Abstract
Nitric oxide (NO) possesses antiinflammatory effects, which may be exerted via its ability to inhibit the transcription factor, NF-κB. A commonly proposed mode of action for inhibition of NF-κBbyNO involves interference with NF-κB binding to DNA. Because activation of inhibitory κB kinase (IKK), the prerequisite enzyme complex necessary to induce NF-κB, is subject to redox regulation, we assessed whether IKK could present a more proximal target for NO to inhibit NF-κB activation. We demonstrate here that S-nitrosothiols (SNO) caused a dose-dependent inhibition of the enzymatic activity of IKK, in lung epithelial cells and in Jurkat T cells, which was associated with S-nitrosylation of the IKK complex. Using biotin derivatization of SNO, we revealed that IKKβ, the catalytic subunit required for NF-κB activation, was a direct target for S-nitrosylation. A mutant version of IKKβ containing a Cys-179-to-Ala mutation was refractory to inhibition by SNO or to increases in S-nitrosylation, in contrast to wild-type IKKβ, demonstrating that Cys-179 is the main target for attack by SNO. Importantly, inhibition of NO synthase activity in Jurkat T cells resulted in activation of IKK, in association with its denitrosylation. Moreover, NO synthase inhibition enhanced the ability of tumor necrosis factor α to activate IKK, illustrating the importance of endogenous NO in regulating the extent of NF-κB activation by cytokines. Collectively, our findings demonstrate that IKKβ is an important target for the redox regulation of NF-κB by endogenous or exogenous NO, providing an additional mechanism for its antiinflammatory properties.
Nitric oxide (NO, nitrogen monoxide) is a pleiotropic shortlived free radical that participates in diverse biological processes such as the regulation of vessel and airway tone, inflammation, neurotransmission, and apoptosis. Although interactions with heme groups (such as in guanylyl cyclase) are the most recognized events associated with the signaling activities of NO, it is increasingly becoming appreciated that nitrosylation of protein sulfhydryl groups represents an important NO-dependent posttranslational modification that impinges on signal transduction cascades (1). Numerous proteins have been identified as targets for S-nitrosylation, including H-Ras (2), caspases (3), c-Jun-N-terminal kinase (JNK) (4), and ornithine decarboxylase (5), among others. In fact, the inhibition of JNK by NO was recently described as a potential antiinflammatory mechanism (4).
NF-κB is a transcription factor that plays a pivotal role in inflammation, cell survival, and proliferation. NF-κB is maintained in a latent form in the cytoplasm via sequestration by inhibitory κB(IκB) proteins. NF-κB-activating stimuli cause the inducible degradation of IκB proteins, unmasking the nuclear localization signal of NF-κB, resulting in its nuclear translocation, binding to NF-κB motifs, and activation of gene transcription (6, 7). The enzyme complex responsible for phosphorylation of IκB on specific serine residues is IκB kinase (IKK), which consists of at least three subunits: IKKα, IKKβ, and IKKγ. Although IKKα and IKKβ are both catalytically active, studies in knockout mice have demonstrated that IKKβ is responsible for degradation of IκB in response to many, but not all, signals (6, 8). In contrast, IKKα plays an important role in transcriptional activation of NF-κB-responsive genes by phosphorylating histone H3 (9, 10). IKKγ is the regulatory subunit responsible for stabilizing the IKK complex and allowing interaction with upstream regulatory proteins (11).
Because the activation of IKK is essential to induce NF-κB, IKK would also be an ideal target for negative regulation to prevent the activation of NF-κB. Indeed, arsenite (12), cyclopentenone prostaglandins (13), hydrogen peroxide (14), and 4-hydroxy-2-nonenal (15) are all capable of inhibiting IKKβ via targeting of cysteine residue(s) of IKKβ, resulting in a failure to activate NF-κB. Numerous studies have reported that NO is capable of modulating the activation of NF-κB (for a review, see ref. 16). The inhibitory effect of NO on NF-κB is believed to play an important role in negative feedback regulation of NO production. The NOS2 gene promoter contains NF-κB regulatory sequences required for maximal gene activation (17, 18), and inhibition of NF-κB therefore decreases NOS2 gene activation (19), decreasing further production of NO. Multiple mechanisms have been described by which NO inhibits NF-κB. For instance, NO has been demonstrated to stabilize IκB (20, 21), induce IκBα mRNA (20), and prevent nuclear translocation of NF-κB (22). Recent emphasis has been focused on S-nitrosylation of Cys-62 of the p50 subunit, which is known to inhibit the ability of NF-κB to bind DNA (23–25).
Despite the current knowledge that the IKK complex is an important regulatory step in the activation of NF-κB by many stimuli (6, 13, 26) and its known sensitivity to redox stress (12, 14, 15), it is unknown whether IKK represents a direct target for inactivation by NO. If NO indeed is an important regulatory molecule in preventing the activation of NF-κB under (patho)physiological conditions, it is plausible that it might inhibit the NF-κB cascade upstream of the degradation of IκB, in addition to inhibiting NF-κB DNA-binding activity, providing more powerful means to prevent NF-κB activation. Therefore, the studies described herein were undertaken to assess whether IKK represents a direct target for S-nitrosylation and inactivation by NO.
Materials and Methods
Cell Culture and Reagents. A line of spontaneously transformed mouse alveolar type II epithelial cells (C10) (27) was propagated in cell culture media-1066 medium containing 50 units/ml penicillin and 50 μg/ml streptomycin (P/S), 2 mM l-glutamine, and 10% FBS, all from GIBCO/BRL. Jurkat T cells were cultured in DMEM high-glucose medium, supplemented with P/S and 10% FBS. At least 1 h before adding the test agents, the cells were switched to phenol red-free DMEM/F12 containing P/S and 0.5% FBS, except for incubations with NG-monomethyl-l-arginine (l-NMMA). Tumor necrosis factor (TNF) α, l-NMMA, S-nitrosoglutathione (GSNO), S-nitroso-N-acetyl-d,l-penicillamine (SNAP), and S-nitrosocysteine antibody were purchased from Calbiochem. The JNK1 and IKKβ and -γ antibodies were purchased from Santa Cruz Biotechnology, the IκBα antibody was purchased from Cell Signaling Technology (Beverly, MA), the phospho-IκBα antibody was purchased from BD Biosciences (Bedford, MA), the phosphoserine antibody was purchased from Zymed, and clasto lactacystin β-lactone was purchased from Sigma. CSNO was prepared fresh before every experiment, as described in ref. 28.
Kinase Assays. Cells were exposed to test agents and, at indicated times, transferred to ice, washed twice with cold PBS, and lysed in buffer containing 50 mM Hepes, 150 mM NaCl, 1 mM EDTA, 2 mM MgCl2, 10 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, 0.1% Nonidet P-40, 10 μg/ml leupeptin, 1% aprotinin, 250 μM DTT, and 100 μM NaF (14). Lysates were cleared by centrifugation at 16,000 × g for 10 min at 4°C. Protein concentrations were determined, and the IKK complex was immunoprecipitated with an IKKγ antibody (Santa Cruz Biotechnology) at 4°C for 1.5 h by using protein G agarose beads. Precipitates were washed once with lysis buffer and twice with kinase buffer (20 mM Hepes/20 mM β-glycerolphosphate/1 mM MnCl2/5 mM MgCl2/2 mM NaF/250 μM DTT). The kinase reaction was performed by using 1 μg of GST-IκBα as a substrate, provided by Rosa Ten (Mayo Clinic, Rochester, MN), and 5 μCi (1 Ci = 37 GBq) of [γ-32P]adenosine triphosphate at 30°C for 30 min. Kinase assays were performed in presence of 250 μM DTT, the minimal concentration necessary to maintain maximal TNFα-stimulated activity (14). Reactions were stopped by the addition of 2× Laemmli sample buffer. Samples were boiled and separated on 15% polyacrylamide gel, and gels were dried and examined by autoradiography. In separate experiments, the immunoprecipitated IKK complex or JNK1 from TNFα-stimulated cells was exposed to SNAP or GSNO for 15 min in lysis buffer in vitro before assessment of kinase activity. The kinase reaction for JNK was performed by using 1 μg of GST-c-Jun as a substrate.
Transfection. C10 cells were transfected (Lipofectamine Plus, Invitrogen) by using 2 μg of plasmid [hemagglutinin-tagged IKKβ wild type (wt HA-IKKβ) or HA-tagged IKKβ C179A; gifts of Michael Karin, University of California at San Diego, La Jolla], for 3 h, washed, and used in experiments 24 h later. The transfection efficiency using this procedure approximates 30% (data not shown). No effects of empty vector were observed.
Detection of S-Nitrosylation Using Biotin Derivatization Coupled to Western Blotting. Detection of S-nitrosylated proteins was performed via the biotin switch method (29) with the following modifications. After treatments, cells were rinsed two times with PBS containing 0.1 mM EDTA and 0.01 mM neocuproine and lysed in HEN buffer (25 mM Hepes, pH 7.7/0.1 mM EDTA/0.01 mM neocuproine) containing 0.5% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate, 0.1% SDS, and 20 mM N-ethylmaleimide (NEM) at 4°C for 30 min to block free thiols. Lysates were cleared by centrifugation at 16,000 × g for 10 min at 4°C, and excess NEM was removed by protein precipitation by using cold acetone. Protein pellets were resuspended in HENS buffer (HEN 1% SDS), SNO bonds were decomposed by adding 20 mM sodium ascorbate, and the resulting free thiols were reacted with 0.05 mM sulfhydryl-specific biotinylating agent, N-(3-malemidylpropionyl)biocytin (MPB, Molecular Probes) for 30 min at room temperature (RT), resulting in biotinylation of SNO. After removal of excess MPB by another protein precipitation using acetone, IKK was immunoprecipitated by using IKKβ or HA (12CA5, Roche Diagnostics) antibodies. Immunoprecipitates were washed three times with HEN buffer and resuspended in 50 μl of HEN containing Laemmli sample buffer, boiled at 95°C for 5 min, loaded on 10% acrylamide gels, and transferred to nitrocellulose. Biotinylated IKKβ was detected on the membrane by using horseradish peroxidase-linked streptavidin. To confirm equal amounts of IKKβ, biotinylated lysates were also subjected to Western blotting for IKKβ or HA. To confirm the specificity of SNO labeling, addition of MPB or reduction by ascorbate was omitted in some samples. All procedures until biotinylation were performed in the dark.
Western Blots. A fraction of the lysates used for in vitro kinase assays, biotin derivatization, or chemiluminescence was mixed with 2× Laemmli sample buffer, and samples were boiled and loaded on a 10% polyacrylamide gel. Proteins were transferred to nitrocellulose and membranes blocked in 5% milk in Tris-buffered saline (TBS). After two washes in TBS containing 0.05% Tween 20 (TBS-Tween), membranes were incubated with primary antibodies against HA, IKKγ, IKKβ, JNK1, IκBα, phospho-IκBα, or phosphoserine for 1 h at RT. Membranes were washed three times for 20 min in TBS-Tween and incubated with a peroxidase-conjugated secondary antibody for 1 h at RT.After three 15-min washes with TBS-Tween, conjugated peroxidase was detected by chemiluminescence according to the manufacturer's instructions (Kirkegaard & Perry Laboratories).
Measurement of SNO Content by Chemiluminescence. The total cellular SNO concentration (protein-bound plus free) was measured in lysates of cells treated with SNO in the presence or absence of l- or d-cys. After three washes with PBS, cells were lysed in the same buffer as was used for the biotin switch method, nitrate was quenched with 0.6% sulfanilamide in 1 M HCl for 10 min at RT, and samples where injected into 5 ml of a solution of 45 mM KI and 10 mM I2 in glacial acetic acid at 60°C, contained within a purge vessel and connected to a NO chemiluminescence analyzer (Ionics, Boulder, CO) (30). The amount of NO released from samples was estimated from a standard curve generated by injection of l-CSNO stock solutions. IKKβ was immunoprecipitated from Jurkat T cell lysates by using a monoclonal IKKβ antibody and protein G agarose beads. After washing the immunoprecipitates three times with HEN buffer (25 mM Hepes, pH 7.7/0.1 mM EDTA/0.01 mM neocuproine) containing 50 mM NaCl to minimize coassociating proteins, antigen–antibody complexes were removed from the beads by three 10-min incubations in 50 μl of 100 mM glycine, pH 3.0, at 4°C. The eluates were treated with 0.6% sulfanilamide before the assessment of the SNO content via chemiluminescence, as described. As a control, some lysates or immunoprecipitates were treated with 4.4 mM HgCl2 for 10 min at RT, followed by 20-min incubation at 4°C and 10-min incubation with sulfanilamide at RT. To confirm that IKKβ was the predominant protein immunoprecipitated under these conditions, Laemmli sample buffer was added to the immunoprecipitate, and samples were boiled and evaluated on a silver-stained gel. All experiments were repeated at least two times, and similar results were obtained.
Results
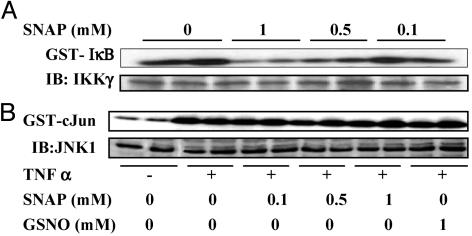
In Vitro Effects of SNO on the Enzymatic Activity of IKK. We first determined whether SNAP or GSNO were capable of inactivating isolated active IKK. For that purpose, C10 cells were stimulated with TNFα for 5 min to induce maximal enzymatic activity (14). The IKK complex was then immunoprecipitated from cell lysates by using an IKKγ antibody and exposed to SNAP or GSNO for 15 min at concentrations ranging from 100 μM to 1 mM before the kinase reaction. Both SNAP (Fig. 1A) and GSNO (data not shown) caused a dose-dependent decrease in IKK enzymatic activity. For comparison, we verified whether these SNO also were capable of inhibiting the activity of JNK, another serine-directed kinase that was recently demonstrated to be sensitive to inhibition by NO (4). In contrast to these previous observations, results in Fig. 1B demonstrate that concentrations of SNAP or GSNO up to 1 mM failed to inhibit JNK enzymatic activity. These results demonstrate that SNO inhibit the enzymatic activity of IKK, but not JNK in vitro.
Fig. 1.
Inactivation of IKK but not JNK by SNAP in vitro. (A) C10 cells were exposed to 10 ng/ml TNFα for 5 min to induce IKK activity. Immunoprecipiated IKK was then exposed to indicated concentrations of SNAP for 15 min, and an in vitro kinase assay was performed by using GST-IκB as a substrate. IB, Western blotting for IKKγ.(B) C10 cells were treated with 10 ng/ml TNFα for 15 min to activate JNK, and JNK1 was immunoprecipitated from lysates. After a 15-min exposure to the indicated concentrations of SNAP or GSNO, an in vitro kinase assay was performed by using GST-c-Jun as a substrate. IB, Western blotting for JNK1.
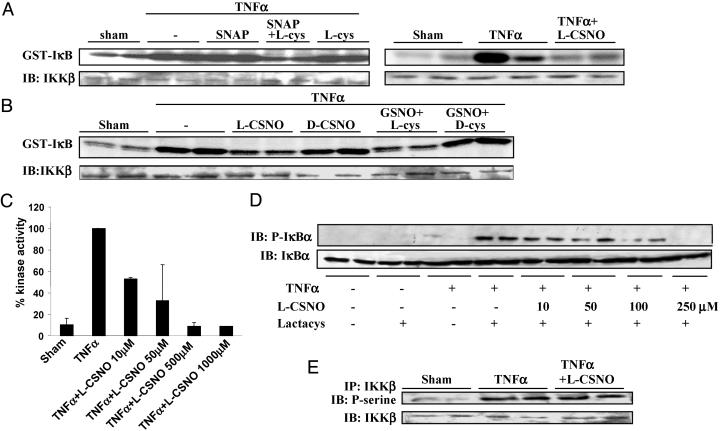
Inhibition of IKK by SNO in Intact Cells. We next established whether SNO are also capable of inhibiting IKK activity in intact cells. TNFα-induced IKK activity was not inhibited after administration of SNAP or GSNO alone, at the doses and time point selected, but was markedly inhibited in intact C10 (Fig. 2A Left) and Jurkat T cells (data not shown) in the presence of l-cys, whereas l-cys itself had no effect. On the other hand, l-CSNO inhibited IKK activity in the absence of extra l-cys in Jurkat T cells (Fig. 2 A Right) and C10 cells (data not shown). In contrast, d-CSNO or GSNO/d-cys failed to inhibit IKK activity (Fig. 2B). Although exposure to GSNO increased the SNO content in TNFα-stimulated cells (TNFα, 0.19 ± 0.01 pmol/μg protein; TNFα + GSNO: 5.78 ± 0.05 pmol/μg protein) this increase was more pronounced in the presence of l-cys (13.65 ± 0.82 pmol/μg protein) but not in the presence of d-cys (8.36 ± 0.66 pmol/μg protein). Collectively, these findings indicate that l-cys mainly facilitates the cellular uptake of NO from SNAP or GSNO through intermediate formation of l-CSNO, thereby promoting inhibition of IKK. Dose-response analyses revealed that inhibition of TNFα-induced activation of IKK (Fig. 2C) or phosphorylation of IκBα (Fig. 2D) was detectable at concentrations of l-CSNO as low as 10 μM, which are believed to reflect pathophysiological amounts of extracellular SNO (31).
Fig. 2.
Repression of IKK activity in intact cells by exposure to SNO. (A Left) C10 cells were treated with 1 mM SNAP in presence or absence of 1 mM l-cys for 15 min before exposure to 10 ng/ml TNFα for 5 min, and an in vitro kinase assay was performed. IB, Western blot of IKKβ. (Right) Jurkat T cells were treated with 500 μM l-CSNO for 30 min and subsequently with 10 ng/ml TNFα for 10 min, and an in vitro kinase assay was performed. (B) C10 cells were treated with 250 μM l-or d-CSNO, or 500 μM GSNO in presence of 500 μMof l-or d-cys for 15 min, before stimulation with 10 ng/ml TNFα for 5 min. The activity of IKK was assessed in an in vitro kinase assay. IB, Western blot of IKKβ.(C) C10 cells were treated with indicated concentrations of l-CSNO for 15 min and subsequently with 10 ng/ml TNFα for 5 min. The activity of IKK was assessed in an in vitro kinase assay. Results were quantified by phospho-image analysis and expressed as the percent kinase activity compared with TNFα-only-treated cells. (D) C10 cells were incubated with 2.5 μM clasto lactacystin β-lactone for 30 min to block proteasomal degradation of proteins and then exposed to l-CSNO for 15 min, followed by a 5-min incubation with 10 ng/ml TNFα. The amount of phosphorylated IκBα (p-IκBα, Upper) and total IκBα (IκBα, Lower) was assessed by Western blotting. (E) C10 cells were treated with 250 μM l-CSNO for 15 min and subsequently with 10 ng/ml TNFα for 5 min. IKKβ was immunoprecipitated from lysates, and phosphoserine content was assessed by Western blot by using a phosphoserine antibody. IB, Western blot of IKKβ.
The possibility exists that SNO inhibit IKK activity by interfering with its phosphorylation, which is required for kinase activity. Results in Fig. 2E demonstrate that exposure of cells to SNO did not interfere with TNFα-induced serine phosphorylation of IKKβ, suggesting that SNO do not act upstream in the activation pathway of IKK but rather inhibit the phosphorylated enzyme directly, consistent with our findings in Fig. 1 A demonstrating that SNO can readily inhibit active IKK in vitro.
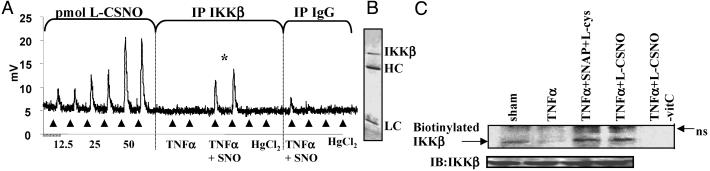
IKKβ Is a Target for S-Nitrosylation. Because the kinase activity of the β subunit of the IKK complex is responsible for phosphorylation of IκBα in response to TNFα, and this was inhibited on treatment with SNO (Figs. 1 A and 2), we next wanted to assess whether IKKβ represents a direct target for S-nitrosylation. For this purpose, we immunopurified IKKβ from TNFα-stimulated Jurkat T cells treated with SNO and performed chemiluminescence analysis to determine the SNO content of IKKβ. Although we could not detect a SNO signal from IKKβ immunopurified from lysates of untreated cells (data not shown) or cells stimulated with TNFα (Fig. 3A), IKKβ obtained from cells exposed to l-CSNO and TNFα demonstrated a marked increase in SNO content. The observed chemiluminescence signal was not due to contaminating nitrite, because samples were treated with sulfanilamide to quench nitrite. In addition, the chemiluminescence signal was completely ablated after HgCl2 treatment, indicating that the measured NO is derived from SNO within the immunopurified protein. Control immunoprecipitations with equal amounts of an isotype-matched control antibody resulted in a barely detectable signal. Analysis of the immunoprecipitate by silver staining (Fig. 3B) revealed that IKKβ was the major detectable protein under the conditions used here, confirming that the measured NO was likely derived from SNO bonds in IKKβ and not from other coimmunoprecipitated proteins.
Fig. 3.
S-nitrosylation of IKKβ. (A) Jurkat T cells were treated with 1 mM l-CSNO for 30 min and subsequently with TNFα for 10 min. IKKβ was immunoprecipitated from lysates containing 2.2 mg of protein by using an IKKβ antibody. A control immunoprecipitation was performed by using an isotype-matched Ig (IgG). After immunoprecipitation, selected samples were treated with HgCl2, and all samples were treated with sulfanilamide to ensure specificity. S-nitrosylation of IKKβ was assessed by chemiluminescence. *, P < 0.05 (Student's t test) compared with NO signal obtained in the TNFα + l-CSNO subjected to immunoprecipitation with the IgG control antibody. (B) IKKβ was immunoprecipitated from untreated Jurkat T cells, samples boiled in sample buffer, separated on SDS/PAGE gel, and silver stained. The location of IKKβ is indicated. HC, antibody heavy chain; LC, antibody light chain. (C) Cells were treated as in A, and lysates were subjected to biotin derivatization. Biotinylation of IKKβ was detected after immunoprecipitation of the IKKβ-containing complex and Western blotting using streptavidin–horseradish peroxidase. In control samples, reduction by ascorbate (–vitC) was omitted. IB, anti-IKKβ immunoblot; ns, nonspecific reactivity.
Alternatively, we used the biotin derivatization method to assess whether the IKKβ subunit is directly targeted by S-nitrosylation. For this purpose, Jurkat T cells were treated as described, and cell lysates were derivatized to selectively biotinylate SNO moieties. IKKβ was subsequently immunoprecipitated from the lysates, and its biotinylation was assessed by using streptavidin–horseradish peroxidase on a Western blot. Results in Fig. 3C demonstrate detectable endogenous S-nitrosylation of IKKβ in untreated cells, which was decreased in response to TNFα. Furthermore, SNAP/l-cys or l-CSNO caused an increase in S-nitrosylation of IKKβ. Biotin labeling was absent when ascorbate was omitted in the derivatization procedure, showing the specificity of biotinylation for detection of SNO.
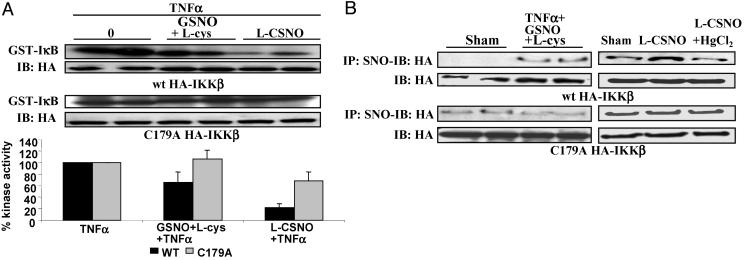
Cysteine Residue 179 of IKKβ Is a Target for S-Nitrosylation. Cys-179 is located between Ser-177 and -181, which are required for activation of IKKβ by TNFα. Because it has been shown that Cys-179 is a target for inhibition by arsenite (12), we wanted to investigate whether this residue was specifically targeted by S-nitrosylation. For this purpose, wt HA-IKKβ or Cys-179-to-Ala mutated HA-tagged IKKβ (C179A HA-IKKβ) were transfected into C10 cells, which were then treated with SNO to assess the extent of inhibition of TNFα-stimulated enzymatic activity. As is apparent from Fig. 4A, both GSNO/l-cys as well as l-CSNO inhibited the activity of wt HA-IKKβ. In contrast, C179A HA-IKKβ was largely refractory to inhibition by SNO. Accordingly, treatment of cells with GSNO/l-cys increased S-nitrosylation of wt HA-IKKβ but not of C179A HA-IKKβ, as revealed by immunoprecipitation using an antibody directed against SNO and Western blotting for HA (Fig. 4B Left). The selectivity of the SNO antibody was demonstrated by incubating lysates of l-CSNO-treated cells with HgCl2 before immunoprecipitation with the SNO antibody, which resulted in a markedly lower amount of wt HA-IKKβ recovered by immunoprecipitation (Fig. 4B Right Upper), whereas HgCl2 did not affect the amount of C179A HA-IKKβ immunoprecipitated with SNO antibody (Fig. 4B Right Lower), illustrating some nonspecific reactivity of this antibody. Collectively, these data demonstrate that Cys-179 of IKKβ is a major target for S-nitrosylation and inhibition by SNO.
Fig. 4.
Cys-179 of IKKβ is target for S-nitrosylation. (A) C10 cells were transfected with wt or C179A HA-IKKβ, treated with 1 mM GSNO/l-cys or l-CSNO for 15 min, before exposure to 10 ng/ml TNFα for 5 min. IKK activity was assessed in an in vitro kinase assay, after immunoprecipitation with an HA antibody. IB, anti-HA immunoblot. (Bottom) Quantitation by phosphoimage analysis. Results are expressed as percentage of IKK activity compared with TNFα-only-treated cells. (B Left)wt(Upper) or C179A HA-IKKβ-transfected C10 cells (Lower) were treated with 1 mM GSNO/l-cys for 15 min before exposure to 10 ng/ml TNFα for 5 min. S-nitrosylated proteins were immunoprecipitated, by using an S-nitrosocysteine antibody (IP SNO) and IKKβ detected by detection of HA by Western blotting. (Lower) HA Western blots on total cell lysates. (B Right) Assessment of specificity of the S-nitrosocysteine antibody. wt (Upper) or C179A HA-IKKβ transfected cells (Lower) were left untreated (left lane), treated with 1 mM l-CSNO for 15 min (middle lane) or treated with 1 mM l-CSNO for 15 min followed by incubation with HgCl2 (right lane) before immunoprecipitation. S-nitrosylated proteins were then immunoprecipitated by using an S-nitrosocysteine antibody (IP: SNO) and IKKβ detected by Western blotting for HA; (Lower) HA Western blots on total cell lysates.
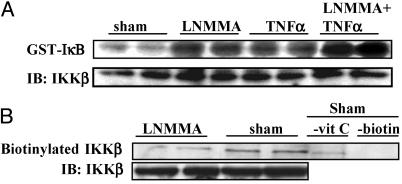
Repression of IKK Enzymatic Activity by Endogenous NOS Activity. If NO is an important negative regulator of IKK in intact cells, inhibition of endogenous NOS activity might relieve this repression, resulting in activation of IKK or enhanced IKK activation by TNFα. To address this question, Jurkat T cells were incubated with 1 mM of the NOS inhibitor, l-NMMA. Results in Fig. 5A demonstrate that 4 h after addition of l-NMMA, IKK activity was markedly enhanced. Furthermore, l-NMMA also potentiated the ability of TNFα to induce IKK activity. Consistent with these findings, inhibition of NOS caused a decrease in SNO-dependent biotinylation of IKKβ, indicating a lesser extent of S-nitrosylation. As described above, omission of ascorbate or the biotin label markedly attenuated biotin reactivity in the sham sample, indicating specific detection of S-nitrosothiols. These results illustrate that endogenous NOS activity is an important repressor of IKK activity in Jurkat T cells.
Fig. 5.
Repression of IKK activity in intact cells by endogenous NOS activity. Jurkat T cells were treated with 1 mM l-NMMA for 4 h followed by stimulation with 10 ng/ml TNFα for 10 min or mock manipulations. Selected dishes were treated with TNFα alone. (A) IKK activity was assessed in an in vitro kinase assay. IB, anti-IKKβ immunoblot. (B) Lysates were subjected to biotin derivatization, and biotinylated IKKβ was detected after immunoprecipitation of IKKβ and Western blotting by using streptavidin–horseradish peroxidase. In control samples, reduction by ascorbate (–vit C) or labeling with N-(3-malemidylpropionyl)biocytin (–biotin) was omitted. IB, anti-IKKβ immunoblot.
Discussion
In the present study, we demonstrate that IKKβ is a direct target for S-nitrosylation. This repression of the NF-κB pathway proximal to DNA binding ensures adequate inhibition of NF-κB activation by NO. The inhibitory action of SNO is not general to all serine-directed kinases. When we evaluated the inhibitory action of SNO on JNK, reported to be inhibited by S-nitrosylation (4), we failed to observe inhibitory effects of SNO. The discrepancy with published work and our observations is puzzling and may be related to the differences in cell types and species investigated. Furthermore, the previous study used anisomycin or UV to activate JNK (4), whereas in the current study, we used TNFα. Nonetheless, our work suggests that the antiinflammatory role of NO previously attributed to inhibition of JNK (4) may also be due to its ability to inhibit IKK.
In cells exposed to SNO, a transnitrosation reaction, i.e., the transfer of NO+ from the SNO to IKK, likely is responsible for the decrease in kinase activity. This explains the relative lack of inactivation of IKK in C10 lung epithelial cells exposed to SNAP and GSNO alone, which cannot readily enter the intact cells (Fig. 2 A and B). In contrast, coincubation of SNAP or GSNO with l-cys results in a transnitrosation reaction to form l-CSNO (28, 32), which is taken up by cells via the l-cys amino acid transport system (33). The transnitrosation mechanism is supported by our observations demonstrating that in the presence of l-cys, but not d-cys, the SNO content of cells exposed to GSNO increased, consistent with inactivation of IKK. Furthermore, direct administration of l-CSNO, but not d-CSNO, inhibited IKK enzymatic activity in intact cells. Although cys may convert S-nitrosothiols to NO extracellularly and thereby promote the formation of N2O3, which would diffuse into the cells and result in S-nitrosylation (34), the observed stereoselective effects of cys argue against such a mechanism and illustrate the importance of cellular SNO uptake in IKK inhibition.
The activity of the IKK complex is markedly inhibited in intact cells by concentrations of SNO as low as 10 μM. Levels of SNO up to 1 μM have been measured in exhaled breath condensate of normal individuals (31). Furthermore, a number of disease states, like pneumonia (35) and inflammatory lung disease (36), have been reported to be accompanied by increased amounts of SNO. The doses of SNO used to detect S-nitrosylation of IKK were higher, likely due to limits of detection imposed by the assays that were used. Importantly, our studies in Jurkat T cells point to a crucial role of endogenous NO in repressing baseline IKK activation and in regulating the magnitude of IKK activation by cytokines. Consequently, inhibition of NOS activity is sufficient to activate IKK in Jurkat T cells. However, incubation of C10 cells with l-NMMA failed to influence IKK enzymatic activity (data not shown), consistent with a recent observation demonstrating that the mode of inhibition of NF-κB by NO is cell type dependent (37). Nonetheless, our observations may have important ramifications in inflammatory conditions where the concentrations of NO, and the sources of its production, are known to be altered. The consumption of NO by peroxidases (38) may lower the concentrations of bioavailable NO and thereby minimize its ability to repress IKK, resulting in chronic activation of NF-κB, which accompanies many inflammatory diseases. Alternatively, direct metabolism of SNO by a recently identified GSNO reductase, which is conserved from bacteria to humans, may lower bioavailable SNO (39). In support of the latter, increased activity of GSNO reductase has recently been demonstrated in allergic airway inflammation (40).
Structural factors that influence the susceptibility to S-nitrosylation include neighboring or surrounding amino acids that affect cysteine reactivity and the presence of a hydrophobic environment that promotes the formation of S-nitrosylating species via the reaction between O2 and NO (34). This “motif” can be apparent from the primary structure of the protein but also as a result of its 3D conformation or protein–protein interactions (41). This makes it difficult to predict the cysteines in IKKβ subject to S-nitrosylation, given that its structure has not been solved. Additionally, the organization of the IKK complex is highly complex and consists of many protein subunits that can associate and dissociate, including Hsp90 and Cdc37 (42, 43). It is furthermore of importance to consider that endothelial NOS associates with Hsp90 (44), thereby directly localizing an endogenous source of NO to the IKK complex.
Although Cys-179 is not an apparent target for S-nitrosylation based on its primary sequence (41), our data demonstrate that this residue is the main target for oxidative modification by SNO. We have shown that the C179A mutant is refractory to inhibition by SNO, and that treatment with SNO did not induce enhanced S-nitrosylation of this form of IKKβ. Cys-179 has previously been shown to be redox sensitive and to be oxidized by arsenite (12) and alkylated by cyclopentenone prostaglandins (13). Cys-179 is strategically located between the two serines that are required for activation of IKK by TNFα. It is conceivable that the TNFα-induced phosphorylation of Ser-177 and -181, which creates a more negative charge surrounding Cys-179, promotes its susceptibility to transnitrosation. However, additional experiments are clearly needed to elucidate the interplay between phosphorylation and S-nitrosylation of IKKβ, as well as the mechanism by which S-nitrosylation impacts on IKK activity.
The identification of IKKβ as a target for S-nitrosylation provides insights into the mechanisms of inhibition of NF-κB by NO, and a proposed model of this mechanism is presented (Fig. 6, which is published as supporting information on the PNAS web site). These observations may provide strategies aimed at enhancing the inhibitory effect of NO at a level before DNA-binding activity, to augment its antiinflammatory mode of action.
Supplementary Material
Acknowledgments
K.C. is supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico-Conselho Nacional de Pesquisas, Brazil (Grant 200538/01-2). This work was supported by National Institutes of Health Grants RO1 HL60014 and HL60812, by Public Health Service Grants P20 RL15557 (National Center for Research Resources Centers of Biomedical Research Excellence) and PO1 HL67004, and by a grant from the Dutch Asthma Foundation.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: IκB, inhibitory κB; IKK, IκB kinase; SNAP, S-nitroso-N-acetylpenicillamine; GSNO, S-nitrosoglutathione; CSNO, S-nitrosocysteine; TNFα, tumor necrosis factor α;NOS, NO synthase; SNO, S-nitrosothiol(s); JNK, c-Jun-N-terminal kinase; wt HA-IKKβ, hemagglutinin-tagged IKKβ wild type; RT, room temperature; C179A HA-IKKβ, Cys-179-to-Ala-mutated HA-tagged IKKβ; l-NMMA, NG-monomethyl-l-arginine.
See Commentary on page 8841.
References
- 1.Stamler, J. S., Lamas, S. & Fang, F. C. (2001) Cell 106, 675–683. [DOI] [PubMed] [Google Scholar]
- 2.Mallis, R. J., Buss, J. E. & Thomas, J. A. (2001) Biochem. J. 355, 145–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mannick, J. B., Hausladen, A., Liu, L., Hess, D. T., Zeng, M., Miao, Q. X., Kane, L. S., Gow, A. J. & Stamler, J. S. (1999) Science 284, 651–654. [DOI] [PubMed] [Google Scholar]
- 4.Park, H. S., Huh, S. H., Kim, M. S., Lee, S. H. & Choi, E. J. (2000) Proc. Natl. Acad. Sci. USA 97, 14382–14387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauer, P. M., Buga, G. M., Fukuto, J. M., Pegg, A. E. & Ignarro, L. J. (2001) J. Biol. Chem. 276, 34458–34464. [DOI] [PubMed] [Google Scholar]
- 6.Karin, M. & Ben-Neriah, Y. (2000) Annu. Rev. Immunol. 18, 621–663. [DOI] [PubMed] [Google Scholar]
- 7.May, M. J. & Ghosh, S. (1998) Immunol. Today 19, 80–88. [DOI] [PubMed] [Google Scholar]
- 8.May, M. J. & Ghosh, S. (1999) Science 284, 271–273. [DOI] [PubMed] [Google Scholar]
- 9.Anest, V., Hanson, J. L., Cogswell, P. C., Steinbrecher, K. A., Strahl, B. D. & Baldwin, A. S. (2003) Nature 423, 659–663. [DOI] [PubMed] [Google Scholar]
- 10.Yamamoto, Y., Verma, U. N., Prajapati, S., Kwak, Y. T. & Gaynor, R. B. (2003) Nature 423, 655–659. [DOI] [PubMed] [Google Scholar]
- 11.Rothwarf, D. M., Zandi, E., Natoli, G. & Karin, M. (1998) Nature 395, 297–300. [DOI] [PubMed] [Google Scholar]
- 12.Kapahi, P., Takahashi, T., Natoli, G., Adams, S. R., Chen, Y., Tsien, R. Y. & Karin, M. (2000) J. Biol. Chem. 275, 36062–36066. [DOI] [PubMed] [Google Scholar]
- 13.Rossi, A., Kapahi, P., Natoli, G., Takahashi, T., Chen, Y., Karin, M. & Santoro, M. G. (2000) Nature 403, 103–108. [DOI] [PubMed] [Google Scholar]
- 14.Korn, S. H., Wouters, E. F., Vos, N. & Janssen-Heininger, Y. M. (2001) J. Biol. Chem. 276, 35693–35700. [DOI] [PubMed] [Google Scholar]
- 15.Ji, C., Kozak, K. R. & Marnett, L. J. (2001) J. Biol. Chem. 276, 18223–18228. [DOI] [PubMed] [Google Scholar]
- 16.Janssen-Heininger, Y. M., Poynter, M. E. & Baeuerle, P. A. (2000) Free Radical Biol. Med. 28, 1317–1327. [DOI] [PubMed] [Google Scholar]
- 17.Xie, Q. W., Kashiwabara, Y. & Nathan, C. (1994) J. Biol. Chem. 269, 4705–4708. [PubMed] [Google Scholar]
- 18.Lowenstein, C. J., Alley, E. W., Raval, P., Snowman, A. M., Snyder, S. H., Russell, S. W. & Murphy, W. J. (1993) Proc. Natl. Acad. Sci. USA 90, 9730–9734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griscavage, J. M., Wilk, S. & Ignarro, L. J. (1996) Proc. Natl. Acad. Sci. USA 93, 3308–3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng, H. B., Libby, P. & Liao, J. K. (1995) J. Biol. Chem. 270, 14214–14219. [DOI] [PubMed] [Google Scholar]
- 21.Katsuyama, K., Shichiri, M., Marumo, F. & Hirata, Y. (1998) Arterioscler. Thromb. Vasc. Biol. 18, 1796–1802. [DOI] [PubMed] [Google Scholar]
- 22.Okamoto, T., Valacchi, G., Gohil, K., Akaike, T. & van der Vliet, A. (2002) Am. J. Respir. Cell Mol. Biol. 27, 463–473. [DOI] [PubMed] [Google Scholar]
- 23.Matthews, J. R., Botting, C. H., Panico, M., Morris, H. R. & Hay, R. T. (1996) Nucleic Acids Res. 24, 2236–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.delaTorre, A., Schroeder, R. A., Bartlett, S. T. & Kuo, P. C. (1998) Surgery 124, 137–141. [PubMed] [Google Scholar]
- 25.Marshall, H. E. & Stamler, J. S. (2001) Biochemistry 40, 1688–1693. [DOI] [PubMed] [Google Scholar]
- 26.Rothwarf, D. M. & Karin, M. (1999) Sci. STKE 1999, RE1. [DOI] [PubMed] [Google Scholar]
- 27.Malkinson, A. M., Dwyer-Nield, L. D., Rice, P. L. & Dinsdale, D. (1997) Toxicology 123, 53–100. [DOI] [PubMed] [Google Scholar]
- 28.Mallis, R. J. & Thomas, J. A. (2000) Arch. Biochem. Biophys. 383, 60–69. [DOI] [PubMed] [Google Scholar]
- 29.Jaffrey, S. R. & Snyder, S. H. (2001) Sci. STKE 2001, L1. [DOI] [PubMed] [Google Scholar]
- 30.Feelisch, M., Rassaf, T., Mnaimneh, S., Singh, N., Bryan, N. S., Jourd'Heuil, D. & Kelm, M. (2002) FASEB J. 16, 1775–1785. [DOI] [PubMed] [Google Scholar]
- 31.Csoma, Z., Bush, A., Wilson, N. M., Donnelly, L., Balint, B., Barnes, P. J. & Kharitonov, S. A. (2003) Chest 124, 633–638. [DOI] [PubMed] [Google Scholar]
- 32.Padgett, C. M. & Whorton, A. R. (1997) Am. J. Physiol. 272, C99–C108. [DOI] [PubMed] [Google Scholar]
- 33.Nemoto, T., Shimma, N., Horie, S., Saito, T., Okuma, Y., Nomura, Y. & Murayama, T. (2003) Eur. J. Pharmacol. 458, 17–24. [DOI] [PubMed] [Google Scholar]
- 34.Hogg, N. (2002) Annu. Rev. Pharmacol. Toxicol. 42, 585–600. [DOI] [PubMed] [Google Scholar]
- 35.Gaston, B., Reilly, J., Drazen, J. M., Fackler, J., Ramdev, P., Arnelle, D., Mullins, M. E., Sugarbaker, D. J., Chee, C., Singel, D. J., et al. (1993) Proc. Natl. Acad. Sci. USA 90, 10957–10961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corradi, M., Montuschi, P., Donnelly, L. E., Pesci, A., Kharitonov, S. A. & Barnes, P. J. (2001) Am. J. Respir. Crit. Care Med. 163, 854–858. [DOI] [PubMed] [Google Scholar]
- 37.Marshall, H. E. & Stamler, J. S. (2002) J. Biol. Chem. 277, 34223–34228. [DOI] [PubMed] [Google Scholar]
- 38.Eiserich, J. P., Baldus, S., Brennan, M. L., Ma, W., Zhang, C., Tousson, A., Castro, L., Lusis, A. J., Nauseef, W. M., White, C. R. & Freeman, B. A. (2002) Science 296, 2391–2394. [DOI] [PubMed] [Google Scholar]
- 39.Liu, L., Hausladen, A., Zeng, M., Que, L., Heitman, J. & Stamler, J. S. (2001) Nature 410, 490–494. [DOI] [PubMed] [Google Scholar]
- 40.Fang, K., Johns, R., Macdonald, T., Kinter, M. & Gaston, B. (2000) Am. J. Physiol. 279, L716–L721. [DOI] [PubMed] [Google Scholar]
- 41.Hess, D. T., Matsumoto, A., Nudelman, R. & Stamler, J. S. (2001) Nat. Cell Biol. 3, E46–E49. [DOI] [PubMed] [Google Scholar]
- 42.Chen, G., Cao, P. & Goeddel, D. V. (2002) Mol. Cell 9, 401–410. [DOI] [PubMed] [Google Scholar]
- 43.Bouwmeester, T., Bauch, A., Ruffner, H., Angrand, P. O., Bergamini, G., Croughton, K., Cruciat, C., Eberhard, D., Gagneur, J., Ghidelli, S., et al. (2004) Nat. Cell Biol. 6, 97–105. [DOI] [PubMed] [Google Scholar]
- 44.Gratton, J. P., Fontana, J., O'Connor, D. S., Garcia-Cardena, G., McCabe, T. J. & Sessa, W. C. (2000) J. Biol. Chem. 275, 22268–22272. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.