Abstract
Poultry are recognized as a main reservoir of Campylobacter spp. However, longitudinal studies investigating the persistence of Campylobacter on commercial meat turkeys are rare. The objectives of this study were to determine the prevalence, antimicrobial susceptibility, and persistence of genotypically related strains of Campylobacter spp. recovered from three commercial turkey farms in Ohio belonging to a single producer. Eight hundred ten samples were collected from birds aged 1 week to slaughter, consisting of 750 fecal droppings and 60 ceca at slaughter. Overall Campylobacter prevalence was 55.9%. Multiplex polymerase chain reaction (PCR) confirmed 72.3% of all isolates as C. coli, 5.3% as C. jejuni, 10.6% as both, and 11.9% as other Campylobacter spp. PCR restriction fragment length polymorphism of the flaA gene subtyping detected 70 types—62 for C. coli and 8 for C. jejuni isolates—with most (80%) of flaA-types constituting farm homogeneous groups. Multilocus sequence typing of 99 selected Campylobacter isolates resulted in 23 sequence types (STs), consisting of 8 STs for C. jejuni and 15 STs for C. coli isolates. Six novel STs—four for C. jejuni and two—for C. coli, were detected. In a subset of isolates (n=98) tested for antimicrobial resistance, the most common resistance was to tetracycline (95%), followed by azithromycin (43%), while 42% and 18% of the isolates were resistant to ciprofloxacin and erythromycin, respectively. All isolates were susceptible to florfenicol. C. coli isolates displayed a higher proportion of resistance than C. jejuni to most antimicrobials. This study highlights the high prevalence, genotypic diversity, and antimicrobial resistance of Campylobacter spp. in commercial turkey from farm to slaughter.
Introduction
Campylobacter is a leading cause of bacterial foodborne diarrheal disease worldwide (Humphrey et al., 2007), with over 1 million cases of campylobacteriosis in the United States annually (Scallan et al., 2011), 48 cases per 100,000 population in European countries (Anonymous, 2012), and affects 5–20% of children in developing regions including Asia, Africa, and Latin America (Oberhelman and Taylor, 2000). Campylobacter spp. in rare cases (1 in 1000) are associated with severe autoimmune-mediated demyelinating neuropathies such as Guillain-Barré and Miller–Fisher syndromes (Skirrow and Blaser, 2000). Antimicrobial therapy is required for severe, prolonged, or systemic infections or to control infections in high-risk groups. Fluoroquinolones (ciprofloxacin), and macrolides (azithromycin and erythromycin) are the antibacterials of choice (Allos and Blaser, 2009). However, antimicrobial use correlates with the emergence of resistant strains (Zhu et al., 2006). Ninety percent of the human Campylobacter infections are attributed to C. jejuni, 6% to C. coli, and the rest are due to C. upsaliensis, C. lari, C. fetus, and C. mucosalis (Friedman et al., 2004). Despite the important public health and socioeconomic impacts of this organism, limited progress has been made in defining routes of infection and reducing associated illness.
Handling and consumption of contaminated raw or undercooked poultry has long been recognized as a major source of human Campylobacter enteritis (Rivoal et al., 2005; Mazick et al., 2006; Lyhs et al., 2010). Though chicken serves as a major source (Horrocks et al., 2009) for human Campylobacter infections, turkey is also an important reservoir of Campylobacter. In recent years there has been a worldwide increase in consumption of turkey products (USDA, 2012). Studies have reported 65–95% prevalence in U.S. turkeys at production (Cox et al., 2000; Luangtongkum et al., 2006; Wesley et al., 2009), with 34.9% from turkey-processing plants (Logue et al., 2003) and 15% in retail turkey meat (Zhao et al., 2010). Considering the significance of these infections, in the United States, mandatory testing of turkey carcasses at slaughter has been instituted by the USDA Food Safety Inspection Services (FSIS, 2010). The current situation prompts the need for strong data on the occurrence and genetic diversity of Campylobacter in the turkey production chain in order to develop effective prevention and control strategies. The purpose of this study was to longitudinally (from farm until slaughter) define the prevalence, genotypes, and antimicrobial resistance of Campylobacter strains isolated from turkeys in order to obtain a better understanding of the epidemiology of the pathogen.
Materials and Methods
Turkey flocks
Between October 2011 and March 2012, three commercial meat turkey farms (A, B, and C) belonging to a single producer were monitored. All farms were located in Ohio, and were within the same climatic zone (within a 10-mile radius from one another). All farms followed similar biosecurity protocols. Farms used an “all in, all out” management system. Each farm had approximately 11,000 turkey males coming from the same hatchery that were reared in 2 barns, each with 5500 turkeys. At the completion of the production cycle (21 weeks), birds from each flock were processed separately in the same processing plant.
Sample collection
Six samplings were made starting with 1-week-old poults to 21-week-old toms. The sample size was calculated according to Fosgate (2009) for a large population size allowing the detection of Campylobacter spp. at a 95% confidence level and considering a within-flock prevalence of 5% or higher. At 1 week of age, 50 fresh fecal droppings from each farm (25/barn) (total of 150 samples/sampling time) were randomly collected from brooder barns. Subsequently, 50 fecal samples were randomly collected in each farm at monthly intervals starting from 4- to 19-week-old birds. At 21 weeks of age, for logistic reasons, 20 viscera from each farm (total 60 samples) were aseptically collected at slaughter from randomly chosen carcasses immediately following evisceration. Samples were placed in sterile polypropylene tubes (fecal droppings) or sterile plastic bags (slaughter materials) and transported to the laboratory on ice and processed within 12 h.
Isolation of thermophilic Campylobacter spp.
Approximately 2 g of feces were suspended with 9 mL of maximum recovery diluent (MRD) (Neogen, USA). One-mL suspension was added to 9 mL of Preston broth containing Campylobacter growth supplements (CM067, SR048, SR117, and SR232; Oxoid, England) and incubated at 42°C for 48 h in a Tri-Gas microaerobic workstation (Microbiology International, USA) (Krause et al., 2006). After incubation, 100 μL of culture was spread onto a modified charcoal cefoperazone deoxycholate agar (mCCDA) plate (CM 0739, Oxoid) containing the selective supplement (SR155E, Oxoid) and incubated for 48 h at 42°C microaerobically (Engberg et al., 2000). Where available, three presumptive Campylobacter colonies from each mCCDA plate were then subcultured onto Muller-Hinton (MH; Difco, MD) agar containing Selective Supplement (SR117, Oxoid) and incubated microaerobically at 42°C for 48 h (Sanad et al., 2011). Pure cultures were stored at −80°C in MH broth supplemented with 30% glycerol (vol/vol) until further identification and characterization. From the slaughter materials, approximately 2 g of cecal homogenates were suspended with 9 mL of MRD and processed for Campylobacter isolation as described above.
DNA extraction and polymerase chain reaction (PCR) amplification
Genomic DNA was extracted using either a Bacterial DNA kit (Epicenter, USA) or by boil-prep method (Dingle et al., 2005). The genomic DNA was quantified using the NanoDrop 1000 (Fisher Scientific, USA) and stored at −20°C until use.
Campylobacter isolates were confirmed by a multiplex PCR as described previously (Yamazaki-Matsune et al., 2007). Primers for simultaneous identification of Campylobacter genus (16S rRNA), C. coli (ceuE), and C. jejuni (mapA) have been previously described (Linton et al., 1997; Denis et al., 1999). C. jejuni 81–176 and C. coli (ATCC 33559) were used as positive controls.
flaA-Restriction fragment length polymorphism (flaA-RFLP) analysis
For flaA-RFLP typing, the PCR product (1.7-kb fragment of the flaA gene) was digested for 5 h at 37°C using DdeI (Promega, USA) (Nachamkin et al., 1996) and the digested products were separated using 4% agarose gel in TAE buffer at 50 V for 5 h at room temperature. The flaA-RFLP profiles were analyzed using BioNumerics V5 (Applied Maths, Belgium). Pairwise comparisons and cluster analysis were made using the Dice correlation coefficient and the unweighted-pair group mathematical average clustering algorithm. The optimization and position tolerance for band analysis were set at 1% and 1.5%, respectively, and a cut-off of 100% was used for the determination of the different flaA-RFLP patterns.
Antimicrobial susceptibility testing
A total of 98 isolates (23 C. jejuni and 75 C. coli) representing different ages, farms, and flaA clusters were tested for antimicrobial susceptibility using Sensititre Campy plates (TREK Diagnostic Systems Inc., USA) as previously described (Sanad et al., 2011, 2013). Minimum inhibitory concentrations (MICs) were determined based on Clinical and Laboratory Standards Institute (CLSI, 2012) breakpoint interpretative criteria for Campylobacteraceae.
Multilocus sequence typing (MLST) analysis
A subset of 99 isolates (19 C. jejuni and 80 C. coli) representing different flaA subtypes, farm source, and ages of turkeys within the farms were analyzed by MLST as described (Dingle et al., 2001; Sanad et al., 2011). PCR products were purified using ExoSAP (Affymetrix Inc., USA) prior to sequencing at GENEWIZ DNA sequencing facility (http://www.genowiz.com). Allele numbers and sequence types (STs) were then generated by comparing the sequences to the Campylobacter MLST database (http://pubmlst.org/Campylobacter).
Statistical analysis
The chi-square test was used to analyze the significance of the difference in prevalence of C. jejuni compared to C. coli between farms, between age groups within the farms, and in antimicrobial resistance between farms. P<0.05 was considered statistically significant for univariate comparisons. In cases of multiple comparisons, the conservative Bonferroni corrected p-value was used as a cut-off for significance.
Simpson's index of diversity was calculated to compare the discriminatory power of flaA and MLST genotyping (Hunter and Gaston, 1988).
The evolutionary relationships of the C. jejuni and C. coli STs were elaborated using minimum spanning tree (Bionumerics V5). Correlations among flaA, MLST, and antimicrobial resistance patterns (R-types) as well as relationships of these profiles to age and farm were examined for each Campylobacter species using cross-tabulation, Fisher's exact test, uncertainty coefficients, and chi-square tests in IBM SPSS statistical software v9 (SPSS Inc., 2010).
Results
Occurrence and distribution of Campylobacter spp. in turkey farms
The overall Campylobacter prevalence was 55.9% (453/810), with 56% (420/750) in turkey feces and 55% (33/60) from cecal contents (Table 1). PCR revealed that the vast majority was C. coli (72.1%; 327/453), whereas 5.3% (24/453) were C. jejuni. In addition, we also identified 10.6% (48/453) isolates that were positive for both ceuE and mapA PCR, and 11.9% (54/453) of isolates were Campylobacter spp., other than C. jejuni or C. coli. The frequency of Campylobacter isolation varied by farm and by age within the farm (Table 1). There was no significant difference in Campylobacter prevalence between farm B and C (p>0.05), but farm A had a significantly higher overall Campylobacter prevalence (p<0.05) than farms B and C. Differences in Campylobacter spp. prevalence were significant between weeks 1 and 4 (p=0.00001), and weeks 19 and 21 (p=0.002) on farm A. Difference in Campylbacter spp. prevalence on farm C varied with age between weeks 1 and 4 (p=0.0317), weeks 4 and 9 (p=0.0110), and between weeks 9 and 13 (p=0.0029). Though these p-values suggest significant differences for all ages, when the stricter Bonferroni-corrected p-value cut-off for significance (0.004) was used, only the weeks 9 and 13 difference was significant with this sample size for farms B and C.
Table 1.
Prevalence of Campylobacter Species Isolated from Turkey Droppings and Viscera That Were Collected from Three Turkey Farms Located in Northwest Ohio, United States
| Farm ID | Age of turkeys (wks) | Total isolates no. (%) | C. coli no. (%) | C. jejuni no. (%) | C. coli/C. jejuni no. (%) | Other Campy. spp no. (%) |
|---|---|---|---|---|---|---|
| A | 1 | 15 (30)a | 12 (24) | 0 | 1 (2) | 2 (4) |
| 4 | 44 (88)b | 27 (54) | 8 (16) | 8 (16) | 1 (2) | |
| 9 | 39 (78)b | 26 (52) | 6 (12) | 4 (8) | 3 (6) | |
| 13 | 42 (84)b | 26 (52) | 5 (10) | 9 (18) | 2 (4) | |
| 19 | 40 (80)b | 31 (62) | 0 | 0 | 9 (18) | |
| 21 | 7 (35)c | 7 (35) | 0 | 0 | 0 | |
| Subtotal | 187 (69.23) | 129 (47.78) | 19 (7.04) | 22 (8.15) | 17 (6.30) | |
| B | 1 | 6 (12)a | 2 (4) | 0 | 0 | 4 (8) |
| 4 | 6 (12)a | 5 (10) | 1 (2) | 0 | 0 | |
| 9 | 14 (28)a | 6 (12) | 1 (2) | 4 (8) | 3 (6) | |
| 13 | 42 (84)b | 24 (48) | 0 | 15 (30) | 3 (6) | |
| 19 | 41 (82)b | 40 (80) | 0 | 0 | 1 (2) | |
| 21 | 12 (60)b | 12 (60) | 0 | 0 | 0 | |
| Subtotal | 121 (44.8) | 89 (32.96) | 2 (0.74) | 19 (7.04) | 11 (4.07) | |
| C | 1 | 2 (4)a | 0 | 0 | 0 | 2 (4) |
| 4 | 9 (18)b | 8 (16) | 0 | 0 | 1 (2) | |
| 9 | 23 (46)c | 19 (36) | 0 | 0 | 4 (4) | |
| 13 | 47 (94)d | 41 (82) | 0 | 3 (6) | 3 (6) | |
| 19 | 50 (100)d | 30 (60) | 3 (6) | 4 (8) | 13 (26) | |
| 21 | 14 (70)d | 11 (55) | 0 | 0 | 3 (15) | |
| Subtotal | 145 (53.70) | 109 (40.37) | 3 (1.11) | 7 (2.59) | 26 (9.63) | |
| Overall prevalence | 1 | 23 (15.3) | 14 (9.3) | 0 | 1 (0.7) | 8 (5.3) |
| 4 | 59 (39.3) | 40 (26.7) | 9 (6.0) | 8 (5.3) | 2 (1.3) | |
| 9 | 76 (50.7) | 51 (34.0) | 7 (4.7) | 8 (5.3) | 10 (6.7) | |
| 13 | 131 (86.7) | 91 (60.7) | 5 (3.3) | 27 (18.0) | 8 (5.3) | |
| 19 | 131 (87.3) | 101 (67.3) | 3 (2.0) | 4 (2.7) | 23 (15.3) | |
| 21 | 33 (55.0) | 30 (60.0) | 0 | 0 | 3 (5.0) | |
| Grand total | 453 (55.9) | 327 (40.37) | 24 (2.96) | 48 (5.92) | 54 (6.67) |
A total of 50 fresh fecal droppings were collected from each farm in the five samplings (week 1, 4, 9, 13, and 19) and 20 viscera/ceca were collected at slaughtering plant from each farm.
For each farm prevalence, numbers in the same column with different letters in the superscript were significantly different (p<0.05), while numbers with the same letters did not differ significantly (chi-square test and Fisher's exact two-tailed test).
At week 1, farm A had higher prevalence (30%) than farms B (12%) and C (4%). Prevalence increased rapidly in farm A to 88% by week 4. In farm B, prevalence was highest (84%) by week 13, while in farm C the prevalence was highest (100%) by week 19. The overall prevalence of Campylobacter spp. declined to 55% at slaughter for all farms combined.
flaA-RFLP polymorphism
The flaA typing was successfully performed on 350/351 (99.7%) isolates (C. jejuni [n=23] and C. coli [n=327]). A total of 7 different banding profiles (∼35–1110 bps) were obtained. Using a similarity cut-off value of 100%, 70 main types were identified (62 for C. coli and 8 for C. jejuni) (Figs. 1 and 2). Overall, flaA types were associated with individual farms (Fisher's exact p-value<0.000; uncertainty coefficient=0.353). This result was true for both C. coli (p-value<0.000) and C. jejuni but slightly less significantly (p-value=0.057), which is reasonable given the differences in sample size. Uncertainty coefficients suggest that flaA type is a good indicator of which farm a sample originated from and predicts between 70% (coefficient=0.709 for C. jejuni) and 80% (coefficient=0.809 for C. coli) of variability in farms.
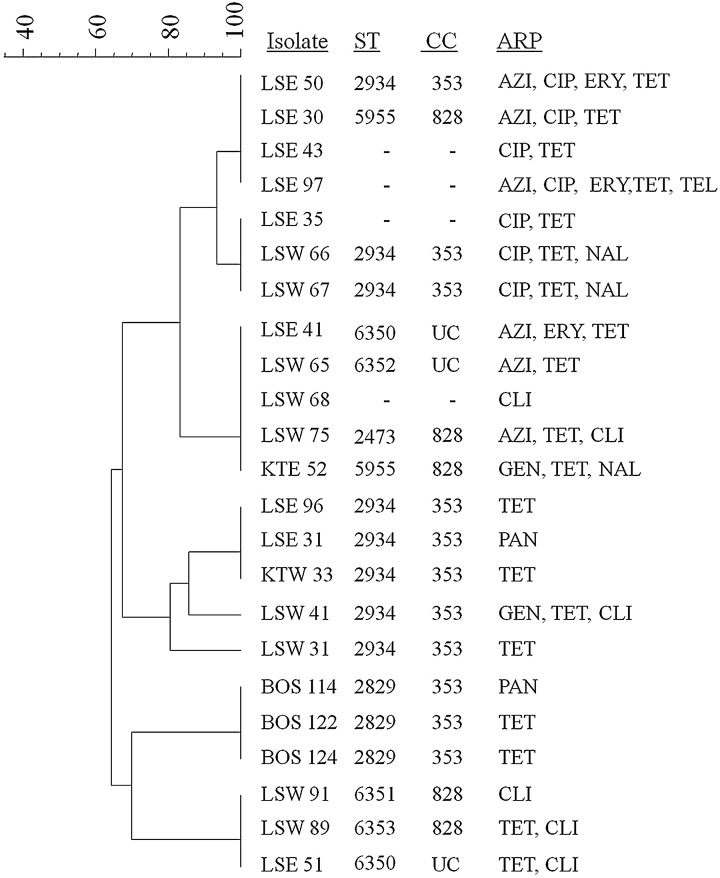
FIG. 1.
Dendrogram showing the relationship of flaA restriction fragment length polymorphism (flaA-RFLP) types of Campylobacter jejuni isolates from turkeys. Similarity and clustering analysis of the polymerase chain reaction–RFLP patterns were performed using the Dice coefficient with unweighted pair-group method with arithmetic averages with optimization of 1% and position tolerance of 1.5%. Multilocus ST and antimicrobial resistance patterns (ARP) of C. jejuni isolates were combined to the tree. UC, undefined clonal complexes; ST, sequence type; CC, clonal complex; ARP, antimicrobial resistance pattern; LSW/LSE, Farm A; KTW/KTE, Farm B; BOS, Farm C; AZI, azithromycin; CIP, ciprofloxacin; ERY, erythromycin; TET, tetracycline; TEL, telithromycin; PAN, pansusceptible; CLI, clindamycin; GEN, gentamicin; NAL, nalidixic acid.
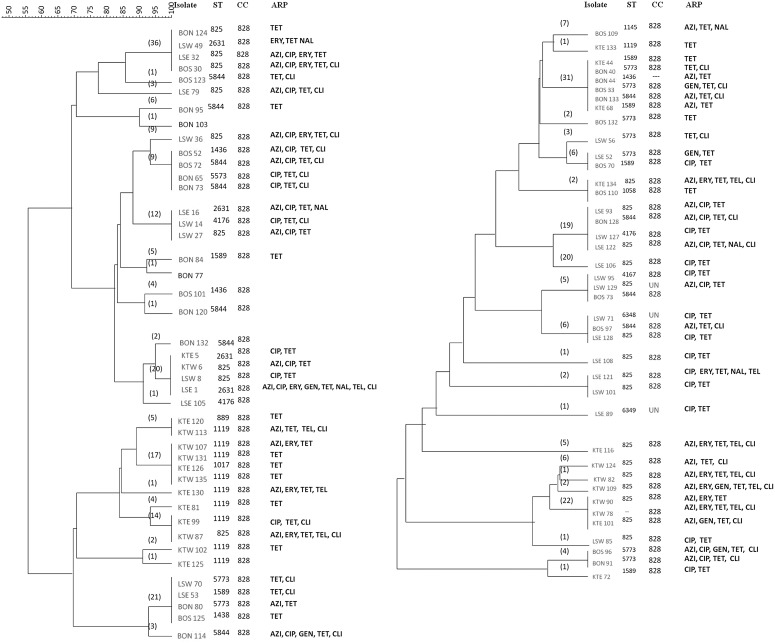
FIG. 2.
Dendrogram showing the relationship of flaA restriction fragment length polymorphism types of Campylobacter coli isolates from turkeys. The cluster analysis was performed as described in Figure 1. Numbers in parentheses denote total number of isolates in each cluster. UC, undefined clonal complexes; ST, sequence type; CC, clonal complex; ARP, antimicrobial resistance pattern; LSW/LSE- Farm A; KTW/KTE- Farm B; BOS/BON-Farm C; NAL, nalidixic acid. For other abbreviations, see Figure 1 legend.
MLST of C. jejuni and C. coli isolates
C. jejuni isolates showed high genetic diversity by MLST analysis (Table 2). A total of 8 different STs were found among 19 C. jejuni isolates, of which 4 (STs 6350, 6351, 6352, and 6353) were identified as novel. ST 2934 was the most predominant, detected in 42.1% (8 of 19) of the isolates. This ST was commonly identified from turkeys of different ages. Six STs were assigned to 2 previously described clonal complexes (CC 353, 11 isolates; and CC 828, 5 isolates), while 2 new STs belonged to undefined CC. Overall, the findings imply the occurrence of spatially diverse strains, while at the same time some clonal strains appear to be limited to specific farms.
Table 2.
Sequence Types and ST-Complexes (ST-CC) of Campylobacter jejuni and C. coli Strains Isolated from Turkeys
| Farms | ||||||
|---|---|---|---|---|---|---|
| ST | ST-CC | No. of isolates | A | B | C | Resistance profiles |
| C. jejuni | ||||||
| 2829 | 353 | 3 | 0 | 0 | 3 | Pan, T |
| 2934 | 353 | 8 | 7 | 1 | 0 | Pan, T, CTNa, ACET |
| 6350 | UC | 2 | 2 | 0 | 0 | AET, TCl |
| 2473 | 828 | 1 | 1 | 0 | 0 | ATCl |
| 5955 | 828 | 2 | 1 | 1 | 0 | ACT, GTNa |
| 6351 | 828 | 1 | 1 | 0 | 0 | Cl |
| 6352 | UC | 1 | 1 | 0 | 0 | ATCl |
| 6353 | 828 | 1 | 1 | 0 | 0 | TCl |
| C. coli | ||||||
| 825 | 828 | 24 | 14 | 9 | 1 | ACT, CT, ACET, ACTCl, ACTNaCl, CETTeCl, AEGTTeCl, AETTeCl |
| 889 | 828 | 1 | 0 | 1 | 0 | T |
| 1017 | 828 | 1 | 0 | 1 | 0 | T |
| 1058 | 828 | 1 | 0 | 0 | 1 | T |
| 1119 | 828 | 10 | 0 | 10 | 0 | T, ATTeCl, AET,CTCl, AETTe |
| 1145 | 828 | 1 | 0 | 0 | 1 | ATNa, |
| 1436 | 828 | 3 | 0 | 0 | 3 | AT, ACTCl |
| 1438 | 828 | 1 | 0 | 0 | 1 | T |
| 1589 | 828 | 6 | 1 | 3 | 2 | T, TCl, CT, AT |
| 2631 | 828 | 4 | 3 | 1 | 0 | CT, ETNa, ETNa, ACEGTNaTeCl |
| 4176 | 828 | 3 | 3 | 0 | 0 | CT, CTCl |
| 5773 | 828 | 10 | 3 | 0 | 7 | T, TCl, AGT, CTCl, GTCl, ACTCl, ACGTCl |
| 5844 | 828 | 12 | 0 | 0 | 12 | T, TCl, ATCl, CTCl, ACTCl, ACGTCl |
| 6348 | 828 | 2 | 2 | 0 | 0 | CTCl, CT |
| 6349 | 828 | 1 | 1 | 0 | 0 | CT |
A, azithromycin; C, ciprofloxacin; Cl, clindamycin; E, erythromycin; G, gentamicin; Na, nalidixic acid; T, tetracycline; Te, telithromycin; Pan, pansusceptible; UC, undefined clonal complex.
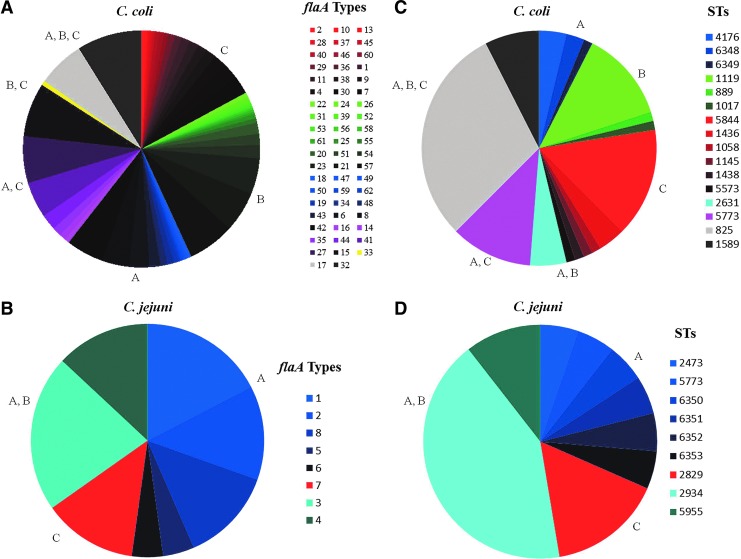
Among 80 C. coli isolates, 15 unique STs were identified. Three isolates assigned to 2 new STs (STs 6348 and 6349). The most common ST was ST 825, which was represented by 24 isolates, with 6 STs occurring only once in the data set. The four most predominant STs (STs 825, 5844, 1119, and 5773) represented more than half of the isolates (56/80 isolates; 70%). All C. coli isolates were assigned to a single previously described CC 828 (Table 2). The discriminatory power of flaA typing (DI=0.78) was greater than MLST (DI=0.51). However, flaA typing did not allow for the clear molecular grouping of the C. jejuni and C. coli strains. The frequency of STs and flaA types are shown in Figure 3.
FIG. 3.
Frequency of flaA types (A, B) and sequence types (C, D) in isolates of Campylobacter coli (n=327) (A, C) and C. jejuni (n=19) (B, D) collected from commercial turkeys. Farm of origin for the isolate is indicated by color: blue (A), green (B), red (C), and mixtures of colors for groups containing isolates from different farms. STs, sequence type. Color images available online at www.liebertpub.com/fpd
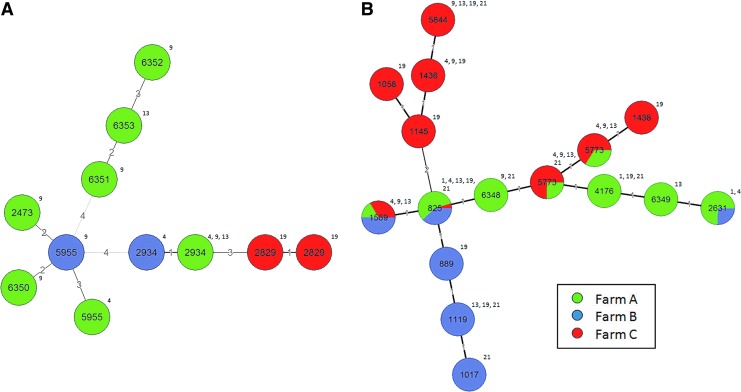
Phylogenetic analysis of C. jejuni and C. coli STs
The evolutionary relationships of the 8 C. jejuni and 15 C. coli STs are represented in 2 minimum spanning trees (Fig. 4). C. jejuni isolates tend to be more clustered by farm, with very limited clonality between farms. Among eight unique STs, five were identified from farm A only, while a single ST was identified from farm C and two STs were common to farms A and B. The majority of the C. jejuni STs were also limited to one age group except ST 2934, which was identified persistently from three different age groups (4, 9, and 13 weeks). On the other hand, for the C. coli, STs showed much more diversity. Though most of the STs were identified from multiple age groups, the STs were found to have multilocus variations, strengthening the highly diverse nature of the C. coli population in turkeys. The founding genotype, ST 828, was identified from all three farms as well as all five age groups.
FIG. 4.
BioNumerics Minimum Spanning Tree for Campylobacter jejuni (A) and C. coli (B). Color codes represent the farm origins of the isolates. The sequence types are numbers in the circles and the numbers outside the circles represent the age of the animals in weeks. The branch length is proportional to the locus variations and as also indicted in numbers on each branch. Color images available online at www.liebertpub.com/fpd
Antimicrobial resistance profiles (ARP) of C. jejuni and C. coli isolates
Campylobacter isolates displayed resistance most frequently to tetracycline (TET) (95%) and all isolates tested were susceptible to florfenicol (FFN) (Table 3). C. coli displayed significantly greater resistance (p=0.0346) than C. jejuni to TET regardless of the source of isolation. The occurrence of the multiple drug resistance isolates varied among C. coli and C. jejuni (Table 3). Approximately 13% (3/23) of C. jejuni isolates were resistant to 4 or more antimicrobials. In contrast, 30.6% (23/75) of C. coli isolates were resistant to 4 or more antimicrobials. A wider range of MICs was observed for some of the antimicrobials, mainly among C. coli isolates (Table 3).
Table 3.
Antimicrobials Resistance and Minimum Inhibitory Concentration (MIC) Distribution of Campylobacter jejuni and C. coli Isolated from Turkeys
| Number of resistant isolates (%) and range of MICs in μg/mL | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Isolates | AZI | CIP | ERY | GEN | TET | FFN | NAL | TEL | CLI | |
| C. jejuni | Total | 6 (26)a | 7 (30.4)a | 3 (13)a | 2 (8.7)a | 19 (82.6)a | 0 | 3 (13.0)a | 1 (4.3)a | 7 (30.4)a |
| (n=23) | 64* | 8–16 | 64 | 8–16 | 32–64 | 64 | 8 | 16 | ||
| Farm A | 5(29.4) | 7 (38.9) | 3 (16.7) | 1 (5.6) | 14 (82.3) | 0 | 1 (5.8) | 1 (5.6) | 6 (35.2) | |
| (n=17) | 64 | 8–16 | 64 | 8 | 32–64 | 64 | 8 | 16 | ||
| Farm B | 1 (33.3) | 0 | 0 | 1 (33.3) | 3 (100) | 0 | 2 (66.7) | 0 | 1 (33.3) | |
| (n=3) | 64* | 16 | 32–64 | 64 | 16 | |||||
| Farm C | 0 | 0 | 0 | 0 | 2 (66.7) | 0 | 0 | 0 | 0 | |
| (n=3) | 64 | |||||||||
| C. coli | Total | 36 (48)a | 34 (45.3)a | 15 (20)a | 7 (9.3)a | 74 (100.0)b | 0 | 6 (8)a | 10 (13.3)a | 33 (44)a |
| (n=75) | 8–64 | 8–32 | 32–64 | 8–32 | 32–64 | 64 | 8 | 8–16 | ||
| Farm A | 10 (38.5)c | 21 (80.8)c | 5 (19.2)c | 2 (7.7)c | 26 (100.0)c | 0 | 5 (19.2)c | 2 (7.7)c | 9 (34.6)c | |
| (n=26) | 8–64 | 8–32 | 32–64 | 16–32 | 32–64 | 64 | 8 | 8–16 | ||
| Farm B | 14 (56.0)c | 4 (16.0)d | 9 (36.0)d | 2 (8.0)c | 25 (100.0)c | 0 | 0d | 8 (32.0)d | 10 (40.0)c | |
| (n=25) | 8–64 | 8–32 | 64 | 8–16 | 32–64 | 8 | 8–16 | |||
| Farm C | 12 (50.0)c | 9 (37.5)e | 1 (4.2)c | 3 (12.5)c | 23 (95.8)c | 0 | 1 (4.2)c | 0c | 14 (58.3)c | |
| (n=24) | 8–64 | 8–32 | 64 | 16 | 32–64 | 64 | 8–16 | |||
MIC ranges of antimicrobials for the resistant isolates.
For each antimicrobial, numbers in the same row with different superscript letters were significantly different (p<0.05), while numbers with the same letters did not differ significantly (chi-square test and Fisher's exact two-tailed test).
Resistance breakpoints: AZI, ≥8 μg/mL; CIP, ≥4 μg/mL; CLI, ≥8 μg/mL; ERY, ≥32 μg/mL; GEN, ≥8 μg/mL; NAL, ≥64 μg/mL; TEL, ≥8 μg/mL; FEN, ≥8 μg/mL; TET, ≥16 μg/mL.
AZI, azithromycin; CIP, ciprofloxacin; CLI, clindamycin; ERY, erythromycin; FFN, florfenicol; GEN, gentamicin; NAL, nalidixic acid; TEL, telithromycin; TET, tetracycline.
Approximately 26% and 30% of C. jejuni strains were resistant to azithromycin (AZI) and ciprofloxacin (CIP), respectively. Similarly, 48% and 45% of C. coli strains were resistant to AZI and CIP, respectively (the difference between the C. coli versus C. jejuni was not significant, Fisher's exact p-value=0.516). In contrast, the majority of both C. jejuni and C. coli strains displayed diversity in resistance within the macrolides group of antimicrobials (AZI, erythromycin [ERY], and telithromycin [TEL]) (Table 3). While about 4% and 13% of C. jejuni and C. coli, respectively, were resistant to TEL (the difference between C. coli versus C. jejuni was not significant, Fisher's exact p-value=0.201), 26% and 13% of C. jejuni strains were resistant to AZI and ERY and about 48% and 20% C. coli strains were resistant to AZI and ERY, respectively (p-values for difference between species 0.197 and 0.516, respectively).
Association of flaA types and STs with antimicrobial resistance pattern (ARP)
Some flaA types for C. coli were more likely to show antibiotic resistance in general (uncertainty coefficient=1.00; Fisher's exact p-value=0.003) and specifically for CIP (uncertainty coefficient=0.80; Fisher's exact p-value<0.000), TET (uncertainty coefficient=1.00; Fisher's exact p-value=0.003), and ERY (uncertainty coefficient=0.728; Fisher's exact p-value=0.049). The relationships between TET and ERY resistance and flaA type were not significant for C. jejuni, but the relationship with CIP resistance was significant (uncertainty coefficient=1.00; Fisher's exact p-value=0.003).
Several C. coli STs showed a good correlation with their antimicrobial resistance profiles (Table 2 and Fig. 2). For instance, the majority of C. coli isolates (18/24; 75%) in ST 825 were resistant to multiple antimicrobials, with the majority (62.5%) of isolates being resistant to CIP. Almost all isolates in ST 1589 (n=6) were resistant to either 1 or 2 antimicrobials, while isolates in ST 5844 showed diverse resistance profiles to a majority of antimicrobials tested, with most isolates (73%) being resistant to clindamycin. All C. coli STs from all farms were pansusceptible to FFN and resistant to TET. In general, STs were significantly associated with ARP (Fisher's exact p-value=0.009) for C. coli, but they accounted for a relatively small portion of the variability in resistance (uncertainty coefficient=0.344).
In general, C. jejuni isolates showed higher antimicrobial resistance (R-type) diversity than C. coli isolates, which is consistent with the MLST findings. With exception of ST 2829, which contained isolates resistant to either one antimicrobial agent or pansusceptible isolates, the majority of STs contained isolates with different resistance profiles (Table 2 and Fig. 1). For example, ST 2934 contained isolates that were pansusceptible, resistant to one, three, and more than three antimicrobials.
Discussion
In the present study, Campylobacter was recovered from the feces starting from 1-week-old birds (14.7%) and continuing to slaughter age (58.3%). The observed timing of colonization of Campylobacter in this study is earlier than what was found in previous reports. Smith et al. (2004) reported that sibling turkey flocks started to be colonized by Campylobacter spp. between weeks 2 and 3 and remained colonized until processing. Similarly, El-Adawy et al. (2012) showed that poults were colonized by thermophilic Campylobacter spp. on week 2. All 3 flocks in our study remained positive throughout, with the highest prevalence after week 13. This confirms the potential rapid bird-to-bird transmission of Campylobacter spp. in commercial meat poultry farms as indicated previously (Newell et al., 2003; Horrocks et al., 2009).
In this study, C. coli was frequently recovered from the turkey fecal droppings. C. coli was also frequently identified previously in turkey flocks in the United States (Logue et al., 2003; Smith et al., 2004; Wesley et al., 2005). Proportional higher prevalence of C. coli than C. jejuni could be due to host adaptation. Resistance to certain antibacterials is reported to be significantly more common in C. coli than in C. jejuni strains (Aarestrup et al., 1997; Ge et al., 2003; Ladely et al., 2007), which could select for one species versus the other. However, studies from both conventional and organic turkey farms in Ohio reported a high prevalence of C. coli (Luangtongkum et al., 2006), so other environmental factors may also contribute to the observed differences.
Eight and 62 different flaA PCR-RFLP profiles were identified for C. jejuni and C. coli, respectively. These results are in line with the previous studies in chickens (Behringer et al., 2011; Miller et al., 2010); however, fewer flaA-types (i.e., 6 for C. jejuni [n=35] and 14 for C. coli [n=65]) isolated from processed turkeys were reported (Lutgen et al., 2009). These discrepancies may be related to animal sources; environmental, management, and husbandry conditions of flocks; primers used; as well as geographic diversity in the distribution of C. jejuni and C. coli isolates.
Of the eight C. jejuni STs, only ST 2934 was previously reported in turkeys (Gu et al., 2009). Therefore, further studies are needed to assess possible host associations of this ST with turkeys. The ST 353 complex, the most common complex seen in C. jejuni strains in this study, has been reported previously in poultry (Manning et al., 2003), and is associated with human gastroenteritis (Dingle et al., 2002; Duim et al., 2003; Manning et al., 2003). Indeed, the ST 353 complex contains a majority of the isolates obtained from human disease. Furthermore, the ST 828 complex, the main group identified in this study for C. coli and some C. jejuni, is associated with strains that are mainly isolated from agricultural and environmental sources, and some from human clinical cases (Sheppard et al., 2010). Also, other researchers have reported the presence of progenitor strains of ST 828 complex in humans, swine, and cattle from different parts of the world (Dingle et al., 2005; Miller et al., 2006). This indicates that ST 828 has potential to infect both humans and animals.
The majority of the isolates (n=77) were resistant to 2 or more antimicrobials, and C. coli displayed significantly more (p=0.0346) resistance than C. jejuni to TET. High occurrences of multidrug-resistant Campylobacter isolated from turkeys have also been reported by other researchers. Lutgen et al. (2009) examined 801 Campylobacter isolates from processed turkey in Midwestern United States and found that C. coli were more resistant to CIP (63%) than C. jejuni (28%), and a subset (n=100) of isolates were resistant to TET (100%) and nalidixic acid (49%). High prevalence of TET resistance in Campylobacter isolates from chickens and turkeys has also been reported (Luangtongkum et al., 2006; Anderson et al., 2006; Gu et al., 2009; Zhao et al., 2010). Studies have also shown that TET resistance is typically transferable (Taylor et al., 1981; Avrain et al., 2004). Since TETs have been used as feed additives for livestock and poultry for both therapeutic and subtherapeutic purposes for a long time (Chopra and Roberts, 2001; Fallon et al., 2003), it is possible that Campylobacter may have become resistant to this class of antimicrobials through selective pressure.
Conclusions
The present study found a high prevalence of Campylobacter spp. in commercial turkey farms and at slaughter, suggesting that on farms high prevalence can be a high risk for carcass contamination. The majority of the Campylobacter isolates examined were resistant to multiple antimicrobials, including ERY and CIP. We observed that turkeys can be colonized by Campylobacter as early as the first week of introduction to the barns; thus, further studies aimed at identifying the sources and vehicles for Campylobacter spp. in turkey farms are needed.
Acknowledgments
We thank Zhe Liu, Mary Drozd, Kshipra Chandrashekhar, Serpil Baspinar, Gokben Ozbey, and Ahmed Hikal for assistance with sample collection and processing. Dr. Kashoma is supported by a National Institutes of Health Fogarty fellowship (D43TW008650-01). Dr. Rajashekara's laboratory is supported by the funds from Ohio Agricultural Research and Development Center (OARDC), and the Agriculture and Food Research Initiative (AFRI) grant# 2012-68003-19679, U.S. Department of Agriculture.
Disclosure Statement
No competing financial interests exist.
References
- Aarestrup FM, Nielsen EM, Madsen M, Engberg J. Antimicrobial susceptibility patterns of thermophilic Campylobacter spp. from humans, pigs, cattle, and broilers in Denmark. Antimicrob Agents Chemother 1997;41:2244–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allos BM, Blaser MJ. Campylobacter jejuni and related species. In: Principles and Practice of Infectious Diseases, 7th Edition. Mandell GL, Bennett JE, Dolin R. (eds.). Philadelphia: Elsevier Publishing Co., 2009, pp. 2793–2802 [Google Scholar]
- Andersen SR, Saadbye P, Shukri NM, Rosenquist H, Nielsen NL, Boel J. Antimicrobial resistance among Campylobacter jejuni isolated from raw poultry meat at retail level in Denmark. Int J Food Microbiol 2006;107:250–255 [DOI] [PubMed] [Google Scholar]
- Anonymous. The European Union Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and Food-borne Outbreaks in 2010. EFSA J 2012;10:2597 [Google Scholar]
- Avrain L, Vernozy-Rozand C, Kempf I. Evidence for natural horizontal transfer of tetO gene between Campylobacter jejuni strains in chickens. J Appl Microbiol 2004;97:134–140 [DOI] [PubMed] [Google Scholar]
- Behringer M, Miller WG, Oyarzabal OA. Typing of Campylobacter jejuni and Campylobacter coli isolated from live broilers and retail broiler meat by flaA-RFLP, MLST, PFGE and REP-PCR. J Microbiol Method 2011;84:194–201 [DOI] [PubMed] [Google Scholar]
- Chopra I, Roberts M. Tetracycline antimicrobials: Mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev 2001;65:232–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [CLSI] Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Second Informational Supplement. CLSI Document M11-S22. Wayne, PA: CLSI, 2012 [Google Scholar]
- Cox NA, Stern NJ, Craven SE, Berrang ME, Musgrove MT. Prevalence of Campylobacter and Salmonella in the cecal droppings of turkeys during production. J Appl Poult Res 2000;9:542–545 [Google Scholar]
- Denis M, Soumet C, Rivoal K, Ermel G, Blivet D, Salvat G, Colin P. Development of a m-PCR for simultaneous identification of Campylobacter jejuni and C. coli. Lett Appl Microbiol 1999;29:406–410 [DOI] [PubMed] [Google Scholar]
- Dingle KE, Colles FM, Wareing DRA, Ure R, Fox AJ. Multilocus sequence typing system for Campylobacter jejuni. J Clin Microbiol 2001;39:14–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingle KE, Colles FM, Ure R, Wagenaar JA, Duim B, Bolton FJ, Fox AJ, Wareing DRA, Maiden MCJ. Molecular characterization of Campylobacter jejuni clones: A basis for epidemiologic investigation. Emerg Infect Dis 2002;8:949–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingle KE, Colles FM, Falush D, Maiden MCJ. Sequence typing and comparison of population biology of Campylobacter coli and Campylobacter jejuni. J Clin Microbiol 2005;43:340–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duim B, Godschalk PCR, Braak NVD, Dingle KE, Dijkstra JR, Leyde E, Plas JVD, Colles FM, Endtz HP, Wagenaar JA, Maiden MCJ, Belkum AV. Molecular evidence for dissemination of unique Campylobacter jejuni clones in Curaçao, Netherlands Antilles. J Clin Microbiol 2003;41:5593–5597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Adawy H, Totzel H, Tomaso T, Neubauer H, Hafez HM. Elucidation of colonization time and prevalence of thermophilic Campylobacter species during turkey rearing using multiplex polymerase chain reaction. Poult Sci 2012;91:454–459 [DOI] [PubMed] [Google Scholar]
- Engberg J, On SLW, Harrington CS, Gerner-Smidt P. Prevalence of Campylobacter, Arcobacter, Helicobacter, and Sutterella spp. in human fecal samples as estimated by a reevaluation of isolation methods for campylobacters. J Clin Microbiol 2000;38:286–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallon R, O'Sullivan N, Maher M, Carroll C. Antimicrobial resistance of Campylobacter jejuni and Campylobacter coli isolates from broiler chickens isolated at an Irish poultry processing plant. Lett Appl Microbiol 2003;36:277–281 [DOI] [PubMed] [Google Scholar]
- [FSIS] Food Safety and Inspection Service. New performance standards for Salmonella and Campylobacter in young chicken and turkey slaughter establishments; new compliance guides. Fed Register 2010;75:27288–27294 [Google Scholar]
- Fosgate GT. Practical sample size calculations for surveillance and diagnostic investigations. J Vet Diagn Invest 2009;21:3–14 [DOI] [PubMed] [Google Scholar]
- Friedman CR, Hoekstra RM, Samuel M, Marcus R, Bende J, Shiferaw B, Reddy S, Ahuja SD, Helfrick DL, Hardnett F, Carter M, Anderson B, Tauxe RV. Risk factors for sporadic Campylobacter infection in the United States: A case-control study in FoodNet sites. Clin Infect Dis 2004;38:S285–S296 [DOI] [PubMed] [Google Scholar]
- Ge B, White DG, McDermott PF, Girard W, Zhao S, Hubert S, Meng J. Antimicrobial-resistant Campylobacter species from retail raw meats. Appl Environ Microbiol 2003;69:3005–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Siletzky RM, Wright S, Islam M, Kathariou S. Antimicrobial susceptibility profiles and strain type diversity of Campylobacter jejuni isolates from turkeys in Eastern North Carolina. Appl Environ Microbiol 2009;75:474–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horrocks SM, Anderson RC, Nisbet DJ, Ricke SC. Incidence and ecology of Campylobacter jejuni and Campylobacter coli in animals. Anaerobe 2009;15:18–25 [DOI] [PubMed] [Google Scholar]
- Humphrey T, O'Brien S, Madsen M. Campylobacters as zoonotic pathogens: A food production perspective. Int J Food Microbiol 2007;117:237–257 [DOI] [PubMed] [Google Scholar]
- Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: An application of Simpson's Index of diversity. J Clin Microbiol 1988;26:2465–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M, Josefsen MH, Lund M, Jacobsen NR, Brorsen L, Moos M, Stockmarr A, Hoorfar J. Comparative, collaborative, and on-site validation of a TaqMan PCR method as a tool for certified production of fresh, Campylobacter-free chickens. Appl Environ Microbiol 2006;72:5463–5468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladely SR, Harrison MA, Fedorka-Cray PJ, Berrang ME, Englen MD, Meinersmann RJ. Development of macrolide-resistant Campylobacter in broilers administered subtherapeutic or therapeutic concentrations of tylosin. J Food Prot 2007;70:1945–1951 [DOI] [PubMed] [Google Scholar]
- Linton D, Lawson AJ, Owen RJ, Stanley J. PCR detection, identification to species level, and fingerprinting of Campylobacter jejuni and Campylobacter coli direct from diarrheic samples. J Clin Microbiol 1997;35:2568–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logue CM, Sherwood JS, Elijah LM, Olah PA, Dockter MR. The incidence of Campylobacter spp. on processed turkey from processing plants in the midwestern United States. J Appl Microbiol 2003;95:234–241 [DOI] [PubMed] [Google Scholar]
- Luangtongkum T, Morishita TY, Ison AJ, Huang S, McDermott PF, Zhang Q. Effect of conventional and organic production practices on the prevalence and antimicrobial resistance of Campylobacter spp. in poultry. Appl Environ Microbiol 2006;72:3600–3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutgen EM, McEvoy JM, Sherwood JS, Logue CM. Antimicrobial resistance profiling and molecular subtyping of Campylobacter spp. from processed turkey. BMC Microbiol 2009;9:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyhs U, Katzav M, Isohanni P, Heiska H, Maijala R. The temporal, PFGE and resistance pattern associations suggest that poultry products are only a minor source of human infections in western Finland. Food Microbiol 2010;27:311–315 [DOI] [PubMed] [Google Scholar]
- Manning G, Dowson CG, Bagnall MC, Ahmed IH, West M, Newell DG. Multilocus sequence typing for comparison of veterinary and human isolates of Campylobacter jejuni. Appl Environ Microbiol 2003;69:6370–6379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazick A, Ethelberg S, Nielsen EM, Mølbak K, Lisby M. An outbreak of Campylobacter jejuni associated with consumption of chicken, Copenhagen, 2005. Euro Surveill 2006;11:137–139 [PubMed] [Google Scholar]
- Miller RS, Miller WG, Behringer M, Hariharan H, Matthew V, Oyarzabal OA. DNA identification and characterization of Campylobacter jejuni and Campylobacter coli isolated from caecal samples of chickens in Grenada. J Appl Microbiol 2010;108:1041–1049 [DOI] [PubMed] [Google Scholar]
- Miller WG, Englen MD, Kathariou S, Wesley IV, Wang G, Pittenger-Alley L, Siletz RM, Muraoka W, Federka-Cray PJ, Mandrell RE. Identification of host-associated alleles by multilocus sequence typing of Campylobacter coli strains from food animals. Microbiol 2006;152:245–255 [DOI] [PubMed] [Google Scholar]
- Nachamkin I, Ung H, Patton CM. Analysis of HL and O serotypes of Campylobacter strains by the Flagellin gene typing system. J Clin Microbiol 1996;34:277–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell DG, Fearnley C. Sources of Campylobacter colonization in broiler chickens. Appl Environ Microbiol 2003;69:4343–4351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhelman RA, Taylor DN. Campylobacter infections in developing countries. In: Campylobacter, 2nd ed. Nachamkin I, Blaser MJ. (eds.). Washington: American Society for Microbiology, 2000, pp. 139–153 [Google Scholar]
- Rivoal K, Ragimbeau C, Salvat G, Colin P, Ermel G. Genomic diversity of Campylobacter coli and Campylobacter jejuni isolates recovered from free-range broiler farms and comparison with isolates of various origins. Appl Environ Microbiol 2005;71:6216–6227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanad YM, Kassem II, Abley M, Gebreyes W, LeJeune JT, Rajashekara G. Genotypic and phenotypic properties of cattle-associated Campylobacter and their implications to public health in the USA. PLoS ONE 2011;6:e25778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanad YM, Closs G, Jr, Kumar A, LeJeune JT, Rajashekara G. Molecular epidemiology and public health relevance of Campylobacter isolated from dairy cattle and European starlings in Ohio, USA. Foodborne Pathog Dis 2013;10:229–236 [DOI] [PubMed] [Google Scholar]
- Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. Foodborne illness acquired in the United States—Major pathogens. Emerg Infect Dis 2011;17:7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard SK, Dallas JF, Wilson DJ, Strachan NJ, McCarthy ND, Jolley KA, Colles FM, Rotariu O, Ogden ID, Forbes KJ, Maiden MC. Evolution of an agriculture-associated disease causing Campylobacter coli clade: Evidence from national surveillance data in Scotland. PLoS ONE 2010;15:e15708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirrow MB, Blaser MJ. Clinical aspects of Campylobacter infection. In: Campylobacter, 2nd ed. Nachamkin I, Blaser MJ. (eds.). Washington, DC: American Society for Microbiology, 2000, pp. 69–88 [Google Scholar]
- Smith K, Reimers N, Barnes HJ, Lee BC, Siletzky R, Kathariou S. Campylobacter colonization of sibling turkey flocks reared under different management conditions. J Food Prot 2004;67:1463–1468 [DOI] [PubMed] [Google Scholar]
- Taylor DE, de Grandis SA, Karmali MA. Transmissible plasmids from Campylobacter jejuni. Antimicrob Agents Chemother 1981;19:831–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [USDA] United States Department of Agriculture. Livestock and Poultry: World Markets and Trades. Foreign Agricultural Services, 2012. Available at: www.fas.usda.gov/psdonline/circular/livestock-poultry.pdf, accessed August15, 2013
- Wesley IV, Muraoka WT, Trampel DW, Hurd HS. Effect of preslaughter events on prevalence of Campylobacter jejuni and Campylobacter coli in market-weight turkeys. Appl Environ Microbiol 2005;71:2824–2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley IV, Rostagno M, Hurd HS, Trampel DW. Prevalence of Campylobacter jejuni and Campylobacter coli in market-weight turkeys on-farm and at slaughter. J Food Prot 2009;72:43–48 [DOI] [PubMed] [Google Scholar]
- Yamazaki-Matsune W, Taguchi M, Seto K, Kawahara R, Kawatsu K, Kumeda Y, Kitazato M, Nukina M, Misawa N, Tsukamoto T. Development of a multiplex PCR assay for identification of Campylobacter coli, Campylobacter fetus, Campylobacter hyointestinalis subsp. hyointestinalis, Campylobacter jejuni, Campylobacter lari and Campylobacter upsaliensis. J Med Microbiol 2007;56:1467–1473 [DOI] [PubMed] [Google Scholar]
- Zhao S, Young SR, Tong E, Abbott JW, Womack N, Friedman SL, McDermott PF. Antimicrobial resistance of Campylobacter isolates from retail meat in the United States between 2002 and 2007. Appl Environ Microbiol 2010;76:7949–7956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Zhang Y, Hua X, Hou J, Jiang Y. Antibiotic resistance in Campylobacter. Rev Med Microbiol 2006;17:107–112 [Google Scholar]