Abstract
Structural studies of myosin have indicated some of the conformational changes that occur in this protein during the contractile cycle, and we have now observed a conformational change in a bound nucleotide as well. The 3.1-Å x-ray structure of the scallop myosin head domain (subfragment 1) in the ADP-bound near-rigor state (lever arm =45° to the helical actin axis) shows the diphosphate moiety positioned on the surface of the nucleotide-binding pocket, rather than deep within it as had been observed previously. This conformation strongly suggests a specific mode of entry and exit of the nucleotide from the nucleotide-binding pocket through the so-called “front door.” In addition, using a variety of scallop structures, including a relatively high-resolution 2.75-Å nucleotide-free near-rigor structure, we have identified a conserved complex salt bridge connecting the 50-kDa upper and N-terminal subdomains. This salt bridge is present only in crystal structures of muscle myosin isoforms that exhibit a strong reciprocal relationship (also known as coupling) between actin and nucleotide affinity.
Myosin is a motor protein that transduces ATP hydrolysis into mechanical work, leading to its translocation along filamentous actin. It is believed that the small conformational changes induced by enzymatic activity in the myosin “motor domain” (MD) are amplified by the motion of the “lever arm.” Three weak actin-binding (actin-detached) states have been identified crystallographically for scallop myosin subfragment 1 (S1) (1). These and other structures reveal that the motor function of myosin S1 is coordinated by three subdomains (50-kDa upper and lower subdomains and the converter) that rotate as rigid bodies around the relatively stable N-terminal subdomain (1–3). These rotations depend on conformational changes in three flexible joints (switch II, relay, and the SH1 helix) between the subdomains. Three-dimensional reconstructions of electron microscopic images of actin decorated by S1 of several myosin isoforms (4–6) have led to the suggestion that actin binds to portions of the 50-kDa upper and lower subdomains, whereas various crystal structures (see, for example, refs. 3 and 7–9) indicate that ATP binds at the opposite side of the myosin head in a pocket between the 50-kDa upper and N-terminal subdomains (Figs. 1 and 2A).
Fig. 1.
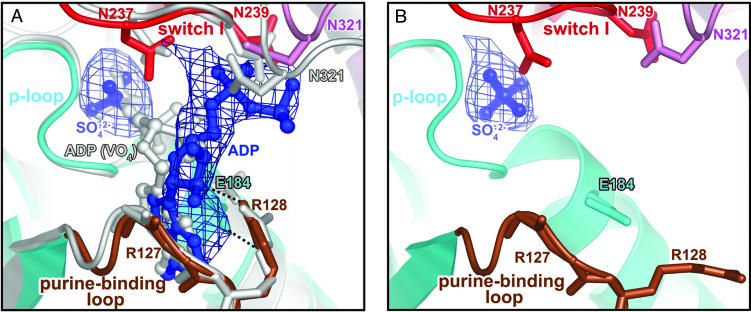
Overview of the nucleotide-binding pocket. (A) ADP (blue) in the scallop myosin near-rigor structure adopts a partially bound conformation at the front door of the nucleotide-binding pocket. For comparison, the fully bound ADP from the S1–ADP·VO4 complex in the scallop prepower-stroke structure (PDB ID code 1QVI) is shown in light gray. Also shown is an Fo – Fc simulated-annealing electron density map at a 1.0 σ contour level, calculated after omitting ADP and sulfate. Residues in switch I (red) and N321 (pink) from the 50-kDa upper subdomain that are involved in weak interactions (see text) with the ADP diphosphate moiety are shown in stick representation. The same residues from the prepower-stroke conformation structure are shown in light gray. The only major difference between the two structures in the nucleotide-binding pocket of the enzyme is that N321 (50-kDa upper subdomain) makes closer contact with the bound nucleotide in the prepower-stroke structure. The side chain of R127 and the hydrogen bond between its main-chain amide and the ADP ribose O2′ are not shown for clarity. The P-loop and purine-binding loop are shown in cyan and brown, respectively. E184 (cyan sticks) and R128 (brown sticks) in the two loops form a salt bridge (dashed lines) that protects one face of the ADP base from solvent. (B) Corresponding view of the ScS1–SO4 structure, with an Fo – Fc simulated-annealing electron density map at a 2.0 σ contour level, calculated after omitting the sulfate. The E184/R128 salt bridge is not formed in the absence of a nucleotide, supporting the role of R128 in nucleotide recruitment.
Fig. 2.
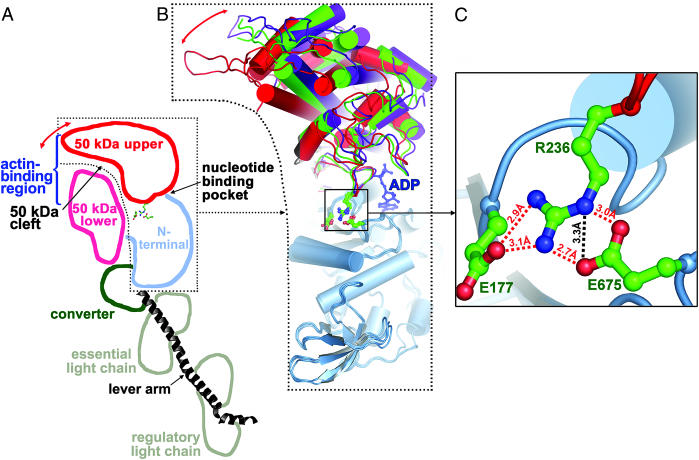
Rotations of the 50-kDa upper subdomain in the three conformational states of scallop myosin. The 50-kDa upper subdomain rotates as a rigid body with respect to the N-terminal subdomain, with the center of rotation near R236 of switch I. (A) Schematic representation of the different subdomains in the ScS1–ADP structure. Red arrow indicates rotation of the 50-kDa upper subdomain, which closes the 50-kDa cleft when myosin binds strongly to actin (5, 6). (B) A superposition of the N-terminal subdomains (light blue) shows the relative positions of the 50-kDa upper subdomains in the present ScS1–ADP near-rigor state (red), the internally uncoupled state (green), and the prepower-stroke state (purple) structures. A red arrow indicates the rotation of the 50-kDa upper subdomain in the plane of the paper. The 50-kDa lower subdomain, the converter, and the lever arm have been omitted for clarity. ADP from the ScS1–ADP structure is shown in a blue ball-and-stick representation. Residues involved in the complex salt bridge (E177, R236, and E675) between the 50-kDa upper and N-terminal subdomains are also shown in a ball-and-stick representation. (C) Close-up view of the complex salt bridge, which is well situated to stabilize the rotation of the 50-kDa upper subdomain. All but one of the salt links have favorable geometry to form hydrogen bonds, and these are shown as red dashed lines. The additional nonhydrogen-bonded salt link is shown as a black dashed line.
One of the three structural conformations, the so-called “prepower stroke,” corresponds biochemically to the ADP·Pi transition state (1, 8, 10–12). Here, S1 displays a primed lever arm (=90° to the actin filament axis), closure of the 50-kDa cleft's base (which is near the nucleotide-binding pocket), and a bent switch II that interacts with both the nucleotide and switch I (a loop in the 50-kDa upper subdomain that coordinates the nucleotide). After this state, the tight binding of myosin to actin results in the so-called “power stroke” (after which the lever arm is =45° to the actin filament), a process believed to be initiated by the near-complete closing of the 50-kDa cleft (4–6). This step is thought to facilitate phosphate release through a “back door,” followed by ADP release through the entrance to the nucleotide-binding pocket (13), here referred to as the “front door.” The strongly actin-bound, nucleotide-free conformation of S1 that results is known as the “rigor state.” Initial binding of MgATP to the rigor S1 at the start of a new ATPase cycle is accompanied by dissociation of myosin from actin (14), possibly because of a partial opening of the 50-kDa cleft (15). Another recently identified state, called “internally uncoupled” (2, 3), is characterized by a disordered SH1 helix, an open 50-kDa cleft, and an extended switch II. The disordered SH1 helix effectively uncouples the S1 MD from the lever arm. Based on this finding and other evidence, it was proposed that this state corresponds to an actin-detached ATP state (2). The third state is called “near-rigor” (3, 16) because it is characterized by the rigor position of the lever arm. Unlike the rigor state, however, this state has an open 50-kDa cleft. Switch II is again extended in this state. The near-rigor state is believed to result when ATP dissociates true rigor myosin from actin in the contractile cycle (7, 9). The structural details of how binding of nucleotide to myosin reduces affinity for actin and of how subsequent rebinding to actin leads to nucleotide release are not fully understood.
Here, we report the crystal structure of scallop myosin S1 in the near-rigor state, this time with a previously unobserved conformation of ADP visualized near the nucleotide-binding pocket. It is unclear whether the ADP provides information about nucleotide entry or exit from the pocket, but the structure suggests a specific sequence for one or both processes. We also have determined a higher-resolution (2.75 Å) near-rigor structure of nucleotide-free scallop S1 that was solved earlier to 3.2 Å (3). In both structures, we identify a complex salt bridge linking switch I, the P-loop, and the N-terminal subdomain. Comparison with kinetic data (17, 18) reveals that the presence or absence of such bridges in the crystal structures of the MD of different muscle myosins correlates with the level of coupling between nucleotide and actin binding in these myosins.
Methods
Crystallization and Data Collection. Purified S1 was obtained from scallop (Argopecten irradians) striated muscle myosin as described in ref. 2. The S1–MgADP complex was crystallized in sitting drops by combining equal volumes (3 μl) of protein solution with a precipitant solution containing 50 mM Mes buffer (pH 6.5), 5 mM MgCl2, 2 mM MgADP, 6–6.5% polyethylene glycol 8K, 50 mM ammonium sulfate, 50 mM trimethylamine N-oxide, and 12–13% glycerol. Nucleotide-free S1 was crystallized with a precipitant solution of 50 mM Na cacodylate (pH 7.0), 10 mM MgCl2, 6–7% polyethylene glycol 8K, 50 mM ammonium sulfate, and 8% glycerol. X-ray data were collected at beamline A1 of the Cornell High Energy Synchrotron Source (S1–MgADP complex) and station ID29 of the European Synchrotron Research Facility (nucleotide-free S1) at 100 K from single crystals cryopreserved in precipitant solution containing final amounts of 25% glycerol and 18% polyethylene glycol 8K. Data were processed with denzo/scalepack (19) (S1–MgADP) or with mosflm (20) and scala (21) (nucleotide-free complex).
Structure Determination and Refinement. Initial phases for both nucleotide-containing and nucleotide-free S1 were determined by molecular replacement and rigid body refinement with the program amore (22) by using the earlier near-rigor scallop S1 structure (3) as an initial search model. The nucleotide-free structure was refined to a 2.75-Å resolution, and the ADP-containing structure was refined to 3.1 Å by iterative model-building with the o graphics package (23), combined with torsional simulated annealing refinement or conjugate-gradient minimization with a bulk solvent correction in cns (24). Unambiguous electron density was observed for the sulfate ion in simulated annealing omit maps (25) of both structures and for the ADP in the nucleotide-containing structure, and these were modeled in at later stages of refinement. Water molecules were added manually in the final stages of refinement and were only built in where justified by hydrogen bonds and Fo – Fc electron density ≥3.0 σ contour level. The crystallographic Rfactor and Rfree for the two structures (Table 1) are within the average values of structures at the given resolutions (26).
Table 1. Data collection and refinement statistics.
| ScS1-ADP | ScS1-SO4 | |
|---|---|---|
| Data collection | ||
| Resolution range, Å | 50.0-3.10 | 38.79-2.75 |
| Unique reflections | 21,662 | 31,951 |
| Multiplicity | 4.5 | 4.0 |
| Average I/σ | 27.2 | 20.5 |
| Rsym, % (all data/outer shell) | 10.2/39.3 | 4.8/38.5 |
| Completeness, % (all data/outer shell) | 89.4/78.8 | 88.0/81.8 |
| Refinement | ||
| σ cutoff | 0.0 | 2.0 |
| Completeness in range, % | 84.6 | 76.3 |
| R factor, % | 23.3 | 24.2 |
| Rfree, % (5% partition) | 26.9 | 28.6 |
| Mean B-factor | 80.5 | 57.1 |
| rms deviations* | ||
| Bond lengths, Å | 0.006 | 0.011 |
| Bond angles,° | 1.6 | 1.4 |
| No. of protein atoms | 8,578 | 8,661 |
| No. of water atoms | 1 | 95 |
| No. of prosthetic atoms | 33 | 8 |
| Cross-validated coordinate error, ņ | 0.47 | 0.5 |
The cell parameters of the two structures are as follows: ScS1-ADP, P21: a = 84.1 Å, b = 50.9 Å, c = 161.6 Å, α = 90.0°, β = 98.4°, and γ = 90.0°; ScS1-SO4, P21: a = 83.6 Å, b = 51.1 Å, c = 162.3 Å, α = 90.0°, β = 98.0°, and γ = 90.0°. Eighty-two percent of the ScS1-SO4 residues and 83% of the ScS1-ADP residues in the final models are in the most favored regions of the Ramachandran plot, with the rest in additionally allowed regions.
See ref. 46.
Results
The ADP-containing structure (ScS1–ADP) and the nucleotide-free structure (ScS1–SO4) are in the near-rigor conformation and both superimpose on the previously reported 3.2-Å scallop near-rigor structure (3) with a rms difference of 0.7 Å for 547 Cα atoms of α-helices and β-strands of the MD and lever arm. In both structures, a sulfate occupies the β-phosphate site in the nucleotide-binding pocket (Fig. 1) and displays the same coordination as observed in previous structures (3, 16). The switch I loop is observed in a constrained conformation that has been identified in many other myosin structures from smooth muscle, skeletal muscle, and Dictyostelium myosin II, usually with nucleotide bound (2, 3, 7–11, 16, 27).
A Partially Bound Conformation of Nucleotide. In the ScS1–ADP structure, the ribose and diphosphate moieties of ADP adopt a previously unobserved conformation at the front door of the nucleotide-binding pocket (Fig. 1). Previous nucleotide-containing structures of the myosin head region have revealed that the nucleotide-binding pocket comprises the purine-binding loop (NPxxxxxxY) on one side and the switch I [AKTxxN(N/D)NSSR]/P-loop (GESGAGKT) pair that sandwich the polyphosphate moiety of the nucleotide on the other side. Interactions between these loops and the nucleotide are strictly conserved in nucleotide-bound near-rigor structures from both Dictyostelium and scallop isoforms, whether the nucleotide bound is ADP, ATP, or an ATP analog (3, 7, 9). In the present ScS1–ADP structure, however, the ribose moiety (to some extent) and the diphosphate moiety are displaced toward solvent and make fewer and more tenuous contacts with N237, N239 (switch I), R127 (purine-binding loop), and N321 (50-kDa upper subdomain) (Fig. 1). The torsional angle between the purine base and the ribose and the angle between the ribose and the C5′ of ADP are different from the corresponding values in the other nucleotide structures mentioned above, so that the present ADP conformation is not simply a rigid-body displacement of the normal ADP structure. The purine base of ADP is in the usual position, held firmly in place by hydrophobic interactions with the purine-binding loop and by the previously observed hydrogen bond to Y132 of the loop (9, 27). Of special interest is R128, which is stacked against the base of ADP. R128 also makes a simple salt bridge with E184, effectively closing off solvent access to the bound ligand on one side of the front door of nucleotide entry (Fig. 1). Similar salt bridges have been observed in a scallop S1 prepower-stroke state structure (12), the scallop internally uncoupled structure (3), and a Dictyostelium myosin II MD structure with MgADP·BeFx bound (27). In the present ScS1–SO4 structure, the salt bridge between R128 and E184 is broken, and R128 is swung out toward solvent in the absence of nucleotide (Fig. 1). The Arg/Glu pair corresponds in skeletal-muscle isoforms to Trp/Val, which can make hydrophobic interactions with the ATP base and with each other, thereby also providing binding interactions and solvent protection to the bound nucleotide. The corresponding W131 in the chicken skeletal myosin structure (16) is also exposed to solvent in the absence of nucleotide. These structural observations imply that R128 in scallop or its equivalent in skeletal muscle myosin is involved in ATP recruitment to the nucleotide-binding pocket. Evidence for this role of R128 is provided by cross-linking studies using photoaffinity ATP analogs (28–30). Moreover, in isoforms lacking a tight contact between the pair of residues equivalent to R128/E184, ATP binding is weaker by one or two orders of magnitude. The actomyosin ATP binding constants are 2.5 μM–1·s–1 and 2.1 μM–1·s–1 for scallop and rabbit skeletal myosin, respectively (17). By contrast, the binding constant is 0.018 μM–·1s–1 for myosin VI (31), in which a pair of negatively charged residues (Asp/Glu) is substituted, and 0.017 μM–1·s–1 for myosin I (32), in which the pair Ser/Glu (whose side chains are too short to form a tight contact) is substituted.
The unusual nucleotide state visualized in the ScS1–ADP structure may provide information about the mode of ADP release from actomyosin before the rigor state and/or the mode of ATP entry to rigor actomyosin before myosin dissociation from actin. The ScS1–ADP structure does not distinguish between these two processes but does suggest that, during either nucleotide entry or exit from the nucleotide-binding pocket, the polyphosphate moiety is the last in and/or first out. A recent kinetic study using a Dictyostelium myosin II mutant containing a single tryptophan near the nucleotide-binding pocket (F129W) revealed at least a three-step nucleotide-binding mechanism (33). They also detected a low-fluorescence binding intermediate that was attributed to an initial unfavorable orientation/conformation of the nucleotide. It is possible that the nucleotide conformation in the ScS1–ADP structure represents one of these steps.
It is not clear why ADP is found in this partially bound conformation in the present structure. One cannot rule out the possibility that the sulfate ion (present at a 50 mM concentration) has displaced the β-phosphate of ADP (present at a 2 mM concentration) in this structure. The ScS1–ADP structure is the only scallop near-rigor structure available with any nucleotide bound. The only other ADP-containing near-rigor structure is that of the Dictyostelium myosin II MD (7), containing a fully bound ADP. But the crystallization conditions of the two studies cannot be compared because the Dictyostelium structure contains a truncated myosin II construct lacking the lever arm and has a low sequence identity (49%) with scallop myosin.
A Connection Between the 50-kDa Upper and N-Terminal Subdomains. The communication between the nucleotide-binding pocket and the lever arm of myosin that leads to the power stroke has been ascribed to coupled rigid-body rearrangements of the MD's four subdomains, which are linked by three flexible joints (2, 3). Recent structural studies have yielded some information about the communication between the nucleotide-binding pocket and the actin-binding 50-kDa cleft. Electron microscopic reconstructions of actin decorated with smooth (5) or skeletal muscle myosin (6) indicate that the 50-kDa upper subdomain rotates away from the N-terminal subdomain when myosin binds strongly to actin. Moreover, the three scallop S1 structures (3) and new crystal structures of myosin V (34) and Dictyostelium myosin II (35) reveal that the 50-kDa upper subdomain rotates about a region comprising switch I and the preceding β-hairpin so that switch I exists in distinctly different conformations or positions during the actomyosin cycle. This rotation is associated with the distortion of the seven-stranded β-sheet that spans the two subdomains (34, 35). Because the nucleotide-binding pocket is located at the interface between the N-terminal and 50-kDa upper subdomains, the rotation of the 50-kDa upper subdomain necessarily opens up the pocket and is believed to lead to the subsequent release of ADP (3, 6, 34). Conversely, in the ATP-bound state, the opening of the 50-kDa cleft (during which the 50-kDa upper subdomain rotates toward the N-terminal subdomain) is accompanied by the closing of the nucleotide-binding pocket (36).
We have now identified a strong linkage between the two subdomains that may strengthen the communication between the nucleotide-binding pocket and the 50-kDa cleft. Including the structures described in our study, we now have eight scallop myosin structures in three different weak actin-binding conformations. Comparison of the 50-kDa upper subdomain positions in these structures reveals that this subdomain rotates as a rigid body with respect to the N-terminal subdomain by as much as 16°, and that the center of rotation is located in the switch I region near R236 (Fig. 2B). The only strong connection between these subdomains that is maintained in all scallop structures (apart from the β-sheet mentioned above) is a complex salt bridge (37) between R236 of switch I in the 50-kDa upper subdomain and two glutamic acid residues in the N-terminal subdomain, E177 and E675 (Fig. 2 B and C). (E177 is a part of the catalytic P-loop in the N-terminal subdomain.) This salt bridge is especially strong because the participating side-chain atoms also form four hydrogen bonds (Fig. 2C) in all three scallop states.
For reasons given below, it is likely that the salt bridge will persist in the strong actin-binding rigor state of scallop myosin, despite the larger rotation of the upper 50-kDa subdomain away from the N-terminal subdomain in that state. This linkage is therefore a possible common feature of all conformational states of certain myosin isoforms in the contractile cycle. The location of the complex salt bridge approximately at the center of rotation of the 50-kDa upper subdomain described above suggests a possible role for the linkage in minimizing the amount of random motion (or “wobble”) that occurs during the rotation of the subdomain. It appears that the strong connection between switch I (part of the 50-kDa upper subdomain) and the N-terminal subdomain provided by the salt bridge helps to direct the rotation of the 50-kDa upper subdomain, thereby improving the communication pathway between the actin-binding 50-kDa cleft and the nucleotide-binding pocket. Efficient communication between the two binding sites could modulate the well documented inverse relationship (coupling) between actin-binding affinity and nucleotide-binding affinity first observed with ATP (14). The affinity of myosin for nucleotide and actin are coupled in that nucleotide weakens actin affinity and actin weakens nucleotide affinity. The ratio of the actin affinities in the absence and presence of nucleotide defines the extent of coupling. With ATP, the coupling is large (=1,000-fold) for all myosin isoforms, whereas for ADP the coupling is variable. A value of 1 means that the two binding sites are independent for that myosin isoform. As the value increases, actin displaces ADP (and ADP displaces actin) more effectively.
This coupling has been investigated in transient kinetic studies in which the dissociation of ADP by actin and of actin by ADP were monitored. These studies indicate that the coupling is significantly stronger in scallop and skeletal muscle myosins than in smooth muscle myosin (17, 18) (Table 2). Correspondingly, the crystal structure of chicken skeletal S1 in the near-rigor state (4) displays the same hydrogen-bonded and arginine-mediated complex salt bridge link between switch I and the N-terminal subdomain as observed in the present structures. Moreover, a recent study (38) places the Drosophila embryonic and indirect flight muscle myosins in the high-coupling category, and sequence alignment reveals that the corresponding residues in the Drosophila muscle myosins are again identical to those in scallop (Table 2). By contrast, the chicken smooth muscle MD prepower-stroke state crystal structure (8) reveals only a weak, lysine-mediated simple salt bridge in this region (Table 2). Thus, a correlation appears to exist between the presence or absence of the complex salt bridge and the degree of coupling between actin and nucleotide binding in different isoforms of muscle myosin. It must be noted that structural features other than the salt bridge might play a role in the coupling in muscle and nonmuscle myosin isoforms of myosin, as indicated in studies with Dictyostelium myosin II (39) and its truncated constructs (40) or engineered mutants (39).
Table 2. The complex salt bridge and kinetic coupling.
| Myosin isoforms | Amino acid at position 177 | Amino acid at position 236 | Amino acid at position 675 | Interactions observed in crystal structure among residues in the three positions (ref.) | Coupling* between ADP/actin affinity KDA/KA = KAD/KD (ref.) |
|---|---|---|---|---|---|
| Scallop (Argopecten irradians) striated muscle | E(177) | R(236) | E(675) | Complex salt bridge (this work) | 45-48 (17) |
| Rabbit skeletal muscle | E(180) | R(240) | E(680) | Complex salt bridge (chicken skeletal muscle S1) (16) | 100 (17) 30-60 (49) |
| Drosophila indirect flight muscle | E(180) | R(237) | E(678) | NA | 55 (38) |
| Drosophila embryonic muscle | E(180) | R(236) | E(678) | NA | 300 (38) |
| Bovine cardiac (α/β) muscle | E(179) | R(237) | E(677) | NA | 15-20 (50) |
| Chicken gizzard smooth muscle | E(177) | K(240) | H(688) | Weak single salt bridge (1BR1, 1BR4) (8) | 4 (18) |
Residues involved in the complex salt bridge or their sequence equivalents from several muscle myosins and the kinetic coupling in the respective isoform. NA, not available.
The coupling is given by the ratio of the actin dissociation constant from myosin in the presence and absence of ADP (KDA/KA) or the ADP dissociation constant from myosin in the presence and absence of actin (KAD/KD).
The complex salt bridge described above might provide a specific mechanism for one aspect of the coupling in high-coupling myosins, namely actin-induced ADP release. The new crystal structure of chicken myosin V (34), which, although not bound to actin, appears to mimic the strong actin-binding rigor state, shows that the 50-kDa upper subdomain along with switch I rotates further away from the N-terminal subdomain than in any previously identified myosin structure. Myosin V lacks one of the residues necessary to form the complex salt bridge observed in scallop (scallop E675 becomes A, D, or S in Myosin V). However, the Myosin V structure suggests that the complex salt bridge could form in the rigor state if a glutamate (with a longer side chain than aspartate) is introduced at that position. (The kinetics of such a construct might reveal the contribution of the salt bridge to the coupling.) While the complex salt bridge is therefore likely to be maintained in the strong actin-bound rigor state in the high-coupling myosins, the large rotation of the 50-kDa upper subdomain would pull switch I away from the nucleotide, breaking important protein–nucleotide interactions and leading to ADP release. Because the rotation of the 50-kDa upper subdomain is well anchored in the high-coupling myosins, this structural feature suggests how increased affinity for actin is strongly coupled with decreased affinity for the nucleotide in these isoforms. By contrast, in low-coupling isoforms such as smooth muscle myosin, less directed movements of the 50-kDa upper subdomain (in the absence of the complex salt bridge) could lead to a slower actin-induced ADP release.
Complex salt bridges have been shown to be involved in the allosteric regulation of the GroEL chaperonin (41) and in promoting intersubdomain contacts in the structure of hemoglobin (42). Although the role of simple salt bridges in providing protein stabilization is controversial (43, 44), the importance of complex salt bridges in providing better stabilization than the sum of two individual salt bridges and the importance of arginine in complex salt bridges have been recognized (37, 45).
Perspective. All high-resolution crystal structures of myosin S1 have been solved in the absence of actin. Nevertheless, some of the most recent x-ray structures and electron microscopic images have begun to reveal the changes in S1 upon binding actin. Two major changes appear to occur: the 50-kDa upper subdomain rotates away from the N-terminal subdomain (thereby closing the actin-binding cleft and opening the nucleotide-binding pocket) and nucleotide (ADP) is released. The center of this rotation is near switch I, leading to the prediction that this rotation triggers the power stroke by pulling switch I away from switch II (3, 35). The current study provides some structural details of both processes: rotation of the 50-kDa subdomain appears to be directed by a complex salt bridge (E177/R236/E675) in myosins of high-coupling muscles, and ADP appears to leave via a front door in agreement with previous kinetic investigations. The unique conformation of ADP near the nucleotide-binding pocket of myosin S1 reported here provides a beginning for understanding the sequence of molecular events leading to nucleotide binding/dissociation from the binding pocket, in which the polyphosphate moiety is the last in and/or first out. Here, a simple salt bridge (R128/E184) in scallop myosin, or equivalent hydrophobic contacts in skeletal muscle myosin, at the front door of the nucleotide-binding pocket appears to play a role in nucleotide recruitment. A high-resolution structure of an acto–S1 complex would be essential to visualize the interactions between these two proteins, to explain how binding to actin produces rotation of the 50-kDa upper subdomain, and to help locate the route for phosphate release.
Acknowledgments
We thank A. Houdusse and P.-D. Coureux (Institut Curie, Paris) for providing the ScS1–ADP data set, which was collected at station ID29 of the European Synchrotron Research Facility (Grenoble, France); A. Houdusse, M. A. Geeves, and J. H. Brown for helpful discussions; E. O'Neall Hennessey for expert technical assistance; and the staff of the Cornell High Energy Synchrotron Source for assistance with data collection. This work was supported by National Institutes of Health Grant AR17346 and a grant from the Muscular Dystrophy Association (to C.C.).
Abbreviations: MD, motor domain; S1, subfragment 1.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 1S5G (ScS1–ADP) and 1SR6 (ScS1–SO4)].
References
- 1.Houdusse, A., Szent-Györgyi, A. G. & Cohen, C. (2000) Proc. Natl. Acad. Sci. USA 97, 11238–11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Houdusse, A., Kalabokis, V. N., Himmel, D., Szent-Györgyi, A. G. & Cohen, C. (1999) Cell 97, 459–470. [DOI] [PubMed] [Google Scholar]
- 3.Himmel, D. M., Gourinath, S., Reshetnikova, L., Shen, Y., Szent-Györgyi, A. G. & Cohen, C. (2002) Proc. Natl. Acad. Sci. USA 99, 12645–12650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rayment, I., Holden, H. M., Whittaker, M., Yohn, C. B., Lorenz, M., Holmes, K. C. & Milligan, R. A. (1993) Science 261, 58–65. [DOI] [PubMed] [Google Scholar]
- 5.Volkmann, N., Hanein, D., Ouyang, G., Trybus, K. M., DeRosier, D. J. & Lowey, S. (2000) Nat. Struct. Biol. 12, 1147–1155. [DOI] [PubMed] [Google Scholar]
- 6.Holmes, K. C., Angert, I., Kull, F. J., Jahn, W. & Schröder, R. R. (2003) Nature 425, 423–427. [DOI] [PubMed] [Google Scholar]
- 7.Gulick, A. M., Bauer, C. B., Thoden, J. B. & Rayment, I. (1997) Biochemistry 36, 11619–11628. [DOI] [PubMed] [Google Scholar]
- 8.Dominguez, R., Freyzon, Y., Trybus, K. M. & Cohen, C. (1998) Cell 94, 559–571. [DOI] [PubMed] [Google Scholar]
- 9.Bauer, C. B., Holden, H. M., Thoden, J. B., Smith, R. & Rayment, I. (2000) J. Biol. Chem. 275, 38494–38499. [DOI] [PubMed] [Google Scholar]
- 10.Smith, C. A. & Rayment, I. (1995) Biochemistry 34, 8973–8981. [DOI] [PubMed] [Google Scholar]
- 11.Smith, C. A. & Rayment, I. (1996) Biochemistry 35, 5404–5417. [DOI] [PubMed] [Google Scholar]
- 12.Gourinath, S., Himmel, D. M., Brown, J. H., Reshetnikova, L., Szent-Györgyi, A. G. & Cohen, C. (2003) Structure (Cambridge, Mass.) 11, 1621–1627. [DOI] [PubMed] [Google Scholar]
- 13.Yount, R. G., Lawson, D. & Rayment, I. (1994) Biophys. J. 68, 44S–49S. [PMC free article] [PubMed] [Google Scholar]
- 14.Lymn, R. W. & Taylor, E. W. (1971) Biochemistry 10, 4617–4624. [DOI] [PubMed] [Google Scholar]
- 15.Rayment, I., Bauer, C. B., Gulick, A. M., Smith, R., Thoden, J. B. & Wesenberg, G. (1999) in Enzymatic Mechanisms, Biomedical and Health Research, eds. Frey, P. A. & Northrop, D. B. (IOS Press, Amsterdam), Vol. 27, pp. 48–60. [Google Scholar]
- 16.Rayment, I., Rypniewski, W. R., Schmidt-Bäse, K., Smith, R., Tomchick, D. R., Benning, M. M., Winkelmann, D. A., Wesenberg, G. & Holden, H. M. (1993) Science 261, 50–58. [DOI] [PubMed] [Google Scholar]
- 17.Kurzawa-Goertz, S. E., Perreault-Micale, C. L., Trybus, K. M., Szent-Györgyi, A. G. & Geeves, M. A. (1998) Biochemistry 37, 7515–7525. [DOI] [PubMed] [Google Scholar]
- 18.Cremo, C. R. & Geeves, M. A. (1998) Biochemistry 37, 1969–1978. [DOI] [PubMed] [Google Scholar]
- 19.Otwinowski, Z. & Minor, W. (1993) in Data Collection and Processing, eds. Sawyer, L., Isaacs, N. & Bailey, S. (CLRC Daresbury Laboratory, Warrington, U.K.), pp. 59–62.
- 20.Leslie, A. G. W., Brick, P. & Wonacott, A. (1986) Daresbury Lab. Inf. Q. Protein Crystallogr. 18, 33–39. [Google Scholar]
- 21.Collaborative Computational Project No. 4. (1994) Acta Crystallogr. D 50, 760–763.15299374 [Google Scholar]
- 22.Navaza, J. (1994) Acta Crystallogr. A 50, 157–163. [Google Scholar]
- 23.Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 24.Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 25.Hodel, A., Kim, S. H. & Brünger, A. T. (1992) Acta Crystallogr. A 48, 851–858. [Google Scholar]
- 26.Kleywegt, G. J. & Brünger, A. T. (1996) Structure (London) 4, 897–904. [DOI] [PubMed] [Google Scholar]
- 27.Fisher, A. J., Smith, C. A., Thoden, J. B., Smith, R., Sutoh, K., Holden, H. M. & Rayment, I. (1995) Biochemistry 34, 8960–8972. [DOI] [PubMed] [Google Scholar]
- 28.Szilagyi, L., Balint, M., Sreter, F. A. & Gergely, J. (1979) Biochem. Biophys. Res. Commun. 87, 936–945. [DOI] [PubMed] [Google Scholar]
- 29.Kerwin, B. A. & Yount, R. G. (1992) Bioconjug. Chem. 3, 328–336. [DOI] [PubMed] [Google Scholar]
- 30.Grammer, J. C., Kuwayama, H. & Yount, R. G. (1993) Biochemistry 32, 5725–5732. [DOI] [PubMed] [Google Scholar]
- 31.De La Cruz, E. M., Ostap, E. M. & Sweeney, H. L. (2001) J. Biol. Chem. 276, 32373–32381. [DOI] [PubMed] [Google Scholar]
- 32.Coluccio, L. M. & Geeves, M. A. (1999) J. Biol. Chem. 274, 21575–21580. [DOI] [PubMed] [Google Scholar]
- 33.Kovacs, M., Malnasi-Csizmadia, A., Woolley, R. J. & Bagshaw, C. R. (2002) J. Biol. Chem. 277, 28459–28467. [DOI] [PubMed] [Google Scholar]
- 34.Coureux, P. D., Wells, A. L., Menetrey, J., Yengo, C. M., Morris, C. A., Sweeney, H. L. & Houdusse, A. (2003) Nature 425, 419–423. [DOI] [PubMed] [Google Scholar]
- 35.Reubold, T. F., Eschenburg, S., Becker, A., Kull, F. J. & Manstein, D. J. (2003) Nat. Struct. Biol. 10, 826–830. [DOI] [PubMed] [Google Scholar]
- 36.Conibear, P. B., Bagshaw, C. R., Fajer, P. G., Kovacs, M. & Malnasi-Csizmadia, A. (2003) Nat. Struct. Biol. 10, 831–835. [DOI] [PubMed] [Google Scholar]
- 37.Musafia, B., Buchner, V. & Arad, D. (1995) J. Mol. Biol. 254, 761–770. [DOI] [PubMed] [Google Scholar]
- 38.Miller, B. M., Nyitrai, M., Bernstein, S. I. & Geeves, M. A. (2003) J. Biol. Chem. 278, 50293–50300. [DOI] [PubMed] [Google Scholar]
- 39.Batra, R., Geeves, M. A. & Manstein, D. J. (1999) Biochemistry 38, 6126–6134. [DOI] [PubMed] [Google Scholar]
- 40.Kurzawa, S. E., Manstein, D. J. & Geeves, M. A. (1997) Biochemistry 36, 317–323. [DOI] [PubMed] [Google Scholar]
- 41.Sot, B., Banuelos, S., Valpuesta, J. M. & Muga, A. (2003) J. Biol. Chem. 278, 32083–32090. [DOI] [PubMed] [Google Scholar]
- 42.Abbasi, A. & Lutfullah, G. (2002) Biochem. Biophys. Res. Commun. 291, 176–184. [DOI] [PubMed] [Google Scholar]
- 43.Hendsch, Z. S. & Tidor, B. (1994) Protein Sci. 3, 211–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waldburger, C. D., Schildbach, J. F. & Sauer, R. T. (1995) Nat. Struct. Biol. 2, 122–128. [DOI] [PubMed] [Google Scholar]
- 45.Kumar, S. & Nussinov, R. (1999) J. Mol. Biol. 293, 1241–1255. [DOI] [PubMed] [Google Scholar]
- 46.Engh, R. A. & Huber, R. (1991) Acta Crystallogr. A 47, 392–400. [Google Scholar]
- 47.Kleywegt, G. J., Bergfors, T., Senn, H., Le Motte, P., Gsell, B., Shudo, K. & Jones, T. A. (1994) Structure (London) 2, 1241–1258. [DOI] [PubMed] [Google Scholar]
- 48.Luzzati, V. (1952) Acta Crystallogr. 5, 802–810. [Google Scholar]
- 49.Ritchie, M. D., Geeves, M. A., Woodward, S. K. & Manstein, D. J. (1993) Proc. Natl. Acad. Sci. USA 90, 8619–8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Siemankowski, R. F. & White, H. D. (1984) J. Biol. Chem. 259, 5045–5053. [PubMed] [Google Scholar]