF50 antibody was used as a control (30). imagequant software was used to quantitate the signals for DmKu relative to the U2AF50 loading control signal.
Effects of Reducing the Levels of DmBLM and DmKu on Recovery of P Element Donor Site Excision and Repair Products. As a first test of how reductions in the levels of DmBLM and DmKu affected P element DNA repair, we quantitated the number of doubly resistant ampicillin and kanamycin colonies under RNAi conditions in which we set the control RNAi activity to 100% (Fig. 2 A). We observed that depletion of the DmBLM protein does not dramatically affect the efficiency of donor plasmid excision and repair. However, depletion of DmKu80 and/or DmKu70 resulted in a decrease to 30–39% of the control efficiency. A more pronounced decrease to 17% of the control level was seen for a double RNAi of DmKu80 and DmKu70. However, when DmBLM and DmKu80 or DmBLM and DmKu70 were knocked down together, the decrease in excision and repair activity was increased relative to the DmKu RNAi alone. Reduction of DmBLM and DmKu together resulted in a repair activity between the expected level for DmBLM or DmKu RNAi treatment alone to 64% and 55%, respectively (Fig. 2 A). This result is intriguing because it shows that a reduction in DmBLM can partially suppress the effects of reducing the DmKu subunits on repair of P element-induced DNA breaks.
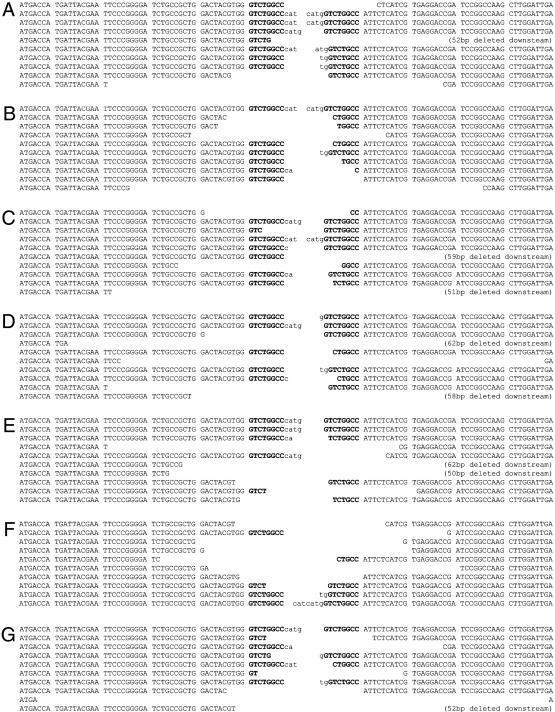
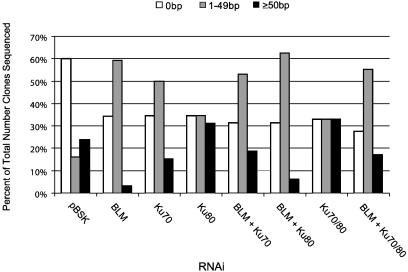
The Quality of DNA Repair at the Donor Site Is Affected by Loss of DmBLM and DmKu. To examine the qualitative nature of the DNA repair events observed under conditions of reduced DmBLM and DmKu, we prepared plasmid DNA from individual bacterial colonies derived from the RNAi transfections described above. This assay requires that the P element interrupting the kanamycin protein-coding sequence is excised and that the kanamycin-resistance gene becomes functional via DNA repair that restores the ORF. We have used this assay previously to show that deletions flanking the P element insertion site occur in mus309 mutant embryos, and thus we know that we can detect defects in repair that do not involve too large a deletion into the downstream gene or the upstream start codon (Fig. 3). The observed repair products were separated into three categories. Those having deletions of the P element are most commonly observed in this assay (0 bp of flanking DNA). Deletions extending into the donor site sequences flanking the P element, including portions of the 8-bp target site duplication, were then categorized by either small deletions (1–49 bp) or large deletions (≥50 bp). About 25 clones were sequenced from each RNAi sample, and the number of clones in each category was plotted as a percentage of the total clones sequenced (a portion of which are shown in Fig. 3). For the control RNAi sample, 60% of the repaired plasmids contained no deletions (0 bp) into the flanking sequence, 16% had deletions of 1–49 bp, and the remaining 24% of the repair products sequenced had deletions ≥50 bp (Fig. 4). In the DmBLM RNAi-treated cells, this high percentage of ≥50-bp deletions decreases to 3%, with a corresponding increase in the 1–49-bp-deletion repair class and a reduction in the 0-bp-deletion repair type (Fig. 4, BLM). Thus, reduction of DmBLM causes reductions in the 0-bp-deletion and ≥50-bp-deletion classes with a concomitant increase in the 1–49-bp-deletion category. In the repair after DmKu70 RNAi alone, a 3-fold increase in 1–49-bp deletions was observed, with a small reduction in the number of large deletions (Fig. 4). In contrast, the DmKu80 RNAi sample behaved more dramatically than the DmKu70 RNAi and resulted in an increase in the number of large deletions, suggesting that reduction of DmKu80 more dramatically affected NHEJ repair (Figs. 3 and 4). Thus, knockdown of DmKu leads to an increase in deletions flanking the site of P element-induced DNA breaks, whereas reduction of DmBLM leads to an increase in small deletions and concomitant reductions in the large and 0-bp deletion classes. Representative sequences of 10 different repair clones from the pBSK control RNAi, DmBLM alone, DmKu70 alone, and DmKu80 alone (Fig. 3 A–D, respectively) were aligned with deletions indicated.
Fig. 3.
Sequences of repaired donor site plasmids. The excision site DNA sequences of repaired pISP-2/Km were aligned by using 10 representative sequences from each RNAi sample. The P element sequence is in lowercase letters. The target site duplication is in bold, and the flanking plasmid sequence is in normal capital type. (A) Repaired plasmids from pBSK control RNAi. (B) DmBLM RNAi-treated L2 cells. (C) RNAi of DmKu70. (D) RNAi of DmKu80. (E) RNAi of DmBLM and DmKu70. (F) RNAi of DmBLM and DmKu80. (G) RNAi of DmBLM and DmKu70 or DmKu80.
Fig. 4.
Summary of deletions and repair quality after depletion of DmBLM and/or DmKu. The total base pair deletion of the plasmid after P element excision was determined, and each sequenced clone was categorized as no deletion (0 bp, open bars) into the flanking donor DNA of pISP-2/Km, small deletions (1–49 bp, gray bars) and large deletions (≥50 bp, black bars). The fraction of total clones sequenced in each category from each RNAi sample was expressed as the percentage of the total clones sequenced.
Loss of both DmBLM and DmKu Results in the DmKu Small and Large Deletion Phenotype. From the results above, it is clear that loss of DmBLM results in a dramatic increase in small (1–49 bp) deletions and a reduction in both the 0-bp and large deletion classes. However, loss of the DmKu subunits caused increases in both the small and the large deletion classes. Thus, it was of interest to examine the pattern of deletions made when both DmBLM and DmKu levels were reduced in a double RNAi experiment. DmBLM RNAi in combination with DmKu70 or both DmKu70 and Ku80 subunits results in the same 2-fold increase in small deletions and an overall decrease in deletions not extending into the flanking donor DNA (0 bp). Conversely, the double DmBLM/Ku80 knockdown resulted in a marked decrease in large deletions and an increase in small deletions compared with the DmKu80 RNAi alone (Fig. 4). This result shows a pattern similar to that of DmBLM RNAi alone. However, the double DmBLM/Ku70 RNAi deletion phenotype is similar to that of DmKu70 RNAi alone (Fig. 4). Our data show that DmBLM and DmKu80 double-RNAi deletion phenotype is similar to DmBLM RNAi alone, whereas DmBlm and DmKu70 double-RNAi deletion phenotype is similar to DmKu70 RNAi alone. These differences may reflect the distinct activities of DmKu70 and DmKu80, or, possibly, these two NHEJ proteins might interact differently with DmBLM helicase. Taken together with the above results, it appears that DmBLM is likely to play a role in the generation of a large-deletion class that is underrepresented in the DmBLM RNAi distribution (Fig. 4). Additionally, loss of the DmKu subunits leads to increases in both small and large deletions. The fact that large deletions are reduced by loss of DmBLM suggests that the direct action of the BLM helicase on the sites of P element DNA breaks could influence how a given repair pathway may be chosen.
Discussion
It is clear that defects in DNA repair pathways can lead to disease phenotypes (36). For Bloom's syndrome helicase, and more generally for the RecQ helicase family, these defects directly lead to cancer predisposition (24, 36). It is thought that these helicases function as genome “caretakers” through their effects on stalled DNA replication forks and other recombination reactions. It is known that mice and flies mutant for BLM show elevated rates of recombination and sister chromatid exchanges, indicating that the fidelity of DNA transactions is impaired. Likewise, components in the NHEJ pathway, particularly the Ku subunits, which function to bind and align DNA ends for repair, also function as genome caretakers, because mice mutant for these proteins exhibit cancer predisposition (37). In Drosophila, there seems to be an interplay between these two classes of molecules because a transgenic copy of DmKu70 can rescue mutations in DmBLM (mus309). In this study, we have shown that large (≥50 bp) deletions flanking the sites of P element-induced DNA breaks are suppressed upon reduction of DmBLM by using RNAi. Moreover, loss of the DmBLM protein partially suppresses the reduction in NHEJ repair efficiency because of reductions in the DmKu subunits. Finally, a higher fraction of deleted molecules and reduced repair efficiency is observed upon reduction of DmKu.
These results may be considered in light of the activities of the BLM and Ku proteins. It is easy to see how reduction in the BLM helicase might result in smaller deletions, if the unwinding of DNA at the break site were impaired. Likewise, the two Ku subunits are known to function in binding and aligning broken DNA ends for repair (38, 39). Thus, reduction of these NHEJ components might be expected to lead to reduced repair and increased deletions flanking the DNA break site. In fact, this sort of phenotype has been observed in x-ray-sensitive mutant mammalian cells defective for Ku70 and Ku80 in tests for faithful DNA repair after variable (diversity) joining recombination (40).
We also note that upon DmBLM depletion, the quality, rather than the efficiency, of repair is affected. The effect of DmBLM loss on deletion size, but not repair efficiency, suggests that DmBLM might interact with the broken DNA ends after DNA cleavage by P element transposase, but before these broken ends are recognized by the Ku proteins for NHEJ repair. However, it is noteworthy that defects in rejoining broken DNA ends, including deletions at the site of the breaks, have been observed in human BLM patient cell lines (41, 42). Biochemical studies should clarify the nature of any possible interactions between DmBLM and DmKu, as well as a possible direct role of DmBLM in NHEJ. Interestingly, in mammals, a protein complex implicated in genome surveillance, termed the BASC complex, was shown by MS to contain both BLM and Ku (43). It should be noted that DmBLM (mus309) mutants display defects in template-directed repair via the SDSA HR pathway, increased sister chromatid exchange, and methyl methanesulfonate sensitivity, as well as chromosome loss and repair defects after a P element-induced DNA break (22, 23, 26, 44). Nonetheless, these genetic studies do not rule out a direct role for DmBLM in the Drosophila NHEJ DNA repair pathway (22, 26). Our results show that DNA repair products from the NHEJ pathway are qualitatively altered under conditions of reduced DmBLM, suggesting that the direct action of the helicase on the broken DNA ends leads to a class of large (≥50 bp) deletions before repair by DmKu and the NHEJ machinery. This suppressive effect is more apparent in the DmBLM + DmKu80 double RNAi data. It is possible that in the Dm Ku70 RNAi, the more functional Ku complex remains, perhaps accounting for the differences seen in the DmKu70 and DmKu80 RNAi experiments. It is also possible that a reduction in DmKu and DmBLM together allows for access of broken DNA ends to other nucleases or processing enzymes, perhaps leading to deletions at the break site. It will be interesting to investigate and clarify the mechanistic role of DmBLM in Drosophila DNA repair pathways.
An alternative view would be a competition between the NHEJ and HR pathways that might involve initial break recognition and recruitment of the DmBLM helicase. Once break recognition occurred, then a decision to enter the NHEJ or HR pathways might be made. This idea is consistent with our observations that whereas DmBLM RNAi causes a qualitative defect in NHEJ products, there is no effect on overall efficiency of NHEJ repair of P element-induced DNA breaks. Biochemical studies of potential interactions of DmBLM and DmKu, as well as tests for the direct involvement of the DmBLM helicase in the Drosophila NHEJ pathway and recognition of DNA breaks, should provide insight on these issues. Our studies also raise the possibility that although the intrinsic biochemical functions among RecQ family members might be conserved across species, their exact roles in distinct DNA repair pathways may differ among different organisms.
Acknowledgments
We thank Eileen Beall (University of California, Berkeley) for the DmKu70 and DmKu80 antibodies and Carla DiGennaro for critical reading of the manuscript. This work was funded by National Institutes of Health Grant R01 GM48862-12.
Abbreviations: DmBLM, Drosophila Bloom's syndrome helicase homolog; DSB, double-strand break; HR, homologous recombination; Ku, Ku autoantigen; NHEJ, nonhomologous end joining; RNAi, RNA interference; SDSA, synthesis-dependent strand annealing.
References
- 1.Craig, N. L. (2002) in Mobile DNA II, ed. Craig, N. L. (Am. Soc. Microbiol., Washington, DC).
- 2.Venter, J. C., Adams, M. D., Myers, E. W., Li, P. W., Mural, R. J., Sutton, G. G., Smith, H. O., Yandell, M., Evans, C. A., Holt, R. A., et al. (2001) Science 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 3.Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. (2001) Nature 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 4.Rio, D. C. (2002) in Mobile DNA II, ed. Craig, N. L. (Am. Soc. Microbiol., Washington, DC), pp. 484–518.
- 5.Kaufman, P. D. & Rio, D. C. (1992) Cell 69, 27–69. [DOI] [PubMed] [Google Scholar]
- 6.Bainton, R., Gamas, P. & Craig, N. L. (1991) Cell 65, 805–816. [DOI] [PubMed] [Google Scholar]
- 7.Benjamin, H. W. & Kleckner, N. (1989) Cell 59, 373–383. [DOI] [PubMed] [Google Scholar]
- 8.Mizuuchi, K. (1992) Annu. Rev. Biochem. 61, 1011–1051. [DOI] [PubMed] [Google Scholar]
- 9.Engels, W. R., Johnson-Schlitz, D. M., Eggleston, W. B. & Sved, J. (1990) Cell 62, 515–525. [DOI] [PubMed] [Google Scholar]
- 10.van Gent, D. C., Hoeijmakers, J. H. & Kanaar, R. (2001) Nat. Rev. Genet. 2, 196–206. [DOI] [PubMed] [Google Scholar]
- 11.Jackson, S. P. (2002) Carcinogenesis 23, 687–696. [DOI] [PubMed] [Google Scholar]
- 12.Nassif, N., Penney, J., Pal, S., Engels, W. R. & Gloor, G. B. (1994) Mol. Cell. Biol. 14, 1613–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engels, W. R., Preston, C. R. & Johnson-Schlitz, D. M. (1994) Science 263, 1623–1625. [DOI] [PubMed] [Google Scholar]
- 14.Formosa, T. & Alberts, B. M. (1986) Cell 47, 793–806. [DOI] [PubMed] [Google Scholar]
- 15.Lieber, M. R., Ma, Y., Pannicke, U. & Schwarz, K. (2003) Nat. Rev. Mol. Cell Biol. 4, 712–720. [DOI] [PubMed] [Google Scholar]
- 16.Gloor, G. B., Moretti, J., Mouyal, J. & Keeler, K. J. (2000) Genetics 155, 1821–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paques, F. & Haber, J. E. (1999) Microbiol. Mol. Biol. Rev. 63, 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taccioli, G. E., Rathbun, G., Oltz, E., Stamato, T., Jeggo, P. A. & Alt, F. W. (1993) Science 260, 207–210. [DOI] [PubMed] [Google Scholar]
- 19.Pergola, F., Zdzienick, M. & Lieber, M. (1993) Mol. Cell. Biol. 13, 3464–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banga, S. S., Velazquez, A. & Boyd, J. B. (1991) Mutat. Res. 255, 79–88. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto, A. H., Brodberg, R. K., Banga, S. S., Boyd, J. B. & Mason, J. M. (1990) Mutat. Res. 229, 17–28. [DOI] [PubMed] [Google Scholar]
- 22.Adams, M. D., McVey, M. & Sekelsky, J. J. (2003) Science 299, 265–267. [DOI] [PubMed] [Google Scholar]
- 23.Kusano, K., Johnson-Schlitz, D. M. & Engels, W. R. (2001) Science 291, 2600–2602. [DOI] [PubMed] [Google Scholar]
- 24.Hickson, I. D. (2003) Nat. Rev. Cancer 3, 169–178. [DOI] [PubMed] [Google Scholar]
- 25.Hickson, I. D., Davies, S. L., Li, J. L., Levitt, N. C., Mohaghegh, P., North, P. S. & Wu, L. (2001) Biochem. Soc. Trans. 29, 201–204. [DOI] [PubMed] [Google Scholar]
- 26.Beall, E. L. & Rio, D. C. (1996) Genes Dev. 10, 921–933. [DOI] [PubMed] [Google Scholar]
- 27.Beall, E. L., Admon, A. & Rio, D. C. (1994) Proc. Natl. Acad. Sci. USA 91, 12681–12685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harlow, E. & Lane, D. (1999) Using Antibodies: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 29.Beall, E. L., Mahoney, M. B. & Rio, D. C. (2002) Genetics 162, 217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rudner, D. Z., Kanaar, R., Breger, K. S. & Rio, D. C. (1998) Mol. Cell. Biol. 18, 1765–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pokrovskaya, I. D. & Gurevich, V. V. (1994) Anal. Biochem. 220, 420–423. [DOI] [PubMed] [Google Scholar]
- 32.Gurevich, V. V., Pokrovskaya, I. D., Obukhova, T. A. & Zozulya, S. A. (1991) Anal. Biochem. 195, 207–213. [DOI] [PubMed] [Google Scholar]
- 33.Cunningham, P. R. & Ofengand, J. (1990) Biotechniques 9, 713–714. [PubMed] [Google Scholar]
- 34.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 35.Clemens, J. C., Worby, C. A., Simonson-Leff, N., Muda, M., Maehama, T., Hemmings, B. A. & Dixon, J. E. (2000) Proc. Natl. Acad. Sci. USA 97, 6499–6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mohaghegh, P. & Hickson, I. D. (2001) Hum. Mol. Genet. 10, 741–746. [DOI] [PubMed] [Google Scholar]
- 37.Roth, D. B. (2003) Nat. Rev. Immunol. 3, 656–666. [DOI] [PubMed] [Google Scholar]
- 38.Jones, J. M., Gellert, M. & Yang, W. (2001) Structure (London) 9, 881–884. [DOI] [PubMed] [Google Scholar]
- 39.Walker, J. R., Corpina, R. A. & Goldberg, J. (2001) Nature 412, 607–614. [DOI] [PubMed] [Google Scholar]
- 40.Lieber, M. R., Grawunder, U., Wu, X. & Yaneva, M. (1997) Curr. Opin. Genet. Dev. 7, 99–104. [DOI] [PubMed] [Google Scholar]
- 41.Onclercq-Delic, R., Calsou, P., Delteil, C., Salles, B., Papadopoulo, D. & Amor-Gueret, M. (2003) Nucleic Acids Res. 31, 6272–6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Runger, T. M. & Kraemer, K. H. (1989) EMBO J. 8, 1419–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, Y., Cortez, D., Yazdi, P., Neff, N., Elledge, S. J. & Qin, J. (2000) Genes Dev. 14, 927–939. [PMC free article] [PubMed] [Google Scholar]
- 44.Boyd, J. B., Golino, M. D., Shaw, K. E. S., Osgood, C. J. & Green, M. M. (1981) Genetics 97, 607–623. [DOI] [PMC free article] [PubMed] [Google Scholar]