Significance
Allosteric sites in protein kinases offer opportunities for developing more selective inhibitors, but these sites are challenging to target because they involve protein–protein interfaces. We designed a site-directed approach to screen for molecules that bind to an allosteric peptide docking site on the protein kinase PDK1. We discovered molecules that structurally mimic the natural peptide ligand and inhibit PDK1 in cells. We also found that combining allosteric and ATP-competitive inhibitors completely blocked the activation of oncogenic kinases downstream of PDK1. This approach could be adapted to target an analogous allosteric site found on many other kinases.
Keywords: allostery, peptide mimicry, drug discovery, protein–protein interaction
Abstract
There is great interest in developing selective protein kinase inhibitors by targeting allosteric sites, but these sites often involve protein–protein or protein–peptide interfaces that are very challenging to target with small molecules. Here we present a systematic approach to targeting a functionally conserved allosteric site on the protein kinase PDK1 called the PDK1-interacting fragment (PIF)tide-binding site, or PIF pocket. More than two dozen prosurvival and progrowth kinases dock a conserved peptide tail into this binding site, which recruits them to PDK1 to become activated. Using a site-directed chemical screen, we identified and chemically optimized ligand-efficient, selective, and cell-penetrant small molecules (molecular weight ∼380 Da) that compete with the peptide docking motif for binding to PDK1. We solved the first high-resolution structure of a peptide docking motif (PIFtide) bound to PDK1 and mapped binding energy hot spots using mutational analysis. We then solved structures of PDK1 bound to the allosteric small molecules, which revealed a binding mode that remarkably mimics three of five hot-spot residues in PIFtide. These allosteric small molecules are substrate-selective PDK1 inhibitors when used as single agents, but when combined with an ATP-competitive inhibitor, they completely suppress the activation of the downstream kinases. This work provides a promising new scaffold for the development of high-affinity PIF pocket ligands, which may be used to enhance the anticancer activity of existing PDK1 inhibitors. Moreover, our results provide further impetus for exploring the helix αC patches of other protein kinases as potential therapeutic targets even though they involve protein–protein interfaces.
Protein kinases are a rich source of targets for the development of chemical probes and therapeutics; however, the remarkable similarity of their ATP-binding pockets presents a formidable challenge for the development of selective ATP-competitive inhibitors. Previous efforts to address these limitations have focused on targeting allosteric sites in kinases. Exquisitely selective allosteric inhibitors of the protein kinases AKT, MEK, and ABL are now in clinical trials for cancer, and various other allosteric kinase inhibitors and activators are in preclinical development (1). Despite these recent successes, finding allosteric modulators remains challenging, because most allosteric opportunities are the sites of protein–protein or protein–peptide interactions, which are very difficult to mimic with small molecules. Moreover, traditional chemical screening approaches most often identify ligands for the more druggable ATP-binding pocket.
The helix αC patch is an ancient allosteric site present on various serine/threonine and tyrosine kinases (2). The binding of effector proteins to the helix αC patch activates some kinases and inhibits others. The helix αC patch is seen most frequently in the AGC family of serine/threonine kinases, where this site is known specifically as the PDK1-interacting fragment (PIF) pocket. A hydrophobic motif (HM) found in the C-terminal tail of most AGC kinases must bind in cis to the PIF pocket for the kinase to be fully active; however, the AGC kinase PDK1 lacks its own HM, and instead uses its PIF pocket as a docking site to recruit, phosphorylate, and thereby activate 23 other AGC kinases, including AKT, S6K, SGK, RSK, and PKC isoforms (3). The known role of PDK1 as a master regulator of these progrowth and prosurvival kinases has motivated the development of numerous PDK1 inhibitors as potential anticancer agents (4). One strategy for inhibiting PDK1 has been to identify compounds that bind to its PIF pocket and disrupt the recruitment of substrates.
Early biochemical studies revealed that PIFtide, a synthetic peptide derived from the HM of the protein kinase PRK2, stimulates PDK1 activity toward a short peptide substrate (5) but disrupts recruitment and phosphorylation of the full-length substrates S6K and SGK (6). Small-molecule mimics of PIFtide have been discovered through pharmacophore modeling (7) and fragment-based approaches (8–10), and some optimized analogs have been characterized structurally (10–13); however, these compounds have limited membrane permeability, which diminishes their utility as chemical probes. Moreover, the lack of a structure of PIFtide bound to PDK1 has impeded the structure-based design of improved analogs that mimic the native allosteric interaction.
We have explored various site-directed methods for targeting the PIF pocket of PDK1. Previously, we used a technique known as disulfide trapping (or tethering) to identify small-molecule fragments (molecular weight <250 Da) that inhibit or activate PDK1 by covalently labeling a cysteine residue that was engineered into the PIF pocket (10). Here we sought to discover noncovalent small molecules that could be used as chemical probes of PIF pocket function in cells. We developed a PIFtide competitive binding assay to perform a site-directed screen of ∼154,000 compounds for new PIF pocket ligands. We discovered a series of diaryl sulfonamides (molecular weight ∼380 Da) that were chemically optimized and then characterized biochemically, structurally, and in cells. We also solved the first structure of PIFtide bound to PDK1, which reveals how small molecules mimic this peptide effector and provides insights into the structure-based design of improved PIF pocket ligands. Remarkably, we found that PIF pocket ligands sensitize PDK1 to inhibition by an ATP-competitive inhibitor, enabling more complete suppression of downstream signaling in cells.
Results
Site-Directed Chemical Screen Identifies Diaryl Sulfonamides as PIF Pocket Ligands.
To identify small molecules that bind to the PIF pocket of PDK1, we developed a fluorescence polarization (FP) competitive binding assay to screen for compounds that disrupt the interaction between PDK1 and PIFtide (Fig. 1A). To achieve optimal sensitivity, FP competitive binding assays require a fluorescent probe that binds tightly to the target (Kd <100 nM) and yields a large change in polarization signal when bound (ΔmP >100 mP) (14). To develop a suitable PIFtide probe for the FP assay, we synthesized a panel of fluorescent PIFtides of various lengths and amino acid content (full-length PIFtide: REPRILSEEEQEMFRDFDYIADW). We found that removal of the first 13 amino acids, substitution of Phe14 and Phe17 with bromo-Phe residues, substitution of Tyr19 with Trp, and conjugation of 6-tetramethylrhodamine to the N terminus resulted in a PIFtide probe with an ideal affinity and dynamic range (Kd = 40 nM; ΔmP = 130 mP; SI Appendix, Fig. S1). We used tetramethylrhodamine, a red-shifted fluorophore relative to fluorescein, to reduce the susceptibility of the assay to interference from autofluorescent compounds (15). The resulting competitive binding assay was suitably robust for a high-throughput screen (Z′ = 0.7).
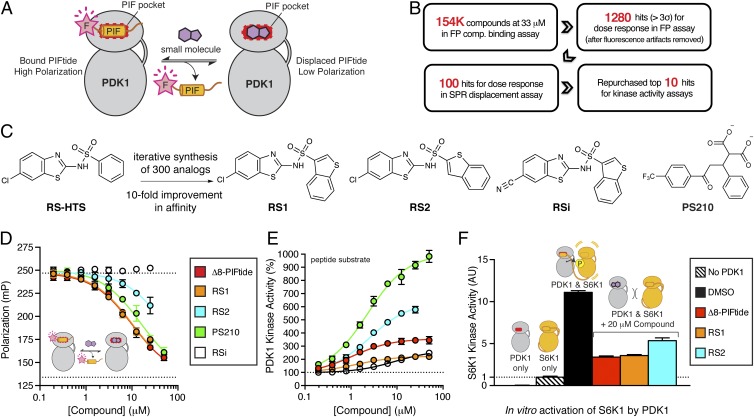
Fig. 1.
Discovery and optimization of diaryl sulfonamides as PIFtide mimics. (A) Schematic of a FP competitive binding assay developed to identify small-molecule mimics of the PIFtide. (B) Overview of the high-throughput screen and triage process. (C) Chemical optimization of the diaryl sulfonamide hit from HTS. RSi is an inactive analog used as a negative control. PS210 is a known PIF pocket ligand used as a positive control. (D) Dose–response curves for PIFtide, the RS compounds, and PS210 in the FP competitive binding assay. (E) Dose–response curves for PIFtide and the RS compounds in a radioactive kinase activity assay monitoring the phosphorylation of T308tide peptide substrate by PDK1. (F) Effect of PIFtide and the RS compounds on the in vitro activation of S6K1 by PDK1. After activation of S6K1 by PDK1 for 30 min, the kinase activity of S6K1 was determined by a radioactive kinase assay using the Crosstide substrate. The activity of S6K1 alone was used for normalization (dotted line). Error bars are ± SD (n = 3).
The high-throughput screen workflow that we used is depicted in Fig. 1B and is discussed in detail in SI Appendix, Materials and Methods. In brief, we screened ∼154,000 compounds at a single dose (33 μM) using the FP assay. We selected hits from this primary screen using a statistical threshold of 3σ (1,460 hits; 0.9% hit rate). We filtered out autofluorescent artifacts by removing compounds that yielded total fluorescence intensities >150% of the untreated control. We tested the remaining 1,280 compounds using the FP assay in dose–response mode. To confirm hits in an orthogonal assay format, we selected the top 100 compounds for dose–response assessment in a surface plasmon resonance (SPR) assay that detects displacement of PDK1 from immobilized PIFtide (7). We confirmed the chemical identity of the top 15 hits by testing repurchased or resynthesized standards in the FP and SPR assays. One of the most potent hits was the diaryl sulfonamide RS-HTS (Fig. 1C), which exhibited a Kd value of 15 μM in the FP assay and an IC50 of 20 μM in the SPR assay. Given the novelty of the diaryl sulfonamide scaffold, we decided to focus further efforts on optimizing and characterizing compounds in this class.
Iterative Synthesis and Characterization of Improved Sulfonamides.
We next sought to improve on the potency of RS-HTS via analog synthesis. We synthesized a panel of 300 diaryl sulfonamides and assessed the potency of each compound in the FP, SPR, and kinase activity assays described above. These efforts yielded the regioisomers RS1 and RS2, as well as the inactive analog RSi (Fig. 1C). RS1 and RS2 bound to PDK1 with a Kd of 1.5 μM and 9 μM, respectively (Fig. 1D), and stimulated the catalytic activity of PDK1 toward a peptide substrate by twofold and sixfold, respectively (Fig. 1E). In comparison, Δ8-PIFtide bound with a Kd of 2 μM and stimulated PDK1 activity by fourfold, whereas the diaryl dicarboxylate PS210 (13) bound with a Kd of 3 μM and stimulated PDK1 activity by 10-fold. In summary, RS1 and RS2 bound to PDK1 with an affinity similar to that of the 15-mer Δ8-PIFtide and the previously described PIF pocket ligand PS210, yet each molecule stimulated the catalytic activity of PDK1 to a different extent (2- to 10-fold).
It is well established that at high concentrations, some small molecules form soluble aggregates, which assemble through a mechanism similar to micelles and can interact nonspecifically with proteins (16). We determined by dynamic light scattering that RS1 and RS2 were monomeric in solution at concentrations <50 μM (SI Appendix, Fig. S2A), which is more than fivefold above their Kd values. Moreover, the stimulation of PDK1 activity by RS2 was not diminished in the presence of Triton X-100 or BSA (SI Appendix, Fig. S2 B and C), additives known to either disperse or mask soluble aggregates of small molecules (17). Thus, RS1 and RS2 do not modulate PDK1 activity through an aggregation-based mechanism.
Although RS1 and RS2 stimulated PDK1 activity toward a short peptide substrate, we expected to find that these PIF pocket ligands prevented PDK1 from phosphorylating most full-length protein substrates (e.g., S6K, SGK, and RSK), because these substrates must dock into the PIF pocket of PDK1 to be efficiently phosphorylated in vitro (6) and in cells (18). Moreover, both a synthetic PIFtide (6) and a small-molecule mimic of PIFtide (7) have been shown to inhibit the phosphorylation of S6K and SGK by PDK1 in vitro. Accordingly, we found that 20 μM Δ8-PIFtide, RS1, and RS2 inhibited the in vitro activation of S6K1 by PDK1 by 75%, 75%, and 60%, respectively (Fig. 1F).
Structures of PDK1 Bound to Diaryl Sulfonamides.
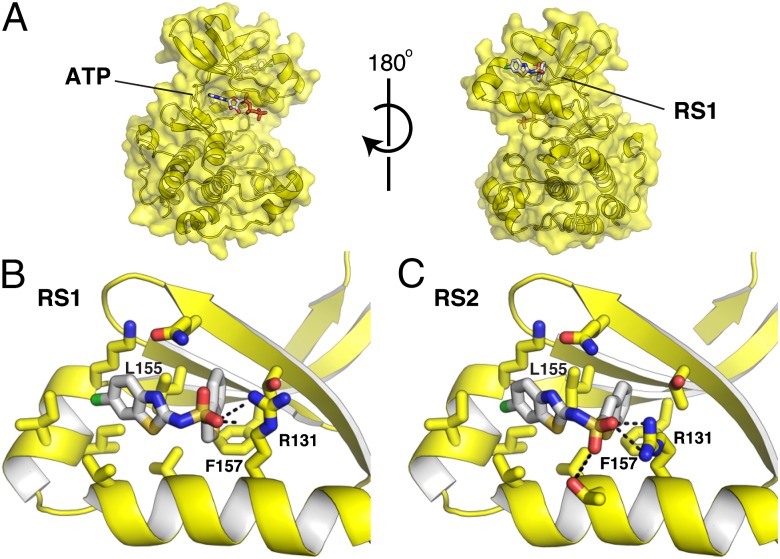
To identify how RS1 and RS2 bind to PDK1, we determined the structures of these compounds in complex with PDK1. We first crystallized PDK1 in complex with ATP, and then soaked these crystals with ligand to obtain the structures in complex with RS1 (1.6-Å resolution; Fig. 2A) or RS2 (1.5-Å resolution; Fig. 2B). Each structure showed unambiguous electron density for the bound compounds (SI Appendix, Fig. S3 A and B). The RS compounds share a binding mode in which the aromatic substituents bind to two adjacent subsites in the PIF pocket. The sulfonyl group of both RS compounds interacts with Arg131 through a salt bridge, because the sulfonamide is likely ionized under the crystallization conditions (pH 7.5; predicted pKa ∼6.5). To confirm the binding mode of the RS compounds, we attempted to disrupt compound binding by mutating Leu155, which resides in the back of PIF pocket and packs against both aromatic substituents. Mutation of Leu155 to Ala or Glu completely abolished the enhancement of PDK1 activity by Δ8-PIFtide, RS1, and RS2 (SI Appendix, Fig. S4 A–C). The Leu155Ala mutation also conferred partial resistance to the RS compounds in the in vitro S6K1 activation assay (SI Appendix, Fig. S4D).
Fig. 2.
Structures of the RS compounds bound to the PIF pocket of PDK1. (A) Structure of the PDK1-RS1 complex. PDK1 is shown as a yellow surface, and both ATP and RS1 are shown as white sticks colored by heteroatom. The relative orientation of the ATP-binding site and the PIF pocket is depicted. (B) Close-up view of the PDK1–RS1 interaction. (C) Close-up view of the PDK1–RS2 interaction.
Molecular Basis of PIFtide Recognition by PDK1.
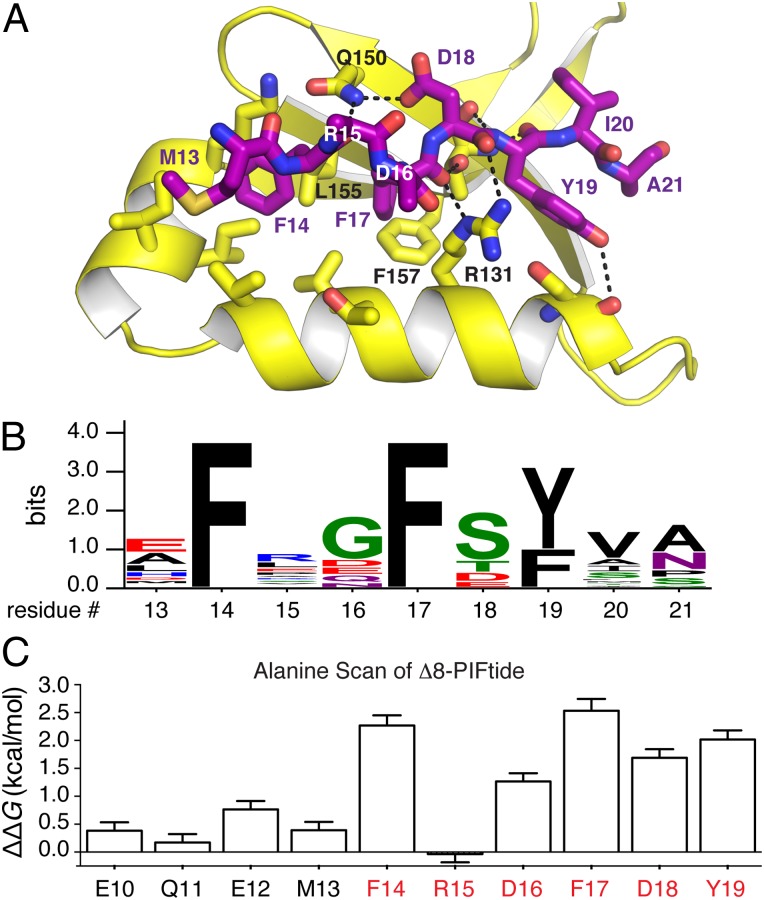
To reveal precisely how PDK1 recognizes a native peptide effector and to gain insight into the mimicry of this peptide by small molecules, we determined the crystal structure of a PDK1-PIFtide complex. To do so, we soaked crystals of PDK1 bound to ATP with PIFtides of varied length and obtained high-quality diffraction data (1.4-Å resolution) with one variant, Δ8-PIFtide (residues 9–23). This structure showed an unambiguous electron density for residues 13–21 of PIFtide (SI Appendix, Fig. S3C) and demonstrates how PDK1 engages the core HM of PIFtide (MFxxFDYIA).
To accommodate PIFtide binding, the side chain of Arg131 swings out to open a hydrophobic channel, and the side chain of Phe157 rotates ∼90° to make room for Phe17 of PIFtide. The conserved aromatic residues of PIFtide (Phe14, Phe17, and Tyr19) occupy three adjacent subsites within the PIF pocket (Fig. 3 A and B). Although the side chain of Asp16 could not be modeled, the Cα-Cβ bond vector for this residue appears to direct the side chain toward Arg131 on PDK1, suggestive of an electrostatic interaction. The conserved negatively charged residue of PIFtide (Asp18) interacts primarily with Gln150, not with Arg131 as was suggested previously (19). This charged binding mode is similar to that observed for the analogous Asp, phospho-Ser, and phospho-Thr in the HM of AKT (20), S6K1 (21), and PKCβII (22), respectively. Finally, Met13, Ile20, and Ala21 of PIFtide occupy small clefts at the periphery of the PIF pocket. A previously reported structure of an AKT chimera bound in cis to PIFtide (20) bears striking resemblance to the PDK1-PIFtide structure (PIFtide all-atom rmsd = 1.6 Å; SI Appendix, Fig. S5). In summary, PIFtide uses a three-pronged hydrophobic plug along with two anionic anchors to engage the PIF pocket of PDK1.
Fig. 3.
Structural and energetic analysis of the PDK1–PIFtide interaction. (A) Structure of the PDK1-PIFtide complex. PDK1 is in yellow, and PIFtide is shown as purple sticks. (B) Sequence logo depicting the consensus HM from 28 AGC kinases known or inferred to interact with PDK1. The consensus sequence is xFxxF[−](Y/F)(V/A/I)x, where [−] indicates a negatively charged residue (Asp, Glu, or phosphorylated Ser/Thr). (C) Hot spot analysis of the PDK1–PIFtide interaction. Residues 10–19 of PIFtide9–23 were subjected to alanine scanning. The red-colored residues red constitute the HM. Energetic contributions were determined from the Ki values for mutant peptides using the formula ΔΔG = 1.4(kcal/mol)*log(Ki,mut/Ki,wt).
To determine the relative energetic contribution of each amino acid within the HM to binding, we individually mutated positions 10–19 of Δ8-PIFtide to alanine and measured the affinities of the mutant peptides using the FP competitive binding assay. Alanine scanning mutagenesis is a proven reliable method for finding binding energy hot spots at protein–protein interfaces (23), and these hot spots often represent ideal small-molecule binding sites (24). We found that the strongly conserved residues at positions 14, 17, 18, and 19 of PIFtide all constituted binding energy hot spots (ΔΔG of 1.5–2.5 kcal/mol), whereas nonconserved residues 10–13 and 15 contributed little to binding affinity (ΔΔG of 0–0.75 kcal/mol) (Fig. 3C; each 1.4-kcal/mol increment in ΔΔG represents a 10-fold loss in affinity). Although Asp16 is not strongly conserved among HMs, mutation to Ala significantly affected binding affinity (ΔΔG of 1.25 kcal/mol), further supporting an electrostatic interaction with PDK1. These quantitative competitive binding data agree with previously reported qualitative immunoprecipitation binding data (25). In summary, the HM of PIFtide contains five amino acids (FxDFDY) that constitute binding energy hot spots (ΔΔG >1.25 kcal/mol).
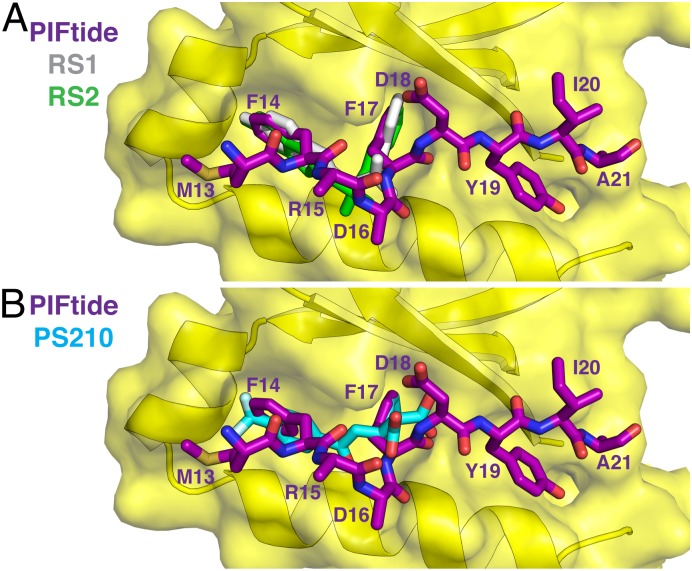
Mimicry of PIFtide by the RS Compounds and PS210.
Comparing the binding modes of PIFtide and its small-molecule mimics revealed that side chains of Phe14 and Phe17 of PIFtide share a nearly identical trajectory with the aromatic substituents of the diaryl sulfonamides RS1 and RS2 (Fig. 4A) and the diaryl carboxylate PS210 (Fig. 4B). In addition, each compound class mimics one native electrostatic interaction; RS1 and RS2 mimic the interaction between Asp16 of PIFtide and Arg131 of PDK1, whereas PS210 mimics the interaction between Asp18 of PIFtide and Gln150 of PDK1. Neither class of compounds engages the hydrophobic pocket occupied by Tyr19/Ile20 of PIFtide. Overall, existing PIF pocket ligands mimic three of the five energetic hotspots of the PDK1–PIFtide interaction.
Fig. 4.
Mimicry of PIFtide by diaryl sulfonamide and diaryl dicarboxylate compounds. (A) Overlay of the binding modes of PIFtide and the RS compounds. This view highlights the similar trajectory of the FxxF motif of PIFtide and the aromatic substituents of the RS compounds. (B) Overlay of the binding modes of PIFtide and the diaryl dicarboxylate PS210 (PDB ID code 4AW1).
RS1 Binds Selectively to PDK1.
To determine whether RS1 binds to PDK1 selectively, we tested the effect of 10 μM RS1 on the catalytic activity of 39 of the 60 AGC family kinases, because these kinases all bind to an HM in their respective PIF pockets. We also tested IGF1 receptor and mTOR, which directly impact the PDK1 signaling pathway that we intended to study, as well as the Aurora kinases. The strongest hits from this independent screen were a 90% stimulation of PDK1 activity and a 33% inhibition of MSK2 activity (SI Appendix, Fig. S6). At 10 μM, RS1 did not inhibit 41 of the 44 kinases by >20%, suggesting that RS1 binds to PDK1 selectively.
Sulfonamides Prevent S6K1 Activation but Permit AKT Activation in Cells.
Having established that RS1 binds selectively to the PIF pocket of PDK1, we wanted to determine its effects on PDK1 signaling in cells. For these cell-based experiments, we used the inactive analog RSi to control for nonspecific effects of RS1, the highly selective ATP-competitive PDK1 inhibitor GSK2334470 (GSK) (26, 27) as a control for pathway modulation, and the diaryl dicarboxylate PS210 and its diester prodrug PS423 as a chemically distinct class of PIF pocket ligands (13).
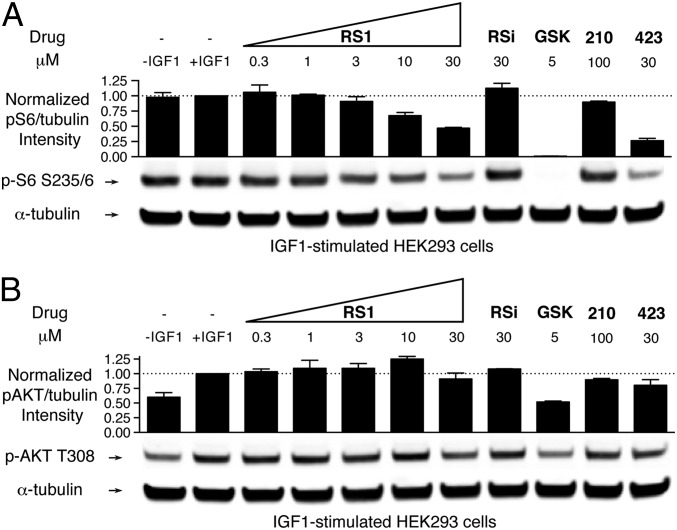
We first tested whether RS1 would inhibit S6K1 activation in cells, as we found in vitro. We serum-starved HEK293 cells, treated them with increasing concentrations of RS1 or control compounds, and then stimulated them with IGF1 for 15 min before lysis. To observe the activation state of S6K1 in the cells, we monitored phosphorylation of its substrate ribosomal protein S6 by quantitative immunoblotting using infrared dyes. Treatment with increasing doses of RS1 led to a dose-dependent but incomplete blockade of S6 phosphorylation (Fig. 5A). At 30 μM, S6 phosphorylation was inhibited by 50% by RS1 and 70% by PS423. Importantly, the inactive analog RSi had no effect on S6 phosphorylation, suggesting that RS1 must specifically bind to PDK1 to exert its effect. The diaryl dicarboxylate PS210 had no effect on substrate phosphorylation at 100 μM, confirming that its carboxylate groups must be masked as esters to be cell active. Thus, both RS1 and PS423 prevent the activation of S6K1 in cells, although this effect did not saturate at the doses tested.
Fig. 5.
PIF pocket ligands block S6K1 activation, but have only a weak effect on AKT activation in cells. (A) Effect of increasing doses of RS1 or control compounds on the phosphorylation of S6. Cell lysates were immunoblotted with antibodies for phospho-S6 S235/6 and α-tubulin. (B) Effect of increasing doses of RS1 or control compounds on the phosphorylation of AKT. The experiment was the same as that depicted in A, except that antibodies against phospho-AKT T308 and α-tubulin were used. The level of phosphorylation was quantified from the infrared signal and normalized to the α-tubulin signal. Error bars are ± SD (n = 2). The following drugs were used: GSK, a selective ATP-competitive inhibitor of PDK1; PS210, a dicarboxylate PIF pocket ligand that does not enter cells; and PS423, a diester prodrug of PS210.
We next assessed the effect of RS1 on the activation of AKT, which does not require binding of its HM to the PIF pocket of PDK1 for efficient activation (6, 18). We treated cells the same as described for monitoring S6K1 activation, but instead monitored the phosphorylation of AKT at Thr308 by PDK1. Treatment with increasing doses of RS1 had little effect on the phosphorylation of AKT (Fig. 5B). At 30 μM, AKT phosphorylation was inhibited by 10% by RS1 and 20% by PS423. At 10 μM, GSK inhibited AKT phosphorylation by only 50%, consistent with previous reports (27, 28). Control compounds RSi and PS210 did not affect AKT activation. In short, these findings indicate that PIF pocket ligands largely permit the activation of AKT by PDK1.
PIF Pocket Ligands Enhance the Ability of an ATP-Competitive Inhibitor to Block PDK1 Signaling.
Previous characterization of GSK in cells revealed that the activation of AKT by PDK1 is much less sensitive to inhibition by this ATP-competitive inhibitor compared with the activation of S6K, SGK, or RSK (27). The insensitivity of AKT activation is not explained by alternative pathways for AKT activation, because PDK1-deficient cells are incapable of activating AKT (18). Disrupting the capacity of AKT to bind to either PIP3 or the PIF pocket of PDK1 markedly sensitized AKT phosphorylation to inhibition by GSK, suggesting that having multiple recruitment mechanisms contributes to insensitivity to GSK (28). Given these data, we wondered whether RS1 would enhance the ability of GSK to block the activation of AKT by disrupting the PIF pocket-dependent recruitment of substrates to PDK1.
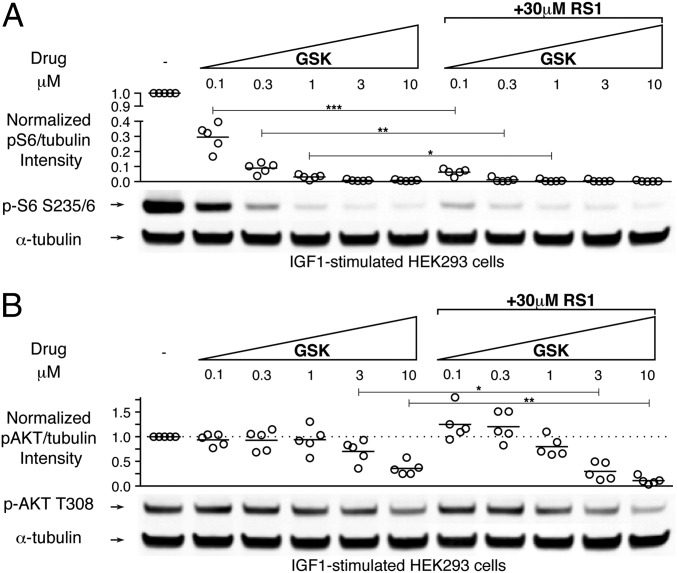
To test whether RS1 enhances the efficacy of GSK, we serum-starved HEK293 cells, treated them with increasing doses of GSK with or without 30 μM RS1, and then stimulated them for 15 min with the growth factor IGF1 before lysis. We monitored both S6K1 activation and AKT activation by immunoblot analysis, as described above. Phosphorylation of S6 was much more sensitive than phosphorylation of AKT to inhibition by GSK, consistent with previous reports (Fig. 6 A and B) (27, 28). Cotreatment with RS1 and GSK resulted in enhanced inhibition of S6 phosphorylation, reflecting the additivity of each compound’s effect (Fig. 6A and SI Appendix, Fig. S7). However, even though RS1 did not block AKT activation by itself (Fig. 5B), it did enhance the ability of GSK to block AKT phosphorylation (Fig. 6B). Thus, combining RS1 with GSK more effectively blocks the activation of both S6K1 and AKT in cells.
Fig. 6.
RS1 enhances the effect of a PDK1 active site inhibitor to more effectively block activation of both S6K1 and AKT. (A) Effect of RS1 on the dose-dependent inhibition of S6 phosphorylation by the PDK1 inhibitor GSK2334470. Cell lysates were immunoblotted with antibodies for phospho-S6 S235/6 and α-tubulin. (B) Effect of RS1 on the dose-dependent inhibition of AKT phosphorylation by the PDK1 inhibitor GSK2334470. The experiment was the same as that depicted in A, except cells were immunoblotted with antibodies for phospho-AKT T308 and α-tubulin. The level of phosphorylation was quantified from the infrared signal and normalized to the α-tubulin signal. Scatterplots represent data pooled from five independent experiments. Statistical significance was calculated using an unpaired two-sided t test. *P < 0.05; **P < 0.01; ***P < 0.001.
Discussion
Using a site-directed chemical screen, we have discovered a series of diaryl sulfonamide compounds that bind to the PIF pocket of PDK1 and disrupt its signaling in cells. Key to our success is the development of a FP competitive binding assay, which we expect could be readily adapted to target the PIF pockets other AGC kinases. Our structures of PDK1 bound to PIFtide or its small-molecule mimics provide insights into the structure-based design of PIF pocket ligands with improved affinity. Unlike previously reported PIF pocket ligands, the diaryl sulfonamide compounds freely diffuse into cells. These compounds are substrate-selective inhibitors of PDK1 as single agents, but in combination with an ATP-competitive inhibitor, they completely suppress the activation of downstream kinases in cells.
We were surprised to find that both RS1 and PS210 bound to PDK1 with an affinity comparable to that of the 15-mer peptide Δ8-PIFtide (Fig. 1D). Alanine scanning mutagenesis revealed that five of the six residues within the HM of Δ8-PIFtide are critical for binding (Fig. 3C), yet RS1 and PS210 each lack a native electrostatic interaction (Asp18 and Asp16, respectively) and a hydrophobic interaction (Tyr19). Thus, RS1 and PS210 make much more efficient contacts with the PIF pocket than PIFtide does (ligand efficiency: 0.35, 0.29, and 0.08 kcal/mol per heavy atom, respectively). Synthesizing analogs of RS1 or PS210 that mimic the native interactions they lack could improve their affinity for PDK1.
The diaryl sulfonamide RS1 and the diaryl dicarboxylate PS210 bind to PDK1 with similar affinity, and both compounds appear to bind to PDK1 selectively. The sulfonamides have a significant advantage over existing carboxylates, however, in that RS1 freely diffuses into cells, whereas PS210 does not. This disparity in cellular permeability may be attributable to the difference in pKa values between the carboxylate and N-arylsulfonamide moieties. At physiological pH, only a negligible fraction of carboxylate is protonated and thus cell-permeable (pKa ∼3), whereas a significant portion of the sulfonamide is protonated (pKa ∼6.5). The carboxylates of PS210 can be masked as esters to create the cell-permeable prodrug PS423, which is hydrolyzed by intracellular esterases and accumulates within cells (29). However, the purpose of an ester prodrug is to improve oral bioavailability of a drug by improving absorption in the gut (30). After its entrance into the circulation, PS423 would likely be rapidly hydrolyzed by esterases in the blood and liver (31), which would limit the delivery of active (cell-permeable) drug to target tissues. In summary, the diaryl sulfonamides represent a promising new chemical scaffold for the development of high-affinity PIF pocket ligands that freely diffuse into cells.
Because PDK1 is an essential mediator of PI3K-AKT growth signaling, numerous PDK1 inhibitors have been developed as potential anticancer therapies (4). Two major findings diminished the enthusiasm for PDK1 as an oncology target, however. First, sustained knockdown of PDK1 levels by ∼90% failed to prevent AKT activation or block tumor formation in PTEN-null mice (32). Second, GSK, the first potent and highly selective PDK1 inhibitor to be extensively characterized, did not significantly impact tumor growth in xenograft models (26). The failure of PDK1 inhibitors as anticancer agents may reside in their inability to effectively block the activation of AKT, even at concentrations that should inhibit >90% of PDK1 within a cell. Here we found that a small molecule that disrupts the recruitment of AKT by the PIF pocket of PDK1 induces sensitivity to an ATP-competitive inhibitor (Fig. 6B). The efficacy of this drug combination is not explained by RS1 increasing the affinity of PDK1 for the active site inhibitor GSK, because binding at the ATP-binding pocket and the PIF pocket are not cooperative in vitro (SI Appendix, Fig. S8). Rather, the ability of the PIF-pocket ligand RS1 to interfere with PDK1’s capacity to recruit substrates likely lowers the threshold of active site occupancy needed to effectively block PDK1 signaling with the ATP-competitive inhibitor GSK.
Taken together, our findings pave the way for the development of potent PIF pocket ligands that freely diffuse into cells. Next-generation analogs can be combined with ATP-competitive inhibitors to determine whether the complete suppression of PDK1 signaling observed here yields favorable outcomes in cancer models. Such a dual-targeting approach also may overcome the emergence of drug resistance, as has been demonstrated previously for targeting Bcr-Abl in chronic myelogenous leukemia (33). Broadly, the site-directed approach described here could be adapted to target the helix αC patch of other protein kinases, which may facilitate the identification of ligands that mimic these challenging protein–protein and protein–peptide interactions.
Materials and Methods
Protein Expression and Purification.
PDK150–359 (WT, Y288G Q292A, L155A, or L155E) and S6K124–421 T412E were expressed in Sf9 insect cells using the Bac-to-Bac system (Invitrogen). Proteins were purified by Ni-NTA affinity chromatography and size-exclusion chromatography. Details are provided in SI Appendix, Materials and Methods.
High-Throughput Screen.
The overall screening strategy is depicted generally in Fig. 1B. Details are provided in SI Appendix, Materials and Methods.
Chemical Synthesis.
Detailed methods for the synthesis and characterization of the RS compounds, PS210, PS423, and the peptides are provided in SI Appendix, Note S1.
Protein Kinase Activity Assays.
The catalytic activity of PDK1 toward the peptide substrate T308tide (10) or the protein substrate S6K1 (6) were measured using radioactivity-based kinase assays as described previously.
Crystallization, Data Collection, and Refinement of PDK1 Complexes.
Crystals were obtained using a PDK150–359 mutant (Y288G Q292A) that disrupts a crystal contact that normally prevents ligands from binding to the PIF pocket (11). PDK1 was crystallized in complex with ATP, and crystals were soaked with ligand overnight before harvesting. Diffraction data were collected at Advanced Light Source beamline 8.3.1. More details on the crystallization conditions and structure solution are provided in SI Appendix, Materials and Methods. Final refinement statistics are summarized in SI Appendix, Table S1.
Cell Culture Studies.
HEK293 cells were cultured in DMEM high-glucose medium supplemented with sodium pyruvate, nonessential amino acids, penicillin/streptomycin, and 10% FBS. Cells were serum-starved for 16 h before drug treatment for 1 h, stimulation with IGF1 for 15 min, and lysis. More details are provided in SI Appendix, Materials and Methods.
Statistics.
All statistical analyses were performed using GraphPad Prism 6. All scatterplot bars and bar graphs depict mean values; all error bars are ± SD. Statistical significance was calculated using an unpaired two-sided t test assuming equal SD. The IC50 and EC50 values were calculated using a sigmoidal dose–response equation with variable slope. IC50 values from the FP competitive binding assay were converted to Kd values using an equation that corrects for ligand depletion (34).
Supplementary Material
Acknowledgments
We thank members of the J.A.W. laboratory for helpful suggestions and critical review of the manuscript. We also thank the staff of Advanced Light Source beamline 8.3.1. This work was supported by National Institutes of Health Grant R01 CA136779-05 (to J.A.W.), a National Institutes of Health Predoctoral Fellowship F31 CA180378-01 (to T.J.R.), a Krevans Fellowship (to T.J.R.), and postdoctoral fellowships from the California Tobacco-Related Disease Research Program [110385 (to J.D.S.)] and the Damon Runyon Cancer Research Foundation [2082-11 (to N.D.T.)].
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4RQK, 4RQV, and 4RRV).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1415365112/-/DCSupplemental.
References
- 1.Fang Z, Grütter C, Rauh D. Strategies for the selective regulation of kinases with allosteric modulators: Exploiting exclusive structural features. ACS Chem Biol. 2013;8(1):58–70. doi: 10.1021/cb300663j. [DOI] [PubMed] [Google Scholar]
- 2.Jura N, et al. Catalytic control in the EGF receptor and its connection to general kinase regulatory mechanisms. Mol Cell. 2011;42(1):9–22. doi: 10.1016/j.molcel.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pearce LR, Komander D, Alessi DR. The nuts and bolts of AGC protein kinases. Nat Rev Mol Cell Biol. 2010;11(1):9–22. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- 4.Medina JR. Selective 3-phosphoinositide–dependent kinase 1 (PDK1) inhibitors: Dissecting the function and pharmacology of PDK1. J Med Chem. 2013;56(7):2726–2737. doi: 10.1021/jm4000227. [DOI] [PubMed] [Google Scholar]
- 5.Biondi RM, et al. Identification of a pocket in the PDK1 kinase domain that interacts with PIF and the C-terminal residues of PKA. EMBO J. 2000;19(5):979–988. doi: 10.1093/emboj/19.5.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biondi RM, Kieloch A, Currie RA, Deak M, Alessi DR. The PIF-binding pocket in PDK1 is essential for activation of S6K and SGK, but not PKB. EMBO J. 2001;20(16):4380–4390. doi: 10.1093/emboj/20.16.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engel M, et al. Allosteric activation of the protein kinase PDK1 with low molecular weight compounds. EMBO J. 2006;25(23):5469–5480. doi: 10.1038/sj.emboj.7601416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stockman BJ, et al. Identification of allosteric PIF-pocket ligands for PDK1 using NMR-based fragment screening and 1H-15N TROSY experiments. Chem Biol Drug Des. 2009;73(2):179–188. doi: 10.1111/j.1747-0285.2008.00768.x. [DOI] [PubMed] [Google Scholar]
- 9.Wei L, et al. Design and synthesis of benzoazepin-2-one analogs as allosteric binders targeting the PIF pocket of PDK1. Bioorg Med Chem Lett. 2010;20(13):3897–3902. doi: 10.1016/j.bmcl.2010.05.019. [DOI] [PubMed] [Google Scholar]
- 10.Sadowsky JD, et al. Turning a protein kinase on or off from a single allosteric site via disulfide trapping. Proc Natl Acad Sci USA. 2011;108(15):6056–6061. doi: 10.1073/pnas.1102376108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hindie V, et al. Structure and allosteric effects of low-molecular-weight activators on the protein kinase PDK1. Nat Chem Biol. 2009;5(10):758–764. doi: 10.1038/nchembio.208. [DOI] [PubMed] [Google Scholar]
- 12.Lopez-Garcia LA, et al. Allosteric regulation of protein kinase PKCζ by the N-terminal C1 domain and small compounds to the PIF-pocket. Chem Biol. 2011;18(11):1463–1473. doi: 10.1016/j.chembiol.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 13.Busschots K, et al. Substrate-selective inhibition of protein kinase PDK1 by small compounds that bind to the PIF-pocket allosteric docking site. Chem Biol. 2012;19(9):1152–1163. doi: 10.1016/j.chembiol.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 14.Huang X, Aulabaugh A. Application of fluorescence polarization in HTS assays. Methods Mol Biol. 2009;565:127–143. doi: 10.1007/978-1-60327-258-2_6. [DOI] [PubMed] [Google Scholar]
- 15.Turek-Etienne TC, et al. Evaluation of fluorescent compound interference in 4 fluorescence polarization assays: 2 kinases, 1 protease, and 1 phosphatase. J Biomol Screen. 2003;8(2):176–184. doi: 10.1177/1087057103252304. [DOI] [PubMed] [Google Scholar]
- 16.Coan KED, Shoichet BK. Stoichiometry and physical chemistry of promiscuous aggregate-based inhibitors. J Am Chem Soc. 2008;130(29):9606–9612. doi: 10.1021/ja802977h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGovern SL, Helfand BT, Feng B, Shoichet BK. A specific mechanism of nonspecific inhibition. J Med Chem. 2003;46(20):4265–4272. doi: 10.1021/jm030266r. [DOI] [PubMed] [Google Scholar]
- 18.Collins BJ, Deak M, Arthur JSC, Armit LJ, Alessi DR. In vivo role of the PIF-binding docking site of PDK1 defined by knock-in mutation. EMBO J. 2003;22(16):4202–4211. doi: 10.1093/emboj/cdg407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biondi RM, et al. High-resolution crystal structure of the human PDK1 catalytic domain defines the regulatory phosphopeptide docking site. EMBO J. 2002;21(16):4219–4228. doi: 10.1093/emboj/cdf437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang J, et al. Crystal structure of an activated Akt/protein kinase B ternary complex with GSK3-peptide and AMP-PNP. Nat Struct Biol. 2002;9(12):940–944. doi: 10.1038/nsb870. [DOI] [PubMed] [Google Scholar]
- 21.Wang J, Zhong C, Wang F, Qu F, Ding J. Crystal structures of S6K1 provide insights into the regulation mechanism of S6K1 by the hydrophobic motif. Biochem J. 2013;454(1):39–47. doi: 10.1042/BJ20121863. [DOI] [PubMed] [Google Scholar]
- 22.Grodsky N, et al. Structure of the catalytic domain of human protein kinase C beta II complexed with a bisindolylmaleimide inhibitor. Biochemistry. 2006;45(47):13970–13981. doi: 10.1021/bi061128h. [DOI] [PubMed] [Google Scholar]
- 23.Clackson T, Wells JA. A hot spot of binding energy in a hormone–receptor interface. Science. 1995;267(5196):383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 24.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein–protein interfaces. Nature. 2007;450(7172):1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 25.Balendran A, et al. PDK1 acquires PDK2 activity in the presence of a synthetic peptide derived from the carboxyl terminus of PRK2. Curr Biol. 1999;9(8):393–404. doi: 10.1016/s0960-9822(99)80186-9. [DOI] [PubMed] [Google Scholar]
- 26.Medina JR, et al. Structure-based design of potent and selective 3-phosphoinositide–dependent kinase-1 (PDK1) inhibitors. J Med Chem. 2011;54(6):1871–1895. doi: 10.1021/jm101527u. [DOI] [PubMed] [Google Scholar]
- 27.Najafov A, Sommer EM, Axten JM, Deyoung MP, Alessi DR. Characterization of GSK2334470, a novel and highly specific inhibitor of PDK1. Biochem J. 2011;433(2):357–369. doi: 10.1042/BJ20101732. [DOI] [PubMed] [Google Scholar]
- 28.Najafov A, Shpiro N, Alessi DR. Akt is efficiently activated by PIF-pocket- and PtdIns(3,4,5)P3-dependent mechanisms leading to resistance to PDK1 inhibitors. Biochem J. 2012;448(2):285–295. doi: 10.1042/BJ20121287. [DOI] [PubMed] [Google Scholar]
- 29.Wilhelm A, et al. 2-(3-Oxo-1,3-diphenylpropyl)malonic acids as potent allosteric ligands of the PIF pocket of phosphoinositide-dependent kinase-1: Development and prodrug concept. J Med Chem. 2012;55(22):9817–9830. doi: 10.1021/jm3010477. [DOI] [PubMed] [Google Scholar]
- 30.Beaumont K, Webster R, Gardner I, Dack K. Design of ester prodrugs to enhance oral absorption of poorly permeable compounds: Challenges to the discovery scientist. Curr Drug Metab. 2003;4(6):461–485. doi: 10.2174/1389200033489253. [DOI] [PubMed] [Google Scholar]
- 31.Liederer BM, Borchardt RT. Enzymes involved in the bioconversion of ester-based prodrugs. J Pharm Sci. 2006;95(6):1177–1195. doi: 10.1002/jps.20542. [DOI] [PubMed] [Google Scholar]
- 32.Ellwood-Yen K, et al. PDK1 attenuation fails to prevent tumor formation in PTEN-deficient transgenic mouse models. Cancer Res. 2011;71(8):3052–3065. doi: 10.1158/0008-5472.CAN-10-2282. [DOI] [PubMed] [Google Scholar]
- 33.Zhang J, et al. Targeting Bcr-Abl by combining allosteric with ATP-binding site inhibitors. Nature. 2010;463(7280):501–506. doi: 10.1038/nature08675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nikolovska-Coleska Z, et al. Development and optimization of a binding assay for the XIAP BIR3 domain using fluorescence polarization. Anal Biochem. 2004;332(2):261–273. doi: 10.1016/j.ab.2004.05.055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.