Significance
Seed behavior is known to be highly dependent on the temperature during seed set, but the mechanism is poorly understood. Here we show that the mother plant plays a central role in the control of progeny seed dormancy, integrating long-term temperature memories in fruit tissues using the same pathway that controls flowering time. Regulation of seed coat properties by maternal flowering time pathways effectively passes timing information across generations, aligning progeny behavior with time of year.
Keywords: seed dormancy, temperature, germination, transgenerational, seed coat
Abstract
Seasonal behavior is important for fitness in temperate environments but it is unclear how progeny gain their initial seasonal entrainment. Plants use temperature signals to measure time of year, and changes to life histories are therefore an important consequence of climate change. Here we show that in Arabidopsis the current and prior temperature experience of the mother plant is used to control germination of progeny seeds, via the activation of the florigen Flowering Locus T (FT) in fruit tissues. We demonstrate that maternal past and current temperature experience are transduced to the FT locus in silique phloem. In turn, FT controls seed dormancy through inhibition of proanthocyanidin synthesis in fruits, resulting in altered seed coat tannin content. Our data reveal that maternal temperature history is integrated through FT in the fruit to generate a metabolic signal that entrains the behavior of progeny seeds according to time of year.
Many organisms use annual changes in temperature to control their phenology, resulting in predictable timing of key life history events, such as flowering, spawning, and migration (1–3). Understanding crop and ecosystem response to climate change requires knowledge of the temperature control of key developmental transitions, but how new generations achieve seasonal orientation is currently unclear. Seed germination is the first step in plant life history and therefore plays a central role in the control of plant phenology (4) and is extremely sensitive to environmental temperature (3–5). Seed dormancy is established during seed maturation and is imposed by control of hormone signaling and the action of the maternal seed coat. Nearly 30 y ago it was found that environmental signaling throughout the whole maternal life history can affect seed dormancy control in wild oats, and that temperature experience in the vegetative phase before flowering affected progeny seed dormancy (6). Here we show that this response is conserved on the model species Arabidopsis. Our data show that fruit tissues carry a memory of past temperature experience and that flowering pathways control a transgenerational metabolic signal of maternal past temperature experience, which modulates progeny dormancy according to time of year.
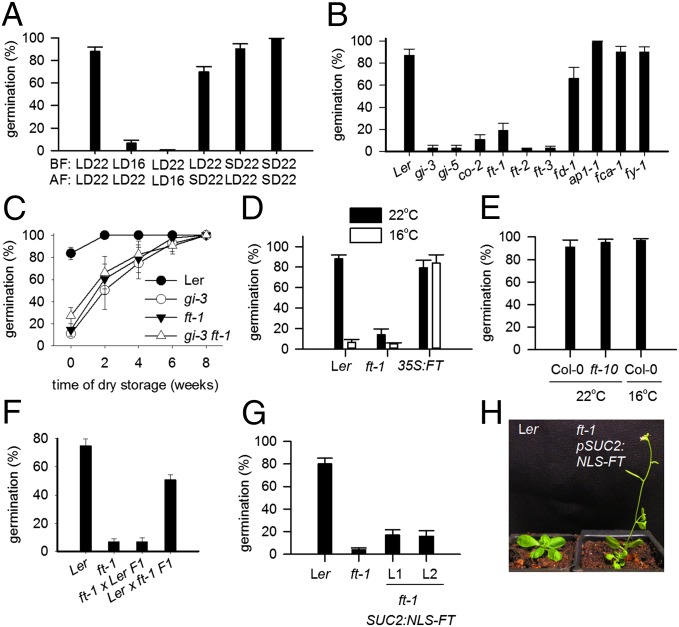
To test whether past parental temperature experience affected progeny dormancy in the model species Arabidopsis thaliana, we grew plants until the first sign of flowering at either 22 °C or 16 °C and then placed plants side by side to set seed at 22 °C in long days (LDs) (Fig. 1A). We found that in Landsberg erecta (Ler) lower temperature during the vegetative phase caused a large increase in the dormancy of seeds produced later on the plants (Fig. 1A). Lower temperatures during seed set also increase progeny dormancy (6), but we observed no effect of photoperiod on dormancy either before or after flowering, as has been reported previously (7, 8). Therefore, temperature signals before seed fertilization are remembered by the parent plant and used to control offspring behavior.
Fig. 1.
FT controls progeny seed dormancy in response to maternal temperature history. (A) Response of wild-type (Ler) progeny seeds to temperature and photoperiod treatments given to the mother plants either before fertilization (BF) or after fertilization during seed set (AF). LD, long days; SD, short days. Figures given are temperatures in degrees Celsius. (B) Germination of late flowering mutant seeds from mother plants grown at 22 °C and long days. (C) GI and FT have an epistatic role in dormancy control. After-ripening of dry-stored seeds of Ler, gi-3, ft-1, and gi-3 ft-1 double mutants. (D) Germination of wild-type, ft-1, and 35S:FT progeny seeds from plants that experienced either 16 °C (white bars) or 22 °C (black bars) until first flowering, and thereafter 22 °C during flowering and seed set. (E) In the Col-0 ecotype, neither FT nor the vegetative temperature experience of the mother plant has an effect on progeny dormancy. Germination of progeny seeds from plants raised under the indicated temperature before fertilization of the first flowers. (F) Reciprocal crosses demonstrate that the ft-1 low-germination phenotype is maternally inherited. (G and H) pSUC2:NLS-FT rescues the flowering time, but not the dormancy phenotype of ft-1 mutants, demonstrating that flowering time itself does not affect progeny seed dormancy. Data points shown are the mean ± SE of a minimum of four biological replicate seed batches for each genotype per experiment.
Previously we have shown that late flowering gigantea (gi) mutants have strongly increased seed dormancy in the Ler and Wassilewskija (Ws) accessions (9). We found that this phenotype was shared by constans (co) and flowering locus t (ft) mutants (10), but not with mutants in meristem-expressed genes flowering locus d (fd-1), or apetala 1 (ap-1), or with autonomous pathway mutants (fca-1 and fy-1), all of which showed similar seed dormancy to wild type (Fig. 1B). The lack of phenotype for the FT-binding partner fd-1 (11) suggests that FT does not act in the shoot apex to control seed dormancy. The role of the canonical photoperiod-sensitive flowering time pathway in seed dormancy inhibition was confirmed by observation of a clear epistatic relationship between gi-3 and ft-1 (Fig. 1C). Therefore, FT has a clear uncharacterized role in seed germination promotion, despite previous reports of a role in germination repression (12).
Because FT is known to play a role in temperature responses, and because the ft-1 mutant phenotype was similar to the effects of low temperature applied before flowering (Fig. 1A), we tested whether mutation or overexpression of FT affected the ability of progeny seeds to respond to temperature treatments given to the mother plant during the vegetative phase (Fig. 1D). Seeds were collected from plants maintained at 22 °C in LDs from first sign of bolting, but which had had experienced either 22 °C or 16 °C before first flowering (BFF). Whereas WT seeds showed a strong decrease in progeny dormancy in response to prior maternal experience of 22 °C, this response was lacking in ft-1. In contrast 35S:FT seeds showed the opposite response and germinated at high levels, regardless of the temperature experience of the parent plant (Fig. 1D). Taken together, these observations reveal that FT is necessary and sufficient for high progeny seed germination in response to experience of warmth before flowering: loss of FT prevents germination in response to warmth, whereas constitutive FT expression results in high germination even after a history of cool temperatures. Reciprocal crosses showed that seed germination responded to the maternal action of the FT gene (Fig. 1F) and that therefore the mother plant can transduce a temperature signal to progeny seeds. Surprisingly, these phenotypes were not observed in Col-0, where loss of FT had no effect on seed dormancy, and Col-0 plants showed no ability to remember temperature BFF (Fig. 1E). However, given that gi alleles in Ws cause increased dormancy (13), that the response was first described in the Poaceae, and that temperature treatments applied to the mother plant affect progeny dormancy in multiple accessions, this response is clearly very general in plants.
The normal seed dormancy of the late flowering fca-1 and fy-1 mutants (Fig. 1B) showed that changes in seed dormancy are unlikely to be a nonspecific consequence of changing flowering time. This finding was further confirmed using transgenic lines overexpressing nuclear-localized FT in phloem tissue (14). These tether FT to the cell of synthesis and do not permit long-range protein movement through the phloem. Unexpectedly these restored early flowering to ft-1 mutants, apparently contradicting an earlier report that this construct could not rescue the ft-10 mutant phenotype (Fig. 1 G and H) (14). This likely arises because position effects permit direct expression of FT in shoot apices, thus negating the need for long-range movement of FT to induce the floral transition. However, these lines did not rescue the increased seed dormancy of ft-1, showing that flowering time and dormancy control by FT can be uncoupled, and confirming that early flowering itself does not affect progeny dormancy. The high prevalence of phenotypes consistent with position effects limited the use of such lines in determining whether FT protein movement is necessary for control of progeny seed behavior.
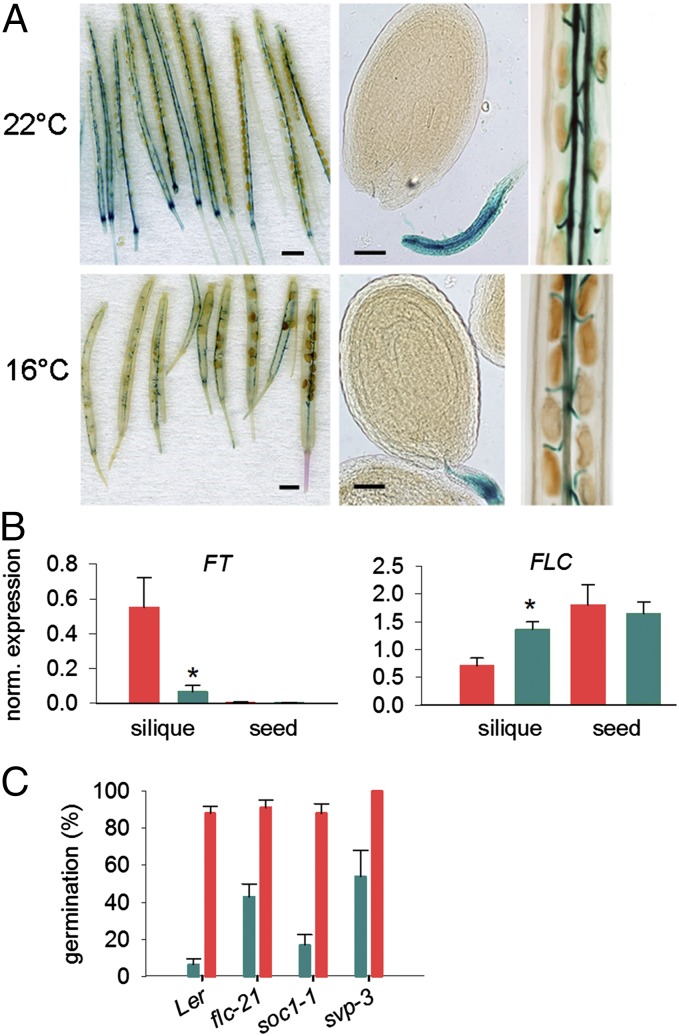
The fact that flowering time and seed dormancy can be uncoupled strongly suggests that FT protein synthesized in leaves does not affect seed germination. In addition, although photoperiod profoundly affects FT levels in leaves and vegetative shoots, maternal photoperiod does not affect seed dormancy (Fig. 1A). Therefore, FT synthesized in leaves does not affect progeny dormancy. Instead, we hypothesized that FT is synthesized in another tissue to control progeny seed behavior, and that in this tissue FT expression should be sensitive to temperature, but not to photoperiod. To localize the tissue in which FT transcriptional control regulates seed dormancy, we used a 5.7-kb FTpro:GUS reporter (15) and sought to identify tissues in which GUS expression was dependent on temperature, but not photoperiod. FTpro:GUS expression in the phloem of silique tissues (16, 17) was sensitive to temperature BFF (Fig. 2A). In siliques, the FT promoter was active in phloem tissue with higher temperatures BFF resulting in higher FT expression. In contrast, no FT promoter activity could be detected in seed tissues at either temperature. GUS assays showed that in siliques, FTpro:GUS activity was responsive to both prior and current temperature, demonstrating that the FT promoter integrates past and present temperature signals in fruits (Fig. S1A). However, in agreement with our physiological observations and previous studies (16, 17), expression of FT was unaffected by daylength in fruits. Note that the FTpro:GUS line is in the Col-0 background, which itself could not increase progeny seed dormancy in response to temperature BFF (Fig. 1): This shows that part of the temperature response is still preserved in Col-0 and that this accession is deficient in signaling downstream rather than upstream of FT. Real-time RT-PCR confirmed that FT expression is indeed high and sensitive to temperature BFF in siliques tissues, but absent from seeds (Fig. 2B and Fig. S1); FLOWERING LOCUS C (FLC) levels were also altered in fruit but not seed tissues (Fig. 2B and Fig. S1). In contrast, no effect of temperature on either CO or GI expression was observed (Fig. S1A). Because FT is expressed in fruits but affects seed behavior, either the movement of FT itself or a downstream consequence of FT activity must pass from fruit to seed to control progeny germination.
Fig. 2.
Maternal temperature experience before fertilization affects FT expression in siliques. (A) FTpro:GUSFTPRO:GUS expression in siliques from plants subjected to either 16 °C or 22 °C in long days before first flowering and both placed at 22 °C in long days for reproduction. (Scale bars, 1 mm for siliques, 50 μm for seeds.) (B) FT and FLC expression in seeds and siliques (without seeds), from plants which had experienced either 16 °C or 22 °C before first flowering and at least 1 wk before analysis. Data represent the mean ± SE of three biological replicates. Black and white bars indicate dark and light periods (plant were grown in long days). Asterisks denote significant differences (P < 0.0.01). (C) The germination of flc-21, soc1-1, and svp-3 seeds from mother plants subjected to either 22 °C (red) or 16 °C (green) before first flowering. Data points indicate mean and SE of four to eight replicate seed batches per genotype.
FLC has previously been shown to carry an epigenetic memory of vernalization (18) and has been shown to affect seed behavior (12), albeit in a manner inconsistent with the phenotypes we report here. FLC cooperates with the floral repressors FLM, SHORT VEGETATIVE PHASE (SVP) and SUPRESSOR OF CONSTANS 1 (SOC1) to control FT expression in response to temperature (19–21). Therefore, we tested the seed dormancy response of svp-3, soc1-1, and a previously uncharacterized Ler transposon insertion allele of FLC, designated flc-21 to maternal temperature experience BFF (Fig. 2C). Analysis of flc-21 revealed it to transcribe undetectable amounts of FLC sense mRNA (Fig. S2), and mutant phenotyping revealed that flc-21 seed dormancy was partly sensitive to temperature BFF (Fig. 2C): Therefore the FLC locus can account for only a fraction of the temperature memory regulating FT in fruits. svp-3 showed a very similar phenotype to flc-21, with a reduced sensitivity to maternal low temperature experience, but retaining a partial response. In contrast, soc1-1 seed germination was similar to wild type. The lower dormancy of flc-21 mutants contrasts with previous reports of low dormancy in high FLC-expressing near isogenic lines (NILs) (12).
To determine the importance of past temperature and FT in controlling gene expression in siliques, we profiled gene expression in fruit tissue only in Ler and ft-1 siliques from plants maintained at 22 °C and compared this expression also to Ler plants reproducing at 22 °C but had been grown until bolting at 16 °C (Fig. S1C). Large changes in gene expression underlined the importance of both FT and past temperature in controlling fruit physiology. Strikingly, there was a large overlap between changes caused by loss of FT and changes caused by prior low-temperature experience, suggesting a major role for FT in transducing temperature signals in fruits.
Next we sought the mechanism through which maternal FT controls progeny seed dormancy. Because seed dormancy is controlled by hormone synthesis and perception, we measured hormone levels in mature Ler and ft-1 mutant seeds (Fig. S3). We found wild-type levels of the germination inhibitors abscisic acid and 12-oxophytodienoic acid in mature ft-1 seeds, alongside increased gibberellin GA4 and jasmonate (JA) levels. Analysis of ft-1 allene oxide synthase-2 (aos-2) double mutants showed that JA was not necessary for the increased dormancy of ft-1 mutants (Fig. S3). Therefore, dormancy changes in ft-1 cannot be explained by control of seed hormone levels.
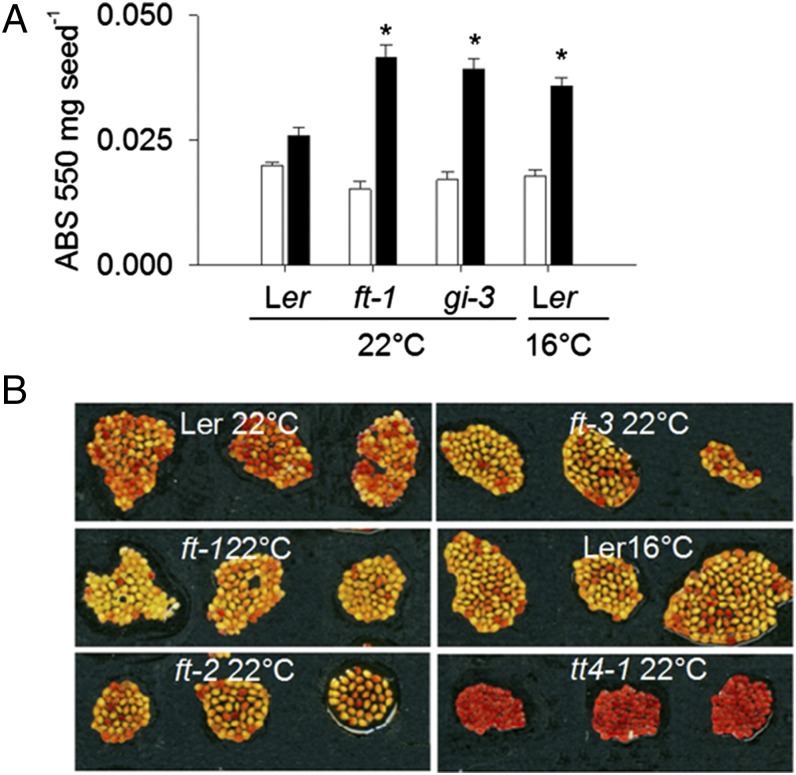
It has previously been observed that seeds of Chenopodium album show changes in seed coat thickness in response to photoperiod during seed set (22). In Arabidopsis the seed coat is dead at maturity, but seed coat tannin biosynthesis during seed maturation is required for normal dormancy (23), as in many other species. Colorimetric assays showed that gi and ft mutant seeds from mother plants grown at 22 °C had an increased condensed tannin content compared with wild type, and that a similar increase in progeny seed tannin content could be obtained by growing wild-type plants at 16 °C BFF (Fig. 3A). This change in seed coat condensed-tannin content was paralleled by decreases in seed permeability to tetrazolium dye (Fig. 3B) showing that the change in tannin content has biophysical consequences for the seed and contrasting with the previously described high permeability of the transparent testa (tt) mutant tt4-1 (23). Therefore, control of seed coat tannin synthesis was a candidate mechanism for the control of progeny dormancy by maternal FT.
Fig. 3.
ft-1 mutant seeds are affected in seed procyanidin content and seed coat permeability. (A) Insoluble (solid bars) and soluble (open bars) procyanidin content measured by acid butanol extraction and absorbance at 550 nm in WT, ft-1, and gi-3 mutant seeds from plants that had experienced the indicated temperature regime before flowering, but all maintained after flowering at 22 °C. Data represent mean and SE of five biological replicate seed batches, with the exception of gi-3, for which only three were available. Astersisks denote significant differences (P < 0.01). (B) Tetrazolium uptake of WT and ft seeds after 48 h from plants raised at 22 °C before fertilization of the first ovules, compared with wild-type seeds from plants that experienced 16 °C. tt4-1 mutants are shown as positive control. All plants were maintained at 22 °C after bolting. Pictures of three biological replicate seed batches are shown.
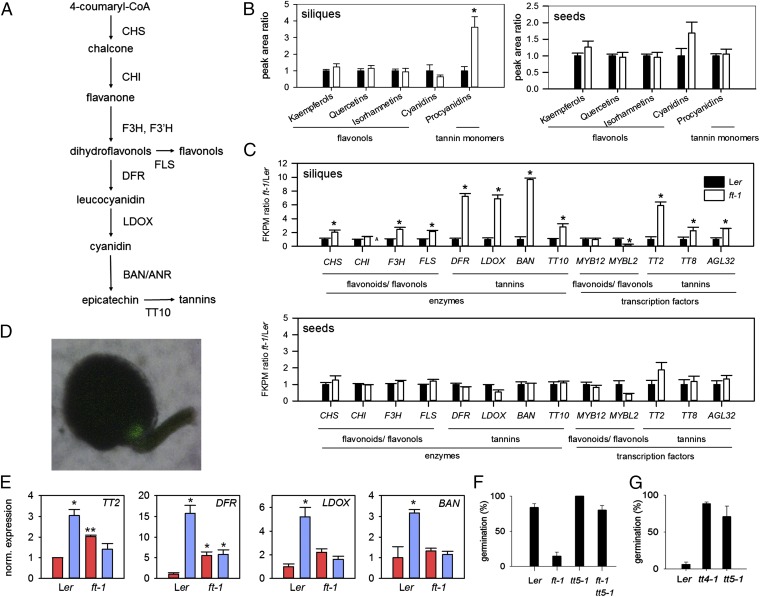
Synthesis of proanthocyanidins (PAs) is believed to take place in the inner integument of the seed coat. We profiled major phenylpropanoid pathway products using liquid chromatography coupled to a triple quadrupole mass spectrometer via an electrospray ionization triple source (24) in wild-type and ft-1 seeds (Fig. 4 A and B). However, we could find no evidence that FT significantly affected the levels of any soluble phenylpropanoid products in seeds. Because FT is expressed in siliques, we also analyzed silique tissues with the seeds removed. Here we could clearly see that PA levels were specifically increased in ft-1 siliques compared with wild type, with levels of alternative flavonoid end products at wild-type levels. Synthesis of PAs was previously believed to be exclusively in the inner integument of Arabidopsis seeds, but it has been detected previously in fruit tissues of grape (Vitis vinifera) (25). To determine how FT affects tannin synthesis during seed maturation, we profiled gene expression in seeds and siliques of wild type and ft-1 (Fig. 4C). We could find no change in phenylpropanoid pathway gene expression in ft-1 seeds compared with wild type. However, in siliques, large increases were observed in the transcript levels of enzymes specifically associated with PA synthesis (Fig. 4C). Therefore, FT specially affects PA synthesis in silique tissues. In seed coats, synthesis of PA is known to be controlled by at least three transcription factors, the MYB/bHLH complex containing TT2 and TT8, and by the MADS box protein Agamous Like 32 (AGL32) (26–28). Whereas modest increases in TT8 and AGL32 levels were observed, ft-1 mutant siliques exhibited large increases in TT2 expression. In contrast, levels of transcriptional regulators of flavonol synthesis were unchanged. TT2 is a known target of Sepallata (SEP) transcription factors (29). SEP transcripts as well as their interacting partners SHATTERPROOF 1 (SHP1), SHP2, and SEEDSTICK (STK) (30, 31) were also up-regulated in ft-1 mutant siliques, suggesting that in fruits, FT is transformed into a repressor of downstream MADS box transcription factor expression, and shp1 shp2 mutants, which have known roles in integument development and also show low dormancy (Fig. S4). Our data, together with previously published work, suggest that FT activity may regulate seed tannin levels via the effects of MADS box transcription factor complexes on TT2 expression. To determine whether the temperature regulation of PA synthesis (32) in siliques is contingent on FT, we analyzed gene expression in torpedo-stage embryo-containing siliques from plants that had experienced either 16 °C or 22 °C BFF (Fig. 4E). Consistent with our previous results, TT gene expression was increased by low-temperature BFF, but in ft-1 the temperature response was severely attenuated. Taken together, these data strongly support the hypothesis that temperature regulation of tannin accumulation in seeds is mediated by FT. Flavonoids have been previously reported in phloem (33). Given that ft-1 seeds accumulate increased condensed PAs (Fig. 3), but that FT affects PA synthesis in siliques, PA synthesized in siliques is likely transported to seeds. In addition, the FT protein itself may move to the seed: Indeed we found that FT–GFP accumulates in the chalazal seed coat when expressed under the phloem-specific SUC2 promoter (34) (Fig. 4D). Both possibilities are not mutually exclusive. To determine whether the regulation of tannin synthesis is sufficient to explain the increased dormancy of ft-1 mutants, we constructed the ft-1 tt5-1 double mutant. The presence of the tannin deficient tt5-1 completely rescued the increased dormancy of ft-1 (Fig. 4F), demonstrating that tannin synthesis is necessary for the increased dormancy of ft-1 mutants. We also compared the ability of tt mutants to increase seed dormancy in response to memory of vegetative low-temperature experience (Fig. 4G). Both tt4-1 and tt5-1 progeny dormancy were insensitive to temperature changes in the maternal vegetative phase, demonstrating that tannin synthesis is necessary for this response.
Fig. 4.
FT controls proanthocyanidin synthesis in siliques. (A) Cartoon demonstrating the main phenylpropanoid pathway active during proanthocyanidin synthesis in Arabidopsis seeds. (B) LC-MS profile of the principle phenylpropanoid end products in wild-type and ft-1 seeds and siliques. Data shown are the mean and SE of five biological replicate samples per genotype. (C) RNA-seq reveals that FT regulates phenylpropanoid pathway gene expression in siliques but not seeds. Data shown are relative RNA levels of enzymes and known transcriptional regulators in wild type and ft-1 mutants. RNA-seq data represent the mean and SE of three biological replicate samples per genotype. Asterisks indicate significant differences (P < 0.01). (D) Overlayed confocal and bright field image of pSUC2:FT-GFP accumulation at the chalazal pole of the seed. (Scale bar, 100 µm.) (E) Real-time PCR was used to analyze the effect of loss of FT on the control of tannin biosynthetic gene expression in siliques. Gene expression was analyzed in siliques containing torpedo-stage embryos 4 h after dawn and data represent the mean and SE of three biological replicates per treatment. Asterisks indicate significant differences from wild type at 22 °C. Plants were grown either at 22 °C (red bars) before fertilization or 16 °C (blue bars). Asterisks denote significant differences from wild type at 22 °C: *P < 0.01, **P < 0.001. (F) The tt5-1 mutant rescues most of the increased dormancy of the ft-1 mutant set at 22 °C. Data represent mean and SE of five biological replicates per genotype. (G) transparent testa mutant seed germination is not inhibited by low temperatures before first flowering. Seeds were collected from Ler and tt mutant plants that had experienced 16 °C BFF. Data represent mean and SE of seven biological replicates per genotype.
Previously we have shown that DELLA proteins and regulation of DELAY OF GERMINATION 1 (DOG1) expression are necessary for the temperature regulation of primary dormancy (5). Analysis of rgl2-1 mutants showed that they had an increase in seed coat permeability, especially if the temperature was 16 °C before seed set (Fig. S5). Analysis of rgl2-1 ft-1 double mutants showed that the low permeability of ft-1 is epistatic to the high permeability of rgl2-1 seeds, showing that maternal RGL2 affects seed coat permeability via FT. These findings are consistent with previous observations that RGL2 affects flowering time via regulation of the FT promoter (35). However, dormancy assays showed that the low dormancy of rgl2-1 mutants was epistatic to the increased dormancy of ft-1 mutants (Fig. S4). Therefore, the zygotic activity of RGL2 is required for seed coat properties to affect dormancy, and RGL2 therefore affects dormancy by both maternal and zygotic mechanisms. In contrast, loss or gain of function of DOG1 did not affect seed coat permeability, or the permeability response to maternal temperature experience (Fig. S5), but like rgl2-1, dog1 mutants could not increase dormancy in response to the maternal experience of low temperature before bolting (Fig. S5). Therefore, we conclude that DOG1 acts exclusively in the zygote to control dormancy and that both maternal and zygotic temperature signaling pathways control primary dormancy levels.
Our data shed new light on the key role of fruit tissues in the control of seed dormancy and show how the mother plant uses temperature history to control progeny behavior. Because FT levels vary with temperature and temperature varies with time of year, it is likely that this process is important in passing seasonal cues to progeny seed. Our data show that FT activity in fruit tissue phloem controls progeny dormancy, and that FT influences PA levels in fruits (Figs. 2 and 4). We also cannot rule out the possibility that in addition, FT itself is transported to seed coats, affecting PA synthesis locally. MADS box complexes directly regulate Arabidopsis TT2 expression (29), the MYB transcription factor known to activate the expression of enzymes required for PA synthesis (27). In kiwifruit SVP overexpression also affects flavonoid synthesis in flowers and seed vigor (36) and can heterodimerize with the SEP proteins as well as control FT expression (19). In seeds, flavonoid pathway gene expression and PA levels are temperature responsive (32). Therefore, it is likely that a complex set of interactions between FT and MADS box transcription factors mediates the control of PA levels in seeds, via TT2.
Previously it has been shown that FLC is also a maternal regulator of Arabidopsis seed dormancy (12), with high FLC levels and late flowering correlating with low seed dormancy. In contrast, our study found that a subset of late flowering mutants predominantly from the canonical photoperiod pathway showed increased seed dormancy, whereas an flc mutant showed reduced levels of dormancy. Because of this discrepancy, we repeated the germination assays under the same conditions used by Chiang et al. (12) (Fig. S6), but still we could not see the previously reported low dormancy of the ft mutant. One possibility is that in the previous study, very high FLC levels in the FLC NILs affects dormancy independently of FT. However, given that in lines with high FLC expression, flowering usually occurs after vernalization, the relevance of high maternal FLC expression to seed dormancy control under natural conditions is unclear.
Instead, our results are more consistent with data from poplar, which associate high FT expression with failure to enter growth cessation and induce bud dormancy (37). In that case and here in this study, high FT levels are associated with the low dormancy state. A recent study of natural variation in Arabidopsis also showed that late flowering is correlated with higher seed dormancy (38). Again this shows that late flowering is associated with more dormant states, in agreement with our observation that some late flowering mutants have increased seed dormancy. Arabidopsis fruits are therefore a good system for the study of flowering pathway plasticity, and given the comparatively late evolution of flowering, might represent a more ancient network topology.
Materials and Methods
Plant Materials.
Arabidopsis mutants were obtained from the European Arabidopsis Stock Centre unless otherwise indicated. The 35S:FT seeds and svp-3 were a gift from Markus Schmidt (Max Planck Institute for Developmental Biology, Tuebingen, Germany); the former was introgressed for three generations from Col-0. The nuclear-localized phloem-expressed FT construct pSUC2:NLS-FT (15) was a gift from Philip Wigge (Sainsbury Laboratory, University of Cambridge, United Kingdom) and was transformed into the ft-1 mutant by Agrobacterium-mediated floral dip. This generated several independent early flowering lines of which two were brought to homozygosity and analyzed for seed dormancy. The 5.7-kb FTpro:GUS reporter line was a gift from Franziska Turck (Max Planck Institute for Plant Breeding Research, Cologne, Germany) (15). pSUC2:FT-GFP plants and soc1-1 mutants were a gift from George Coupland (Max Planck Institute for Plant Breeding Research, Cologne, Germany) (20, 34). flc-21 seeds were obtained from the Cold Spring Harbor gene trap collection.
Seed Dormancy Assays.
Seeds were harvested from mature brown siliques from plants watered consistently grown in Sanyo MLR growth chambers under 16 h of white light (long days) at 100 µmol⋅m−2⋅sec−1 at 22 °C, except where indicated. Germination frequency was measured after 7 d from freshly harvested seed batches placed in 12 h white light/12 h dark at 22 °C on 0.9% water agar plates. Batches harvested from separate individual plants were used as biological replicates. Where the effect of growth temperature before fertilization on progeny dormancy is addressed, plants were moved to the seed set conditions at first visible sign of the floral transition. Seed produced 2–4 wk after the temperature shift were analyzed, and this included seed on primary inflorescence, including branches, and so was not limited to the analysis of seeds from ovules differentiating before the temperature shift.
Seed Coat and Metabolite Analysis.
Tetrazolium staining of Arabidopsis seeds has been described (23) and took place in the dark at 30 °C for 48 h. PA analysis was performed as described (39, 40) using freshly harvested flash frozen seeds. LC-MS analysis of phytohormones and phenylpropanoids has been described previously (24, 41). Significance was tested using the T statistic.
RNA Analysis.
RNA was extracted from siliques and seeds using sodium tetraborate extraction as described previously (42). cDNA was synthesized using M-MLV reverse transcriptase (Promega) and PCR carried out under standard conditions for 40 cycles on an Agilent Mx3005P qPCR system. The control gene used was At5g46630. Primers for real time RT-PCR are given in Table S1. RNA-seq was carried out using three biological replicate samples for WT and ft-1 seeds and siliques using an Illumina Hi-sEq. 2500. Preliminary data analysis was performed using Tophat and Cufflinks (43), using the TAIR10 Arabidopsis genome annotation. The cuffcompare and cuffdiff components of Cufflinks were used to quantify gene and isoform differential expression. Further analysis was performed using the cummeRbund package (43). The RNA-seq data are available at ArrayExpress, accession no. E-MTAB-2869.
GUS Assays.
For GUS staining, developing siliques were cut from plants and vacuum infiltrated in the GUS staining solution as described previously (44). Quantitative GUS assays were performed as described previously (44). The fluorescent intensity was measured using an Infinite 200 plate reader (Tecan; excitation, 365 nm; emission, 455 nm). Protein concentrations were determined using the Qubit Protein Assay kit (Invitrogen) following the manufacturer’s instructions.
Supplementary Material
Acknowledgments
We thank Franziska Turk for supplying FTpro:GUS seeds, Philip Wigge for the gift of the pSUC2:NLS-FT construct, George Coupland for the pSUC2:FT-GFP and soc1-1 seeds, and Markus Schmid for svp-3. This work was supported by Biotechnology and Biological Sciences Research Council Grants BB/J000949/1 (to S.P.) and BB/J00216X/1 (to I.A.G.), a Royal Society University Research Fellowship (to S.P.), and a Wellcome Trust Institutional Strategic Support Award (WT097835MF).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The RNAseq data reported in this paper have been deposited in the ArrayExpress database (accession no. E-MTAB-2869).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1412274111/-/DCSupplemental.
References
- 1.Parmesan C, Yohe G. A globally coherent fingerprint of climate change impacts across natural systems. Nature. 2003;421(6918):37–42. doi: 10.1038/nature01286. [DOI] [PubMed] [Google Scholar]
- 2.Fitter AH, Fitter RS. Rapid changes in flowering time in British plants. Science. 2002;296(5573):1689–1691. doi: 10.1126/science.1071617. [DOI] [PubMed] [Google Scholar]
- 3.Fenner M. The effects of the parent environment on seed germinability. Seed Sci Res. 1991;1(2):75–84. [Google Scholar]
- 4.Chiang GC, et al. Pleiotropy in the wild: The dormancy gene DOG1 exerts cascading control on life cycles. Evolution. 2013;67(3):883–893. doi: 10.1111/j.1558-5646.2012.01828.x. [DOI] [PubMed] [Google Scholar]
- 5.Kendall SL, et al. Induction of dormancy in Arabidopsis summer annuals requires parallel regulation of DOG1 and hormone metabolism by low temperature and CBF transcription factors. Plant Cell. 2011;23(7):2568–2580. doi: 10.1105/tpc.111.087643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sawhney R, Quick WA, Hsiao AI. The effect of temperature during parental vegetative growth on seed-germination of wild oats (Avena-Fatua L) Ann Bot (Lond) 1985;55(1):25–28. [Google Scholar]
- 7.Munir J, Dorn LA, Donohue K, Schmitt J. The effect of maternal photoperiod on seasonal dormancy in Arabidopsis thaliana (Brassicaceae) Am J Bot. 2001;88(7):1240–1249. [PubMed] [Google Scholar]
- 8.Donohue K, et al. Environmental and genetic influences on the germination of Arabidopsis thaliana in the field. Evolution. 2005;59(4):740–757. [PubMed] [Google Scholar]
- 9.Penfield S, Hall A. A role for multiple circadian clock genes in the response to signals that break seed dormancy in Arabidopsis. Plant Cell. 2009;21(6):1722–1732. doi: 10.1105/tpc.108.064022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koornneef M, Hanhart CJ, van der Veen JH. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet. 1991;229(1):57–66. doi: 10.1007/BF00264213. [DOI] [PubMed] [Google Scholar]
- 11.Wigge PA, et al. Integration of spatial and temporal information during floral induction in Arabidopsis. Science. 2005;309(5737):1056–1059. doi: 10.1126/science.1114358. [DOI] [PubMed] [Google Scholar]
- 12.Chiang GC, Barua D, Kramer EM, Amasino RM, Donohue K. Major flowering time gene, flowering locus C, regulates seed germination in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2009;106(28):11661–11666. doi: 10.1073/pnas.0901367106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmuths H, Bachmann K, Weber WE, Horres R, Hoffmann MH. Effects of preconditioning and temperature during germination of 73 natural accessions of Arabidopsis thaliana. Ann Bot (Lond) 2006;97(4):623–634. doi: 10.1093/aob/mcl012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaeger KE, Wigge PA. FT protein acts as a long-range signal in Arabidopsis. Curr Biol. 2007;17(12):1050–1054. doi: 10.1016/j.cub.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 15.Adrian J, et al. cis-Regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell. 2010;22(5):1425–1440. doi: 10.1105/tpc.110.074682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hiraoka K, Yamaguchi A, Abe M, Araki T. The florigen genes FT and TSF modulate lateral shoot outgrowth in Arabidopsis thaliana. Plant Cell Physiol. 2013;54(3):352–368. doi: 10.1093/pcp/pcs168. [DOI] [PubMed] [Google Scholar]
- 17.Liu L, Farrona S, Klemme S, Turck FK. Post-fertilization expression of FLOWERING LOCUS T suppresses reproductive reversion. Front Plant Sci. 2014;5:164. doi: 10.3389/fpls.2014.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bastow R, et al. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature. 2004;427(6970):164–167. doi: 10.1038/nature02269. [DOI] [PubMed] [Google Scholar]
- 19.Lee JH, et al. Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 2007;21(4):397–402. doi: 10.1101/gad.1518407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Samach A, et al. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science. 2000;288(5471):1613–1616. doi: 10.1126/science.288.5471.1613. [DOI] [PubMed] [Google Scholar]
- 21.Posé D, et al. Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature. 2013;503(7476):414–417. doi: 10.1038/nature12633. [DOI] [PubMed] [Google Scholar]
- 22.Pourrat Y, Jacques R. The influence of photoperiodic conditions received by the mother plant on morphological and physiological characteristics of Chenopodium polyspermum L. seeds. Plant Sci Lett. 1975;4(4):273–279. [Google Scholar]
- 23.Debeaujon I, Léon-Kloosterziel KM, Koornneef M. Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol. 2000;122(2):403–414. doi: 10.1104/pp.122.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Page M, Sultana N, Paszkiewicz K, Florance H, Smirnoff N. The influence of ascorbate on anthocyanin accumulation during high light acclimation in Arabidopsis thaliana: Further evidence for redox control of anthocyanin synthesis. Plant Cell Environ. 2012;35(2):388–404. doi: 10.1111/j.1365-3040.2011.02369.x. [DOI] [PubMed] [Google Scholar]
- 25.Gagne S, Lacampagne S, Claisse O, Geny L. Leucoanthocyanidin reductase and anthocyanidin reductase gene expression and activity in flowers, young berries and skins of Vitis vinifera L. cv. Cabernet-Sauvignon during development. Plant Physiol Biochem. 2009;47(4):282–290. doi: 10.1016/j.plaphy.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 26.Nesi N, et al. The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell. 2000;12(10):1863–1878. doi: 10.1105/tpc.12.10.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L. The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell. 2001;13(9):2099–2114. doi: 10.1105/TPC.010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nesi N, et al. The TRANSPARENT TESTA16 locus encodes the ARABIDOPSIS BSISTER MADS domain protein and is required for proper development and pigmentation of the seed coat. Plant Cell. 2002;14(10):2463–2479. doi: 10.1105/tpc.004127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaufmann K, et al. Target genes of the MADS transcription factor SEPALLATA3: Integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol. 2009;7(4):e1000090. doi: 10.1371/journal.pbio.1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pinyopich A, et al. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature. 2003;424(6944):85–88. doi: 10.1038/nature01741. [DOI] [PubMed] [Google Scholar]
- 31.Favaro R, et al. MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell. 2003;15(11):2603–2611. doi: 10.1105/tpc.015123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacGregor DR, et al. Seed production temperature regulation of primary dormancy occurs through control of seed coat phenylpropanoid metabolism. New Phytol 2014 doi: 10.1111/nph.13090. , in press. [DOI] [PubMed] [Google Scholar]
- 33.Grayer RJ, Harborne JB, Kimmins FM, Stevenson PC, Wijayagunasekera HNP. Phenolics in rice phloem sap as sucking deterrents to the Brown Planthopper, Nilaparvata lugens. Acta Hortic. 1994;381(2):691–694. [Google Scholar]
- 34.Corbesier L, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316(5827):1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- 35.Galvão VC, Horrer D, Küttner F, Schmid M. Spatial control of flowering by DELLA proteins in Arabidopsis thaliana. Development. 2012;139(21):4072–4082. doi: 10.1242/dev.080879. [DOI] [PubMed] [Google Scholar]
- 36.Wu R, et al. Overexpression of the kiwifruit SVP3gene affects reproductive development and suppresses anthocyanin biosynthesis in petals, but has no effect on vegetative growth, dormancy, or flowering time. J Exp Bot. 2014;65(17):4985–4995. doi: 10.1093/jxb/eru264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Böhlenius H, et al. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science. 2006;312(5776):1040–1043. doi: 10.1126/science.1126038. [DOI] [PubMed] [Google Scholar]
- 38.Debieu M, et al. Co-variation between seed dormancy, growth rate and flowering time changes with latitude in Arabidopsis thaliana. PLoS ONE. 2013;8(5):e61075. doi: 10.1371/journal.pone.0061075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Routaboul JM, et al. Flavonoid diversity and biosynthesis in seed of Arabidopsis thaliana. Planta. 2006;224(1):96–107. doi: 10.1007/s00425-005-0197-5. [DOI] [PubMed] [Google Scholar]
- 40.Routaboul JM, et al. Metabolite profiling and quantitative genetics of natural variation for flavonoids in Arabidopsis. J Exp Bot. 2012;63(10):3749–3764. doi: 10.1093/jxb/ers067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dave A, et al. 12-oxo-phytodienoic acid accumulation during seed development represses seed germination in Arabidopsis. Plant Cell. 2011;23(2):583–599. doi: 10.1105/tpc.110.081489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Penfield S, et al. Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr Biol. 2005;15(22):1998–2006. doi: 10.1016/j.cub.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 43.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6(13):3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.