Abstract
Alterations in the expression and activity of the centrosomal kinase, Aurora-A/serine/threonine kinase 15 (STK15), affect genomic stability, disrupt the fidelity of centrosome duplication, and induce cellular transformation. Here, we provide evidence that p160ROCK, a Rho-associate serine/threonine kinase, associates with Aurora-A in a protein complex with other STK15-associated factors. Suppression of Aurora-A by small interfering RNA in HeLa cells blocks the ability of centrosomes to organize normal mitotic spindles, induces G2/M cell cycle arrest, and promotes accumulation of tetraploid cells. In many cases, one outcome of such abnormalities is apoptosis. Introduction of a second genetic lesion, suppression of p160ROCK by RNA interference, can rescue abnormal mitotic spindle formation, release the G2/M cell cycle arrest, and alleviate apoptosis, leading to a greater accumulation of aneuploid cells. These results suggest that Aurora-A and p160ROCK act in a common genetic pathway that promotes and monitors progression through G2/M.
The eukaryotic centrosome is the microtubule-organizing center and is composed of two perpendicularly positioned centrioles and surrounding amorphous pericentriolar materials (for reviews, see refs. 1–6). Four groups of protein kinases are found to regulate the centrosome replication cycle: cyclin-dependent kinases (7–9), Polo-like kinases (10), NIMA kinases (11), and Aurora kinases. In mammalian cells, the Aurora kinase family has three members: Aurora-A, -B, and -C. Alterations in the expression and activity of Aurora-A/serine/threonine kinase 15 (STK15) affect genomic stability, disrupt the fidelity of centrosome duplication, and induce cellular transformation (12–15). Aurora-A is regulated by ubiquitin pathways and protein phosphorylation (16–19). A kinesin-related protein (Eg5), cytoplasmic polyadenylation element-binding protein (CPEB), and transforming acidic coiled-coil protein number 3 (TACC3) have been found to be substrates of Aurora-A (20–23).
Recently, p160ROCK has been identified as a centrosome component and functions in centrosome positioning and centrosome-dependent exit from mitosis (24). p160ROCK is a member of Rho-associated serine-threonine protein kinases. p160ROCK/Rho-kinase/ROK kinases are involved in various cellular functions downstream of Rho, such as smooth muscle contraction, stress fiber formation, and cytokinesis (25). p160ROCK phosphorylates LIM kinase and mDia, which then phosphorylates cofilin to regulate the Rho-induced reorganization of cytoskeleton (26). p160ROCK has also been reported to interact with CDC25A in transforming growth factor β-induced cell cycle arrest (27) and to be involved in tumorigenesis (28, 29), and apoptosis (30). Here we provide biochemical and genetic evidence that Aurora-A and p160ROCK associate physically and functionally in a protein complex. These proteins interact genetically, with suppression of p160ROCK alleviating the G2/M cell cycle arrest induced by inactivation of Aurora-A.
Materials and Methods
Cell Culture. Normal human foreskin fibroblast IMR90 cells were immortalized by hTERT expression, and early passage HeLa and LinXA (293Tt derivative as in ref. 31) were cultured as in ref. 32. To assess mitotic spindle reassembly capacity in vivo, microtubules were disassembled with nocodazole and cold treatment (32). The cells were incubated at 37°C for 10 min to promote spindle reassembly. This process was monitored by anti-β-tubulin indirect immunofluorescence. p160ROCK specific inhibitor Y27632 (Calbiochem) was used at 100 μM in Fig. 2b.
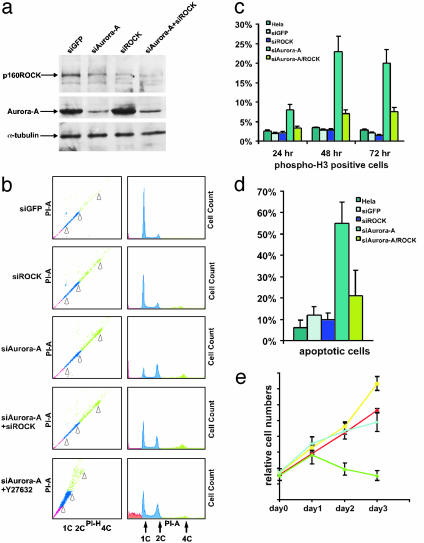
Fig. 2.
Depletion of Aurora-A in HeLa cells induces G2/M cell cycle arrest and apoptosis, but further deletion of p160ROCK alleviates this phenotype. HeLa cells with siGFP, siAurora-A, siROCK, or both, were used at 48 h after transfection. (a) Cell lysates (25 μg) were subjected to immunoblot assay by anti-Aurora-A, -p160ROCK, and -tubulin. The proteins are indicated on the left. (b) Cells treated with various siRNAs and/or Y-27632 were collected, fixed, and analyzed by flow cytometry. The blue portion indicates the diploid cells. The red portion indicates apoptotic cells, and the green portion indicates tetraploid/aneuploid cells. 1C, 2C, and 4C DNA contents are also indicated by arrows. (c, complete haploid DNA content). (c) Cells were stained for mitotic marker by using anti-phospho-H3 antibody, counterstained by Hoechst 33342 to visualize nuclei, and subjected to immunofluorescence microscopy. (d) Cells were collected including the floating cells in the media. Precipitated cells were resuspended in mounting media supplemented with Hoechst 33342 to stain DNA in the nuclei, and checked under a microscope for apoptotic cells. (e) HeLa cells were plated in six-well plates and transfected with siRNAs. At the indicated time, cells were collected and counted. Purple line, siGFP; yellow line, siROCK; green line, siAurora-A; blue line, siAurora-A+siROCK. In c–e, three independent experiments were performed and in each experiment, >300 cells were counted. Error bars are labeled on the top.
Antibodies. Affinity-purified anti-Aurora-A, anti-GST-Aurora-A sera (32), rabbit polyclonal anti-γ-tubulin, mouse monoclonal anti-α-tubulin, anti-β-tubulin (Sigma), rabbit anti-phospho-H3 (Ser-10, Upstate Biotechnology, Lake Placid, NY), mouse monoclonal anti-BrdUrd (Amersham Pharmacia), rabbit polyclonal anti-MAD2 (Covance, Berkeley, CA), anti-p160ROCK serum (a kind gift from Shuh Narumiya, Kyoto University, Kyoto), anti-BubR1 and anti-Bub1 (kind gifts from S. Taylor, Manchester, U.K.), anti-DDX3 and DDX6 (kind gifts from J. Sommerville, University of St. Andrews, St. Andrews, Scotland and A. H. Patel, Medical Research Council, Institute of Virology, Glasgow, U.K.) were used. Another rabbit polyclonal anti-p160ROCK antibody against the C-terminal 9 aa (VKNTS-GKTS) of p160ROCK was affinity-purified as in ref. 32.
Immunoprecipitation, Immunofluorescence, Immunoblot, and Aurora-A Depletion Assay. Immunoprecipitation, immunofluorescence, and immunoblotting were as described in ref. 32. Denaturing immunoprecipitation was done as in ref. 33. To measure how much p160ROCK associates with Aurora-A in vivo, we used increasing amounts of anti-GST-Aurora-A serum, and found that Aurora-A in 1 × 106 immortalized IMR90 cells can be 90% depleted by using 50 μl of anti-GST-Aurora-A serum. In Fig. 1c, the same amount of anti-Aurora-A antibody was used and images on immunoblot were captured with Alphaimager 2200 and quantified by chemilmager v5.5 program (Alpha Innotech, San Leandro, CA).
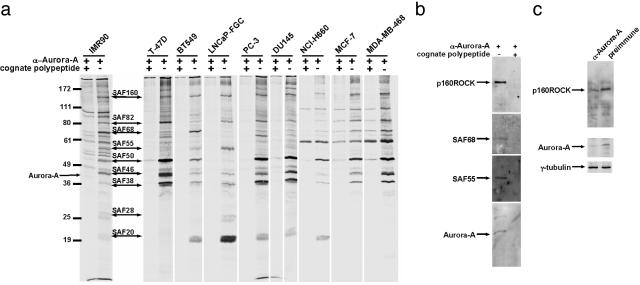
Fig. 1.
Aurora-A and p160ROCK associate in a protein complex. (a) IMR90 cells, several prostate (LNCaP-FGC, PC-3, DU145, and NCI-H660), and breast (T-47D, BT549, MCF-7, and MDA-MB-468) cancer cell lines were labeled with [35S]methionine. Cell lysates were immunoprecipitated with affinity-purified anti-Aurora-A antibody (αAurora-A+ lanes) or anti-Aurora-A plus cognate antigen polypeptide (αAurora-A+/cognate polypeptide+ lanes) and were resolved on SDS-10% polyacrylamide gel and subjected to autoradiography. SAFs with numbers indicate the apparent molecular mass on the gel, starting from the largest component (160 kDa). Molecular mass markers are indicated on the left. (b) Anti-p160ROCK, -SAF68, -SAF55, and -Aurora-A immunoblotting on anti-Aurora-A immunoprecipitates from IMR90 cells. (c) IMR90 cell lysates that were either titrated with anti-GST-Aurora-A to remove Aurora-A protein, or with same amount of preimmune sera as a control were subjected to immunoblotting with anti-p160ROCK and Aurora-A. Tubulin was used as a loading control.
35S-Labeling of Cells. IMR90 or cancer cell lines (2 × 106 cells) were labeled with 35S-translabeling mixtures (Easy tag express [35S] protein-labeling mix, 1,175 Ci/mmol; NEN) for 4 h. The cells were lysed, and immunoprecipitations were as in ref. 32.
Peptide Sequencing, MS Analysis, and Sequence Identification. Preparative immunoprecipitates from 2.5 × 108 IMR90 cells by using 1.5 mg of affinity-purified anti-Aurora-A antibody were resolved in 10% or 12% SDS-polyacrylamide gel and stained by Bio-Safe Coomassie G250 (Bio-Rad) or sequencing-grade Coomassie blue G250 (Sigma). Individual bands were excised and digested with trypsin, and peptides were fractionated by RP-HPLC. The MS sequencing was done in the Bio-medical Mass Spectrometry Facility at Washington University, St. Louis, as in ref. 34.
RNA Interference (RNAi) in Transient Transfection. To design target-specific small interfering RNA (siRNA) duplexes, AA(N19)dTdT type sequences were selected from the ORFs of the p160ROCK (AAGGTGATTGGTAGAGGTGCA), Aurora-A (AAGCACAAAAGCTTGTCTCCA), and GFP (TCAGCGTGTCCGGCGAGGGCGA) (35). The selected sequences were submitted to blast searches against the nonredundant protein database in National Center for Biotechnology Information to make sure that only the selected genes were targeted. The 21-nt RNAs purchased form Dharmacon (Lafayette, CO). In brief, 100 nM siRNA was used with transfection reagent Oligotransfectamine (Invitrogen) in HeLa cell transfections as in ref. 35. The 24-, 48-, and 72-h time points had one, two, and three rounds of transfections, respectively. Cells were assayed 24 h after the last transfection.
Flow Cytometry. Transfected HeLa cells were trypsinized and fixed for flow cytometry on Becton Dickinson FACSCalibur or LSRII as in ref. 36. The data were processed for cell cycle distribution by modfit v.2 (Macintosh) or v.3 (PC).
Apoptosis Assay. HeLa cells transfected in six-well cell-culture plates were trypsinized. The cells together with the culture media were spun down at 1,000 rpm in a tabletop centrifuge, and cells were stained with Hoechst 33342 to visualize the nuclei DNA morphology. Cells with intact uniform nuclei were counted as live cells and cells with fragmented nuclei, a signature of apoptosis cell, were counted as apoptotic cells (37). Annexin V-FITC apoptosis detection kit II (BD Pharmingen) and terminal deoxynucleotidyltransferase-mediated dUTP nick end-labeling assay (In Situ cell death detection kit, Roche Applied Science) were used according to the manufacturers' instructions.
Results and Discussions
To identify regulators and substrates of Aurora-A kinase, immortalized human IMR90 fibroblast cells were labeled with 35S and Aurora-A was immunoprecipitated from cell lysates with affinity purified polyclonal anti-Aurora-A antibodies. Nine protein bands associated specifically with the antibody (Fig. 1a). We named the immunoprecipitated Aurora-A-bound protein STK15-associate factors (SAFs; see below).
The integrity of STK15 complexes is compromised in various breast and prostate cancer cell lines tested. However, SAF160 appears in every cell line examined, pointing to the potential importance of its association with Aurora-A (Fig. 1a). To identify components of the Aurora-A complex, Aurora-A-associated protein bands were excised and analyzed by matrix-assisted laser desorption ionization–time-of-flight MS and electrospray ionization MS/MS. Mass fingerprinting indicated that the 46-kDa protein was Aurora-A and that SAF160 was p160ROCK. Immunoblotting Aurora-A immunoprecipitates showed that anti-Aurora-A specifically immunoprecipitated Aurora-A, and p160ROCK, SAF68, and SAF55 (also identified as DDX3 and DDX6 by matrix-assisted laser desorption ionization–time-of-flight) (Fig. 1b). Immunoblot analysis demonstrated that γ-tubulin is not present at detectable levels in the SAF immunocomplex (data not shown). Therefore, it is unlikely that the purified SAF complex represents part of the centrosome that is indirectly associated with Aurora-A through γ-tubulin. TPX2 has been found to interact with Aurora-A by immunoprecipitation (38). However, we did not see a band of 100 kDa corresponding to TPX2 (Fig. 1a). Use of different anti-Aurora-A antibodies, cell lines, and labeling methods may account for this discrepancy in the immunoprecipitation results.
To examine whether SAFs coexist with Aurora-A in a large multiprotein complex, IMR90 lysates were fractionated by FPLC. All the identified proteins cofractionated with Aurora-A on Mono S and Mono Q ion-exchange columns and on a third Superose 6 gel filtration column (Fig. 5a, which is supporting information on the PNAS web site). On Superose 6, Aurora-A split into two distinct peaks, indicating that Aurora-A exists in multiple protein complexes (Fig. 5b and ref. 32). Immunodepletion with anti-GST-Aurora-A antibody in IMR90 cell lysates suggested that roughly half of endogenous p160ROCK associates with Aurora-A (Fig. 1c), either at the centrosome or in the cytoplasm (24).
We used RNAi to investigate consequences of suppressing Aurora-A or p160ROCK in HeLa cells. siRNAs dramatically reduced the expression of targeted proteins (Fig. 2a). Immunofluorescence stains with anti-Aurora-A and -p160ROCK also confirmed the depletions. The depletion of Aurora-A also significantly reduced the p160ROCK protein level (Fig. 2a), consistent with the notion that two proteins interact in a complex in vivo (39).
Twenty-four hours after transfection of siAurora-A, cells became rounder and grew more slowly than the control siGFP-transfected cells. Flow cytometry showed a significant increase in G2/M phase cells in comparison with that observed in the control siGFP- or siROCK-treated cells (Fig. 2b Right). Notably, this cell cycle phenotype could be alleviated by simultaneous cotransfection of siRNAs directed against Aurora-A and p160ROCK (Fig. 2b Right, siAurora-A+siROCK). Correspondingly, a ROCK kinase specific inhibitor, Y27632, also showed a dosage-dependent suppression of the G2/M arrest phenotype in siAurora-A-transfected cells (Fig. 2b Right, siAurora-A+Y27632). These results demonstrate an epistatic relationship between Aurora-A and p160ROCK and indicate that they function in the same genetic pathway.
The observed phenotypes were also confirmed by immunofluorescence staining of cells with anti-phospho-H3 antibody, a mitotic marker that specifically labels cells from the beginning of prophase to the end of anaphase (Fig. 2c). Quantitation of anti-phospho-H3 staining by flow cytometry supported interpretations from the immunofluorescence studies (data not shown). BrdUrd incorporation to assess the S-phase population did not show a noticeable difference in the transfected cells at any time point (data not shown). The significantly prolonged mitotic phase correlated with an increased frequency of apoptosis (Fig. 2d), which was also confirmed by annexin V staining and terminal deoxynucleotidyltransferase-mediated dUTP nick end-labeling assays (data not shown) and a decrease in surviving cell numbers as measured by plotting growth curves (Fig. 2e). Again, cotransfection of siROCK together with siAurora-A partially rescued the apoptotic phenotype (Fig. 2 d and e).
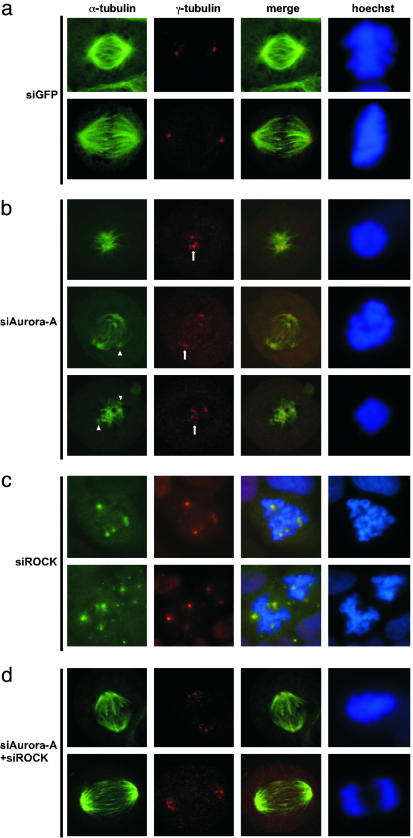
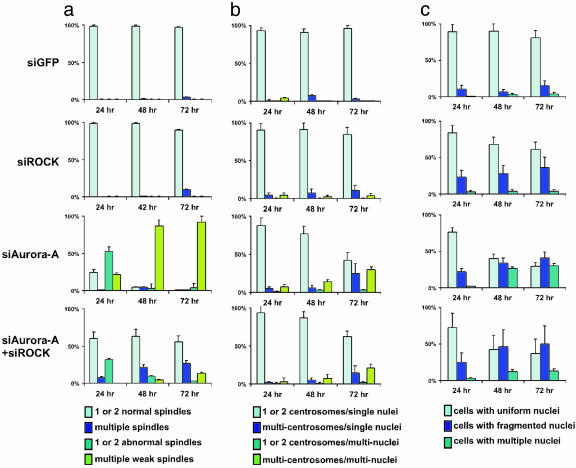
Because Aurora-A is a centrosomal kinase, we next assessed the integrity of mitotic spindles and examined centrosome and nuclear morphologies in siAurora-A-transfected cells (Fig. 3). Most mitotic cells contained abnormal spindles with disorganized arrays of microtubules and fewer and shorter spindles than in normal control cells (Fig. 3b, anti-α-tubulin columns in siAurora-A images). In addition, comparison of the spindle lengths, intensities, and levels of the anti-α-tubulin stains in siAurora-A-treated cells with those of siGFP control cells showed that the spindle microtubule networks in Aurora-A-depleted cells did not extend to the condensed prometaphase chromosomes (Fig. 3 a and b, anti-α-tubulin columns). Some of the abnormal spindles still apparently radiated from centrosomes (Fig. 3b). Others had multiple origins that did not always colocalize with centrosomes or concentrations of γ-tubulin (Fig. 3b, anti-α-tubulin stains indicated by triangles). The portion of cells with abnormal spindles increased dramatically over the time course of the experiment (Fig. 4a). This outcome is the most prominent defect in the siAurora-A-transfected cells. Anti-γ-tubulin staining showed that with abnormal mitotic spindles, multiple concentrated spots of γ-tubulin occurred in the cytoplasm (Fig. 3b, anti-γ-tubulin spots indicated with arrows). Suppression of Aurora-A also induced centrosome amplification in interphase cells (Fig. 4b). Although chromosomes condensed at prometaphase in arrested mitotic cells, virtually no normal metaphase and anaphase mitotic figures were found in these transfected cells, suggesting cells arrested at prometaphase (Fig. 3b). Live cell imaging over a 4-h time course showed no progression of the arrested cells to metaphase, whereas control cells progressed from prometaphase through anaphase in ≈50 min (data not shown). siRNAs to Aurora-A also resulted in a significant accumulation of binucleated cells (Fig. 4c). Many nuclei had satellite micronuclei around the major interphase nucleus. In addition, many nuclei showed irregular undefined edges and shapes, a phenotype that often accompanies genomic instability and aneuploidy. The cell population with these unusual nuclear morphologies increased dramatically over the time course of the experiment (Figs. siAurora-A, 4C portion 2b Left, and 4c), indicating an increase in genomic instability of the cells. The mitotic spindle defects we observed here are closely resemble to those recently observed in Aurora-A-depleted HeLa cells (38, 40).
Fig. 3.
Depletion of Aurora-A in HeLa cells causes abnormal mitotic spindles and split centrosomes in mitotic arrested cells, but further deletion of p160ROCK alleviates the phenotype. HeLa cells transfected with siGFP (a), siAurora-A (b), siROCK (c), or both (d) were subjected to immunofluorescence analysis with anti-α-tubulin to visualize mitotic spindles, anti-γ-tubulin to visualize centrosomes, and Hoechst 33342 to visualize nuclei DNA morphologies. The merge lane combines the anti-α- or anti-γ-tubulin images together for colocalization. In c cells were treated with nocodazole and cold to dissipate the microtubule networks. The mitotic spindles were allowed to reassemble for 10 min in 37°C.
Fig. 4.
Depletion of Aurora-A, p160ROCK, or both induces genomic instability. HeLa cells were analyzed 24, 48, and 72 h after transfection with siRNAs for mitotic spindle (a), centrosome (b), and nuclear (c) morphologies. (a) Spindles that can organize arrays of extended microtubules were counted as normal, either with one or two spindles, or with multiple (>2) spindles. Disorganized, distinctively fewer, and shorter spindles were counted as weak spindles (either one or two, or multiple). (b) Centrosomes were grouped according to their numbers in either single or multiply (≥2) nucleated cells. (c) Nuclei with free chromosome(s) (satellite nuclei), lagging chromosomes between two cells, string-like (abnormal chromosomes that were fragmented or unraveled), misplaced (positions in the cells dramatically deviated form wild-type controls), were counted as fragmented nuclei. The nuclei with perfectly round, smooth edges were counted as uniform nuclei. Cells with more than two nuclei were counted as cells with multiple nuclei. Three independent experiments were conducted and in each experiment, at least 100 mitotic (a) or 200 (b and c) cells were examined.
In siROCK-transfected cells, the number of cells with multiple mitotic spindles and the number of cells with apparent centrosome amplification in interphase cells increased significantly in comparison with siGFP control cells (Fig. 4 a and b). A dramatic increase in mitotic spindle reassembly capacity was also observed in siROCK-transfected cells (see Materials and Methods). Eighty percent of mitotic cells showed multiple, reassembled mitotic spindle networks after transient treatment with nocodazole (Fig. 3c, anti-α-tubulin columns in siROCK images), in comparison with 20% of control cells. The result showed the siROCK-transfected cells are highly prone to reassembling inappropriate mitotic spindles using nascent γ-tubulin spots. In accordance with this result, we observed an increase in aneuploid cells (Figs. siROCK, 4C portion 2b Left, and 4c). Also, siROCK transfection caused nuclear fragmentation (Fig. 4c). However, a dramatic increase did not occur in multinucleated cell populations (Fig. 4c). This result suggests that depletion of p160ROCK alone can give rise to aneuploid cells, although to a lesser degree than does loss of Aurora-A.
Again, we found the mitotic arrest caused by suppression of Aurora-A was alleviated by coincident depletion of p160ROCK (Fig. 2 b Right, siAurora-A+siROCK, and c). Cotransfection of both siAurora-A and siROCK led to assemble of functional mitotic spindles (Fig. 3d, anti-α-tubulin columns in siAurora-A+siROCK images), completion of mitosis, regrouping of split centrosomes into two functional groups for organizing bipolar spindles (Fig. 3d, anti-γ-tubulin columns in siAurora-A+siROCK images), and greater accumulation of aneuploid cells (Figs. siAurora-A+siROCK, 4C portion 2b Left, and 4c).
The immunoprecipitated Aurora-A not only phosphorylated an artificial substrate, myelin basic protein, but also phosphorylated a p160 in the SAF complex (Fig. 6 a and b, which is published as supporting information on the PNAS web site). This finding was confirmed by denaturing IP to be p160ROCK (Fig. 6c). Aurora-A also phosphorylated another protein, p82, in purified centrosomes (Fig. 6 a and b). Phosphorylation of p160ROCK in the immunoprecipitated Aurora-A complex can be specifically increased in cells transfected with constructs encoding Aurora-A, but not a mutant Aurora-A K162M, which lacks kinase activity (41) (Fig. 6d). In fact, [32P]orthophosphate labeling indicated that phosphorylation of p160ROCK in the cells expressing a kinase dead Aurora-A K162M mutant appreciably decreased, suggesting that Aurora-A is responsible, at least in part, for p160ROCK phosphorylation in vivo (Fig. 6e, comparing P-p160ROCK band intensity in K162M lane with those in other control lanes). Phosphoamino acid analysis of excised 32P-labeled p160ROCK band showed that only serine residue(s) are phosphorylated (Fig. 7a, which is published as supporting information on the PNAS web site). In vitro kinase assays with various GST-p160ROCK fusion proteins as substrates for Aurora-A (Fig. 7b, all fusion proteins started from N terminus of p160ROCK and ended at the indicated amino acids) showed GST-p160ROCK1034 was significantly labeled, but not GST-p160ROCK726 or other fusion proteins (Fig. 7c), suggesting the phosphorylated serine residues are located between 726 and 1,034 aa of p160ROCK.
A G2/M delay phenotype had been reported after overexpression of Aurora-A in HeLa cells (42). A similar mitotic defect is also observed on Aurora-A loss-of-function in Drosophila melanogaster and in S2 cells (ref. 43 and data not shown) and most recently in HeLa cells (40, 44). These results together with our observation indicate that balance of Aurora-A protein in the cell is critical and that either down- or up-regulation of its protein level will compromise its function and lead to mitotic defects. Suppression of aurora-A (aurora-1) in Caenorhabditis elegans by RNAi does not affect centrosome separation, but mitotic spindle formation (45), similar to what we observed here in HeLa cells. However, in D. melanogaster syncytial embryos, aurora mutations cause centrosome separation defects, which result in monopolar spindles and abnormal mitoses. The differences between these observed phenotypes could be due to cell type-specific functions of Aurora kinases. However, we favor the notion that different genetic lesions (e.g., point mutation versus nearly, but not complete suppression by RNAi versus null mutations) leads to different phenotypic outcomes. Although a causal relationship between phosphorylation of p160ROCK by Aurora-A and suppression of mitotic arrest has not been firmly established here, our results indicate that p160ROCK acts downstream of Aurora-A in a common genetic pathway and phosphorylation of p160ROCK by Aurora-A may mimic the depletion of p160ROCK to bypass the mitosis arrest caused by Aurora-A inactivation.
Supplementary Material
Acknowledgments
We thank M. and C. Crankshaw (Washington University, St. Louis) for their expert help on MS, D. Helfman for his insight on the Y27632 and p160ROCK experiments, S. Narumiya (Kyoto, Japan) for his kind gift of the p160ROCK plasmid and antibody, S. Taylor, J. Sommerville, and A. H. Patel for kind gifts of antibodies, M. Mayer (Cold Spring Harbor Laboratory) for the help with the phosphoamino acid assay, G. Nourjanova for help on 2D protein gel, S. Prasanth for advice on HeLa cell transfection, P. Bubulya for help in live cell imaging, M. McCurrach for advice in BrdUrd labeling, A. Denli and A. Caudy for their critical reading of the manuscript and helpful discussion, and J. Duffy in the graphic arts department and S. Hearn in the microscope core facility (Cold Spring Harbor Laboratory) for their help. J.D. is supported by Postdoctoral Fellowship DAMD17-02-1-0344 from the Department of Defense Breast Cancer Research Program. This work is also supported by National Institutes of Health Grant P01-CA13106 (to G.J.H.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: STK15, serine/threonine kinase 15; SAF, STK15-associate factor; RNAi, RNA interference; siRNA, small interfering RNA.
References
- 1.Zheng, Y., Jung, M. K. & Oakley, B. R. (1991) Cell 65, 817–823. [DOI] [PubMed] [Google Scholar]
- 2.Hinchcliffe, E. H. & Sluder, G. (2001) Curr. Biol. 11, R698–R701. [DOI] [PubMed] [Google Scholar]
- 3.Bornens, M. (2002) Curr. Opin. Cell Biol. 14, 25–34. [DOI] [PubMed] [Google Scholar]
- 4.Rieder, C. L., Faruki, S. & Khodjakov, A. (2001) Trends Cell Biol. 11, 413–419. [DOI] [PubMed] [Google Scholar]
- 5.Doxsey, S. (2001) Nat. Rev. Mol. Cell Biol. 2, 688–698. [DOI] [PubMed] [Google Scholar]
- 6.Stearns, T. (2001) Cell 105, 417–420. [DOI] [PubMed] [Google Scholar]
- 7.Hinchcliffe, E. H., Li, C., Thompson, E. A., Maller, J. L. & Sluder, G. (1999) Science 283, 851–854. [DOI] [PubMed] [Google Scholar]
- 8.Matsumoto, Y., Hayashi, K. & Nishida, E. (1999) Curr. Biol. 9, 429–432. [DOI] [PubMed] [Google Scholar]
- 9.Meraldi, P., Lukas, J., Fry, A., Bartek, J. & Nigg, E. A. (1999) Nat. Cell Biol. 1, 88–93. [DOI] [PubMed] [Google Scholar]
- 10.do Carmo Avides, M., Tavarest, A. & Glover, D. M. (2001) Nat. Cell Biol. 3, 421–424. [DOI] [PubMed] [Google Scholar]
- 11.Fry, A. M. (2002) Oncogene 21, 6184–6194. [DOI] [PubMed] [Google Scholar]
- 12.Zhou, H., Kuang, J., Zhong, L., Kuo, W., Grey, J., Sahin, A., Brinkley, B. & Sen, S. (1998) Nat. Genet. 20, 189–193. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka, T., Kimura, M., Matsunaga, K., Fukada, D., Mori, H. & Okano, Y. (1999) Cancer Res. 59, 2041–2044. [PubMed] [Google Scholar]
- 14.Miyoshi, Y., Iwao, K., Egawa, C. & Noguchi, S. (2001) Int. J. Cancer 92, 370–373. [DOI] [PubMed] [Google Scholar]
- 15.Sen, S., Zhou, H., Zhang, R. D., Yoon, D. S., Vakar-Lopez, F., Ito, S., Jiang, F., Jounston, D., Grossman, H. B., Ruifrok, A. C., et al. (2002) J. Natl. Cancer. Inst. 94, 1320–1329. [DOI] [PubMed] [Google Scholar]
- 16.Farruggio, D. C., Townsley, F. M. & Ruderman, J. V. (1999) Proc. Natl. Acad. Sci. USA 96, 7306–7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walter, A., Seghezzi, O., Korver, W., Sheung, J. & Lees, E. (2000) Oncogene 19, 4906–4916. [DOI] [PubMed] [Google Scholar]
- 18.Honda, K., Mihara, H., Kato, Y., Yamaguchi, A., Tanaka, H., Yasuda, H., Furukawa, K. & Urano, T. (2000) Oncogene 19, 2812–2819. [DOI] [PubMed] [Google Scholar]
- 19.Katayama, H., Zhou, H., Li, Q., Tatsuka, M. & Sen, S. (2001) J. Biol. Chem. 276, 46219–46224. [DOI] [PubMed] [Google Scholar]
- 20.Giet, R., Uzbekov, R., Cubizolles, F., Le Guellec, K. & Prigent, C. (1999) Biol. Cell 91, 461–470. [PubMed] [Google Scholar]
- 21.Mendez, R., Hake, L. E., Andresson, T., Littlepage, L. E., Ruderman, J. V. & Richter, J. D. (2000) Nature 404, 302–307. [DOI] [PubMed] [Google Scholar]
- 22.Groisman, I., Huang, Y. S., Mendea, R., Cao, Q., Theurkauf, W. & Richter, J. D. (2000) Cell 103, 435–447. [DOI] [PubMed] [Google Scholar]
- 23.Giet, R., McLean, D., Descamps, S., Lee, M. J., Raff, J. W. & Prigent, C. (2002) J. Cell. Biol. 156, 437–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chevrier, V., Piel, M., Collomb, N., Saoudi, Y., Frank, R., Paintrand, M., Narumiya, S., Bornens, M. & Didier, J. (2001) J. Cell. Biol. 157, 807–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amano, M., Fukata, Y. & Kaibuchi, K. (2000) Exp. Cell Res. 261, 44–51. [DOI] [PubMed] [Google Scholar]
- 26.Maekawa, M., Ishizaki, T., Boku, S., Watanabe, N., Fujita, A., Iwamatsu, A., Obinata, T., Ohashi, K., Mizuno, K. & Narumiya, S. (1999) Science 285, 895–898. [DOI] [PubMed] [Google Scholar]
- 27.Bhowmick, N. A., Ghiassi, M., Aakre, M., Brown, K., Singh, V. & Moses, H. L. (2003) Proc. Natl. Acad. Sci. USA 100, 15548–15553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watnick, R. S., Cheng, Y. N., Rangarajan, A., Ince, T. A. & Weinberg, R. A. (2003) Cancer Cell 3, 199–200. [DOI] [PubMed] [Google Scholar]
- 29.Sahai, E. I. T., Narumiya, S. & Treisman, R. (1999) Curr. Biol. 9, 136–145. [DOI] [PubMed] [Google Scholar]
- 30.Coleman, M. L., Sahai, E. A., Yeo, M., Bosch, M., Dewar, A. & Olson, M. F. (2001) Nat. Cell Biol. 3, 339–345. [DOI] [PubMed] [Google Scholar]
- 31.Hannon, G. J., Sun, P., Carnero, A., Xie, L. Y., Maestro, R., Conklin, D. S. & Beach, D. (1999) Science 283, 1129–1130. [DOI] [PubMed] [Google Scholar]
- 32.Du, J. & Hannon, G. (2002) Nucleic Acids Res. 30, 5465–5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herbst, A. & Tansey, W. P. (2000) Mol. Biol. Rep. 27, 203–208. [DOI] [PubMed] [Google Scholar]
- 34.Grant, G. A. & Crankshaw, M. W. (2003) Methods Mol. Biol. 211, 247–268. [DOI] [PubMed] [Google Scholar]
- 35.Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K. & Tuschl, T. (2001) Nature 411, 494–498. [DOI] [PubMed] [Google Scholar]
- 36.Du, J., Nasir, I., Benton, B. K., Kladde, M. P. & Laurent, B. C. (1998) Genetics 150, 987–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lassus, P., Optiz-Araya, X. & Lazebnik, Y. (2002) Science 297, 1352–1354. [DOI] [PubMed] [Google Scholar]
- 38.Kufer, T. A., Sillje, H. H., Korner, R., Gruss, O. J., Meraldi, P. & Nigg, E. A. (2002) J. Cell Biol. 158, 617–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laurent, B. L. & Carlson, M. (1992) Genes Dev. 6, 1707–1715. [DOI] [PubMed] [Google Scholar]
- 40.Hirota, T., Kunitoku, N., Sasayama, T., Marumoto, T., Zhang, D., Nitta, M., Hatakeyama, K. & Saya, H. (2003) Cell 114, 585–598. [DOI] [PubMed] [Google Scholar]
- 41.Bischoff, J. R., Anderson, L., Zhu, Y., Mossie, K., Ng, L., Souza, B., Schryver, B., Flanagan, P., Chairvoyant, F., Ginther, C., et al. (1998) EMBO. J. 17, 3052–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meraldi, P., Honda, R. & Nigg, E. A. (2002) EMBO, J. 21, 483–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Glover, D. M., Leibowitz, M. H., McLean, D. A. & Parry, H. (1995) Cell 81, 95–105. [DOI] [PubMed] [Google Scholar]
- 44.Kunitoku, N., Sasayama, T., Marumoto, T., Zhang, D., Honda, S., Kobayashi, O., Hatakeyama, K., Ushio, Y. H. S. & Hirota, T. (2003) Dev. Cell 5, 853–864. [DOI] [PubMed] [Google Scholar]
- 45.Schumacher, J. M., Ashcroft, N., Donovan, P. J. & Golden, A. (1998) Development (Cambridge, U.K.) 125, 4391–4402. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.