Significance
Long noncoding RNA (lncRNA) has been implicated in carcinogenesis and regarded as an emerging alternative target for cancer therapy. This study provides, to our knowledge, the first case of lncRNA that serves as a coactivator of c-Myc and a master regulator of tumor metabolism. This lncRNA, prostate cancer gene expression marker 1 (PCGEM1), is overexpressed in prostate cancer and implicated in castration resistance. It also forms an independent complex with androgen receptor (AR). Being a dual coactivator for AR and c-Myc, PCGEM1 reprograms the transcriptional network of metabolic genes and androgen-responsive genes, making it an ideal therapeutic target for prostate cancer. The present study provides significant insights into lncRNA’s role in c-Myc activation, tumor metabolism, and prostate carcinogenesis.
Keywords: lncRNA, tumor metabolism, c-Myc coactivator, prostate cancer
Abstract
Long noncoding RNAs (lncRNAs) have been implicated in a variety of physiological and pathological processes, including cancer. In prostate cancer, prostate cancer gene expression marker 1 (PCGEM1) is an androgen-induced prostate-specific lncRNA whose overexpression is highly associated with prostate tumors. PCGEM1’s tumorigenic potential has been recently shown to be in part due to its ability to activate androgen receptor (AR). Here, we report a novel function of PCGEM1 that provides growth advantages for cancer cells by regulating tumor metabolism via c-Myc activation. PCGEM1 promotes glucose uptake for aerobic glycolysis, coupling with the pentose phosphate shunt to facilitate biosynthesis of nucleotide and lipid, and generates NADPH for redox homeostasis. We show that PCGEM1 regulates metabolism at a transcriptional level that affects multiple metabolic pathways, including glucose and glutamine metabolism, the pentose phosphate pathway, nucleotide and fatty acid biosynthesis, and the tricarboxylic acid cycle. The PCGEM1-mediated gene regulation takes place in part through AR activation, but predominantly through c-Myc activation, regardless of hormone or AR status. Significantly, PCGEM1 binds directly to target promoters, physically interacts with c-Myc, promotes chromatin recruitment of c-Myc, and enhances its transactivation activity. We also identified a c-Myc binding domain on PCGEM1 that contributes to the PCGEM1-dependent c-Myc activation and target induction. Together, our data uncover PCGEM1 as a key transcriptional regulator of central metabolic pathways in prostate cancer cells. By being a coactivator for both c-Myc and AR, PCGEM1 reprograms the androgen network and the central metabolism in a tumor-specific way, making it a promising target for therapeutic intervention.
Long noncoding RNAs (lncRNAs) have recently drawn increasing attention as important players in physiological and pathological processes. In cancer, aberrant expression and mutations of lncRNAs can contribute to tumor development and progression by promoting proliferation, invasion, metastasis, and survival (1–3). LncRNAs thus may serve as diagnostic biomarkers and therapeutic targets for cancer. LncRNAs function at several levels of cellular processes, and the majority thus far studied are involved in gene regulation either at the transcriptional or posttranscriptional level (4). At the transcriptional level, lncRNA can serve as a chaperon or scaffold to deliver transcriptional factor to the chromatin site, to modulate the chromatin conformation by recruiting histone-modifying complexes, and to connect distal gene-regulatory elements together to effectively modulate the transcription of the targeted loci (4).
Prostate cancer gene expression marker 1 (PCGEM1) is a prostate tissue-specific lncRNA highly associated with prostate cancer (5). Over 80% of patient specimens show elevated levels (5), and the occurrence of PCGEM1 overexpression seems to be significantly higher in African-American patients, whose population has the highest prostate-cancer incidence in the world (6). The clinical evidence thus strongly indicates the tumorigenic potential of PCGEM1 in prostate-cancer development. The oncogenic property of PCGEM1 is further demonstrated by its ability to promote cell proliferation and increase colony formation upon overexpression (6), as well as conferring resistance to doxorubicin-induced apoptosis via attenuation of p53 and p21 responses (7). Despite being a hormone-regulated malignancy at early stages, the majority of prostate cancer develops into hormone independence during progression, resulting in disease relapse, and makes the current hormone-deprivation therapy ineffective. Recent research has therefore emphasized mechanisms underlying cancer progression to hormone independence or castration resistance. In a systematic transcriptome analysis using a human prostate cancer cell line (LNCaP) mouse xenograft model to identify genes differentially expressed during tumor progression, PCGEM1 was found significantly up-regulated in the castration-recurrent stage, implicating its role in the development of hormone-refractory cancer (8). The recent work of Yang et al. (9) elegantly demonstrated that PCGEM1 is associated with and activates androgen receptor (AR), which contributes to the development of castration-resistant prostate cancer. Although a recent report does not support the role of PCGEM1 or PRNCR1 in AR activation (10), evidence from other studies described above suggests PCGEM1 as a potentially useful biomarker as well as a therapeutic target for prostate cancer. In the present study, we define a previously unidentified role of PCGEM1 in prostate carcinogenesis. It functions as a master regulator of tumor metabolism that facilitates the biosynthesis of cellular building materials, providing proliferative advantages for cancer cells. PCGEM1 regulates metabolic programming by enhancing activation of c-Myc and AR, which, in turn, control the expression profiles of multiple key metabolic pathways. We found that PCGEM1 directly binds c-Myc, enhances c-Myc transactivation potency, and facilitates the recruitment of c-Myc to the chromatin target sites. These functions are independent of its association with AR. However, being a coactivator of both c-Myc and AR, PCGEM1 reprograms both metabolic and AR genes and represents an ideal target for therapy.
Results
PCGEM1 Regulates Prostate Cancer Cell Growth and Tumor Metabolism.
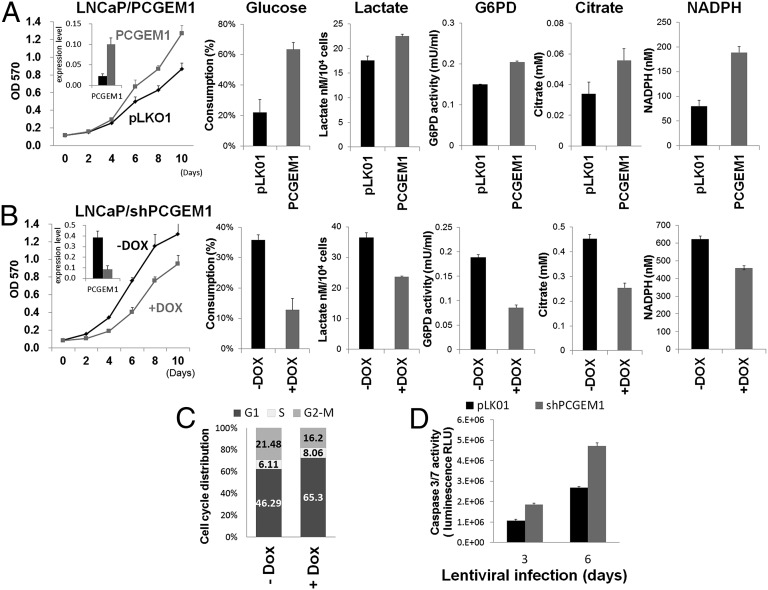
PCGEM1 is an androgen-inducible gene whose expression is reported to associate exclusively with AR-positive cell lines (5). To further explore its physiological role, we used the androgen-responsive, androgen-dependent LNCaP prostate cancer cell line and developed derivatives with either constitutive overexpression of PCGEM1 (LNCaP/PCGEM1) or with doxycycline (DOX)-inducible knockdown of PCGEM1 (LNCaP/shPCGEM1). Consistent with a previous report (6), PCGEM1 overexpression led to accelerated cell growth (Fig. 1A). By contrast, knockdown of PCGEM1 by DOX treatment to LNCaP/shPCGEM1 cells resulted in retarded proliferation (Fig. 1B) and G1 arrest (Fig. 1C). Prolonged PCGEM1 knockdown also gave rise to increased caspase 3/7 activity, indicating induced apoptosis in the knockdown cells (Fig. 1D). Given that tumor cells often develop altered metabolism to cope with the demand of cell-mass increase during growth, we next examined whether the PCGEM1-dependent proliferation involves metabolic reprogramming. Indeed, LNCaP/PCGEM1 showed increased glucose uptake and lactate production, indicating elevated glycolysis (Fig. 1A). PCGEM1 overexpression also increased the activity of glucose-6-phosphate dehydrogenase (G6PD), the rate-limiting enzyme of pentose phosphate pathway (PPP) shunting the carbon flow from glucose to ribose-5-phosphate, through which generating the reducing agent NADPH that is essential for maintaining cellular redox status. The subsequent increase of ribose-5-phosphate presumably further accelerated nucleotide synthesis. Citrate, the intermediate for fatty-acid synthesis, was also increased in LNCaP/PCGEM1 cells (Fig. 1A). These results suggest that PCGEM1 overexpression leads to increased glucose uptake and glycolysis that facilitate macromolecule biosynthesis and ensures the supply of intracellular reducing energy. Knockdown of PCGEM1 in contrast, resulted in decreased glucose consumption and lactate production, as well as decreased G6PD activity and citrate level, indicating reduced glycolysis and anabolism (Fig. 1B). Together, our data suggest that PCGEM1 plays an important role in regulating metabolism essential for cell-cycle progression, proliferation, and survival of LNCaP cells.
Fig. 1.
PCGEM1 regulates proliferation and metabolism. (A and B) MTT proliferation assay and metabolic profiles of PCGEM1-overexpressing cells (A) and DOX-inducible PCGEM1 knockdown cells (B). Cells were collected for metabolic enzyme reactions on day 3 after lentiviral transduction or DOX treatment and were monitored for proliferation to 10 d. The measurement of glucose consumption, lactate production, G6PD activity, and citrate and NADPH levels were normalized by cell number. SD was derived from biological triplicates. Inset graphs illustrate PCGEM1 expression level. (C) The DNA content of LNCaP/shPCGEM1 cells was detected by propidium iodide staining and flow-cytometry analysis. Population of each cell-cycle stage is quantified and plotted, as a percentage of the total cell population, in the stacked bar graph. (D) Caspase 3/7 activity was measured in LNCaP cells transduced with pLKO.1or shPCGEM1 for the indicated days.
PCGEM1 Regulates Metabolic Genes.
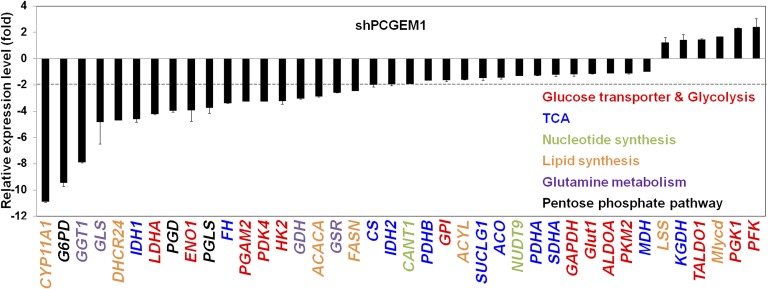
To investigate how PCGEM1 regulates cellular metabolism, we surveyed expression of metabolic enzymes in PCGEM1 knockdown cells (LNCaP/shPCGEM1 +DOX) and found that the PCGEM1-mediated metabolic alterations take place at the transcriptional level. Several enzymes involved in glucose uptake, glycolysis, PPP, lipid synthesis, glutamine metabolism, and the TCA cycle were prominently down-regulated in PCGEM1 knockdown cells (Fig. 2). These data strongly suggest that PCGEM1 functions as a key regulator of multiple metabolic genes, whose expression alterations in turn lead to metabolic outcomes beneficial to tumor growth.
Fig. 2.
PCGEM1 regulates expression of metabolic enzymes in multiple pathways. Expression of the metabolic genes in PCGEM1 knockdown LNCaP cell was examined by qRT-PCR. Positive and negative values indicate up and down-regulation compared with control cell, respectively.
PCGEM1 Regulates Metabolic Genes in both AR-Dependent and -Independent Manners.
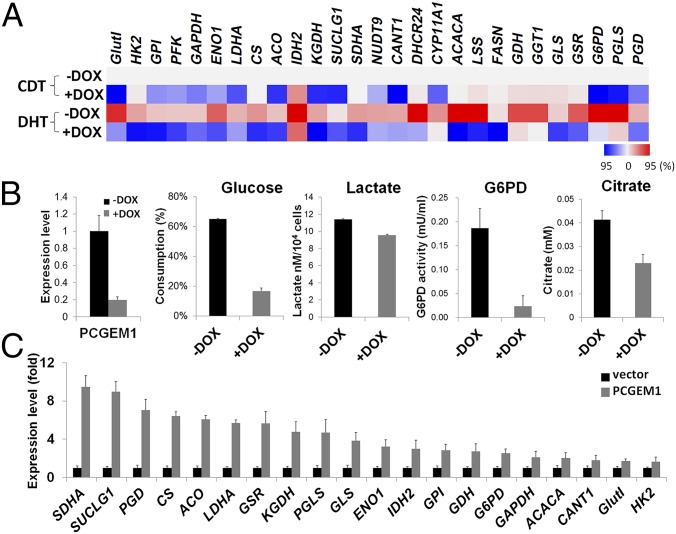
In prostate-cancer cells, androgen signaling and AR play an important role in regulating cellular metabolism (11). Consistent with the report of Massie et al. (11), our data showed that androgen (DHT) significantly induced genes in multiple metabolic pathways, including glucose uptake, glycolysis, PPP, lipid, and nucleotide synthesis. Additionally, we found that several enzymes in glutamine metabolism and the TCA cycle were also induced upon DHT treatment (Fig. 3A). Strikingly, when knocking down PCGEM1 (LNCaP/shPCGEM1 + DOX), the DHT-dependent gene induction was drastically compromised, suggesting that PCGEM1 is a key regulator for androgen-dependent metabolic gene expression (Fig. 3A). PCGEM1 has been recently shown to be a coactivator of AR (9). In agreement with Yang et al. (9), we independently confirmed the association of AR with PCGEM1 and extended the results to demonstrate PCGEM1’s ability to transactivate the prostate-specific antigen (PSA) enhancer and FK506 binding protein 5 promoter, two well-known AR targets (Fig. S1 A and B). We also showed that AR was recruited to a subset of the PCGEM1-regulated gene promoters and that the chromatin recruitment was partially reduced in PCGEM1 knockdown cells (Fig. S1C). These results indicate that PCGEM1 may facilitate AR binding to some of the metabolic gene promoters. However, given the limited reduction of AR chromatin occupancy in the knockdown cell observed, we anticipated that the PCGEM1-mediated AR activation and target transcription may rely on other mechanisms, such as the formation of androgen-dependent chromatin looping reported by Yang et al. (9). Together, our results suggest that PCGEM1 plays an essential role in the androgen-induced metabolic gene regulation via AR.
Fig. 3.
Metabolic gene regulation in androgen-dependent and -independent manners. (A) Metabolic expression profiles of LNCaP/shPCGEM1 cells cultured in hormone-deprived (CDT) or androgen-treated (DHT) conditions. The expression levels of control cells in CDT medium (−DOX CDT) were used as baseline to compare with other treatment. Relative expression fold changes are illustrated by heat map (red, up-regulation; blue, down-regulation). Color scale indicates the 95th percentile of either up- or down-regulated expression range. (B) Metabolic profiles of the PCGEM1 knockdown cell under hormone deprivation. The procedures for PCGEM1 knockdown and enzymatic reactions are as described in Fig. 1B. (C) Metabolic gene expression in PC3 cells overexpressing control vector or PCGEM1 was detected by qRT-PCR. The normalized expression levels were compared using vector control as the baseline (shown by fold, P < 0.05).
Interestingly, in addition to its role in androgen response, PCGEM1 seemed to be essential for the tumor metabolic regulation in hormone-deprived conditions as well. Under hormone deprivation (CDT), knockdown of PCGEM1 caused down-regulation of multiple metabolic genes (Fig. 3A, +DOX CDT) and further resulted in significant metabolic alterations indicative of down-regulated glycolysis and macromolecule biosynthesis (Fig. 3B). These results revealed PCGEM1’s critical role in tumor metabolism independent of androgen signaling and suggest that, in addition to AR, another transcription factor may be involved in PCGEM1’s metabolic regulatory role. This assumption was strongly supported by the observation that, in AR-negative prostate cancer cell PC3, whose growth and survival do not require androgen, overexpression of PCGEM1 was capable of widely inducing the metabolic genes (Fig. 3C). Our data support the notion that, as an androgen-inducible gene, PCGEM1 enhances AR activity and contributes to the androgen-induced metabolic reprogramming. Moreover, we also revealed PCGEM1’s critical role in metabolic regulation independent of androgen and AR.
PCGEM1 Functions as a Coactivator of c-Myc.
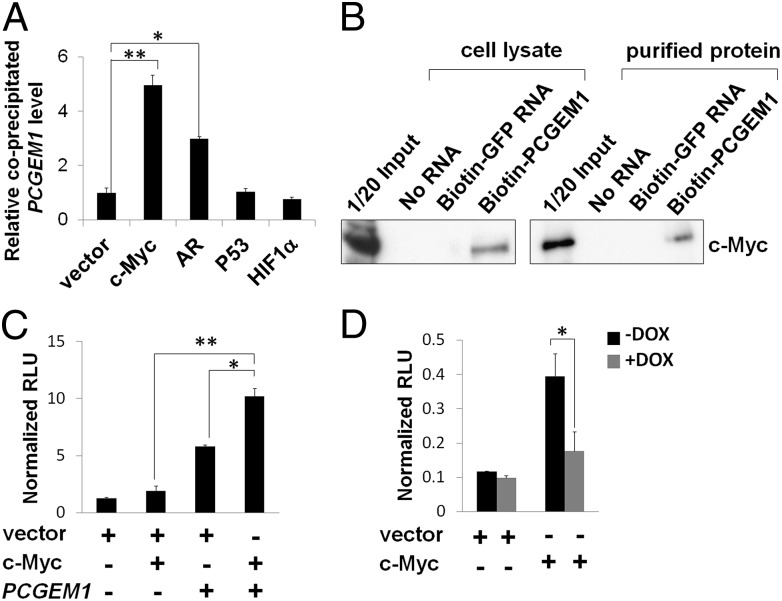
Several transcription factors, such as c-Myc, p53 and HIF-1α, are implicated in regulating cancer-cell metabolism (12, 13). To identify the transcription factors involved in PCGEM1-mediated metabolic regulation, we performed RNA immunoprecipitation (RIP) to examine whether they form complexes with PCGEM1. As shown in Fig. 4A, we found that, in addition to AR, ectopically expressed PCGEM1 predominantly interacted with c-Myc whereas no significant association with p53 or HIF-1α was detected. The PCGEM1 and c-Myc interaction was also detected at the endogenous level, and the specificity was confirmed by c-Myc exclusively binding with PCGEM1 but not PRNCR1, another prostate cancer-associated lncRNA (Fig. S2). Consistently, in RNA pull-down assays using purified recombinant c-Myc or LNCaP cell lysate, we showed that the in vitro transcribed biotinylated PCGEM1 associated with c-Myc through direct binding (Fig. 4B). Given the evidence of physical interaction, we further tested whether PCGEM1 directly enhances c-Myc transactivation potency. Using a Myc-responsive luciferase construct, we showed that overexpression of PCGEM1 itself was capable of inducing c-Myc transactivation activity whereas the combination of PCGEM1 and c-Myc overexpression synergistically enhanced the promoter activity (Fig. 4C). In a reciprocal experiment, the c-Myc activity was compromised when knocking down endogenous PCGEM1 (+DOX), indicating that PCGEM1 functions as a natural coactivator of c-Myc in prostate cancer cells (Fig. 4D). In conclusion, we identified c-Myc as a novel binding partner of PCGEM1, which positively regulates the transactivity of c-Myc.
Fig. 4.
PCGEM1 functions as a c-Myc coactivator. (A) Association of ectopically expressed PCGEM1 with HA-tagged c-Myc, AR, p53, or HIF-1α was detected by RIP assay. The relative levels of coimmunoprecipitated PCGEM1 were calculated as fold difference compared with vector control (*P < 0.05, **P < 0.01). (B) RNA pull-down of the in vitro transcribed biotinylated PCGEM1 incubated with LNCaP cell lysate (Left) or purified recombinant c-Myc protein (Right). The biotinylated GFP mRNA served as a negative control. (C) Myc responsive luciferase was cotransfected with empty vector, c-Myc, PCGEM1, or both into PC3 cells for the reporter assay. Coexpression of PCGEM1 and c-Myc significantly enhanced the luciferase activity. (D) The luciferase activity (same as C) was measured in LNCaP/shPCGEM1 cell cultured with or without DOX treatment. Knockdown of the endogenous PCGEM1 significantly reduced c-Myc transactivity. The relative luciferase activity was calculated by normalization against Renilla-Luc activity (*P < 0.05, **P < 0.01).
c-Myc Is the Major Effector of PCGEM1-Dependent Metabolism Regulation.
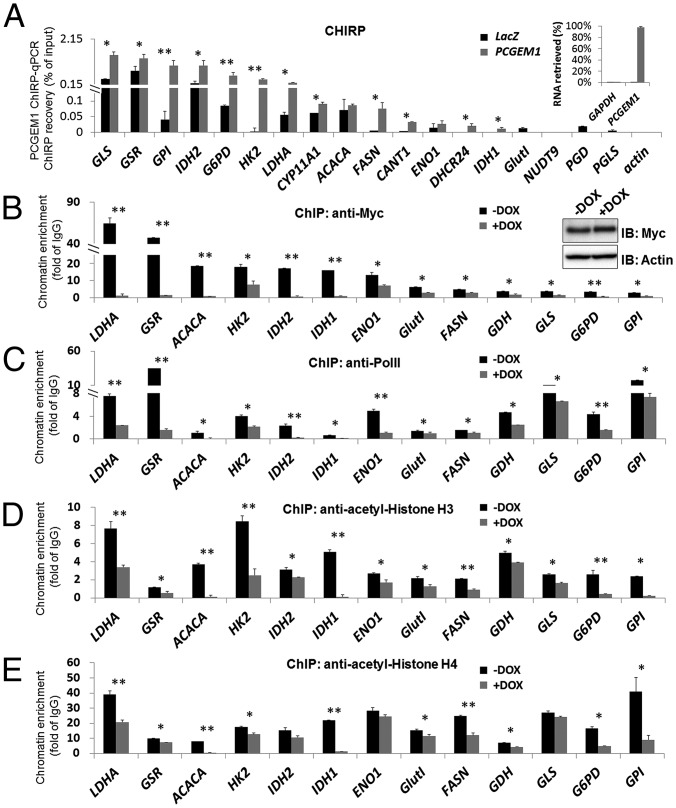
Global metabolic programming is one of the most prominent c-Myc properties in tumorigenesis (14). Our findings that PCGEM1 formed complexes with and coactivated c-Myc strongly suggest c-Myc as a key effector of PCGEM1 in metabolic gene regulation. This hypothesis was supported by the following evidence. First, several metabolic enzymes showing more than twofold expression alteration upon PCGEM1 knockdown have been documented as c-Myc targets (Fig. 2; summarized in Fig. S3) (14, 15). Second, chromatin isolation by RNA purification (ChIRP) analysis showed that PCGEM1 physically associated to a subset of the metabolic gene promoters (Fig. 5A) that are c-Myc binding sites identified in The Encyclopedia of DNA Elements (ENCODE) ChIP-seq data (also confirmed by our c-Myc ChIP assay) (Fig. 5B). Therefore, the DNA loci that were detected in the captured PCGEM1 complex suggest overlapped chromatin occupancy of PCGEM1 with c-Myc, indicating lncRNA-transcriptional complex formation on these targets. The few c-Myc binding loci failing to show PCGEM1 enrichment could be due to various reasons, including the assay stringency or the possibility that PCGEM1 associates with the target at regions distal to the c-Myc binding site.
Fig. 5.
PCGEM1 associates with the target chromatin and promotes c-Myc recruitment and active transcription. (A) Chromatin occupancy of PCGEM1 to the target loci was detected by ChIRP assay. The PCGEM1-associated metabolic gene promoters were detected by qPCR, and the recovered DNA level was estimated as the percentage of input chromatin. Actin promoter was used as negative control. The Inset graph illustrates the efficiency and specificity of RNA retrieved from streptavidin-bound probes. GAPDH mRNA was used to evaluate nonspecific binding of the biotinylated probes. (B–E) ChIP analysis of c-Myc (B) and PolII (C) recruitment, and acetylated-histone H3 (D), -histone H4 (E) on metabolic gene promoters. Recovered DNA in the precipitated complexes was analyzed by qPCR, and the DNA levels were normalized against nonspecific IgG-bound DNA to obtain the enrichment folds, indicated by fold of IgG. Significantly reduced chromatin enrichment in PCGEM1 knockdown cell is indicated by asterisks (*P < 0.05, **P < 0.01). The Inset Western blots show similar c-Myc protein level in –DOX and +DOX cells.
To further understand the mechanism of how PCGEM1 regulates c-Myc, we first tested whether it increases the protein stability of c-Myc. We found that no significant changes in the protein turnover rate were detected (Fig. S4A) and that no detectable difference of total c-Myc protein level was observed in the PCGEM1 knockdown cell either (Fig. 5B, Inset). We next tested whether PCGEM1 promotes c-Myc loading to the chromatin. Significantly, the ChIP assay confirmed c-Myc occupancy on a significant number of PCGEM1-dependent metabolic gene promoters and showed that its recruitment was drastically impaired by the knockdown of PCGEM1 (+DOX) (Fig. 5B). Concurrently, the recruitment of RNA polymerase II (PolII) was generally diminished in PCGEM1 knockdown cells (Fig. 5C), indicating a suppressed transcriptional status of these metabolic genes. c-Myc-mediated transcription in vivo involves several histone modification events at the target promoters (16). In particular, once bound to the target, c-Myc induces local hyperacetylation of both histone H3 and H4, further maintaining the active chromatin status (16, 17). Given the reduced c-Myc occupancy upon PCGEM1 knockdown, we examined whether the histone hyperacetylation on the target loci is decreased as well. Consistently, we found that PCGEM1 knockdown (+DOX) resulted in decreased H3 and H4 acetylation on most of the targets (Fig. 5 D and E), indicating inactive chromatin and compromised transcriptional status. Together, these data suggest that PCGEM1 forms complexes with c-Myc on their target loci and functions in promoting the recruitment of c-Myc and presumably other transcriptional regulators to enhance c-Myc transcriptional activity.
Structural and Functional Domain Mapping of PCGEM1.
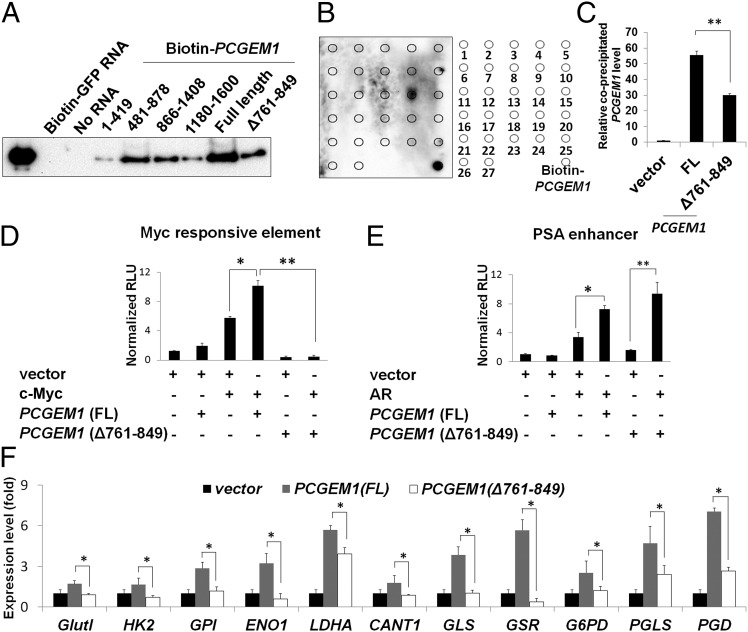
To map the c-Myc binding domain on PCGEM1, various in vitro transcribed and biotinylated PCGEM1 truncates were incubated with recombinant c-Myc protein for RNA pull-down assays. The results showed that, compared with others, the RNA fragment containing nucleotides 481–878 associated to c-Myc with the highest affinity (Fig. 6A). Subsequently, we used synthesized tiling oligos that are complementary to the PCGEM1 transcript to probe the c-Myc–bound PCGEM1 fragments. The fourteenth probe complementary to nucleotides 781–840 exhibited the strongest intensity (Fig. 6B). This region falls within the fragment 481–878 identified in Fig. 6A. Conversely, both RIP assays and in vitro RNA pull-down assays showed significantly reduced association of c-Myc with the mutant PCGEM1 ∆761–849 (Fig. 6 A and C), strongly suggesting that nucleotides 781–840 represent the major c-Myc binding site on PCGEM1. Importantly, the c-Myc binding site seemed to be essential for PCGEM1’s role in c-Myc transactivation. We found that the synergistic effect of PCGEM1 on c-Myc transactivation was significantly diminished when expressing the mutant PCGEM1 (∆761–849) (Fig. 6D) whereas its efficacy on AR transactivation was not affected (Fig. 6E). Given that AR binds to PCGEM1 on a distinct site locating on nucleotides 421–480 (9) (Fig. S1D), our data suggest that the c-Myc binding domain on PCGEM1 is functionally distinct from that of AR. Finally, using the AR-negative PC3 cell, we tested whether the c-Myc binding domain on PCGEM1 is essential for the downstream target induction. As shown in Fig. 6F, compared with full-length PCGEM1, the ability of the ∆761–849 mutant to induce metabolic genes was considerably weakened. A similar gene regulation pattern was observed when overexpressing the full-length and deletion mutant PCGEM1 in LNCaP cells although the presence of AR and the endogenous PCGEM1 may have masked some of the c-Myc effects (Fig. S5). Together, we identified the c-Myc binding domain on PCGEM1 and showed that the structural domain for c-Myc binding is consistent as the functional domain for c-Myc target-gene regulation.
Fig. 6.
c-Myc binding domain on PCGEM1. (A) In vitro binding of c-Myc recombinant protein with full-length and various PCGEM1 constructs. Equal amount of biotinylated PCGEM1 RNA was incubated with c-Myc in the pull-down assay. The streptavidin-RNA–bound c-Myc was analyzed by Western blotting. (B) The recombinant c-Myc and PCGEM1 RNA pull-down assay was coupled with a dot blot assay. Twenty-seven probes, each containing 60 bases of the complementary PCGEM1 sequence, were dotted on membrane for c-Myc binding domain detection. Probe 14 complementary to PCGEM1 781–840 exhibited the strongest signal. The biotinylated full-length PCGEM1 was dotted at right lower corner as a positive control for streptavidin-HRP chemiluminescent reaction. (C) Association of HA-tagged c-Myc with full-length (FL) or mutant (∆761–849) PCGEM1 was detected by RIP assay. (D) Myc-responsive element reporter and (E) PSA enhancer reporter assays were carried out to determine the transactivities of c-Myc and AR in PCGEM1 (FL and ∆761–849) overexpressing PC3 cell, as described in Fig. 4C. (F) Metabolic gene expression in PC3 cells overexpressing control vector, full-length, or mutant (∆761–849) PCGEM1. The relative expression levels were calculated as described in Fig. 3C. Asterisks indicate statistical significance (*P < 0.05, **P < 0.01).
Discussion
Distinct from normal cells, cancer cells acquire alterations in central metabolic pathways to fulfill their high demands on biomass and energy production, while maintaining appropriate redox. These metabolic changes are critical for cancer cells to sustain rapid proliferation and adapt to a dynamic tumor microenvironment (12, 18, 19). In this report, we identified the lncRNA PCGEM1 as a key regulator of metabolic pathways that provide proliferating advantages for prostate-cancer cells. We found that PCGEM1-overexpressing cells showed significantly enhanced glucose uptake and lactate production, indicative of an increased glycolysis rate, as well as an increased cellular level of citrate, G6PD activity, and NADPH, indicating elevated biosynthesis of fatty acid and nucleotide and redox control. Conversely, knockdown of PCGEM1 resulted in opposite metabolic outcomes that further arrested cell-cycle progression and induced apoptosis. These findings indicate an essential role of PCGEM1 in tumor metabolic regulation, critical for cancer-cell proliferation and survival. By enhancing aerobic glycolysis, the most common metabolic phenotype, known as the Warburg effect in cancer (20), overexpression of PCGEM1 facilitates anabolism to produce cellular building materials (21, 22) and consequently drives the cell to higher proliferation potential.
Our study has uncovered PCGEM1 lncRNA as a key transcriptional regulator of metabolic genes that profoundly affects the gene expression profiles in several pathways linked to tumor metabolism. The PCGEM1-mediated tumor metabolic regulation can largely be attributed to its dual role as c-Myc and AR coactivators. In prostate-cancer cells, AR has been shown to regulate metabolic genes involved in glycolysis, PPP, fatty acid, and nucleotide synthesis (11). On the other hand, as one of the most highly amplified oncogenes in many cancers (23), c-Myc emerges as a master regulator of global metabolism, including those regulated by AR, as well as glutamine metabolism and the TCA cycle (14, 15). We showed, in both cases, that PCGEM1 associated with the transcription factors, enhanced their transactivation activities, enriched their recruitments to the target promoters, and up-regulated the target genes in all of the pathways described above. Our data indicate that the complex formation of PCGEM1 with c-Myc and AR is mutually independent. First, PCGEM1 associated with c-Myc in the absence of AR in vitro, and vice versa. Moreover, the PCGEM1–c-Myc association was detected in AR-negative cells. Second, c-Myc did not interact with lncRNA PRNCR1, which is required for AR and PCGEM1 complex formation (9). Third, PCGEM1 enhanced c-Myc activity and up-regulated c-Myc–targeted metabolic genes in cells without AR expression. Finally, c-Myc and AR bind to PCGEM1 on distinct domains, and deletion of the c-Myc binding site did not affect the PCGEM1-induced AR transcriptional activity. Therefore, we propose that the PCGEM1-mediated transactivation of c-Myc and AR is functionally and structurally distinct. We cannot completely rule out that, under certain conditions, c-Myc and AR may be embedded in the same complex and coregulate certain targets. We have used ChIP-reChIP to test whether there is PCGEM1-mediated corecruitment of c-Myc on the AR-targeted loci. At least for ENO1, HK2, GLS, GSR, and LDHA promoters, such corecruitment was not observed (Fig. S6A), indicating the formation of two independent PCGEM1 complexes. On the other hand, even without coexisting in the same complex, PCGEM1-dependent activation of c-Myc and AR can enhance the cross talk between these two transcriptional programs. It has previously been reported that AR is able to induce the expression of c-Myc, in turn reinforcing amplification of the AR-transcriptional signals (24). The c-Myc induction by AR can also confer hormone-independent growth of prostate cancer cells (25, 26). Our data showed that, upon androgen stimulation, the metabolic gene induction was drastically diminished when knocking down PCGEM1 (Fig. 3A) or c-Myc (Fig. S6B), indicating that the androgen-induced metabolic reprogramming is dependent on the action of PCGEM1 and c-Myc. Together, we propose a model for the androgen-stimulated metabolic regulation that is through AR-dependent transcriptional up-regulation of the key regulators PCGEM1 and c-Myc, as well as the formation of a dual PCGEM1 complex with AR and c-Myc (Fig. S6C).
Regarding the mechanisms as to how PCGEM1 serves as a coactivator of transcriptional factors, Yang et al. demonstrated that the PCGEM1-induced AR activation involves looping of the chromatin enhancer to the promoter for subsequent transcriptional activation (9). The data from Prensner et al. (10), however, indicated otherwise. In our system, we were able to detect PCGEM1-enhanced AR activation, but we also identified an AR-independent function of PCGEM1 through c-Myc. At present, we know little about how PCGEM1 enhances the transcriptional potential of c-Myc. We observed that the chromatin recruitment of c-Myc is compromised when knocking down PCGEM1, indicating that PCGEM1 facilitates binding of c-Myc to the targeted promoters tested. Because heterodimerization of c-Myc and Max is sufficient to bind the E-box containing DNA in vitro (27, 28), it is unlikely that the DNA binding of c-Myc per se in prostate cancer cell requires PCGEM1. In fact, we found that PCGEM1 did not affect the interaction of c-Myc with Max or c-Myc protein stability (Fig. S4), indicating a mechanism independent of facilitating active Myc–Max dimerization or increasing the c-Myc protein level for enhanced transactivation. On the other hand, because the target recognition and binding for c-Myc in vivo depends on preexisting chromatin modification, such as histone methylation and acetylation (29), it is possible that PCGEM1 binds to epigenetic modifiers or specific histone marks of active chromatin, and in turn, promotes and stabilizes c-Myc enrichment on the target chromatin. The detailed mechanism requires further investigation.
Given the kinship of lncRNA to gene expression, the roles of lncRNAs in regulating metabolism are anticipated but have not been extensively studied. Recently, it was shown that lncRNA-p21 associates with HIF-1α and modulates glycolysis under hypoxia (30). As to c-Myc regulation, two recent reports identified lncRNA CCAT1-L and GHET1 involved in modulating transcription and RNA stability of c-Myc (31, 32). To our knowledge, the present study is the first report of an lncRNA that binds c-Myc and functions as a coactivator of c-Myc that modulates metabolic programming. Targeting tumor metabolism and its key regulators has emerged as an alternative strategy to complement the conventional genotoxic stress-based cancer therapy (19, 33, 34). One rationale is that “starvation” therapy may use a different cellular death mechanism to overcome the resistance to apoptosis often developed after cancer therapy (35). However, despite some success, the challenge of targeting individual metabolic enzymes in tumor, while avoiding toxic effects on normal proliferating cells, remains due to their essential housekeeping roles. Because PCGEM1 is prostate tissue-specific (5) and is overexpressed in cancer cells (6), it may serve as a unique target of metabolic regulation for prostate-cancer therapy. The specific role of PCGEM1 in prostate cancer metabolism and AR activation ( ref. 9 and this study) makes it an ideal therapeutic target for prostate cancer.
Materials and Methods
Detailed materials and methods are provided in SI Materials and Methods. The sequences of all primers and probes used in this study are listed in Tables S1 and S2.
LNCaP cells stably expressing Tet repressor (TR) were used to generate the inducible PCGEM1 knockdown cell line (LNCaP/shPCGEM1). Briefly, lentiviral particles carrying the shPCGEM1 construct that was driven by Tet-operating H1 promoter were generated according to the manufacturer’s instructions (Invitrogen) and were subsequently infected to LNCaP/TR cells. After 48 h of transduction, drug-resistant clones were selected by zeocin. The obtained stable cell lines were cultured and maintained in blasticidin (10 ng/mL) and zeocin (100 ng/mL; Invitrogen). PCGEM1 expression levels in the isolated zeocin-resistant clones were monitored by quantitative RT (qRT)-PCR to confirm knockdown efficiency. To induce knockdown, doxycycline (DOX) (100 ng/mL) was added to culture medium for 3 d. PCGEM1-overexpressing LNCaP and PC3 cells were also generated by the lentiviral system, following the same procedure.
Supplementary Material
Acknowledgments
This work was supported by National Science Council of Taiwan Grant NSC102-2320-B-400-018. We also acknowledge US National Institutes of Health Grants CA114575, CA165263, and DK065977, as well as National Health Research Institutes of Taiwan Grant 03A1-MGPP18-014.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1415669112/-/DCSupplemental.
References
- 1.Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: New links in cancer progression. Cancer Res. 2011;71(1):3–7. doi: 10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: A new frontier of translational research? Oncogene. 2012;31(43):4577–4587. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geisler S, Coller J. RNA in unexpected places: Long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14(11):699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Srikantan V, et al. PCGEM1, a prostate-specific gene, is overexpressed in prostate cancer. Proc Natl Acad Sci USA. 2000;97(22):12216–12221. doi: 10.1073/pnas.97.22.12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petrovics G, et al. Elevated expression of PCGEM1, a prostate-specific gene with cell growth-promoting function, is associated with high-risk prostate cancer patients. Oncogene. 2004;23(2):605–611. doi: 10.1038/sj.onc.1207069. [DOI] [PubMed] [Google Scholar]
- 7.Fu X, Ravindranath L, Tran N, Petrovics G, Srivastava S. Regulation of apoptosis by a prostate-specific and prostate cancer-associated noncoding gene, PCGEM1. DNA Cell Biol. 2006;25(3):135–141. doi: 10.1089/dna.2006.25.135. [DOI] [PubMed] [Google Scholar]
- 8.Romanuik TL, et al. LNCaP Atlas: Gene expression associated with in vivo progression to castration-recurrent prostate cancer. BMC Med Genomics. 2010;3:43. doi: 10.1186/1755-8794-3-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang L, et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500(7464):598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prensner JR, et al. The IncRNAs PCGEM1 and PRNCR1 are not implicated in castration resistant prostate cancer. Oncotarget. 2014;5(6):1434–1438. doi: 10.18632/oncotarget.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Massie CE, et al. The androgen receptor fuels prostate cancer by regulating central metabolism and biosynthesis. EMBO J. 2011;30(13):2719–2733. doi: 10.1038/emboj.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11(2):85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 13.Ward PS, Thompson CB. Metabolic reprogramming: A cancer hallmark even Warburg did not anticipate. Cancer Cell. 2012;21(3):297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dang CV. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harb Perspect Med. 2013;3(8):a014217. doi: 10.1101/cshperspect.a014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morrish F, Isern N, Sadilek M, Jeffrey M, Hockenbery DM. c-Myc activates multiple metabolic networks to generate substrates for cell-cycle entry. Oncogene. 2009;28(27):2485–2491. doi: 10.1038/onc.2009.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinato F, Cesaroni M, Amati B, Guccione E. Analysis of Myc-induced histone modifications on target chromatin. PLoS ONE. 2008;3(11):e3650. doi: 10.1371/journal.pone.0003650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 2001;15(16):2069–2082. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dang CV. Links between metabolism and cancer. Genes Dev. 2012;26(9):877–890. doi: 10.1101/gad.189365.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schulze A, Harris AL. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature. 2012;491(7424):364–373. doi: 10.1038/nature11706. [DOI] [PubMed] [Google Scholar]
- 20.Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 21.Newsholme EA, Crabtree B, Ardawi MS. The role of high rates of glycolysis and glutamine utilization in rapidly dividing cells. Biosci Rep. 1985;5(5):393–400. doi: 10.1007/BF01116556. [DOI] [PubMed] [Google Scholar]
- 22.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463(7283):899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ni M, et al. Amplitude modulation of androgen signaling by c-MYC. Genes Dev. 2013;27(7):734–748. doi: 10.1101/gad.209569.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernard D, Pourtier-Manzanedo A, Gil J, Beach DH. Myc confers androgen-independent prostate cancer cell growth. J Clin Invest. 2003;112(11):1724–1731. doi: 10.1172/JCI19035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao L, et al. Androgen receptor promotes ligand-independent prostate cancer progression through c-Myc upregulation. PLoS ONE. 2013;8(5):e63563. doi: 10.1371/journal.pone.0063563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papoulas O, Williams NG, Kingston RE. DNA binding activities of c-Myc purified from eukaryotic cells. J Biol Chem. 1992;267(15):10470–10480. [PubMed] [Google Scholar]
- 28.Littlewood TD, Amati B, Land H, Evan GI. Max and c-Myc/Max DNA-binding activities in cell extracts. Oncogene. 1992;7(9):1783–1792. [PubMed] [Google Scholar]
- 29.Guccione E, et al. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol. 2006;8(7):764–770. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- 30.Yang F, Zhang H, Mei Y, Wu M. Reciprocal regulation of HIF-1α and lincRNA-p21 modulates the Warburg effect. Mol Cell. 2014;53(1):88–100. doi: 10.1016/j.molcel.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 31.Xiang JF, et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014;24(5):513–531. doi: 10.1038/cr.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang F, et al. Long non-coding RNA GHET1 promotes gastric carcinoma cell proliferation by increasing c-Myc mRNA stability. FEBS J. 2014;281(3):802–813. doi: 10.1111/febs.12625. [DOI] [PubMed] [Google Scholar]
- 33.Tennant DA, Durán RV, Gottlieb E. Targeting metabolic transformation for cancer therapy. Nat Rev Cancer. 2010;10(4):267–277. doi: 10.1038/nrc2817. [DOI] [PubMed] [Google Scholar]
- 34.Vander Heiden MG. Targeting cancer metabolism: A therapeutic window opens. Nat Rev Drug Discov. 2011;10(9):671–684. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- 35.Kim RH, Bold RJ, Kung HJ. ADI, autophagy and apoptosis: Metabolic stress as a therapeutic option for prostate cancer. Autophagy. 2009;5(4):567–568. doi: 10.4161/auto.5.4.8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.