Significance
Small noncoding RNA molecules have been found in eukaryotes and prokaryotes and are also encoded by viruses. Wolbachia are endosymbiotic bacteria that are widespread in invertebrate populations. Significantly, certain strains of Wolbachia inhibit replication of mosquito-borne pathogens, such as dengue viruses, the malaria parasite, and filarial nematodes. Our results demonstrate that Wolbachia encode conserved small RNAs of approximately 30 nt in Aedes aegypti mosquito and Drosophila melanogaster. We show that the small RNAs may regulate bacterial and host genes, providing a means of communication across two kingdoms.
Keywords: Aedes aegypti, mosquito, microRNA
Abstract
In prokaryotes, small noncoding RNAs (snRNAs) of 50–500 nt are produced that are important in bacterial virulence and response to environmental stimuli. Here, we identified and characterized snRNAs from the endosymbiotic bacteria, Wolbachia, which are widespread in invertebrates and cause reproductive manipulations. Most importantly, some strains of Wolbachia inhibit replication of several vector-borne pathogens in insects. We demonstrate that two abundant snRNAs, WsnRNA-46 and WsnRNA-49, are expressed in Wolbachia from noncoding RNA transcripts that contain precursors with stem-loop structures. WsnRNAs were detected in Aedes aegypti mosquitoes infected with the wMelPop-CLA strain of Wolbachia and in Drosophila melanogaster and Drosophila simulans infected with wMelPop and wAu strains, respectively, indicating that the WsnRNAs are conserved across species and strains. In addition, we show that the WsnRNAs may potentially regulate host genes and Wolbachia genes. Our findings provide evidence for the production of functional snRNAs by Wolbachia that play roles in cross-kingdom communication between the endosymbiont and the host.
microRNAs (miRNAs) are small noncoding RNAs (snRNAs) of ∼22 nt that regulate gene expression at the transcriptional and posttranscriptional levels (reviewed in ref. 1). They have been reported from animals, plants, viruses, and potentially bacteria. Evidence shows that miRNAs are directly or indirectly involved in regulation of almost all cellular pathways and, consequently, affect various biological processes such as development, differentiation, immunity, cancer, and host–pathogen interactions. Recently, it was shown that miRNAs from sickle cell erythrocytes are translocated and enriched in the malaria parasite, Plasmodium falciparum, when they infect the cells. Consequently, the translocated miRNAs suppress the cAMP-dependent protein kinase (PKA-R), resulting in inhibition of translation in the parasite, contributing to the resistance of the sickle cells to malaria infection (2). This example demonstrates the role of snRNAs in regulation of gene expression across kingdoms.
Wolbachia are obligate intracellular alphaproteobacteria (family Anaplasmataceae, order Rickettsiales) transmitted maternally and infect more than 40% of terrestrial arthropod species, suggesting that they may be the most prevalent vertically transmitted endosymbiont worldwide (3). The bacteria’s impacts on the host are variable, but they are best known for their reproductive manipulations of the host and, more recently, because of inhibition of replication of several vector-borne pathogens after their transinfections; these pathogens include arboviruses such as dengue and chikungunya viruses in Aedes aegypti (4) and the human malaria parasite Plasmodium falciparum in an important malaria vector, Anopheles stephensi (5). However, these effects seem to be strain and host specific (reviewed in ref. 6). The effects on vector-borne pathogens place Wolbachia as a potential agent for the control of insect-transmitted diseases. Despite the significance of these discoveries, the molecular mechanisms behind these effects are largely unknown and need to be elucidated. To date, all of the studies on small RNAs in Wolbachia-infected insects have focused on the impact of the endosymbiont on the host miRNAs/genes. For instance, microarray and deep sequencing analyses of A. aegypti mosquitoes infected with the wMelPop-CLA strain of Wolbachia revealed substantial changes in the mosquito’s miRNA profile and its localization within the cell (7–9). One of these miRNAs, aae-miR-2940, is exclusively up-regulated in Wolbachia-infected mosquitoes positively regulating the metalloprotease ftsh (7) and negatively regulating the metalloprotease AaDnmt2 (10) genes; both of the effects shown to benefit the endosymbiont’s persistence in the host. However, how Wolbachia manipulates the host miRNA profile is still unknown.
The majority of snRNAs found in pathogenic and nonpathogenic bacteria are of 50–500 nt, which play key roles in bacterial virulence and response to the environment (11). However, recent studies suggest that bacteria could also produce functional miRNA or miRNA-like snRNAs (12, 13). In this study, we found that Wolbachia, as endosymbiotic bacteria, are able to encode snRNAs that act as effectors to modulate the expression of Wolbachia and mosquito host genes.
Results and Discussion
Wolbachia Expresses Small RNAs.
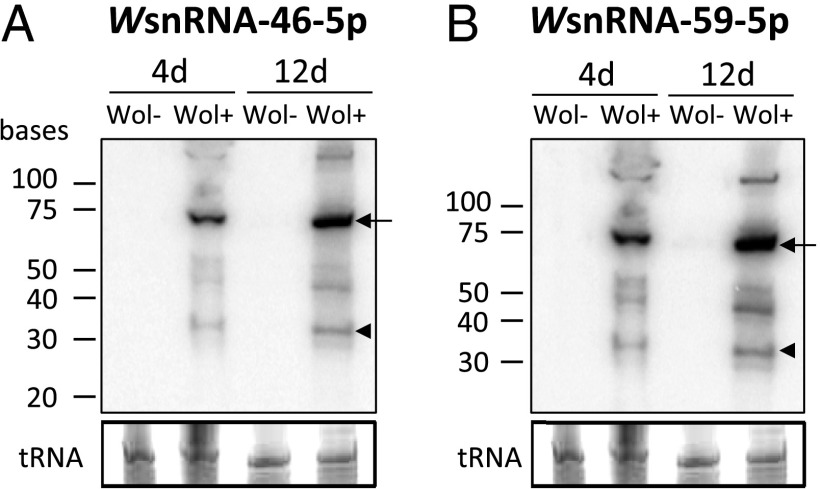
To explore whether Wolbachia expresses snRNA molecules, we deep sequenced small RNAs (less than 35 nucleotides) of two A. aegypti Aag2 cell lines, one infected with the obligate endosymbiont Wolbachia strain wMelPop-CLA and another without it. After filtering the RNA reads, 87 sequences mapped to the genome of Wolbachia, potentially originating from stem-loop structures (14). The 10 most abundant sequences were selected (Fig. S1) and analyzed by Northern blotting from 4- and 12-d-old A. aegypti adult female mosquitoes with Wolbachia-wMelPop-CLA (Wol+) and without (Wol−). Two snRNAs, WsnRNA-46-5p and WsnRNA-59-5p, were highly expressed in Wol+ mosquitoes (Fig. 1), whereas the other eight could not be detected. Northern blot analysis of mosquito RNAs showed two fragments of ∼30 bp; deep-sequencing data from the mosquito cell line showed sequences of 27 and 30 nt, respectively. Both deep-sequencing and Northern blot analyses showed that WsnRNA-46 and WsnRNA-59 are expressed exclusively in insects or cells infected with Wolbachia. An intense band of ∼65–70 bp was detected for each snRNA, which corresponded to the size of precursor miRNAs (premiRNAs) found in eukaryotes. There were more of the mature and presnRNA forms in 12-d-old mosquitoes compared with 4-d-old ones, which could either be due to up-regulation of expression or simply because of the higher density of Wolbachia with age. We designed antisense probes for WsnRNA-46-3p and WsnRNA-59-3p (also known as star strands) and analyzed their expression by using Northern blot analysis in 4-d and 12-d-old mosquitoes. In both cases, a strong band corresponding to their presnRNAs was detected, but not their mature forms as seen in Fig. 1 (Fig. S2). Star strands are found in relatively low numbers compared with the mature miRNA.
Fig. 1.
Adult female mosquitoes infected with Wolbachia express small RNAs. WsnRNA-46 (A) and WsnRNA-59 (B) detected by Northern blotting from 4- and 12-d-old A. aegypti mosquitoes with Wolbachia-wMelPop-CLA (Wol+) and without (Wol−). Arrow and arrowhead indicate the precursor and mature RNAs, respectively. tRNA is shown as loading control.
WsnRNAs were found located apart from each other in the Wolbachia genome; WsnRNA-46 mapped to the positive strand of the intergenic section in between the rod shape-determining MreB protein gene (WD1258) and the tRNA-specific 2-thiouridylase MnmA gene (WD1250). WsnRNA-59 mapped to the negative strand of the intergenic region in between the predicted phage major capsid protein E (WD0604) and the small hypothetical protein (WD0605) (Fig. S3). Interestingly, transcripts of preWsnRNA-59 were sequenced in a small RNA library of Aedes albopictus infected with Wolbachia strain wMelPop-CLA (15). Two peaks of antisense expression were observed in a region of the Wolbachia genome corresponding to two hypothetical proteins and annotated as phage major capsid protein E. One of these two structures matched the location, sequence, and length of preWsnRNA-59. The results from Darby et al. (15) are therefore consistent with our results. Secondary structure prediction revealed a stem and hairpin loop structure for both WsnRNAs (Fig. S4). In addition, we were able to clone the presnRNAs from Wolbachia-infected cells, which matched the stem-loop structures.
In comparison with eukaryotes, there is only one gene coding for RNA polymerase in bacteria; all RNA is transcribed from DNA by the product of this one gene (including noncoding RNAs). Using reverse transcription and PCR, we investigated the biogenesis of WsnRNAs. Both WsnRNA-46 and WsnRNA-59 are on an intergenic area close to adjacent genes (Fig. S5 B and D); WsnRNA-46 is located 57 bp after the stop codon of the rod shape-determining protein mreb, and WsnRNA-59 overlaps with the last 5 bp at the 3′ end of the phage major capsid protein E transcript (in the negative strand). Yang et al. (16) described four miRNAs in the fungus Neurospora crassa with the primary miRNAs (pri-miRNAs) transcribed together with their adjacent genes, two tRNA-Ala genes. Subsequently, at least for one of them, they were trimmed and processed by RNase Z releasing the premiRNA. The possibility that WsnRNA-46 and WsnRNA-59 are transcribed together with one or both of the two adjacent genes was tested by RT-PCR. Expression of preWsnRNA-46 and the downstream gene tRNA-specific 2-thiouridylase were high but very different from each other; transcript levels of the rod shape-determining protein were substantially lower (Fig. S5A). Differential expression levels of these genes suggest expression of the genes under different promoters. Furthermore, we could not amplify any amplicons when internal primers of neighboring genes and primers inside the preWsnRNA sequences were used (Fig. S5B). Similar results were obtained for preWsnRNA-59 and its flanking genes; preWsnRNA-59 and capsid protein E transcripts were highly expressed and no expression was detected for the hypothetical protein gene WD0605 (Fig. S5C). No amplicon was obtained when internal primers for preWsnRNA-59 and any of the flanking genes were used. In addition, using the bioinformatics tools “The Terminator” (17) and ARNold (18), we examined whether the preWsnRNA-46 stem loop could be a Rho-independent transcription terminator for mrbe gene (Fig. S5B) but none were predicted. The results indicate that WsnRNA-46 and WsnRNA-59 are not transcribed together with any of the flanking genes, and therefore, they are independently expressed and regulated. Analysis of the genomic area upstream of the two presnRNA sequences revealed conserved prokaryote promoter regions and boxes −35 and −10 were identified (Fig. S6A). Bioinformatics analyses predicted that the transcription initiation site is 1 nt before the beginning of the experimental sequence obtained for preWsnRNA-59, and the initiation site for preWsnRNA-46 was predicted to be 17 nt from the beginning of the presnRNA sequence (Fig. S6A). Furthermore, transcription factors that may have an important role in the activation of these snRNAs in diverse environmental conditions were found in their promoter regions. i.e., for WsnRNAs-46, the sigma factor RpoD19 that controls the regulation of the fec gene in the iron transport pathway (Fig. S6B). The presence of conserved transcription factors in the promoter region reinforces the hypothesis that WsnRNA-46 and WsnRNA-59 may be independently regulated through their own promoters, and that may play an important role in the maintenance of the fitness of the bacteria.
Mosquitoes used for the experiments as “Wolbachia free” were previously infected with Wolbachia but treated with the antibiotic tetracycline (Tet). This exposure to the endosymbiont could have caused DNA fragments from Wolbachia to be integrated into the mosquito genome. In this scenario, mosquitoes would be Wolbachia free after the antibiotic treatment, but they may still conserve those Wolbachia DNA fragments integrated into their genomic DNA. After certain stress factors or environmental condition, those DNA fragments could be transcribed into RNA. To rule out this possibility, we used PCR and specific primers for preWsnRNA-46 and preWsnRNA-59 by using genomic DNA of Tet mosquitoes and mosquitoes infected with Wolbachia (Pop). Amplicons were obtained in mosquitoes infected with Wolbachia but not from those treated with tetracycline (Fig. S7 A and B). These results confirm that WsnRNA-46 and WsnRNA-59 are transcribed from the genome of Wolbachia.
To further elucidate the origin of these snRNAs (prokaryotic or eukaryotic), a fragment of ∼400 bp containing the preWsnRNA sequences of -46 and -59 were inserted into the vector pIZ/V5 and pGEM-T-Easy. The vectors containing the fragments were transfected into Aag2 and Pop mosquito cells (with pIZ/V5) or transformed into Escherichia coli (with pGEM-T-Easy). Mosquito cells transcribed the fragments cloned into pIZ/V5 compared with the control with empty pIZ/V5, and endogenous slicer activity of the cells produced a band of the right size for preWsnRNA-46 and preWsnRNA-59; however, very little WsnRNA was detected (Fig. S8A). It is worth mentioning that despite the presence of the WsnRNAs in the deep sequencing, their concentrations in Pop cells were considerably lower than that in whole mosquitoes or flies and, therefore, not as detectable as in Pop cells. This observation could be due to lower density of Wolbachia in cells compared with whole mosquitoes. In addition, in the blot shown in Fig. S8A only 10 μg of total RNA was loaded, which would not show any signal with mosquito RNA either. Remarkably, two well-defined bands corresponding to mature WsnRNA-46 and WsnRNA-59 were detected when they were expressed in E. coli overnight (Fig. S8B). A less abundant but noticeable fragment corresponding to the preWsnRNA size was also detected; this reduction in intensity of preWsnRNA is likely due to its processing to its mature form by the endogenous slicer machinery of E. coli. Slightly different sizes of mature WsnRNA were obtained in this experiment with E. coli if compared with their size when they were detected in A. aegypti mosquitos infected with Wolbachia (Fig. 1). The large phylogenetic distance between Wolbachia and E. coli, and therefore their processing machineries, could justify this difference. These results suggest that WsRNA-46 and WsRNA-59 are most likely synthesized and processed inside of Wolbachia.
Wolbachia-Encoded snRNAs Are Conserved in Different Strains.
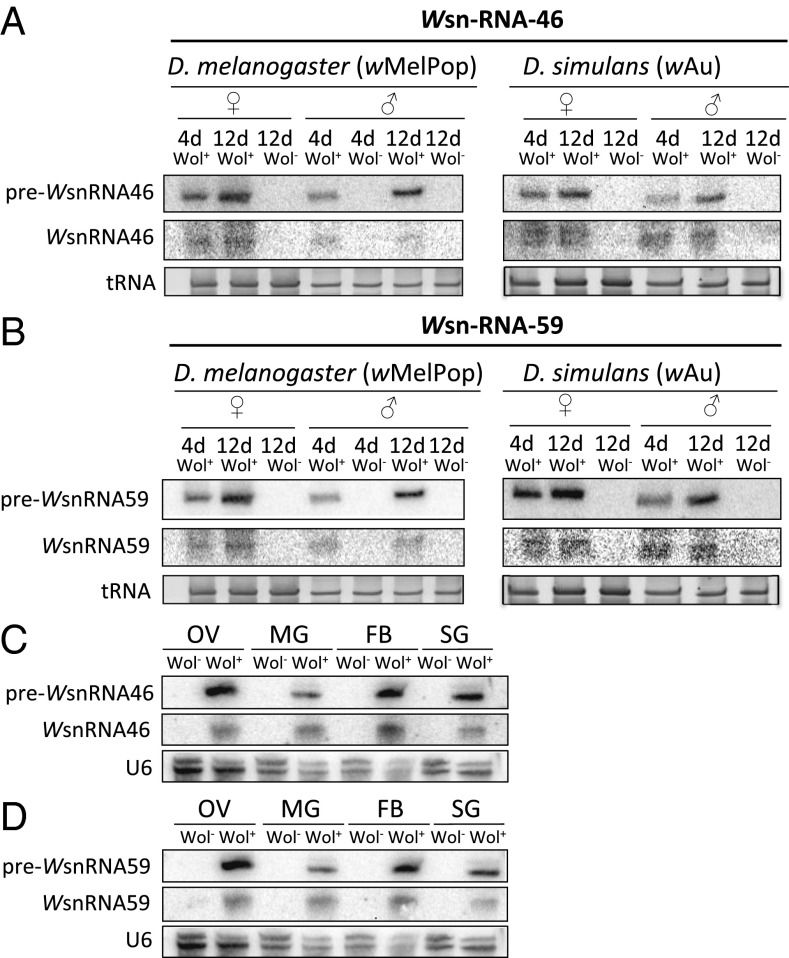
To test whether WsnRNA-46 and WsnRNA-59 are expressed in other hosts and by other Wolbachia strains, 4- and 12-d-old noninfected Drosophila melanogaster and Drosophila simulans and flies infected with wMelPop and wAu Wolbachia strain, respectively, were analyzed by using Northern blot. WsnRNA-46 and WsnRNA-59 were both detected in flies infected with both strains of bacteria and with a similar overall pattern as the one obtained in mosquitoes infected with wMelPop-CLA (Fig. 2 A and B). WsnRNA-46 and WsnRNA-59 were expressed in both females and males, and they were up-regulated in 12-d-old flies compared with 4-d-old ones, consistent with the results obtained from mosquitoes. No expression was detected in noninfected flies of the same sex, age, and physiological conditions (Fig. 2 A and B). To fit all of the samples in one gel, we only included 12-d-old noninfected flies for female samples as negative control because obviously the small RNAs are not produced in noninfected flies and also not detected in 4-d-old female mosquitoes (Fig. 1).
Fig. 2.
WsnRNA-46 and WsnRNA-59 are expressed in females and males of D. melanogaster and D. simulans infected with Wolbachia strains wMelPop and wAu, respectively, and in the reproductive and nonreproductive tissues of A. aegypti mosquito females. Expression of WsnRNA-46 (A) and WsnRNA-59 (B) in D. melanogaster and D. simulans infected with wMelPop and wAu strains (Wol+), respectively, and without Wolbachia (Wol−) were examined by Northern blot analysis. WsnRNA-46 (C) and WsnRNA-59 (D) expression in ovaries (OV), Malpighian tubules (MG), fat bodies (FB), and the salivary glands (SG) of the adult A. aegypti female Wol+ and Wol− mosquitoes. tRNA and U6 are shown as loading controls.
We further carried out a sequence homology search of the available sequences of different Wolbachia strains to find out how conserved the sequences are. The results showed that the whole preWsnRNA sequences of miRNA-59 and miRNA-46 are conserved in most strains belonging to the supergroup A Wolbachia (Table S1). Unfortunately, the sequence for wAu was not available to include in the analysis, but the strong hybridization (Fig. 2) indicates that the sequence must be highly conserved. Consistently, wAu also belongs to supergroup A. These results indicated that WsnRNA-46 and WsnRNA-59 are expressed in at least three host species and by different strains of Wolbachia. Furthermore, their up-regulation in 12-d-old mosquitoes and flies were consistent across the species.
Wolbachia can successfully infect reproductive and nonreproductive tissues in females and males of a variety of insect hosts, i.e., Drosophila, Aedes, and Culex (19). Depending on the host and the strain, Wolbachia is more abundant in some tissues than in others. To study the tissue specificity of WsnRNA-46 and WsnRNA-59 by using Northern blot analysis, we examined their expression in ovaries, Malpighian tubules, fat body, and salivary glands of adult female mosquitoes. All these tissues are known to be infected by Wolbachia at different levels. A good expression of WsnRNA-46 and WsnRNA-59 was detected in all of the tissues tested (Fig. 2 C and D); however, the expression in ovaries and fat body was considerably higher than in the other two tissues. Ovaries are known for being a preferred target of this intracellular symbiont and for affecting its physiology (i.e., cytoplasmic incompatibility). Midgut was the tissue with the lowest expression, which is in agreement with the low levels of Wolbachia reported from this tissue (19). The expression pattern was similar for both snRNAs.
Wolbachia Genes Interact with Wolbachia-Encoded snRNAs.
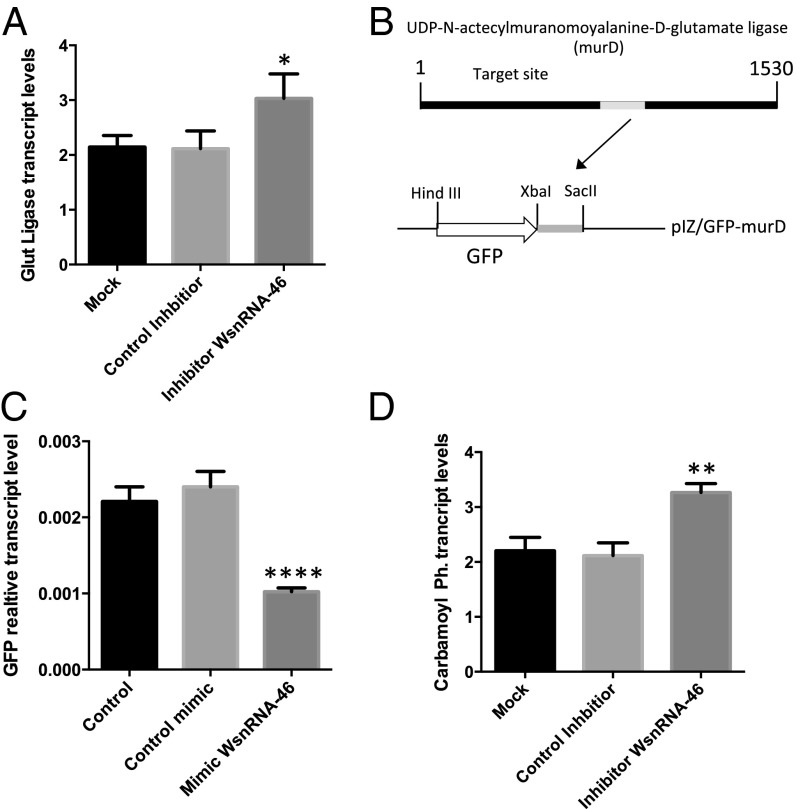
The induction of WsnRNAs-46 and WsnRNAs-59 in 12-d-old mosquitos and flies infected with Wolbachia led us to investigate their possible function as transcriptional regulators. Using NCBI and RNAhybrid, we found that WsnRNAs-46 and WsnRNAs-59 could potentially target several bacterial genes (Table S2). Predicted target sites of WsnRNA-46 in the target genes were within the coding DNA sequence (CDS) of those proteins. WsnRNAs-46 targets palindromic repeat sequences inserted in frame for all those target genes (Fig. S9 A–F). However, the insertion of the targeted palindromic sequences in those genes was not consistent for all Wolbachia strains. To test whether these palindromic sequences act as regulatory points of the genes we studied, mRNA levels of UDP-N-acetylmuramoyalanine-d-glutamate ligase (murD) were monitored after the transfection of WsnRNA-46 inhibitor into mosquito cells infected with wMelPop-CLA. Transcript levels of murD were significantly higher in the presence of the inhibitor compared with the mock or control inhibitor sequences (Fig. 3A). An additional experiment was performed to confirm the interaction of WsnRNA-46 with murD; a 177-bp DNA fragment containing the target region was cloned downstream of the GFP gene in pIZ/V5 expression vector (Fig. 3B). The construct pIZ/GFP-murD was cotransfected into Aag2 mosquito cells along with WsnRNA-46 mimic, a control mimic or a mock. Using quantitative RT-PCR (RT-qPCR) assay, significantly lower GFP transcript levels were found when the pIZ/GFP-murD construct was cotransfected with WsnRNA-46 mimic compared with the other control treatments (Fig. 3C). The results confirmed that WsnRNA-46 is able to interact with murD. Importantly the interaction between WsnRNA-46 and murD resulting in a down-regulation was confirmed. In addition to murD, in the presence of WsnRNA-46 inhibitor, the transcript levels of another target, carbamoyl-phosphate synthase, was significantly increased (Fig. 3D).
Fig. 3.
WsnRNA-46 interacts with Wolbachia genes. (A) RT-qPCR analysis of the Wolbachia UDP-N-acetylmuranomoyalanine-d-glutamate ligase gene (murD) relative to Wolbachia surface protein (wsp) in Pop cells 48 h after transfection with mock, control inhibitor, and inhibitor of WsnRNA-46. (B) Cloning strategy of the murD target sequence complementary to WsnRNA-46 under the GFP reporter gene ORF in the pIZ/V5 vector. (C) RT-qPCR analysis of RNA from Aag2 cells 72 h after cotransfection of pIZ-GFP-murD with control, control mimic, and WsnRNA-46 mimic. Control cells were mock transfected with only the pIZ/GFP vector. (D) RT-qPCR analysis of the Wolbachia gene carbomoyl-phosphatase synthase (carA) relative to wsp in Pop cells 48 h after transfection with mock, control inhibitor, and inhibitor of WsnRNA-46. *P < 0.05, **P < 0.01, ****P < 0.0001; ANOVA.
Wolbachia-Encoded snRNAs Are Exported into the Host Cell and Regulate Host Genes.
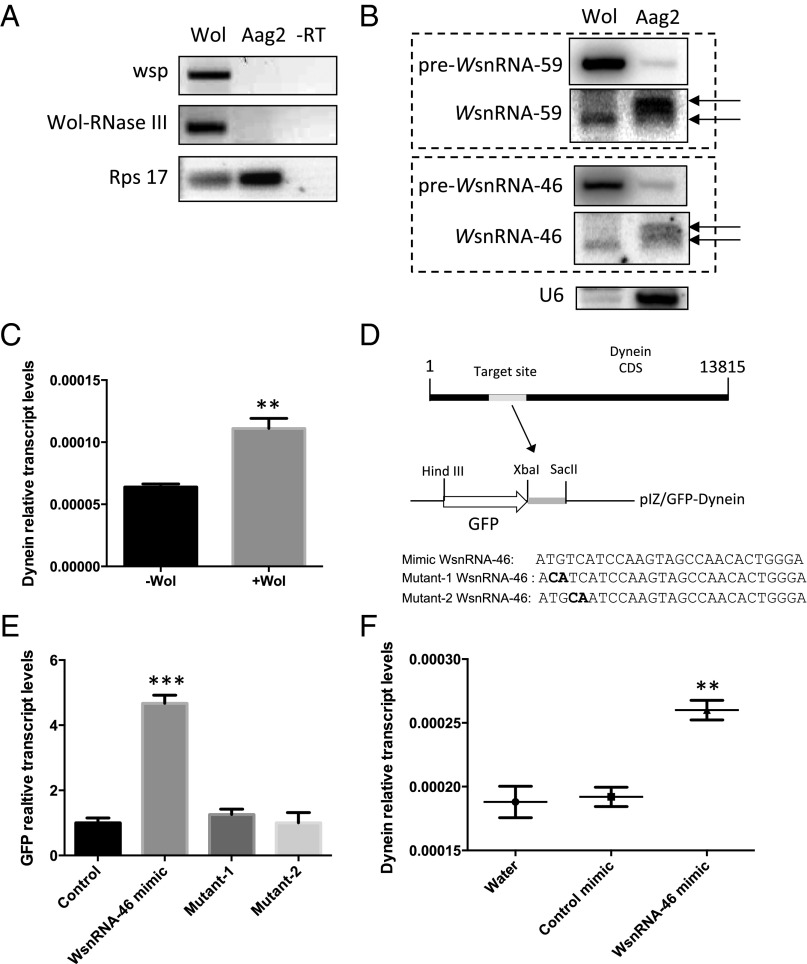
The presence of WsnRNAs, their transport out into the cytoplasm of the mosquito cells, and their potential cross-kingdom effect modulating host genes was assessed. Wolbachia was isolated to purity from Pop cells (Fig. 4A), RNA extractions were performed, and the presence of WsnRNAs in the mosquito cell fraction was evaluated. Using Northern blot, WsnRNA-46 and WsnRNA-59 were detected in Wolbachia but were also detected in the mosquito fraction (Fig. 4B). Although it is likely that during the purification some Wolbachia cells may have lysed and, therefore, contents were released in the host cell fraction (Fig. 4B), the considerable amount of the detectable Wolbachia small RNAs present in the purified cellular fraction, their slightly larger size in the cellular fraction, and that using PCR no wsp or RNase III from Wolbachia were amplified from the purified cellular fraction (Fig. 4A) strongly suggest that these small RNAs most likely have been exchanged between Wolbachia and the host cell.
Fig. 4.
WsnRNA-46 is exported out of Wolbachia and target mosquito genes. (A) Confirmation of seperation of the Wolbachia fraction from Aag2 mosquito cells by RT-PCR using Wolbachia-specific primers, Wolbachia surface protein (wsp) and wRNase III, and the ribosomal host-specific primers Rps17. –RT, negative control. (B) Detection of precursor and mature WsnRNA-46 and WsnRNA-59 in Wolbachia and Aag2 fractions after Northern blot analysis with their respective probes. Arrows indicate mature RNAs. The small nuclear RNA U6 was used as control. (C) Expression levels of Dynein heavy chain in A. aegypti mosquitoes with wMelPop-CLA (Wol+) and without (Wol−). (D) Cloning strategy of Dynein heavy chain target sequence complementary to WsnRNA-46 under the GFP reporter gene ORF in the pIZ/V5 vector, and the mimic and mimic-mutated sequences transfected into the Aag2 cells in the experiment. Mutated residues are in bold. (E) Validation of Dynein–WsnRNA-46 interaction by using a GFP reporter assay. RT-qPCR was used to analyze RNA from Aag2 cells 72 h after cotransfection of pIZ-GFP-dynein with control, WsnRNA-46 mimic, Mutant-1 WsnRNA-46, and Mutant-2 WsnRNA-46. Control cells were mock transfected with only the pIZ/GFP vector. (F) mRNA levels of Dynein heavy chain in A. aegypti mosquitoes collected 3 d after the injection with water, control mimic, or mimic WsnRNA-46. **P < 0.01, ***P < 0.001.
To find out whether the WsnRNAs are functional, target genes for WsnRNA-46 and WsnRNA-59 were searched in the genome of A. aegypti by using a stringent target prediction, and several targets were predicted in a broad array of pathways (Table S3). We focused on the candidate target gene Dynein heavy chain (Dhc) because it has a complete sequence complementarity to the WsnRNA-46 seed region (nucleotides 2–8 from the 5′ end) in the coding region of the gene, the interaction shows a low minimum free energy (mfe) of −30.3 kcal/mol, and that it is a well-characterized gene. Using RT-qPCR, we found that transcript levels of Dhc were significantly higher in Wol+ mosquitoes compared with Wol− mosquitoes (Fig. 4C). The increase was consistent with the up-regulation of WsnRNA-46 in Wol+ mosquitoes compared with Wol− mosquitoes (Fig. 1A), suggesting that by binding to the CDS region of the target gene, WsnRNA-46 could enhance mRNA transcript levels and/or the stability of the mRNA as shown in many other examples (reviewed in ref. 20).
Two additional independent experiments were performed to confirm the specific interaction of WsnRNA-46 with Dhc and the resulting transcript up-regulation. First, we cloned a fragment of 181 nt including the target sequence of Dhc downstream of the GFP gene in the pIZ expression vector (pIZ/GFP-Dynein; Fig. 4D). The construct was transfected into Aag2 cells together with WsnRNA-46 mimic, and also another set of cells were independently transfected with pIZ/GFP-Dynein and two WsnRNA-46 sequences mutated in two different nucleotides in the seed region. After 48 h, significantly higher GFP transcript levels were detected in cells transfected with pIZ/GFP-Dynein and WsnRNA-46 mimic than those transfected with pIZ/GFP-Dynein and any of the two mutated sequences (Fig. 4E).
In a second experiment, A. aegypti Wol− mosquitoes were injected with WsnRNA-46 mimic. Controls were injected with water or a control mimic that consisted of a random sequence with the same length as the mimic. After 72 h, we observed higher transcript levels of the Dhc gene in mosquitoes injected with WsnRNA-46 mimic compared with mosquitoes injected with water or the control mimic (Fig. 4F). All these results together confirm that WsnRNA-46 targets Dhc and enhances mRNA transcript levels and/or the stability of the mRNA. Ferree et al. (21) described how Wolbachia uses the host microtubule network for their placement at the anterior pole of the oocyte in Drosophila. These microtubules facilitate the inward transport of maternal mRNA and protein necessary for proper oocyte and embryonic development. By associating with microtubules through Dynein, Wolbachia may intercept membrane vesicles or other host factors transported along microtubules, thus facilitating its own replication. This idea is supported by the findings in other studies that other bacterial endoparasites, such as Salmonella typhimurium, require host microtubules and Dynein for their replication (21). If Wolbachia localization and replication depend on Dynein expression, it is to its best interest to have a molecular mechanism to modulate its expression.
Conclusions
Here, we revealed the expression of snRNAs (∼30 bp) in Wolbachia that are conserved in different strains of Wolbachia and act as effector molecules regulating both bacterial and mosquito host genes. miRNA and siRNAs have been reported to have cross-kingdom effects in a fungal plant pathogen and also in Plasmodium, the malaria parasite (2, 22). The discovery of these regulatory small RNAs that can affect bacterial and host gene expression opens up an exciting avenue to better understand and manipulate host–pathogen interactions, in particular, the epigenetic effect endosymbionts may have on vector-borne pathogens, leading to better control of diseases that have global impact on human and animal health.
Materials and Methods
Mosquitoes, Flies, and Insect Cells.
A. aegypti infected with the wMelPop-CLA strain of Wolbachia (Wol+) and a Wolbachia-free tetracycline-treated line (Wol−) generated previously (23) were used for the experiments. Insects were reared at 26 °C with 60% relative humidity and a 12-h light regime and fed on a 10% (wt/vol) sucrose solution ad libitum. A. aegypti Aag2 cells were infected with the wMelPop-CLA strain of Wolbachia (Pop) as described for the C6/36.wMelPop-CLA cell line (24). Cells were maintained in growth media in a 1:1 mixture of Mitsuhashi–Maramorosch and Schneider’s insect media (Invitrogen), supplemented with 10% FBS. D. melanogaster and D. simulans naturally infected with wMelPop and wAu strains of Wolbachia, respectively, were maintained in the laboratory at 25 °C with 80% relative humidity and a 12-h light regime.
Deep Sequencing Analysis of Aag2 and Pop Cells.
A small RNA library was generated from total RNA extracted from Aag2 and Pop cells by using TRI Reagent (Molecular Research Centre) using the Illumina Truseq Small RNA Preparation kit according to Illumina’s TruSeq Small RNA Sample Preparation Guide. The purified cDNA library was used for cluster generation on Illumina’s Cluster Station and then sequenced on Illumina GAIIx by following the vendor’s instructions for running the instrument. Raw sequencing reads (40 nt) were obtained by using Illumina’s Sequencing Control Studio software version 2.8 (SCS v2.8) following real-time sequencing image analysis and base-calling by Illumina's Real-Time Analysis version 1.8.70 (RTA v1.8.70). The extracted sequencing reads were then used for data analysis. Reads with less than 15 bp, low quality reads, and reads without a 3′ adaptor were removed. Reads mapped to Rfam and Repbase were discarded before any further analysis. Read were mapped to the Wolbachia genome, and a sequence extended from the mapped locations was folded to predict the secondary structure by using UNAFold (25). The reads located in the sequence that can form a potential hairpin were selected as potential candidates. Further, RNAfold (26) was used to predict the secondary structures of the preWsnRNA and their minimum folding energy. Deep sequencing data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession no. GSE55210.
Northern Blot Hybridizations.
Total RNA was extracted from mosquitoes, flies, or female mosquito tissues by using the TRI Reagent (Molecular Research Centre). RNA samples were run on 15% urea denaturing polyacrylamide gels, electroblotted to nylon membranes by a semidried Western blotting apparatus (Bio-Rad), and UV cross-linked. DNA oligonucleotides with reverse complementarity to specific WsnRNA sequences were labeled with [α-32P]dCTP by using terminal nucleotide transferase. All probe hybridizations and washes were done at 50 °C. Blots were exposed to a phosphorimager screen overnight, and radioactive signals were detected by using a phosphorimager scanner. In cases when a blot was used multiple times, the probe was removed by washing the blot with boiling 0.1% SDS twice for 30 min each time. Stripping of the probe was confirmed by scanning the blots as described above, and most hybridizations were repeated at least three times to ensure specificity of the probes.
Expression of preWsnRNA in E. coli and Mosquito Cells.
A fragment of 428 and 439 bp containing the preWsnRNA-46 and preWsnRNA-59 sequences, respectively, were inserted into the vector pIZ/V5 (Invitrogen) and pGEM-T-Easy (Promega). After the transformation of the vector pGEM-T-Easy-preWsnRNA into E. coli, expression was carried out in an incubator for 16 h at 37 °C and horizontal shaking. A sequence of 420 bp from a random Wolbachia gene cloned into pGEM-T-Easy was used as a control. After the incubation, cells were collected by centrifugation, and RNA was extracted by using TRI Reagent (Molecular Research Centre) and analyzed by Northern blot. Probes against WsnRNA-46 and WsnRNA-59 were used, and a probe matching the 3′ end of the control gene sequence was used as control. A total of 2 μg of preWsnRNA sequences into pIZ/V5 were transfected into Aag2 and Pop cells by using the Cellfectin reagent (Invitrogen). Cells transfected with the empty pIZ/V5 vector were used as control. Cells were kept at 27 °C in an incubator, and collected 72 h after transfection for RNA isolation by using TRI Reagent.
qPCR Studies.
Total RNA of Wol+ and Wol− mosquitoes or Aag2 and Pop cells were isolated by using Tri-Reagent and subsequently treated with DNase I before being used for reverse transcription. A total of 2 µg of RNA for each sample was reverse transcribed by using oligo dT in a total volume of 20 μL. qPCR with gene-specific primers was performed to determine their mRNA levels in infected versus uninfected mosquitoes or cells. Platinum SYBR Green Mix (Qiagen) with 1 μL of the first-strand cDNA reaction was used in a Rotor-Gene thermal cycler (Qiagen) under the following conditions: 95 °C hold for 30 s, then 40 cycles of 95 °C for 15 s, 50 °C for 15 s, and 72 °C for 20 s, followed by the melting curve (68–95 °C). The RPS17 gene was used for normalization of the RNA samples by using mosquito genes, and wsp gene was used for normalization of the RNA samples by using Wolbachia genes. Student’s t test was used to compare the differences in means. For qPCR, each biological sample had two technical replicates. Inhibitor and control inhibitor experiments (Fig. 3) had three biological replicates each, with two technical replicates per biological sample.
WsnRNA Target Analysis.
NCBI BLAST, RNAHybrid, and RNA22 software (IBM) were used to find potential targets of WsnRNA-46 and WsnRNA-59 in the A. aegypti genome and in the Wolbachia wMelPop-CLA strain. A fragment of ∼150 bp for each target tested containing the target sequences complementary to WsnRNA-46 were cloned into pIZ/V5-His vector (Invitrogen) downstream of GFP by using XbaI and SacII restriction sites, resulting in pIZ/GFP target (Fig. 3B). Mimic, mutant mimics, and control inhibitors were synthesized by Genepharma and used in transfection studies at a concentration of 100 μM/mL. The expression levels of the GFP gene was analyzed 72 h after transfection by RT-qPCR by using three biological replicates, each with three technical replicates.
Inhibition of WsnRNA-46 in Wolbachia-Infected Cells.
An inhibitor for WsnRNA-46 (5′-UCCCAGUGUUGGCUACUUGGAUGACAU-3′) and a control inhibitor (a random sequence; 5′-CAGUACUUUUGUGUAGUACAA-3′) were synthesized by Genepharma. One hundred nanograms of the WsnRNA-46 inhibitor or the control inhibitor was transfected into Pop cells by using the Cellfectin transfection reagent (Invitrogen). Cells were collected at 72 h after transfections, total RNA was extracted, RT-PCR was conducted, and qPCR analyses were performed as described above.
Wolbachia Isolation from Aag2 Cells.
Wolbachia isolation from host cells was conducted by following the protocol described (27) with some modifications. Briefly, Wolbachia was isolated from Aag2 cells infected with Wolbachia wMelPop-CLA. The cells were homogenized in a glass Dounce homogenizer by using a 40-mL cold sucrose-phosphate-glutamate (SPG) buffer (218 mM sucrose, 3.8 mM KH2PO4, 7.2 mM K2HPO4, 4.9 mM l-glutamate, pH 7.2). The extract was split into four Falcon tubes containing another 20 mL of SPG buffer each and centrifuged at 3,200 × g for 15 min, twice. The supernatant was sequentially filtered through 5, 2.7 and 1.2 μm syringe filters, and Wolbachia were pelleted at 18,000 × g for 20 min and resuspended in cold SPG buffer in Eppendorf tubes. The supernatant fraction containing the host RNA was filtered through a 0.22-μm syringe filter to remove any Wolbachia left, and the fraction was saved for later RNA extraction. Intact Wolbachia in SPG buffer were treated with 20 μL of DNase I (Roche) (30 μg/mL) for 30 min at 37 °C to remove host DNA contamination without disrupting the cells and with 5 μL of RNase (Fermentas) for 15 min at 37 °C. RNA from isolated Wolbachia and from the host fraction were extracted separately by using the TRI Reagent (Molecular Research Centre).
Mosquito Injections with WsnRNA-46.
Wolbachia-free 3-d-old mosquitoes were CO2-anesthetized and injected into the thorax with 20 μM WsRNA-46 mimic (5′-UCCCAGUGUUGGCUACUUGGAUGACAU-3′) in 69 μL of sterile water. Control mosquitoes were injected with sterile water or control mimic (5′-UUCUCCGAACGUGUCACGUTT-3′), also in 69 μL of water. All mosquitoes were collected 3 d after injection. qPCRs were performed in triplicates with cycling conditions detailed above. The ANOVA test was used to compare differences in means between different treatments.
Promoter Predictions.
Promoters and transcription factors upstream of the WsnRNAs were predicted by using the Softberry software BPROM (28).
Supplementary Material
Acknowledgments
This project was funded by Australian Research Council Discovery Grant DP110102112 (to S.A.), DECRA Fellowship DE120101512 (to M.H.), and a grant from the Foundation of the National Institutes of Health through the Grand Challenges in Global Health Initiative of the Bill and Melinda Gates Foundation (to S.L.O.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE55210).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1420131112/-/DCSupplemental.
References
- 1.Asgari S. MicroRNA functions in insects. Insect Biochem Mol Biol. 2013;43(4):388–397. doi: 10.1016/j.ibmb.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 2.LaMonte G, et al. Translocation of sickle cell erythrocyte microRNAs into Plasmodium falciparum inhibits parasite translation and contributes to malaria resistance. Cell Host Microbe. 2012;12(2):187–199. doi: 10.1016/j.chom.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zug R, Hammerstein P. Still a host of hosts for Wolbachia: Analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS ONE. 2012;7(6):e38544. doi: 10.1371/journal.pone.0038544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moreira LA, et al. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell. 2009;139(7):1268–1278. doi: 10.1016/j.cell.2009.11.042. [DOI] [PubMed] [Google Scholar]
- 5.Bian G, et al. Wolbachia invades Anopheles stephensi populations and induces refractoriness to Plasmodium infection. Science. 2013;340(6133):748–751. doi: 10.1126/science.1236192. [DOI] [PubMed] [Google Scholar]
- 6.Bourtzis K, et al. Harnessing mosquito-Wolbachia symbiosis for vector and disease control. Acta Trop. 2014;132(Suppl):S150–S163. doi: 10.1016/j.actatropica.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 7.Hussain M, Frentiu FD, Moreira LA, O’Neill SL, Asgari S. Wolbachia uses host microRNAs to manipulate host gene expression and facilitate colonization of the dengue vector Aedes aegypti. Proc Natl Acad Sci USA. 2011;108(22):9250–9255. doi: 10.1073/pnas.1105469108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hussain M, O’Neill SL, Asgari S. Wolbachia interferes with the intracellular distribution of Argonaute 1 in the dengue vector Aedes aegypti by manipulating the host microRNAs. RNA Biol. 2013;10(12):1868–1875. doi: 10.4161/rna.27392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayoral JG, Etebari K, Hussain M, Khromykh AA, Asgari S. Wolbachia infection modifies the profile, shuttling and structure of microRNAs in a mosquito cell line. PLoS ONE. 2014;9(4):e96107. doi: 10.1371/journal.pone.0096107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang G, Hussain M, O’Neill SL, Asgari S. Wolbachia uses a host microRNA to regulate transcripts of a methyltransferase, contributing to dengue virus inhibition in Aedes aegypti. Proc Natl Acad Sci USA. 2013;110(25):10276–10281. doi: 10.1073/pnas.1303603110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caldelari I, Chao Y, Romby P, Vogel J. RNA-mediated regulation in pathogenic bacteria. Cold Spring Harb Perspect Med. 2013;3(9):a010298. doi: 10.1101/cshperspect.a010298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shmaryahu A, Carrasco M, Valenzuela PD. Prediction of bacterial microRNAs and possible targets in human cell transcriptome. J Microbiol. 2014;52(6):482–489. doi: 10.1007/s12275-014-3658-3. [DOI] [PubMed] [Google Scholar]
- 13.Furuse Y, et al. Search for microRNAs expressed by intracellular bacterial pathogens in infected mammalian cells. PLoS ONE. 2014;9(9):e106434. doi: 10.1371/journal.pone.0106434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedländer MR, et al. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol. 2008;26(4):407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 15.Darby AC, et al. Integrated transcriptomic and proteomic analysis of the global response of Wolbachia to doxycycline-induced stress. ISME J. 2014;8(4):925–937. doi: 10.1038/ismej.2013.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Q, et al. Transcription of the major neurospora crassa microRNA-like small RNAs relies on RNA polymerase III. PLoS Genet. 2013;9(1):e1003227. doi: 10.1371/journal.pgen.1003227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor L, Meador S, Richeson K. 2010 The Terminator: A terminator identification tool. Available at gcat.davidson.edu/Spring2010/terminators/index.html. Accessed December 2, 2014.
- 18.Naville M, Ghuillot-Gaudeffroy A, Marchais A, Gautheret D. ARNold: A web tool for the prediction of Rho-independent transcription terminators. RNA Biol. 2011;8(1):11–13. doi: 10.4161/rna.8.1.13346. [DOI] [PubMed] [Google Scholar]
- 19.Dobson SL, et al. Wolbachia infections are distributed throughout insect somatic and germ line tissues. Insect Biochem Mol Biol. 1999;29(2):153–160. doi: 10.1016/s0965-1748(98)00119-2. [DOI] [PubMed] [Google Scholar]
- 20.Vasudevan S. Posttranscriptional upregulation by microRNAs. Wiley Interdiscip Rev RNA. 2012;3(3):311–330. doi: 10.1002/wrna.121. [DOI] [PubMed] [Google Scholar]
- 21.Ferree PM, et al. Wolbachia utilizes host microtubules and Dynein for anterior localization in the Drosophila oocyte. PLoS Pathog. 2005;1(2):e14. doi: 10.1371/journal.ppat.0010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weiberg A, et al. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science. 2013;342(6154):118–123. doi: 10.1126/science.1239705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McMeniman CJ, et al. Stable introduction of a life-shortening Wolbachia infection into the mosquito Aedes aegypti. Science. 2009;323(5910):141–144. doi: 10.1126/science.1165326. [DOI] [PubMed] [Google Scholar]
- 24.Frentiu FD, Robinson J, Young PR, McGraw EA, O’Neill SL. Wolbachia-mediated resistance to dengue virus infection and death at the cellular level. PLoS ONE. 2010;5(10):e13398. doi: 10.1371/journal.pone.0013398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Markham NR, Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005;33(Web Server issue):W577–W581. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31(13):3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iturbe-Ormaetxe I, Woolfit M, Rancès E, Duplouy A, O’Neill SL. A simple protocol to obtain highly pure Wolbachia endosymbiont DNA for genome sequencing. J Microbiol Methods. 2011;84(1):134–136. doi: 10.1016/j.mimet.2010.10.019. [DOI] [PubMed] [Google Scholar]
- 28.Solovyev V, Salamov A. Automatic annotation of microbial genomes and metagenomic sequences. In: Li R, editor. Metagenomics and its Applications in Agriculture, Biomedicine and Environmental Studies. Nova Science Publishers; Hauppauge, NY: 2011. pp. 61–78. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.