Significance
mAbs are increasingly being used for treatment of chronic diseases wherein the subcutaneous delivery route is preferred to enable self-administration and at-home use. To deliver high doses (several hundred milligrams) through a small volume (∼1 mL) into the subcutaneous space, mAb solutions need to have low viscosity. Concomitantly, acceptable chemical stability is required for adequate shelf life, and normal in vivo clearance is needed for less frequent dosing. We propose in silico tools that provide rapid assessment of atypical behavior of mAbs (high viscosity, chemical degradation, and fast plasma clearance), which are simply predicted from sequence and/or structure-derived parameters. Such analysis will greatly improve the probability of success to move mAb-based therapeutics efficiently into clinical development and ultimately benefit patients.
Keywords: monoclonal antibodies, viscosity, degradation, prediction, pharmacokinetics
Abstract
For mAbs to be viable therapeutics, they must be formulated to have low viscosity, be chemically stable, and have normal in vivo clearance rates. We explored these properties by observing correlations of up to 60 different antibodies of the IgG1 isotype. Unexpectedly, we observe significant correlations with simple physical properties obtainable from antibody sequences and by molecular dynamics simulations of individual antibody molecules. mAbs viscosities increase strongly with hydrophobicity and charge dipole distribution and decrease with net charge. Fast clearance correlates with high hydrophobicities of certain complementarity determining regions and with high positive or high negative net charge. Chemical degradation from tryptophan oxidation correlates with the average solvent exposure time of tryptophan residues. Aspartic acid isomerization rates can be predicted from solvent exposure and flexibility as determined by molecular dynamics simulations. These studies should aid in more rapid screening and selection of mAb candidates during early discovery.
Treatment of certain chronic ailments, e.g., rheumatoid arthritis, using mAb-based therapies require delivery via the s.c. route for patients’ at-home use, self-administration, and compliance (1). To deliver several hundred milligrams of the active drug in a small volume (∼1 mL) into the s.c. space, a liquid formulation containing high concentrations of mAb is required (1, 2). Therefore it is essential that the lead clinical mAb candidate meets the following criteria: (i) an injectable solution of low viscosity (higher viscosity solutions are difficult to manufacture and administer, and could be painful to inject) (2); (ii) minimal chemical/physical degradation in solution such that the efficacy and safety is maintained; and (iii) a normal in vivo clearance profile to avoid multiple injections and/or more frequent dosing (3, 4).
Effort must be spent early on to screen for lead mAb candidates, which exhibit desirable physicochemical and biological attributes. Inclusion of computational in silico tools along with experimental approaches will enable rapid screening of a larger number of mAb candidates (5–7).
In this report, we describe in silico screening approaches that aid in selection of mAb candidates with respect to viscosity, in vivo clearance in Cynomolgus monkeys (a relevant preclinical model for human clearance), Trp oxidation, and Asp isomerization. We show that using the optimal parameters extracted from sequence and/or structure including molecular dynamic simulations, the desirable attributes of mAbs can be predicted to enable lead candidate selection.
Viscosity
mAbs, differing largely in the complementarity determining region (CDR) sequence or in the Fv domain, exhibit a variety of viscosity-concentration profiles under similar conditions of shear rate (Fig. 1A). Recognizing that for mAbs of similar isotype (IgG1 in the present case), the variable domain Fv (and the CDRs within) presumably plays a critical role in defining intermolecular interactions leading to differences in viscosity (8, 9), we set out to determine what parameters can be extracted to capture the contributing hydrophobic and electrostatic elements (10, 11). It has been postulated recently that such interactions may produce molecular entanglements, leading to increased viscosity (12). We focused on sequence only because this provided the simplest means of data generation and analysis. However, we note that any of the parameters discussed below and calculated from the sequence can be readily calculated from structure as well (Figs. S1 and S2). As shown (Figs. S1 and S2), the sequence-based calculations correlate with structure-based calculations.
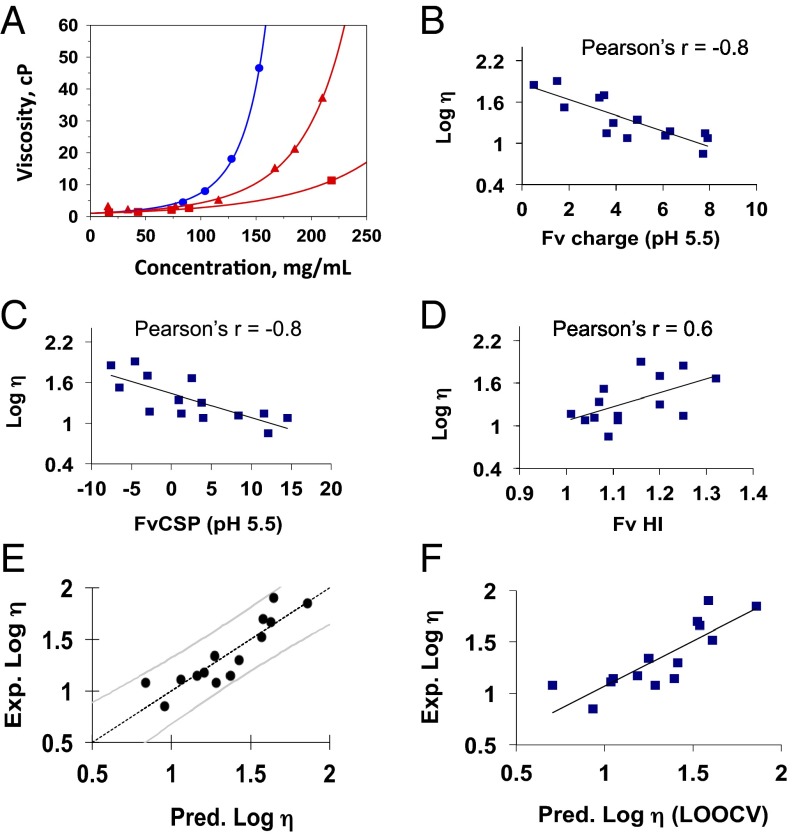
Fig. 1.
(A) Viscosity-concentration profiles of three monoclonal antibodies of the IgG1 isotype in a buffered solution at pH 5.5 and 200 mM arginine-HCl. The points represent the experimental data. The lines are used as a guide to the eye and were generated using the equation of the exponential form y = a + becx, where y is viscosity, x is protein concentration, and a, b, and c are fitting parameters. Correlation of log viscosity with the calculated sequence-based parameters of (B) charge at pH 5.5, (C) Fv charge symmetry parameter (FvCSP) at pH 5.5, and (D) Fv hydrophobicity index (HI). The viscosity values were obtained in buffered solution at pH 5.5 and 200 mM arginine-HCl. (E) Principal component regression analysis plot showing the predicted viscosity values against the experimental viscosity values for 180 mg/mL mAb concentration. The observed viscosity values are the experimental values obtained in buffered solution at pH 5.5 and 200 mM arginine-HCl. The predicted viscosity values are the output values from PCR analysis and are described by Eq. 1. Each data point represents an mAb, and the curved lines represent the 90% CIs. (F) Scatterplot between the predicted values obtained using the LOOCV approach through PCR analysis and the experimental viscosity values.
The parameters calculated were (i) net charge of Fv at a given pH (pH 5.5 in this study), (ii) Fv charge symmetry parameter (FvCSP) between the VH and VL domain at pH 5.5, and (iii) the hydrophobicity index (HI) of the Fv. The net charge potentially contributes to repulsive interactions, whereas FvCSP and HI can potentially contribute to attractive interactions. The FvCSP parameter represents similarity in the net charge states of the VH and VL domains. We hypothesized that a lack of charge symmetry, i.e., opposite net charges (negative FvCSP), provides an opportunity for the Fv domain to interact with another Fv domain through a dipole-like interaction or with another charge patch present on the mAb (11, 13). Therefore, a greater negative FvCSP value is expected to lead to stronger attractive interactions. We recognize that the actual structural conformation could distribute the charge asymmetry in a way that may not be captured as defined above by the sequence. However, our sequence-based calculations provide a reasonable first approximation to the lack of charge symmetry that is relatively easy to compute.
We examined correlations between these three parameters and experimental viscosity values (measured at 180 mg/mL in a 200 mM arginine-based buffer at pH 5.5) for a set of 14 different mAbs (Fig. 1 B–D). A fair correlation is observed between the Fv charge and viscosity (Pearson’s r = −0.8) and between FvCSP and viscosity (Pearson’s r = −0.8); however, the correlation somewhat weakens between HI and viscosity (Pearson’s r = 0.6). Evidently, electrostatic interactions play a dominant role in modulating viscosity, whereas hydrophobicity contributes to the overall viscosity of these mAbs to a less extent under these solution conditions. A stronger correlation between the FvCSP and viscosity points to the fact that the charge asymmetry between the VH and VL domain potentially plays a role in modulating viscosity.
Next, we used principal component regression (PCR) analysis for providing a predictive model for viscosity. Viscosity at 180 mg/mL at 25 °C was used as the independent variable. Fv charge represented as q, FvCSP represented as qsym, and HI represented as ϕ were used as dependent variables. The details of the calculated parameters are shown in Table S1. The observed experimental viscosity values at 180 mg/mL for various mAbs are plotted against the predicted viscosity values as obtained through the best-fit equation, together with a 90% CI (Fig. 1E). The best-fit equation is described as
| [1] |
The coefficients are specific to this buffer system and the respective protein concentration. The SEs associated with the coefficients are indicated in parentheses next to the coefficients. Overall, a strong correlation (Pearson’s r = 0.9) and a mean absolute error of 7 ± 9 cP at 180 mg/mL between observed and predicted values demonstrates that the model works well in predicting viscosity values. To further test the validity of the model, we used the leave-one-out cross-validation (LOOCV) approach. PCR analysis was performed while leaving out one mAb and using the remaining mAbs as the training set; the resulting best-fit equation was then used to predict the viscosity of the left-out mAb; the steps were then repeated for each mAb. A strong correlation is observed (Pearson’s r = 0.8) with a mean absolute error of 9 ± 10 cP between the predicted and the observed viscosity values (Fig. 1F and Table S1). Based on this analysis, we show that the current training set and the resulting output model (Eq. 1) enables prediction of the viscosity values for mAbs of the IgG1 isotype.
The model equation obtained through PCR regression analysis, using the sequence-derived theoretical parameters, is effective in predicting the viscosity for this protein-buffer system involving the antibodies of the IgG1 isotype. This approach can be extended to other buffer systems, as well as other IgG subclasses, as long as the critical theoretical parameters contributing to viscosity are identified. Based on the type of buffer systems and solution conditions, it is likely that other parameters, for example, related to ion binding, may need to be included to generate an effective predictive model.
Clearance
Antibodies within a similar isotype exhibit vast differences in plasma clearance in humans and in Cynomolgus (Cyno) monkeys (an established preclinical model) (14). A few studies have shown such differences to be correlated to pI or specific mutations in the sequence (15, 16); however, no clear trend is reported. The underlying cause for faster clearance has been attributed to off-target/nonspecific binding of the mAbs in vivo (14), presumably through hydrophobic and/or electrostatic interactions. We set out to explore whether any of the sequence properties would predict the differences in Cyno clearance. We hypothesized that any extremes of such properties in the variable domain, such as pI, charge, or hydrophobicity, would translate into the antibody exhibiting a faster Cyno clearance. Based on data previously published, a clearance value of ≥10 mL/kg per day (i.e., amount of drug cleared from plasma volume in a given unit of time for a given body weight) in Cyno monkeys was designated as faster clearance, and a value of <10 mL/kg per day was designated as normal clearance (14).
A large set of IgG1 mAbs (61 mAbs) was evaluated for their Cyno clearance at the maximum administered dose (ranging from 10 to 100 mg/kg). As reported previously (14), no clear correlation was observed between the calculated mAb pI or HI and clearance (Fig. S3 A and B). Rather than evaluating the molecular properties of the whole mAb, our next step was to examine if differences in the variable domain, Fv, would be more discriminating with respect to clearance rates. To this end, Fv charge in a certain pH range (pH 5.0–7.5) was calculated and compared against the clearance values. A broader pH range was covered to include physiological and the endosomal pH (antibody clearance involves FcRn salvation through the endosomal environment that has low pH, pH 5–6) (3). Furthermore, it was examined if hydrophobicity of certain CDRs would correlate better with clearance rather than the overall CDR or Fv hydrophobicity.
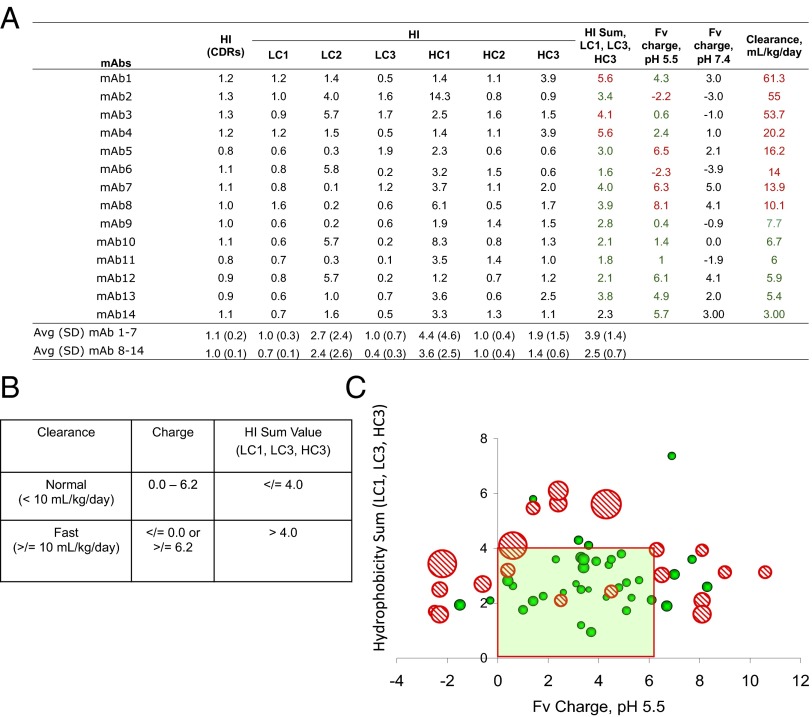
To simplify our analysis, a test set of 14 mAbs, similar to that used in ref. 14, was first used to cover the full range of clearance values (Fig. 2A). The mAbs in the training set were arranged in decreasing order of clearance values. We evaluated criteria that would allow us to separate out the two groups of mAbs (≥10 vs. <10 mL/kg per day). Examination of the calculated parameters revealed that faster clearing mAbs tend to have higher hydrophobicity among three specific CDRs (LC CDR1, LC CDR3, and HC CDR3). A calculated sum of the HI values of these three CDRs was used to simplify further analysis. The averaged HI sum for the mAbs in the faster clearing group is significantly higher that those in the normal clearing mAbs (3.9 ± 1.4 vs. 2.5 ± 0.7, respectively, P = 0.045, unpaired t test). With respect to the Fv charge, we noticed that at pH 5.5 (which coincidentally happens to be in the endosomal pH range), all six mAbs with normal clearance values tend to have charge values between 0.4 and 6.1, whereas five of eight mAbs that clear faster have charge values outside this range. We noticed that the charge and selective HI of CDRs are complementary to each other in differentiating faster clearing mAbs, i.e., those mAbs with charge values between 0.4 and 6.1 in the fast clearance group have a relatively higher HI sum value. This data analysis indicates that either high hydrophobicity of certain CDRs or extremes of Fv charge values (either high negative or high positive) could be predictive of mAbs with faster clearance.
Fig. 2.
(A) Clearance values in Cynomolgus monkeys and the calculated sequence parameters for a training set of 14 mAbs. (B) Assigned criteria of the sequence-derived parameters based on the training set of 14 mAbs to differentiate between mAbs of fast clearance and normal clearance. (C) Distribution of a set of 61 mAbs based on their respective theoretical Fv domain charge at pH 5.5 and a calculated HI sum of CDRs for LC1, LC3, and HC3. Each data point on the grid represents a given mAb, and the size of each data point is proportional to the clearance values by area. Solid green circles represent mAbs with normal clearance (<10 mL/kg per day), and red dashed circles represent mAbs with fast clearance (≥10 mL/kg per day) in Cynomolgus monkeys.
The above analysis led to developing the criteria to differentiate faster clearing mAbs from those with normal clearance (Fig. 2B). In the training set, if we set arbitrary criteria that mAbs with the HI sum value of >4.0 and/or an Fv charge value of either ≤0 or ≥6.2 are at risk for exhibiting faster clearance, whereas those with the hydrophobicity sum value of ≤4.0 and Fv charge values within 0–6.2 will exhibit normal clearance, then faster clearing mAbs (colored red) are clearly separated compared with the normal clearing mAbs (colored green).
We extended these criteria to a set of 61 mAbs to test its validity. To facilitate visualization of such analysis, we converted the data into a graphical format using the The Fv charge and HI sum as the two axes (Fig. 2C). Each mAb is plotted on the graph based on their respective charge, and HI sum values and the size of each data point are proportional to the clearance value of that mAb (by area). All Cyno clearance values of ≥10 mL/kg per day are represented by red dashed circles, whereas the remaining mAbs with normal clearance are represented by green solid circles. The solid line square box represents the charge-HI sum criteria based on the training set. It is evident that the criteria hold up well for the complete set of 61 mAbs including the test mAbs. Based on either high HI sum or Fv charge extremes, we were able to correctly predict faster Cyno clearance for 10 of 13 (77%) mAbs (86% including the training set mAbs) and normal Cyno clearance for 24 of 34 (70%) mAbs (75% including the training set mAbs). Table S2 provides a presentation of the basis of this analysis. If the analysis is done differently, that is, if false positives and false negatives are examined by looking at the predicted outcome, it also becomes clear that probability of assigning normal clearance based on charge and hydrophobicity is much higher (only 3 mAbs are fast clearing that were predicted to be normal) compared with assigning faster clearance (10 mAbs are normal clearing that were predicted to be fast). Overall, our analysis essentially establishes the ability of using fundamental molecular properties of charge and hydrophobicity in assessing the clearance of mAbs in Cynomolgus monkeys.
Trp Oxidation and Asp Isomerization
Along with low viscosity and normal clearance, a liquid formulation is paramount to enable at-home administration because of its ease of use with a device. Chemical stability (prevention of amino acid side chain modification) is required to enable a liquid formulation for prolonged shelf-life. We used all-atom molecular dynamics (MD) simulations with explicit water to enable ranking the relative liability of Trp oxidation and Asp isomerization, common degradation pathways that can limit the viability of an aqueous mAb solution for injection. A homology model-based approach has been recently reported to assess Asp isomerization (17).
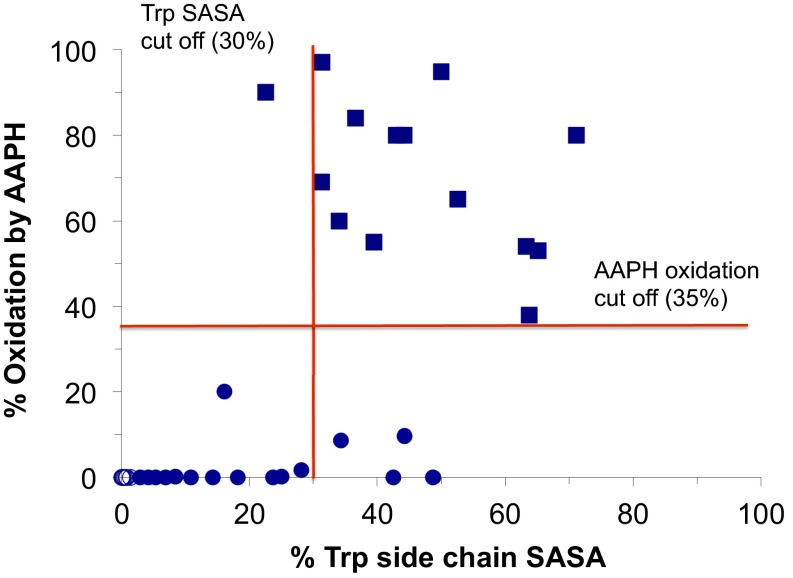
For Trp oxidation, we examined the correlation between MD generated time-averaged solvent-accessible surface area (SASA) and the extent of oxidation of Trp residues by 2,2′-azobis (2-amidinopropane) dihydrochloride (AAPH) (18) (Fig. 3). We evaluated 38 Trp residues in 17 different mAbs, with most of these Trps present in CDRs except for 8 Trps that were present in the constant region of Fab domain (shown as open circles in Fig. 3). Examination of the plot between % SASA and AAPH-induced oxidation clearly demonstrates a binary dependence on % SASA, wherein most Trps below a certain % SASA do not undergo oxidation, whereas most Trps with higher % SASA show significant oxidation. Historically we observed that AAPH-induced oxidation of Trps in mAbs is correlative of thermal-induced Trp oxidation. Based on our historical data available between % oxidation by AAPH and thermal-induced oxidation on a set of six mAbs, we defined a criterion where Trps with >35% oxidation were designated as oxidation labile Trps, and Trps with less than 35% oxidation were designated nonlabile Trps. Based on this criteria, we examined % SASA of Trp residues against reactive and nonreactive sites. This analysis revealed that a cutoff value of 30% SASA of Trp side chains (i.e., 80 Å2) was largely sufficient to distinguish between reactive and nonreactive Trp sites. This selected criteria correctly identified 13 of 14 (93%) labile Trp residues and 20 of 24 (83%) nonlabile Trp residues (Fig. 3 and Table S3). Therefore, we conclude that the time-averaged SASA of Trp side chain is sufficient to be able to differentiate between the labile and the nonlabile Trp residues to enable risk ranking against oxidation risk. It should be noted that the current model addresses organic free radical-induced Trp oxidation and may need to be modified to expand its utility toward other oxidation pathways such as light-induced or metal-induced oxidation.
Fig. 3.
Trp oxidation prediction using the time-averaged solvent accessible surface area of a number of Trps on various mAbs. A total of 38 Trps in 17 mAbs were used for the analysis, of which 8 were present in the constant domain of the Fab (indicated by open circles) and the rest were present in the CDRs. A plot compares the AAPH-induced Trp oxidation and % Trp SASA. The lines represent the % SASA cutoff (horizontal) and the % AAPH-induced oxidation cutoff (vertical), respectively, to differentiate between the reactive and the nonreactive Trps in various mAbs. Each data point on the plot is a Trp residue in a given mAb. Based on historical data between AAPH-induced and thermal-induced oxidation of Trps in mAbs, a 35% oxidation cutoff was used to differentiate between reactive (squares) and nonreactive (circles) Trps. Reactive Trp were predicted based on a >30% average SASA of the Trp side chain obtained through MD simulations.
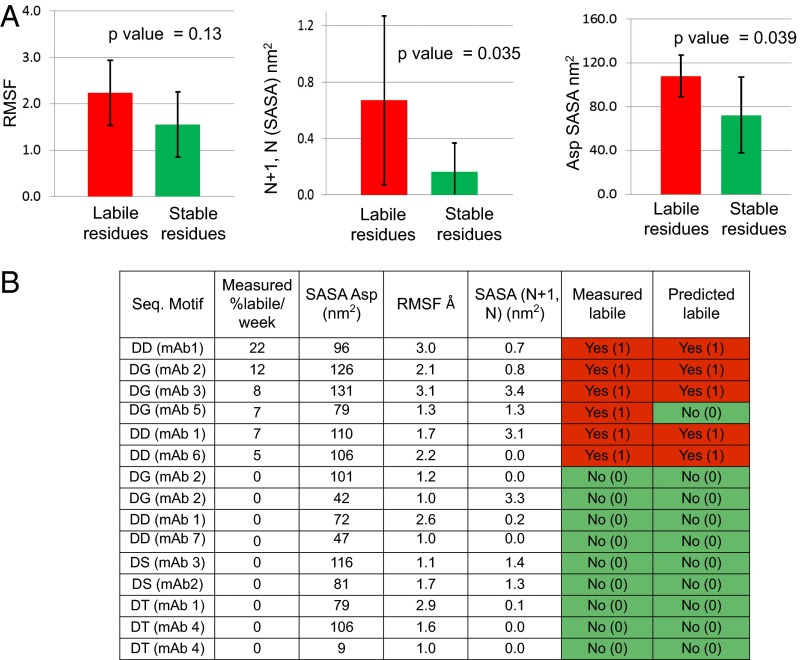
For Asp isomerization, multiple variables related to Asp residues were generated from MD trajectories. Consistent with the Asp isomerization mechanism (19), we examined the following properties: time average SASA of Asp side chains, the peptide backbone N atom (−NH) of the n + 1 residue [SASA (n + 1), N], the peptide backbone H atom associated with –NH of the n + 1 residue [SASA (n + 1), H], intraresidue mutual information (MI) for Asp residues, Shannon entropy for φ-ψ distributions, and root-mean-square fluctuations for Cα atoms (RMSF). A number of Fabs, which contained both known labile and stable Asp residues, were chosen for MD calculations. The calculated output from MD simulations along with Asp sites is detailed in Table S4. We focused on motifs, which have been previously demonstrated, to isomerize on timescales that impact shelf-life (DG, DS, DT, DD, DA) (20, 21) and excluded the remaining Asps.
The MD-derived parameters were compared against the experimental isomerization degradation rates at pH 5.5 and 40 °C. For the mAbs studied, all CDR Asp residues were included with the exception of non-CDR Asp residues (framework residues) that were only listed for a single mAb. Data analysis was twofold. First, we separated labile residues (≥2.5%/wk) from stable residues (<2.5%/wk) and compared the average value of each of the properties among the two groups (labile and stable) of Asp residues. Three properties, namely the SASA, RMSF, and SASA (n + 1, N), showed significant differences (80% CI) among these two groups of Asp residues (Fig. 4A). We next evaluated if a binary correlation could be established between experimental rates and MD-derived properties; i.e., can we simply differentiate sites with rates > 2.5%/wk, i.e., labile residues from ones with <2.5%/wk, i.e., stable residues. To this end, we assigned a value of 1 (labile) to rates of >2.5%/wk and a value of 0 (stable) to rates <2.5%/wk (stable) (Fig. 4B). We then performed logistic regression using SASA, RMSF, and SASA (n + 1, N) as independent variables and the binary rate output as the dependent variable. The equation output as a result of this regression is shown as
| [2] |
The output of this equation was rounded off to one significant figure to deliver a result of either 1 (labile) or 0 (stable) and is shown in Fig. 4B.
Fig. 4.
(A) Comparison of the average and SD of various properties, i.e., SASA, RMSF, and SASA (n + 1, N), extracted from MD simulations for labile group (red) vs. stable group of Asp residues (green). Calculated P values are shown on each plot. (B) Outcome of the logistical regression to enable prediction of labile vs. stable Asp sites. Logistic regression was performed using SASA, RMSF, and SASA (n + 1, N) as independent variables and the binary rate output as the dependent variable. All sites were assigned a value of 1 or 0 for labile and stable sites, respectively, based on a cutoff value of Asp degradation rate of 2.5%/wk at pH 5.5. Labile residues are shown in red and stable residues are shown in green. The predicted outcome was generated using the binary logistic model as described in the text. The model was validated using the LOOCV approach.
The logistic regression predicted five of six labile sites and all nine of nine nonreactive sites correctly. Essentially, the equation generated through logistic modeling enables us to use the three parameters [SASA, RMSF and SASA (n + 1, N)] to predict the susceptibility of an Asp residue to degrade at a rate greater than 2.5%/wk under the experimental conditions tested. We tested the validity of the model using the LOOCV approach similar to that of viscosity analysis, wherein we predicted five of six labile sites and seven of nine nonreactive sites. Although the number of sites correctly predicted was reduced a little, a total of 12 of 15 sites (80%) were still correctly predicted and therefore the model was deemed satisfactory. Finally we note that, although this modeling approach is specific to the current experimental conditions, the underlying approach can be likely extended to any given experimental condition as long as the experimental rates are known for a set of Asp residues.
Discussion
Monoclonal antibodies are an important class of therapeutic agents for treating diseases including cancers, autoimmune disorders, and infections. For manufacturability, ease of use, patient convenience, and less-frequent dosing, it is critical to develop low-viscosity formulations having adequate shelf-life using mAb candidates that exhibit normal clearance, and an expected long plasma half-life. Here, we show surprisingly good predictive power for these properties from simple physical properties—such as hydrophobicity, net charge, charge distribution, flexibility, and solvent accessibility—that are readily obtainable from amino acid sequence and MD simulations of individual mAbs molecules.
Hydrophobicity and dipolar charge distribution increase the solution viscosity, whereas protein charge decreases it. Previous experimental work has demonstrated that viscosity of mAb solutions is modulated through protein-protein interactions involving hydrophobic and electrostatic forces (10, 11). Our findings through use of in silico tools remarkably align well with these experimental observations. For in vivo clearance of mAbs, the correlation between normal vs. fast clearance and fundamental mAb properties is indeed striking. Fast clearance of mAbs has been attributed to nonspecific antigen-independent binding of mAb in vivo such that it becomes undetectable in plasma (14). At the fundamental level, these nonspecific interactions (and most biological protein-protein interactions) are driven by the same physical forces of electrostatics, hydrophobic, van der Waal’s, etc. We demonstrated that these forces/interactions can be extracted from simple sequence-based analysis and used to differentiate between faster clearing and normal clearing mAbs. A sequence-based analysis was likely effective because of the use of a single isotype of mAbs, i.e., IgG1 in this study. However, the overall approach can be extended to other isotypes/subtypes and a structure based analysis could be included to expand the overall scope. For site-specific properties such as Trp oxidation or Asp isomerization, our work is consistent with a previously published mechanism regarding which sites are reactive or nonreactive, affecting the shelf-life of the product. For Trp oxidation, time-averaged SASA can differentiate between reactive and nonreactive sites. For Asp isomerization, the mechanism is more complex. In summary, the correlations we observe here will be useful for more efficient selection of lead mAbs candidates as therapeutics.
Methods
All mAbs are of the IgG1 isotype expressed in Chinese Hamster Ovary (CHO) cells. It is noted that across different datasets used in this study, mAbs are represented by numbers, i.e., mAb1 and mAb2; however, a given mAb with the same number designation may not represent the same actual original mAb across different datasets.
Sequence-Based Parameters.
The net charge for a given sequence at a given pH was calculated by adding up the contribution from all charged amino acids using the known pKas of the side chains (22) and the Henderson–Hasselbalch equation. The FvCSP was calculated by obtaining the product between the net charge on the VH domain and the VL domain at a given pH. A positive product is a result of either net positive charge or net negative charge on each domain. A negative product represents lack of charge symmetry between the two domains, i.e., one of the domain with a net negative charge and the other with a net positive charge. The hydrophobicity index (HI) was calculated as , where i represents the hydrophobic amino acids, i.e., A, C, F, I, L, P, V, W, and Y, and j represents the hydrophilic amino acids, i.e., D, E, G, H, K, M, N, Q, R, S, and T; n is the number of each amino acid, and E is the Eisenberg scale value of each amino acid (23). The calculation can be easily transformed to a 3D structure of a protein where the parameter n is replaced by S, defined as the solvent accessible surface area of each amino acid (Fig. S2).
Viscosity.
Viscosity measurements were performed as described previously using an Anton Paar Physica MCR 501 cone and plate rheometer (Anton Paar) (24).
Clearance.
Clearance values in Cynomolgus monkeys used in this study were obtained from previously published data and additional data generated in-house (14).
MD Simulations.
MD starting structures.
The structures of the Fabs were obtained either from the 3D crystal structure (if available) or a homology model generated using a local adaption of Modeler (25). The Fab domain was used as the starting structure for MD before addition of ions (where needed) and explicit solvent molecules.
MD analysis.
All SASAs are calculated using g_sas of GROMACS (26); mutual information calculation is implemented locally similar to Lange and Grubmüller (27). g_rsmf, g_hbond, and dssp of GROMACS were used to calculate the root mean square fluctuations, the hydrogen bonds, and the secondary structure status, respectively. Shannon entropy and mutual information were calculated using previously published methods (28). MD simulations for aspartate isomerization and analysis followed that described previously (28), with solvation using TIP3P water (29). MD simulations for tryptophan oxidation were conducted in a similar fashion except using Amber 11 (FF99SB fixed-charge force field; SASA was calculated using areaimol (30); 100-ns trajectories were used as they provided sufficient data within available computational power.
AAPH-Induced Trp Oxidation.
AAPH-induced oxidation was carried out by mixing the mAb solution with AAPH at final concentrations of 1 mg/mL and 1 mM, respectively (18). The solutions were incubated at 40 °C for 16 h. The reaction was quenched by addition of 20 mM Met following by buffer exchange into a 20 mM buffer at pH 5.5 using PD-10 desalting columns. The solutions were then analyzed using tryptic digest followed by LC-MS/MS for site-specific Trp oxidation.
Experimental Determination of Asp Degradation Rates.
mAb solutions were buffer exchanged using Centricon ultrafiltration tubes with a final formulation of 5 mg/mL protein in a 20 mM buffered solution at pH 5.5, 240 mM sucrose. Samples were placed at 40 °C and withdrawn at t = 0, 14, and 28 d.
Thermal stressed samples were analyzed using tryptic peptide digest followed by LC-MS/MS. Protein samples were digested following published protocols (31), and peptide mapping was performed on an Agilent 1200 HPLC system coupled to a Thermo Fisher LTQ Orbitrap mass spectrometer. The degradation level at each site was determined by extracted ion chromatography (EIC) using Xcalibur software (31).
Regression Analysis.
Principal component regression and logistic regression analysis were carried out using XLSTAT (Addinsoft).
Supplementary Material
Acknowledgments
We thank Lisa Bernstein, Andrew Kosky, Steve Shire, Paul Carter, Saileta Prabhu, Louie Naumovski, Thomas Scherer, and Relly Brandman for helpful discussions and Robert Ovadia, Michael Kim, William Galush, and Ankit Patel for assistance with data collection. We also thank Stacey Ma and Jamie Moore for support of this project.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1421779112/-/DCSupplemental.
References
- 1.Eisenstein M. Something new under the skin. Nat Biotechnol. 2011;29(2):107–109. doi: 10.1038/nbt.1768. [DOI] [PubMed] [Google Scholar]
- 2.Shire SJ, Shahrokh Z, Liu J. Challenges in the development of high protein concentration formulations. J Pharm Sci. 2004;93(6):1390–1402. doi: 10.1002/jps.20079. [DOI] [PubMed] [Google Scholar]
- 3.Wang W, Wang EQ, Balthasar JP. Monoclonal antibody pharmacokinetics and pharmacodynamics. Clin Pharmacol Ther. 2008;84(5):548–558. doi: 10.1038/clpt.2008.170. [DOI] [PubMed] [Google Scholar]
- 4.Zheng Y, et al. Minipig as a potential translatable model for monoclonal antibody pharmacokinetics after intravenous and subcutaneous administration. MAbs. 2012;4(2):243–255. doi: 10.4161/mabs.4.2.19387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buck PM, et al. Computational methods to predict therapeutic protein aggregation. Therapeutic Proteins: Methods and Protocols. 2012 doi: 10.1007/978-1-61779-921-1_26. eds Voynov V, Caravella JA (Springer, New York), Vol 899, pp 425–451. [DOI] [PubMed] [Google Scholar]
- 6.Chennamsetty N, Voynov V, Kayser V, Helk B, Trout BL. Design of therapeutic proteins with enhanced stability. Proc Natl Acad Sci USA. 2009;106(29):11937–11942. doi: 10.1073/pnas.0904191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li L, et al. Concentration dependent viscosity of monoclonal antibody solutions: Explaining experimental behavior in terms of molecular properties. Pharm Res. 2014;31(11):3161–3178. doi: 10.1007/s11095-014-1409-0. [DOI] [PubMed] [Google Scholar]
- 8.Kanai S, Liu J, Patapoff TW, Shire SJ. Reversible self-association of a concentrated monoclonal antibody solution mediated by Fab-Fab interaction that impacts solution viscosity. J Pharm Sci. 2008;97(10):4219–4227. doi: 10.1002/jps.21322. [DOI] [PubMed] [Google Scholar]
- 9.Liu J, Nguyen MDH, Andya JD, Shire SJ. Reversible self-association increases the viscosity of a concentrated monoclonal antibody in aqueous solution. J Pharm Sci. 2005;94(9):1928–1940. doi: 10.1002/jps.20347. [DOI] [PubMed] [Google Scholar]
- 10.Du W, Klibanov AM. Hydrophobic salts markedly diminish viscosity of concentrated protein solutions. Biotechnol Bioeng. 2011;108(3):632–636. doi: 10.1002/bit.22983. [DOI] [PubMed] [Google Scholar]
- 11.Yadav S, Liu J, Shire SJ, Kalonia DS. Specific interactions in high concentration antibody solutions resulting in high viscosity. J Pharm Sci. 2010;99(3):1152–1168. doi: 10.1002/jps.21898. [DOI] [PubMed] [Google Scholar]
- 12.Schmit JD, et al. Entanglement model of antibody viscosity. J Phys Chem B. 2014;118(19):5044–5049. doi: 10.1021/jp500434b. [DOI] [PubMed] [Google Scholar]
- 13.Yadav S, et al. Establishing a link between amino acid sequences and self-associating and viscoelastic behavior of two closely related monoclonal antibodies. Pharm Res. 2011;28(7):1750–1764. doi: 10.1007/s11095-011-0410-0. [DOI] [PubMed] [Google Scholar]
- 14.Hötzel I, et al. A strategy for risk mitigation of antibodies with fast clearance. MAbs. 2012;4(6):753–760. doi: 10.4161/mabs.22189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Igawa T, et al. Reduced elimination of IgG antibodies by engineering the variable region. Protein Eng Des Sel. 2010;23(5):385–392. doi: 10.1093/protein/gzq009. [DOI] [PubMed] [Google Scholar]
- 16.Wu H, et al. Development of motavizumab, an ultra-potent antibody for the prevention of respiratory syncytial virus infection in the upper and lower respiratory tract. J Mol Biol. 2007;368(3):652–665. doi: 10.1016/j.jmb.2007.02.024. [DOI] [PubMed] [Google Scholar]
- 17.Sydow JF, et al. Structure-based prediction of asparagine and aspartate degradation sites in antibody variable regions. PLoS ONE. 2014;9(6):e100736. doi: 10.1371/journal.pone.0100736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ji JA, Zhang B, Cheng W, Wang YJ. Methionine, tryptophan, and histidine oxidation in a model protein, PTH: Mechanisms and stabilization. J Pharm Sci. 2009;98(12):4485–4500. doi: 10.1002/jps.21746. [DOI] [PubMed] [Google Scholar]
- 19.Wakankar AA, et al. Aspartate isomerization in the complementarity-determining regions of two closely related monoclonal antibodies. Biochemistry. 2007;46(6):1534–1544. doi: 10.1021/bi061500t. [DOI] [PubMed] [Google Scholar]
- 20.Radkiewicz JL, Zipse H, Clarke S, Houk KN. Neighboring side chain effects on asparaginyl and aspartyl degradation: An ab initio study of the relationship between peptide conformation and backbone NH acidity. J Am Chem Soc. 2001;123(15):3499–3506. doi: 10.1021/ja0026814. [DOI] [PubMed] [Google Scholar]
- 21.Yi L, et al. Isomerization of Asp-Asp motif in model peptides and a monoclonal antibody Fab fragment. J Pharm Sci. 2013;102(3):947–959. doi: 10.1002/jps.23423. [DOI] [PubMed] [Google Scholar]
- 22. Berg JM, Tymoczko JL, Stryer L (2002) Biochemistry (Springer, New York)
- 23.Eisenberg D, Schwarz E, Komarony M, Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol. 1984;179(1):125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- 24.Connolly BD, et al. Weak interactions govern the viscosity of concentrated antibody solutions: High-throughput analysis using the diffusion interaction parameter. Biophys J. 2012;103(1):69–78. doi: 10.1016/j.bpj.2012.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 26.Eisenhaber F, Lijnzaad P, Argos P, Sander C, Scharf M. The double cube lattice method: Efficient approaches to numerical integration of surface area and volume and to dot surface contouring of molecular assemblies. J Comput Chem. 1995;16(3):273–284. [Google Scholar]
- 27.Lange OF, Grubmüller H. Full correlation analysis of conformational protein dynamics. Proteins. 2008;70(4):1294–1312. doi: 10.1002/prot.21618. [DOI] [PubMed] [Google Scholar]
- 28.Kortkhonjia E, et al. Probing antibody internal dynamics with fluorescence anisotropy and molecular dynamics simulations. MAbs. 2013;5(2):306–322. doi: 10.4161/mabs.23651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79(2):926–935. [Google Scholar]
- 30.Bailey S. Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 31.Yu XC, et al. Accurate determination of succinimide degradation products using high fidelity trypsin digestion peptide map analysis. Anal Chem. 2011;83(15):5912–5919. doi: 10.1021/ac200750u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.