Abstract
Peroxisome proliferator-activated receptor γ (PPARγ) coactivator 1α (PGC-1α) is a transcriptional coactivator that is a key component in the regulation of energy production and utilization in metabolic tissues. Recent work has identified PGC-1α as a strong coactivator of the orphan nuclear receptor estrogen-related receptor α (ERRα), implicating ERRα as a potential mediator of PGC-1α action. To understand the role of ERRα in PGC-1α signaling, a parallel approach of high-throughput screening and gene-expression analysis was used to identify ERRα small-molecule regulators and target genes. We report here the identification of a potent and selective ERRα inverse agonist that interferes effectively with PGC-1α/ERRα-dependent signaling. This inverse agonist inhibits the constitutive activity of ERRα in both biochemical and cell-based assays. Also, we demonstrate that monoamine oxidase B is an ERRα target gene whose expression is regulated by PGC-1α and ERRα and inhibited by the ERRα inverse agonist. The discovery of potent and selective ERRα modulators and their effect on PGC-1α signaling provides mechanistic insight into gene regulation by PGC-1α. These findings validate ERRα as a promising therapeutic target in the treatment of metabolic disorders, including diabetes and obesity.
Peroxisome proliferator-activated receptor γ (PPARγ) coactivator 1α (PGC-1α) is a critical regulator of genes controlling many aspects of energy metabolism (1). PGC-1α is a coactivator for many different transcription factors through which it regulates adaptive thermogenesis in brown fat and skeletal muscle and stimulates mitochondrial biogenesis and respiration in muscle cells (2, 3). PGC-1α is induced in mouse liver under conditions of fasting or diabetes and has been shown to regulate expression of several genes encoding gluconeogenic enzymes (4, 5). In skeletal muscle, PGC-1α expression increases with exercise, and PGC-1α is involved in mediating muscle fiber-type determination (6–8). PGC-1α has also been linked to expression of genes involved in oxidative phosphorylation (OX/PHOS) in muscle (9, 10). Importantly, expression of PGC-1α and OX/PHOS enzymes are reduced in human type 2 diabetic muscle, indicating a role for PGC-1α and its transcriptional regulators in diabetes and insulin resistance (9, 10). One potential mediator of PGC-1α activity is the orphan nuclear receptor estrogen-related receptor α (ERRα). PGC-1α binds to ERRα and functions as a potent ligand-independent coactivator of this orphan receptor (11–13).
The ERRs are a family of orphan nuclear hormone receptors identified initially based on their homology to the estrogen receptor ERα (14). The ERR family consists of three members, designated ERRα (NR3B1), ERRβ (NR3B2), and ERRγ (NR3B3), that share similarity with ER; however, they are not activated by natural estrogens and do not bind 17β-estradiol (E2) (14–16). The ERRs generally are considered to be constitutively active receptors that interact with coactivator proteins in the absence of exogenous ligands. Although natural estrogens do not activate ERRs, the synthetic estrogens 4-hydroxytamoxifen, tamoxifen, and diethylstilbestrol (DES) can bind and function as inverse agonists of ERRβ and ERRγ (17, 18). DES has also been shown to affect the activation and coactivator interactions of ERRα (19). In addition to binding selective estrogen-receptor modulators, the ERs and ERRs also may share common target genes and thereby control overlapping transcriptional regulatory networks (20–23).
ERRα is expressed in many tissues in developing and adult animals, with highest expression in tissues with increased metabolic demands, including skeletal muscle, heart, kidney, liver, and adipose tissue (14, 24). There are several lines of evidence that indicate a potential role for ERRα in the regulation of metabolic homeostasis. Importantly, genetic deletion of the ERRα gene in mice results in a lean phenotype with decreased white adipose tissue deposits (25). ERRα–/– mice are also resistant to high-fat diet-induced obesity and have reduced lipogenesis in adipose tissues (25). In addition to genetic evidence, experimental evidence in cultured cells indicates that ERRα is involved in both brown and white adipocyte development (24, 26). The involvement of ERR in controlling lipid metabolism is supported also by the identification of the gene encoding medium-chain acyl CoA dehydrogenase, an enzyme involved in mitochondrial β-oxidation of fatty acids, as an ERRα target gene (11, 24, 26).
A role for ERRα in metabolic control is supported further by recent data (11–13) that demonstrate physical and functional interactions between ERRα and PGC-1α. In addition to having similar expression patterns in mouse and human tissues, PGC-1α and ERRα are also induced in skeletal muscle and adipose tissue of mice that are exposed to cold and in the liver of fasted mice (2, 4, 5, 12, 13). Furthermore, PGC-1α can strongly induce ERRα mRNA expression in several cell lines, suggesting that ERRα induction in vivo may be mediated by PGC-1α (13). Because PGC-1α is also a potent coactivator of ERRα, it is likely that ERRα is an important transcriptional regulator of PGC-1α-induced genes, implicating ERRα as a potential therapeutic target for the treatment of diseases such as obesity, diabetes, and the metabolic syndrome.
Given the potential involvement of ERRα in various pathologic conditions including metabolic disorders, cancer, and osteoporosis (22, 25, 27–29), we set out to identify synthetic ERRα-specific ligands to elucidate the therapeutic relevance of ERRs in human disease. We have identified and characterized an ERRα ligand that inhibits PGC-1α signaling, suggesting that ERRα functions in concert with PGC-1α as a critical regulator of energy homeostasis.
Materials and Methods
Fluorescence Polarization (FP) Assay. N-terminal His-tagged ERRα ligand-binding domain (LBD) protein (amino acids 145–423) was expressed in bacteria and purified with Ni-NTA resin according to the manufacturer's instructions (Qiagen, Valencia, CA). We incubated 2.5 μM protein and 10 nM peptide (fluor-ILRKLLQE) at room temperature in FP buffer {20 mM KH2PO4, pH 7.3/150 mM NaCl/2 mM 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate/2 mM EDTA/10 mM DTT} containing XCT790 or DMSO. Readings were taken with the excitation wavelength at 485 nm and emission at 530 nm.
Plasmids and Reagents. GAL4-ERRα encodes amino acids 174–423 of human ERRα fused to the DNA-binding domain (DBD) of GAL4 in the pCMX plasmid (30). CMX-hERRα was cloned from human kidney cDNA, and it matched the ERRα sequence in the GenBank database (accession no. NM_004451). CMX-ERRαΔAF2 was generated by site-directed mutagenesis to change amino acid 410 to a stop codon. The pGL3-MAOB-Luc reporter contains 1.68 kb of the human monoamine oxidase B (MAOB) promoter, including the TATA box cloned into pGL3 basic (Promega). Mutations in this promoter were generated by using the QuikChange mutagenesis kit (Stratagene). TK-MAOBx3-Luc reporter contains the sequence AGTCCTAGGTGACCTCTCCGCCCAGAAGTCCTAGGTGACCTCTCCGCCCAGAAGTCCTAGGTGACCTCTCCGCCCA, which is cloned into the HindIII and SalI sites of TK-Luc. pcDNA-PGC-1α contains full-length mouse PGC-1α, which is cloned into pcDNA3.1/myc-His A (Invitrogen). pGL3-mSHP contains nucleotides –435 to +20 of the mouse short heterodimer partner (mSHP) promoter cloned into pGL3-basic (Promega). All clones were verified by sequencing.
Cell Culture. MDA-MB-231 cells (American Type Culture Collection) were maintained in DMEM (without phenol red) supplemented with 10% FBS/2 mM GlutaMAX I (GIBCO)/1 mM sodium pyruvate/0.1 mM nonessential amino acids/20 mM Hepes/1% penicillin–streptomycin. GP-293 cells (Clontech) were grown in DMEM supplemented with 10% FBS. CV-1 cells were maintained in DMEM (without phenol red) with 10% charcoal/dextran-treated FBS. HeLa cells were maintained in DMEM (without phenol red) with 10% heat-inactivated FBS. C2C12 cells were cultured in DMEM supplemented with 10% FBS, 4 mM l-GlutaMAX I (GIBCO)/1 mM MEM sodium pyruvate/0.5% chicken embryo extract (GIBCO)/50 μg/ml gentamicin. All cells were maintained at 37°C and 5% CO2.
Transient Transfections. We plated 7,500 cells per well in 96-well plates and incubated them at 37°C at 5% CO2 overnight. Media was changed to fresh DMEM (without phenol red) containing 5% charcoal/dextran-treated FBS, and cells were transfected by using FuGENE6 (Roche). Compounds were added 5 h after transfection, and cells were lysed and assayed for luciferase and β-galactosidase activity 18 h later. Transfections were performed in triplicate, and luciferase values were normalized to β-galactosidase activity. Experiments were repeated several times to assure reproducibility.
Retroviral Constructs. Human ERRα (encoding amino acids 2–423) was PCR amplified from kidney cDNA and cloned into a pLNCX2 retroviral expression vector (Clontech) containing two copies of the Flag epitope tag at the 5′ end. The QuikChange mutagenesis kit (Stratagene) was used to generate the ERRα AF2 deletion by changing amino acid 410 to a stop codon. The mouse PGC-1α retrovirus encodes amino acids 6–797 with an N-terminal Flag tag in the pLNCX2 expression vector. All clones were verified by sequencing.
Generation of Stable Retroviral Cell Lines. GP-293 cells were transfected with retroviral DNA and pVSV-G plasmid (Clontech) by using FuGENE6 (Roche). After 48 h, virus-containing media were filtered, concentrated by centrifugation according to the manufacturer's instructions (Clontech), and resuspended in TNE (50 mM Tris·HCl, pH 7.8/130 mM NaCl/1 mM EDTA) at 4°C overnight. Aliquoted virus was stored at –80°C. MDA-MB-231 cells were grown to 50–70% confluency in six-well plates and infected with 100 μl of virus stock per well for 24 h at 37°C and 5% CO2. Media were then replaced with fresh media containing 900 μg/ml G418 sulfate, and stable cell populations were selected for 10 days. C2C12 cells (≈30–40% confluent) were infected with mPGC-1α or empty pLNCX2 vector virus for 24 h, after which the media were replaced with fresh media containing 600 μg/ml G418 sulfate, and stable cell populations were selected for 10 days.
Short-Term Infection with mPGC-1α Retrovirus. MDA-MB-231 wild-type or stable cell lines were plated and allowed to reach 50–70% confluence overnight. Media were changed to fresh media, containing 8 μg/ml hexadimethrine bromide (Sigma) and either mPGC-1α retrovirus or control LNCX2 retrovirus (expressing the empty vector), and incubated at 37°C and 5% CO2 for 24 h. Media were then replaced with fresh media containing 900 μg/ml G418 sulfate. RNA was collected at various times after infection, as indicated in figure legends.
Real-Time Quantitative RT-PCR. Stable cell lines were plated in 10-cm plates and grown to 80–90% confluence, and RNA was purified by using RNeasy mini-columns (Qiagen). Real-time RT-PCR analysis was performed as described (31), with the following modifications. RT-PCR was performed in a 384-well assay plate by using the Prism 7900HT sequence detector (Applied Biosystems) with 400 nM reverse and forward primer (each) and 100 nM probe. PCRs used 0.5 units of Taq polymerase (Invitrogen) per 20-μl reaction and an initial incubation at 95°C for 1 min. A six-point standard curve was run in parallel. Data were normalized to cyclophilin mRNA.
Northern Blot Analysis. We separated 10 μg of total RNA in a 1% agarose gel containing 1× Mops buffer (20 mM Mops [3-(N-morpholino)propanesulfonic acid]/5mM EDTA, pH 7.0) and 6% formaldehyde. RNA was transferred to a Zeta-Probe membrane (Bio-Rad) and UV cross-linked. Blots were prehybridized and hybridized at 65°C with Rapid-hyb buffer (Amersham Biosciences) and washed with 0.5× SSC (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7)/0.1% SDS at 65°C. Blots were stripped with 0.1× SSC/0.1% SDS at 100°C before reprobing. DNA probes corresponded to the following regions: mouse PGC-1α amino acids 7–314, human ERRα amino acids 1–97, and human MAOB amino acids 133–453.
Results
To identify small-molecule modulators of ERRα, a high-throughput biochemical FP coactivator interaction assay was used to screen ≈340,000 synthetic compounds. The initial screen identified several different structural classes of compounds that inhibited the constitutive interaction between the ERRα LBD and a small LXXLL peptide. These compounds were validated further in a transient transfection assay by using a GAL4-ERRα LBD fusion and a luciferase reporter containing GAL4 binding sites (32). As in the FP assay, GAL4-ERRα exhibits constitutive transcriptional activity (≈20-fold, relative to the reporter alone) that is inhibited in a concentration-dependent manner by the inverse agonists identified in the FP assay. Compounds displaying activity in both biochemical and transcriptional assays were analyzed further in various cell-based assays to test for potency, efficacy, and selectivity for ERRα. The prototype compound from one chemical series with an IC50 value of ≈1–2 μM was modified chemically to yield XCT790 (Fig. 1A). This compound is an ERRα inverse agonist with an IC50 value of ≈300–500 nM in transient transfection assays using GAL4-ERRα LBD (Fig. 1B) or full-length ERRα with the mSHP promoter (Fig. 1C). In these assays XCT790 demonstrates ≈90–100% inhibition of ERRα constitutive activity.
Fig. 1.
XCT790 acts as a selective inverse agonist of ERRα. (A) Chemical structure of XCT790. (B) GAL4-ERRα transient transfection assay in CV-1 cells treated with increasing concentrations of XCT790 from 3 nM to 10 μM. (C) CV-1 cell transfections with full-length human ERRα and a luciferase reporter containing the mSHP promoter. Cells were treated with XCT790 as in B. (D) Selectivity assays in CV-1 cells with GAL4-ERRα, GAL4-ERRγ, or GAL4-ERα in the presence of 1 nM DES (ERα antagonist assay) treated with XCT790 as in B. Percentage of activity is shown relative to 100%, which represents the normalized activated state of each receptor. (E) FP analysis using His-tagged ERRα LBD protein and a fluorescein-labeled peptide (ILRKLLQE), demonstrating that the constitutive interaction between ERRα and the peptide is disrupted specifically by increasing concentrations of XCT790. (F) Transient transfections in CV-1 cells demonstrating that XCT790 acts as an agonist of the mutant receptor ERRαF232A in the presence of the coactivator GRIP-1. Assays were performed in 96-well plates, and they included 50 ng of reporter, 40 ng of receptor, and 20 ng of GRIP-1. RLU, relative light units.
XCT790 is selective and shows no significant antagonist activity on related nuclear receptors, such as ERRγ or ERα, at concentrations below 10 μM (Fig. 1D). Additionally, XCT790 has no agonist activity on either receptor (data not shown). The antagonist activity of XCT790 was tested also on various other GAL4-chimeric receptors, including ROR, RXR, PPARα, PPARδ, and PPARγ, and it showed no significant activity (see Fig. 6, which is published as supporting information on the PNAS web site). Potential agonist activity of XCT790 was tested further on several nuclear receptors, including LXRα, LXRβ, FXR, PPARα, PPARδ, PPARγ, Nurr1, RARα, RORα, and RXRα. No significant activity was detected in these receptor assays, with the exception of PPARγ, which was weakly activated by 10 μM XCT790 to a level ≈10% of the activity of the PPARγ agonist rosiglitazone (data not shown).
To demonstrate a specific interaction between XCT790 and the ERRα LBD, the activity of XCT790 was tested also in a biochemical protein–protein interaction assay. As shown in Fig. 1E, XCT790 specifically disrupts the interaction between ERRα and an LXXLL peptide in a dose-dependent manner, whereas the ERRγ ligand 4-hydroxytamoxifen has no effect. Furthermore, XCT790 has no effect on ERRγ–peptide interactions in an ERRγ FP assay (data not shown).
To rule out the possibility that XCT790 inhibits coactivator interactions independent of binding to the ligand-binding pocket, a mutation was introduced into the ligand-binding pocket of ERRα, and the mutant receptor was used to test XCT790 activity. ERRαF232A has the phenylalanine at position 232 replaced with an alanine, resulting in a mutant receptor lacking constitutive activity and capable of binding the synthetic ERα ligands DES and 4-hydroxytamoxifen (17, 33). Also, this mutant receptor is activated by the weak ERRα antagonist toxaphene (33). In transient transfections with ERRαF232A and GRIP-1, treatment with XCT790 increased transcription of the mSHP promoter, demonstrating that XCT790 is an agonist ligand for ERRαF232A (Fig. 1F). Together, data from the biochemical assay and ERRαF232A experiments indicate that XCT790 most likely binds in the ligand-binding pocket of ERRα.
In addition to identification of ERRα ligands, microarray experiments were done to identify ERRα target genes in MDA-MB-231 breast cancer cells overexpressing ERRα or a dominant-negative form of ERRα lacking the activation function 2 (AF2) domain. Two genes strongly induced by overexpression of ERRα are MAOA and MAOB. These enzymes are expressed in neurologic and metabolic tissues and are critical regulators of biogenic amine metabolism. Also, MAOA and MAOB are the pharmacologic targets of monoamine oxidase inhibitors used for therapeutic intervention in various neurological disorders, including depression, anxiety, and Parkinson's disease (34). Validation of the microarray results by using quantitative real-time RT-PCR demonstrated that expression of MAOB mRNA increased ≈10-fold when ERRα was overexpressed and that it decreased ≈4-fold upon overexpression of the ERRα AF2 deletion (Fig. 2A).
Fig. 2.
MAOB is an ERRα target gene. (A) Quantitative RT-PCR of human MAOB mRNA expression in MDA-MB-231 cells overexpressing a retroviral vector, ERRα, or ERRαΔAF2. mRNA levels are expressed relative to the amount of cyclophilin mRNA. (B) VP16-ERRα and ERRγ transcriptional activation of the human MAOB promoter in transient transfections in HeLa cells. (C) Region of the human MAOB promoter containing the ERR response element (ERRE). The sequence of the MAOB ERRE, cloned as three copies into the TK-Luc reporter, is underlined. *, Nucleotides mutated from C to A in mutagenesis experiment. The TATA box is capitalized. (D) VP16-ERRα activation of wild-type and mutant forms of the human MAOB promoter in HeLa cells. (E) Transient transfection in HeLa cells with full-length ERRα and a luciferase reporter containing three copies of the MAOB ERR binding site (TK-MAOBx3-Luc) treated with various concentrations of XCT790 for 18 h. RLU, relative light units.
To determine whether the effect of ERRα overexpression on MAOB mRNA expression is direct, 1,680 bp of the human MAOB promoter was cloned into a reporter and tested for activation by ERRα. In transient transfection experiments, ERRα promotes a 2-fold increase in transcriptional activation of the MAOB promoter (data not shown), whereas a hyperactive form of the receptor containing the VP16 activation domain fused to the N terminus of the full-length receptor (VP16-ERRα) produces a 60-fold increase in transcription from the MAOB promoter (Fig. 2B). Transfections using several different VP16-activated receptors demonstrated that the MAOB promoter is activated specifically by ERRα and ERRγ but not by the VP16-activated forms of two other orphan receptors, RORα and Nurr1 (Fig. 2B).
Several potential ERR binding sites were identified in the 1.68-kb human MAOB promoter. These sites were mutated individually, and the promoters were tested for activation by VP16-ERRα in transient transfection assays. Results from these experiments identified a putative ERR binding site 133 bp from the TATA box as the site affected most severely by mutagenesis, resulting in an ≈10-fold reduction of ERRα transactivation (Fig. 2 C and D). To validate these findings further, three copies of this binding site were cloned into the TK-Luc reporter (TK-MAOBx3-Luc) and tested for activity by using the wild-type receptor. As shown in Fig. 2E, this putative ERR binding site is able to confer strong activation by ERRα. Furthermore, XCT790 inhibits ERRα activity on the TK-MAOBx3-Luc reporter in a dose-dependent manner with an IC50 value of ≈500 nM (Fig. 2E).
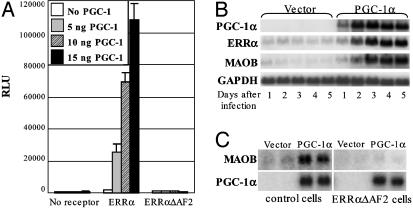
Because of the known interaction between ERRα and the coactivator PGC-1α (11–13), transient transfections were repeated by using the human MAOB promoter and wild-type ERRα in the presence or absence of PGC-1α. The relatively weak activation by ERRα is enhanced up to 120-fold by addition of increasing amounts of PGC-1α (Fig. 3A). This activation is highly specific and not observed in the absence of receptor or with the ERRα AF2 deletion that does not interact with PGC-1α (11).
Fig. 3.
PGC-1α and ERRα induce target gene expression cooperatively. (A) Transient transfections in HeLa cells, demonstrating that PGC-1α specifically coactivates ERRα on the MAOB promoter in an AF2-dependent manner. (B) Northern blot of total RNA from MDA-MB-231 cells collected 1–5 days after infection with a retrovirus expressing PGC-1α. The blot was probed with PGC-1α, ERRα, MAOB, and GAPDH (loading control). (C) Northern blot analysis of total RNA from MDA-MB-231 stable cell lines overexpressing the ERRαΔAF2 retrovirus or the empty retroviral vector and infected subsequently with a PGC-1α retrovirus for 4 days. The blot was probed with PGC-1α and MAOB. Cyclophilin mRNA was used to check for equal RNA loading (data not shown).
Because ERRα and PGC-1α can regulate target gene expression cooperatively, the effect of PGC-1α overexpression on ERR target genes in MDA-MB-231 cells was investigated also. Cells were infected with a retrovirus overexpressing PGC-1α, and RNA was collected at various times after infection. Northern blot analysis of total RNA shows a clear induction of both ERRα and MAOB mRNAs beginning 2 days after PGC-1α infection (Fig. 3B). To determine whether ERRα is involved in the PGC-1α-mediated induction of MAOB expression, PGC-1α was subsequently overexpressed in MDA-MB-231 stable cell lines overexpressing the ERRα AF2 deleted receptor or a control retroviral vector. In this experiment, if ERRα is required for PGC-1α induction of MAOB expression, overexpression of the dominant-negative form of ERRα would be expected to block MAOB mRNA induction by PGC-1α. The results shown in Fig. 3C demonstrate that overexpression of ERRαΔAF2 does indeed prevent induction of MAOB gene expression by PGC-1α, and they suggest further that PGC-1α initiates a cascade involving ERRα regulation of MAOB expression. Together, the data in Figs. 2 and 3 indicate that MAOB is an ERR target gene that is regulated by ERRα and PGC-1α predominantly through an ERR binding site in the 5′ promoter region of the human MAOB gene.
Our data suggest that PGC-1α induces ERRα expression and that together they regulate transcription of target genes coordinately. We reasoned, therefore, that the ERRα inverse agonist XCT790 should affect the PGC-1α-mediated activation of gene expression. In transient transfection experiments, XCT790 inhibits activation of the MAOB promoter by the combination of ERRα and PGC-1α in a dose-dependent manner (Fig. 4A).
Fig. 4.
The ERRα inverse agonist XCT790 inhibits PGC-1α regulation of ERRα and ERRα target gene expression. (A) Transient transfection in CV-1 cells, demonstrating ERRα and PGC-1α activation of the human MAOB promoter reporter. At 6 h after transfection, cells were treated for 18 h with the indicated concentrations of DMSO or XCT790. (B–D) Quantitative RT-PCR of ERRα (B), MAOB (C), and medium-chain acyl CoA dehydrogenase (MCAD) (D). mRNA expression in MDA-MB-231 cells infected with a control retrovirus or a PGC-1α retrovirus for 24 h, followed by treatment with 10 μM XCT790 for 4 days before RNA analysis, is shown. mRNA levels are expressed relative to the amount of cyclophilin mRNA. RLU, relative light units.
To determine the effect of XCT790 on PGC-1α/ERRα-induced target genes in cells, a PGC-1α retrovirus was used to infect MDA-MB-231 cells that were treated subsequently with 10 μM XCT790 for 4 days before RNA analysis by quantitative RT-PCR. The data shown in Fig. 4 B and C demonstrate that PGC-1α overexpression induces ERRα and MAOB mRNA expression ≈5-fold and 9-fold, respectively. As predicted, addition of XCT790 inhibits PGC-1α-mediated induction of both genes by ≈90%. Furthermore, expression of medium-chain acyl CoA dehydrogenase, an ERRα target gene involved in fatty acid β-oxidation, is also down-regulated by XCT790 treatment (Fig. 4D). In addition to experiments in breast cancer cells, XCT790 inhibits expression of ERRα and carnitine palmitoyl transferase 1 (CPT-1m) in C2C12 myotubes overexpressing PGC-1α (see Fig. 7, which is published as supporting information on the PNAS web site). These data suggest a strong link between this orphan receptor and coactivator, and they further support a role for ERRα in the transcriptional control of PGC-1α-regulated genes involved in energy metabolism.
Discussion
Recent evidence (25) suggests that the orphan receptor ERRα is one of several nuclear receptors involved in controlling lipid and energy metabolism. To further examine the role of ERRα in metabolism and metabolic disease, we used a combined approach consisting of both ligand and target gene identification. High-throughput screening and medicinal chemistry coupled with cell-based transfection assays were used to identify and characterize synthetic ERRα ligands. By using this approach, we identified several classes of compounds that function as inverse agonists of ERRα. One compound, XCT790, is an efficacious and selective ERRα inverse agonist that inhibits ERRα constitutive activity with an IC50 of ≈400 nM in various cell-based and biochemical assays. By using XCT790, it is possible to specifically inhibit ERRα activity without affecting ER signaling, thereby allowing one to address specific questions regarding the role of ERRα and its mechanism of action in endocrine signaling.
In addition to ligand identification, transcriptional profiling in ERRα overexpressing cells was used to identify and characterize MAOB as an ERR target gene whose expression is regulated by both ERRα and the coactivator PGC-1α. MAOB is expressed in various metabolic tissues, including liver, kidney, skeletal muscle, and heart (data not shown), and its induction by PGC-1α/ERRα suggests an increased need for oxidative deamination with increased energy metabolism. Additionally, oxidative deamination may be a means to activate or inactivate signaling molecules or endogenous ligands involved in regulating ERRα and/or PGC-1α activity.
PGC-1α regulates the expression of many genes involved in energy production and utilization by means of its interactions with various transcription factors, including nuclear receptors. Studies demonstrating a role for PGC-1α in regulating expression of gluconeogenic enzymes in the liver served as the first link between this coactivator and the pathogenesis of type 2 diabetes (4, 5). More recently, genetic evidence has linked PGC-1α expression in muscle to type 2 diabetes and insulin resistance (9, 10). In muscle cells, PGC-1α regulates the expression of many genes involved in energy production by means of OX/PHOS. Interestingly, the expression of many of these OX/PHOS genes is lower in the muscle of individuals with type 2 diabetes, in which PGC-1α levels are also decreased. These results indicate a strong correlation among PGC-1α activity, OX/PHOS gene expression, and type 2 diabetes. As a coactivator, PGC-1α does not control gene expression directly but, rather, acts through specific DNA binding transcription factors at the promoters of target genes. Therapeutic intervention aimed at affecting the activity of the transcription factors that recruit PGC-1α could, therefore, prove to be useful in treating metabolic diseases.
ERRα and PGC-1α are expressed in tissues with high metabolic demands, including liver, adipose tissue, and skeletal muscle, and they are induced by many of the same environmental conditions, such as fasting and exposure to cold (2, 4, 5, 12, 13). Importantly, PGC-1α can induce ERRα expression in cultured cells, and it acts as a potent ligand-independent coactivator of ERRα at target promoters (11, 13). Together, these data suggest that ERRα mediates a subset of the many effects of PGC-1α on gene expression in metabolic tissues.
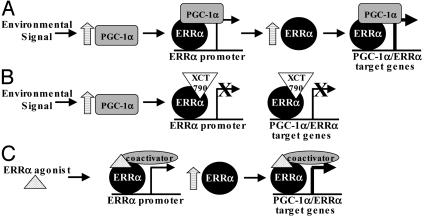
To investigate the functional relationship between ERRα and PGC-1α signaling further, we used the ERRα inverse agonist XCT790 to examine the regulation of MAOB, a model ERRα/PGC-1α target gene. Importantly, XCT790 inhibits PGC-1α induction of ERRα and MAOB gene expression, indicating a central role for ERRα in this signaling pathway. The observation that XCT790 also inhibits induction of the ERRα gene by PGC-1α implies that ERRα may be involved in regulating its own expression (Fig. 5A). This autoregulatory model suggests that the transcriptional activity of endogenous ERRα will increase significantly when its coactivator PGC-1α is induced under conditions such as fasting, exercise, or exposure to cold. Together, ERRα and PGC-1α then stimulate transcription of ERRα through putative binding sites in the ERRα promoter, and, when the levels of both receptor and coactivator have increased significantly, they can regulate the expression of various downstream ERRα/PGC-1α target genes coordinately. In support of this idea, microarray experiments indicate that overexpression of ERRα in breast cancer cells induces endogenous ERRα gene expression (data not shown). This model predicts that inhibitors of ERRα will prevent ERRα and target gene expression (Fig. 5B), and it is supported by our data (Figs. 4 and 7).
Fig. 5.
Schematic model for regulation of ERRα and PGC-1α target genes. (A) PGC-1α expression increases with environmental signals. PGC-1α then interacts with ERRα to activate the ERRα promoter, resulting in increased ERRα expression. ERRα and PGC-1α then cooperatively induce expression of target genes. (B) In the presence of the ERRα inverse agonist XCT790, expression of ERRα and target genes is blocked. (C) Activity of a hypothetical ERRα agonist that would increase expression of ERRα/PGC-1α target genes by means of the recruitment of other coactivators in the absence of PGC-1α.
It will be important to determine whether ERRα ligands can be used to treat metabolic diseases. Given the decrease in PGC-1α expression observed in the muscle of type 2 diabetics, one could speculate that ERRα agonists may counteract the loss of PGC-1α by increasing ERRα activity and subsequently expression of PGC-1α target genes, leading to increased energy production and insulin sensitivity in diabetic muscle (Fig. 5C). However, if ERRα mediates the effects of PGC-1α in the liver, an ERRα inverse agonist may have therapeutic use in preventing gluconeogenesis associated with diabetes.
Under certain conditions, ERRα is considered to be a constitutively active receptor; however, it has also been characterized as a transcriptional repressor, leading to the suggestion that its activity may depend on the cell type and promoter context in which it is assayed (23, 35, 36). As suggested for the related receptor ERRγ, the constitutive activity of ERRα may arise from a native conformation that promotes coactivator association in the absence of a bound ligand (37). In this view, expression and availability of the appropriate coactivators would control the “constitutive” activity of the receptor (38). Another possibility is that ERRα constitutive activity comes from an unidentified endogenous agonist ligand. Alternatively, if ERRα is constitutively active in the apo state, an endogenous antagonist ligand could regulate its activity in a tissue-specific manner. In either case, ERRα activity would be regulated by factors controlling expression and/or activity of the endogenous ERRα ligand.
Importantly, the ligand- and coactivator-regulated models for ERRα activity need not be mutually exclusive. For example, in tissues where PGC-1α expression is regulated by environmental stimuli, ERRα activity would be high when PGC-1α levels increase, and this coactivator regulation would cease with the removal of the environmental stimulus and subsequent loss of PGC-1α.
Experiments and structural analyses using selective ligands should shed light on the true mechanism of ERRα activity. The identification of potent and selective ERR modulators now enables one to move forward and explore ERRα function pharmacologically in normal endocrine signaling and metabolic disease.
Supplementary Material
Acknowledgments
We thank Griffin Macondray, Susan Zimmerman, Rosa Moreno, Lynn Wheeler, Nhin Lu, and Robin Summers for excellent technical assistance with various aspects of cell culture, high-throughput screening, and follow-up assays. We also thank Mary Petrowski, Janet Moritsugu, and Amy Liu for RT-PCR data and analysis; Jennifer Kitto for microarray experiments; Parinaz Pircher for help with retroviral experiments; Dr. David Moore for the gift of the mSHP promoter; and Dr. David Mangelsdorf for the peptide used in FP assays.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PPARγ, peroxisome proliferator-activated receptor γ; AF2, activation function 2; DES, diethylstilbestrol; ER, estrogen receptor; ERR, estrogen-related receptor; FP, fluorescence polarization; MAOB, monoamine oxidase B; mSHP, mouse short heterodimer partner; OX/PHOS, oxidative phosphorylation; PGC-1α, PPARγ coactivator 1α; LBD, ligand-binding domain.
References
- 1.Puigserver, P. & Spiegelman, B. M. (2003) Endocr. Rev. 24, 78–90. [DOI] [PubMed] [Google Scholar]
- 2.Puigserver, P., Wu, Z., Park, C. W., Graves, R., Wright, M. & Spiegelman, B. M. (1998) Cell 92, 829–839. [DOI] [PubMed] [Google Scholar]
- 3.Wu, Z., Puigserver, P., Andersson, U., Zhang, C., Adelmant, G., Mootha, V., Troy, A., Cinti, S., Lowell, B., Scarpulla, R. C. & Spiegelman, B. M. (1999) Cell 98, 115–124. [DOI] [PubMed] [Google Scholar]
- 4.Yoon, J. C., Puigserver, P., Chen, G., Donovan, J., Wu, Z., Rhee, J., Adelmant, G., Stafford, J., Kahn, C. R., Granner, D. K., Newgard, C. B. & Spiegelman, B. M. (2001) Nature 413, 131–138. [DOI] [PubMed] [Google Scholar]
- 5.Herzig, S., Long, F., Jhala, U. S., Hedrick, S., Quinn, R., Bauer, A., Rudolph, D., Schutz, G., Yoon, C., Puigserver, P., et al. (2001) Nature 413, 179–183. [DOI] [PubMed] [Google Scholar]
- 6.Goto, M., Terada, S., Kato, M., Katoh, M., Yokozeki, T., Tabata, I. & Shimokawa, T. (2000) Biochem. Biophys. Res. Commun. 274, 350–354. [DOI] [PubMed] [Google Scholar]
- 7.Baar, K., Wende, A. R., Jones, T. E., Marison, M., Nolte, L. A., Chen, M., Kelly, D. P. & Holloszy, J. O. (2002) FASEB J. 16, 1879–1886. [DOI] [PubMed] [Google Scholar]
- 8.Lin, J., Wu, H., Tarr, P. T., Zhang, C. Y., Wu, Z., Boss, O., Michael, L. F., Puigserver, P., Isotani, E., Olson, E. N., et al. (2002) Nature 418, 797–801. [DOI] [PubMed] [Google Scholar]
- 9.Mootha, V. K., Lindgren, C. M., Eriksson, K. F., Subramanian, A., Sihag, S., Lehar, J., Puigserver, P., Carlsson, E., Ridderstrale, M., Laurila, E., et al. (2003) Nat. Genet. 34, 267–273. [DOI] [PubMed] [Google Scholar]
- 10.Patti, M. E., Butte, A. J., Crunkhorn, S., Cusi, K., Berria, R., Kashyap, S., Miyazaki, Y., Kohane, I., Costello, M., Saccone, R., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 8466–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huss, J. M., Kopp, R. P. & Kelly, D. P. (2002) J. Biol. Chem. 277, 40265–40274. [DOI] [PubMed] [Google Scholar]
- 12.Ichida, M., Nemoto, S. & Finkel, T. (2002) J. Biol. Chem. 277, 50991–50995. [DOI] [PubMed] [Google Scholar]
- 13.Schreiber, S. N., Knutti, D., Brogli, K., Uhlmann, T. & Kralli, A. (2003) J. Biol. Chem. 278, 9013–9018. [DOI] [PubMed] [Google Scholar]
- 14.Giguere, V., Yang, N., Segui, P. & Evans, R. M. (1988) Nature 331, 91–94. [DOI] [PubMed] [Google Scholar]
- 15.Hong, H., Yang, L. & Stallcup, M. R. (1999) J. Biol. Chem. 274, 22618–22626. [DOI] [PubMed] [Google Scholar]
- 16.Heard, D. J., Norby, P. L., Holloway, J. & Vissing, H. (2000) Mol. Endocrinol. 14, 382–392. [DOI] [PubMed] [Google Scholar]
- 17.Coward, P., Lee, D., Hull, M. V. & Lehmann, J. M. (2001) Proc. Natl. Acad. Sci. USA 98, 8880–8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tremblay, G. B., Bergeron, D. & Giguere, V. (2001) Endocrinology 142, 4572–4575. [DOI] [PubMed] [Google Scholar]
- 19.Tremblay, G. B., Kunath, T., Bergeron, D., Lapointe, L., Champigny, C., Bader, J. A., Rossant, J. & Giguere, V. (2001) Genes Dev. 15, 833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang, N., Shigeta, H., Shi, H. & Teng, C. T. (1996) J. Biol. Chem. 271, 5795–5804. [DOI] [PubMed] [Google Scholar]
- 21.Vanacker, J. M., Pettersson, K., Gustafsson, J. A. & Laudet, V. (1999) EMBO J. 18, 4270–4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu, D., Kiriyama, Y., Lee, K. Y. & Giguere, V. (2001) Cancer Res. 61, 6755–6761. [PubMed] [Google Scholar]
- 23.Kraus, R. J., Ariazi, E. A., Farrell, M. L. & Mertz, J. E. (2002) J. Biol. Chem. 277, 24826–24834. [DOI] [PubMed] [Google Scholar]
- 24.Sladek, R., Bader, J. A. & Giguere, V. (1997) Mol. Cell. Biol. 17, 5400–5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo, J., Sladek, R., Carrier, J., Bader, J. A., Richard, D. & Giguere, V. (2003) Mol. Cell. Biol. 23, 7947–7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vega, R. B. & Kelly, D. P. (1997) J. Biol. Chem. 272, 31693–31699. [DOI] [PubMed] [Google Scholar]
- 27.Ariazi, E. A., Clark, G. M. & Mertz, J. E. (2002) Cancer Res. 62, 6510–6518. [PubMed] [Google Scholar]
- 28.Bonnelye, E., Vanacker, J. M., Dittmar, T., Begue, A., Desbiens, X., Denhardt, D. T., Aubin, J. E., Laudet, V. & Fournier, B. (1997) Mol. Endocrinol. 11, 905–916. [DOI] [PubMed] [Google Scholar]
- 29.Vanacker, J. M., Delmarre, C., Guo, X. & Laudet, V. (1998) Cell Growth Differ. 9, 1007–1014. [PubMed] [Google Scholar]
- 30.Umesono, K., Murakami, K. K., Thompson, C. C. & Evans, R. M. (1991) Cell 65, 1255–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muscat, G. E., Wagner, B. L., Hou, J., Tangirala, R. K., Bischoff, E. D., Rohde, P., Petrowski, M., Li, J., Shao, G., Macondray, G. & Schulman, I. G. (2002) J. Biol. Chem. 277, 40722–40728. [DOI] [PubMed] [Google Scholar]
- 32.Kang, T., Martins, T. & Sadowski, I. (1993) J. Biol. Chem. 268, 9629–9635. [PubMed] [Google Scholar]
- 33.Chen, S., Zhou, D., Yang, C. & Sherman, M. (2001) J. Biol. Chem. 276, 28465–28470. [DOI] [PubMed] [Google Scholar]
- 34.Yamada, M. & Yasuhara, H. (2004) Neurotoxicology 25, 215–221. [DOI] [PubMed] [Google Scholar]
- 35.Wiley, S. R., Kraus, R. J., Zuo, F., Murray, E. E., Loritz, K. & Mertz, J. E. (1993) Genes Dev. 7, 2206–2219. [DOI] [PubMed] [Google Scholar]
- 36.Johnston, S. D., Liu, X., Zuo, F., Eisenbraun, T. L., Wiley, S. R., Kraus, R. J. & Mertz, J. E. (1997) Mol. Endocrinol. 11, 342–352. [DOI] [PubMed] [Google Scholar]
- 37.Greschik, H., Wurtz, J. M., Sanglier, S., Bourguet, W., van Dorsselaer, A., Moras, D. & Renaud, J. P. (2002) Mol. Cell 9, 303–313. [DOI] [PubMed] [Google Scholar]
- 38.Kamei, Y., Ohizumi, H., Fujitani, Y., Nemoto, T., Tanaka, T., Takahashi, N., Kawada, T., Miyoshi, M., Ezaki, O. & Kakizuka, A. (2003) Proc. Natl. Acad. Sci. USA 100, 12378–12383. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.