Significance
Acetaminophen toxicity is significantly influenced by the hepatocyte circadian clock through its control of xenobiotic metabolizing systems. We have found that, although the central circadian clock can influence detoxification through glutathione biosynthesis, the autonomous hepatocyte circadian clock also controls major aspects of acetaminophen (APAP) bioactivation. One mechanism by which APAP bioactivation is controlled is through the clock’s regulation of cytochrome P450-dependent activity through NADPH-cytochrome P450 oxidoreductase.
Keywords: circadian clock, acetaminophen toxicity, NADPH-cytochrome P450 oxidoreductase, chronotoxicity, Bmal1
Abstract
The diurnal variation in acetaminophen (APAP) hepatotoxicity (chronotoxicity) reportedly is driven by oscillations in metabolism that are influenced by the circadian phases of feeding and fasting. To determine the relative contributions of the central clock and the hepatocyte circadian clock in modulating the chronotoxicity of APAP, we used a conditional null allele of brain and muscle Arnt-like 1 (Bmal1, aka Mop3 or Arntl) allowing deletion of the clock from hepatocytes while keeping the central and other peripheral clocks (e.g., the clocks controlling food intake) intact. We show that deletion of the hepatocyte clock dramatically reduces APAP bioactivation and toxicity in vivo and in vitro because of a reduction in NADPH-cytochrome P450 oxidoreductase gene expression, protein, and activity.
Acetaminophen (APAP) is generally regarded as safe at recommended doses, but at higher doses it is well-reported to cause significant hepatotoxicity in humans (1). APAP overdose is the number one cause of acute liver failure in many western countries including the United States, Canada, and Great Britain, with up to half of these cases being accidental (2–4). At recommended doses, the large proportion of APAP is detoxified by direct enzymatic conjugation to sulfate or glucuronic acid, but a small percentage is bioactivated to the toxic metabolite N-acetyl-p-benzoquinone imine (NAPQI) by cytochromes P450. When glutathione levels are high, small amounts of NAPQI can be detoxified efficiently in the hepatocyte by conjugation to glutathione. When glutathione is low, NAPQI forms direct macromolecular adducts and also generates reactive oxygen and nitrogen species that damage cellular proteins and disrupt cellular processes (1).
Acetaminophen, like many pharmaceutical agents, is thought to exhibit chronotoxicity because it displays variation in toxicity over a 24-h (circadian) period (5, 6). Although the mechanisms underlying APAP chronotoxicity are unclear, past research shows that toxicity is inversely correlated with liver glutathione levels that rise and fall with the daily phases of feeding and fasting. That is, high glutathione levels after feeding correlate with protection, whereas low glutathione levels after fasting correlate with susceptibility (6). The importance of food intake in chronotoxicity is supported by data showing that shifting the food intake of mice to their typical fasting phase also shifts glutathione rhythms and APAP chronotoxicity (7–9). Although the importance of glutathione is often emphasized, it also has been proposed that variations in cytochrome P450 activity may be important in chronotoxicity (10). In this regard, ketones generated during the fasting phase have been proposed to increase levels of a specific APAP-metabolizing cytochrome P450, such as CYP2E1, presumably via ligand stabilization (11, 12). Collectively, these data have led to the idea that feeding- and fasting-driven oscillations in glutathione and the cytochromes P450 are major determinants of APAP chronotoxicity in the liver.
In mammals, circadian physiology is directed by the transcription factors CLOCK and BMAL1 (aka MOP3, ARNTL) (13). These molecular clock components control transcriptional and translational feedback loops that guide the circadian expression of as much as 15% of the mammalian transcriptome (14, 15). Emerging models of circadian biology suggest that the majority of cells throughout the body have their own molecular clocks and that a central clock in the suprachiasmatic nucleus (SCN) of the brain, entrained by environmental cues such as light, plays a role in synchronizing peripheral cellular clocks through its influence over activity/resting periods that determine food intake (16, 17). In fact, food intake may be a dominant synchronizer of peripheral clocks, because restricting food intake in the presence of normal light cues shifts the phase of peripheral cellular clocks (18).
We propose that APAP chronotoxicity is controlled by two distinct molecular clocks, the central clock within the SCN and a peripheral clock in the hepatocyte. We propose that the central clock influences toxicity through feeding/fasting-related metabolic products, such as glutathione or ketones, and the hepatocyte clock influences toxicity through its control of cytochromes P450. If this model is correct and the hepatocyte clock is an important player in APAP chronotoxicity, then we should see differential toxicity in an animal model in which only the hepatocyte circadian clock has been disrupted and the central clock and feeding rhythms are intact. In contrast, if food-induced oscillations are the primary drivers of toxicity, then we would anticipate that the ablation of the hepatocyte clock would have little effect on chronotoxicity. To test this hypothesis, we created a conditional null allele of Bmal1, an essential component of the circadian clock (13), to disrupt the molecular clock in hepatocytes while keeping the central and other peripheral clocks intact. Using this model, we demonstrate that the hepatocyte clock, through regulation of NADPH-cytochrome P450 oxidoreductase (Por), independently controls bioactivation of APAP.
Results
Bmal1fx/fxCreAlb Mice Exhibit Normal Feeding and Locomotor Activity Patterns.
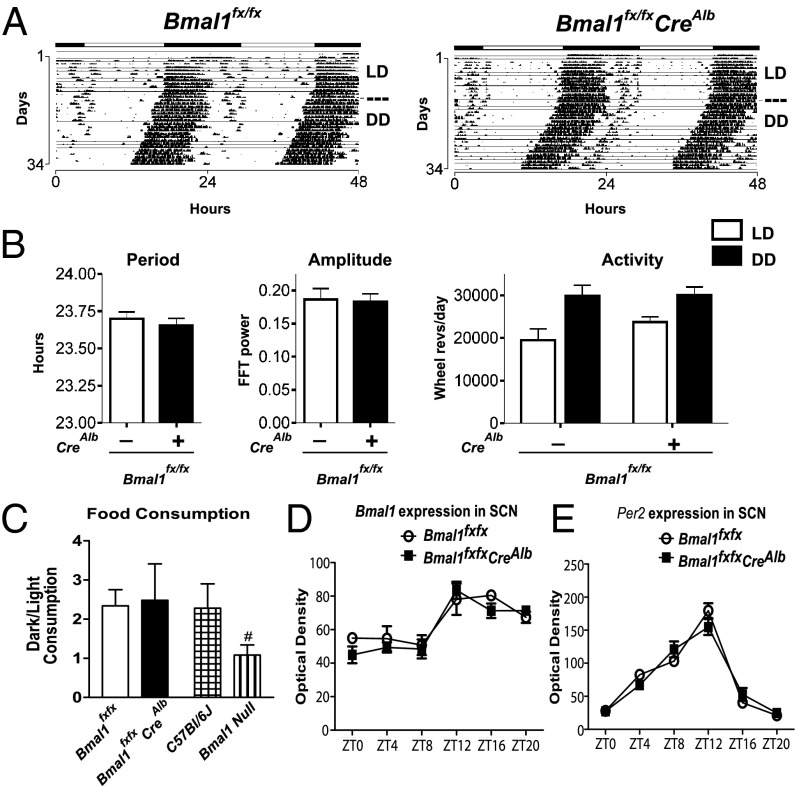
Details on the creation of the Bmal1fx allele and subsequent crosses are provided in SI Experimental Procedures and Fig. S1. Deletion of Bmal1 in hepatocytes occurred through a cross of mice homozygous for the Bmal1fx allele (i.e., Bmal1fx/fx) with mice heterozygous for the Bmal1fx allele (i.e., Bmal1fx/wt) that also harbored a separate Cre transgene under the control of the Albumin promoter (i.e., CreAlb) (19). To characterize the effects of hepatocyte-specific Bmal1 deletion on locomotor activity, we examined the wheel-running activity of Bmal1fx/fxCreAlb and WT littermate Bmal1fx/fx controls. Animals were entrained to a 12-h light/dark (LD 12:12) schedule for 14 d and then were transferred to constant darkness (DD). Zeitgeber time 0 (ZT0) corresponds to the time the entrainment signal initiates, which here is light onset. WT and Bmal1fx/fxCreAlb mice expressed indistinguishable circadian oscillations in activity (Fig. 1 A and B): average free-running periods of WT and Bmal1fx/fxCreAlb mice were 23.7 ± 0.06 h and 23.7 ± 0.05 h, respectively. Spectral analysis using fast Fourier transforms detected high-amplitude periodicity in the circadian range for both genotypes (Fig. 1B). Activity levels, measured as wheel revolutions, were similar for WT and Bmal1fx/fxCreAlb mice, with higher activity in the dark phase than during the light period (Fig. 1B). The ratio of food consumed during the subjective night/day (i.e., dark/light phase) showed no difference between WT and Bmal1fx/fxCreAlb mice, with both genotypes showing a twofold increase in food consumption during the dark phase (P = 0.88). In contrast, the dark/light ratios of food consumption by global Bmal1-null animals were significantly different from those of C57BL/6J controls (P < 0.05) (Fig. 1C).
Fig. 1.
Bmal1fx/fxCreAlb mice exhibit normal extrahepatic circadian physiologies. (A) Actograms of wheel-running activity in WT and Bmal1fx/fxCreAlb mice. Animals were entrained to a schedule of 12:12 LD and then were transferred to DD. (B) WT and Bmal1fx/fxCreAlb mice expressed robust circadian rhythms of activity with no significant differences in amplitude, periodicity, or total activity (P > 0.05). (C) Normal dark/light ratios of food consumption in Bmal1fx/fxCreAlb mice, in contrast to Bmal1-null mice (P = 0.88 and 0.012, respectively, compared with matched controls). (D and E) Bmal1 and Per2 expression in the SCN by in situ hybridization. WT and Bmal1fx/fxCreAlb mice showed diurnal expression of Bmal1 (D) and Per2 (E) over a 24-h period in the SCN (P < 0.0001 for both genes with respect to time; P = 0.33 and P = 0.76 for Bmal1 and Per2, respectively, with respect to genotype; two-way ANOVA). All data represent means ± SEM. Also see Fig. S1.
Bmal1 and the Hepatocyte Clock Drive the Majority of Transcriptional Oscillations in the Liver.
To confirm that CreAlb-mediated excision of the conditional Bmal1fx allele did not affect the central circadian clock, we examined the expression of two prototype molecular clock-regulated genes, Per2 and Bmal1, in the SCN using in situ hybridization. We observed that WT and Bmal1fx/fxCreAlb mice showed similar diurnal expression of both Bmal1 and Per2 in the SCN over a 24-h period (P < 0.05 with respect to time, P > 0.05 with respect to genotype) (Fig. 1 D and E).
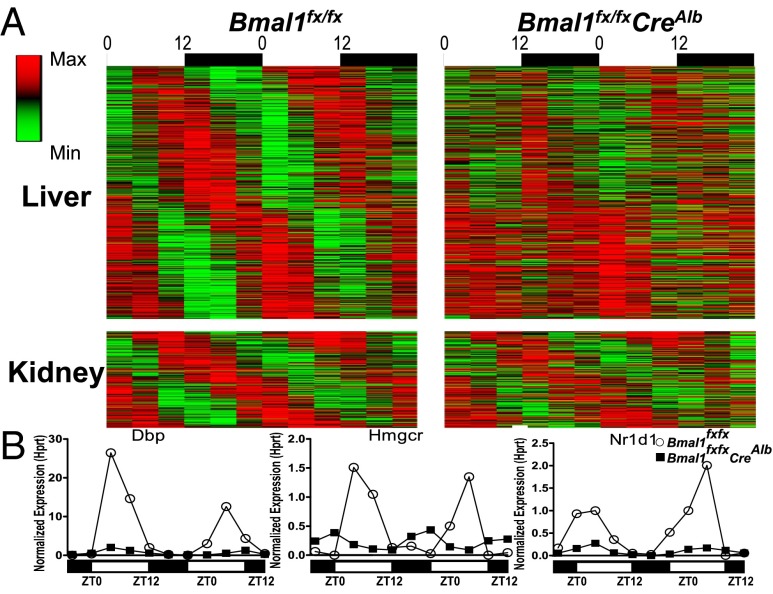
To determine if circadian oscillations in transcription were disrupted in hepatocytes and to examine the effect on other peripheral sites, we performed expression microarrays on liver and kidney from WT and Bmal1fx/fxCreAlb mice. Animals were killed every 4 h in two separate 24-h experiments, one using 10-wk-old mice and one using 20-wk-old mice. We performed COSOPT analysis on the microarray results and found 5.6% of the expressed transcripts in liver and 1.8% of the expressed transcripts in kidney oscillated with a circadian periodicity [20–28 h, mean measure corrected P value (pMMC-β) < 0.05] in WT mice (Fig. 2A, Left). Of these cycling transcripts, 90% of cycling liver transcripts showed significantly different expression in Bmal1fx/fxCreAlb mice (P ≤ 0.05), but only 16% of kidney transcripts showed a significant difference (Fig. 2A, Right). Quantitative PCR analysis supported the array data showing that the cycling of known output genes regulated by Bmal1, such as Dbp, Nr1d1 (Rev-Erb-a), and Hmgcr, showed substantially dampened oscillations in the livers of Bmal1fx/fxCreAlb mice (Fig. 2C).
Fig. 2.
Microarray analysis reveals the independence of the Bmal1-driven hepatocyte clock. (A) Heatmap of cycling transcripts in liver (Upper) and kidney (Lower). WT and Bmal1fx/fxCreAlb mice were killed every 4 h over two different days. (Left) Transcripts with corrected P value < 0.03 in WT liver and kidney are shown and ordered by peak phase. (Right) Ninety percent of cycling transcripts in liver and 16% of cycling transcripts in kidney were significantly different in Bmal1fx/fxCreAlb and WT mice. (B) Validation of arrays by multiplex PCR of cycling transcripts Dbp, Nr1d1 (Reverb-a), and Hmgcr internally normalized to Hprt in WT mice (open circles) and Bmal1fx/fxCreAlb mice (filled squares). Also see Fig. S3 and Table S1.
Lack of APAP Chronotoxicity in Bmal1fx/fxCreAlb Mice.
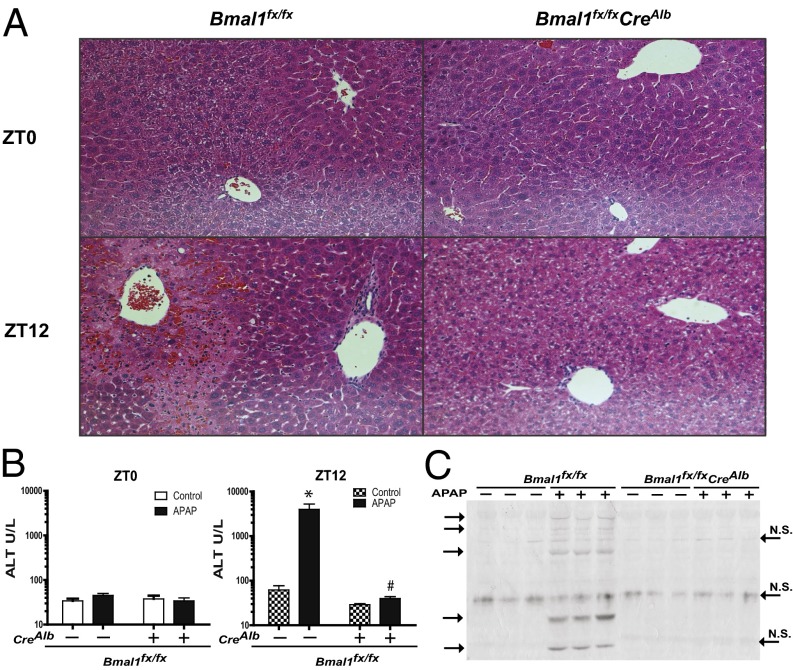
To study the effects of the hepatocyte circadian clock on APAP chronotoxicity, we dosed WT and Bmal1fx/fxCreAlb mice with 300 mg/kg APAP at ZT0 and ZT12. We observed that when mice were given APAP at ZT0 and were killed 6 h later, both WT and Bmal1fx/fxCreAlb mice displayed normal liver histology and normal serum ALT levels (Fig. 3 A and B). In contrast, we observed significant differences between genotypes when mice were administered the same dose of APAP at ZT12 and were killed 6 h later. APAP-treated WT mice displayed extensive multifocal centrilobular necrosis, extending to the midzonal region, fragmented hepatic plates, hepatocellular dropout, dilated sinusoids, and neutrophil inflammation (Fig. 3A). Consistent with the histopathological findings, serum ALT levels in WT mice were markedly elevated compared with saline-treated controls (Fig. 3B). In contrast, the livers from APAP-treated Bmal1fx/fxCreAlb mice displayed no histological evidence of centrilobular necrosis or fibrosis, normal hepatocellular architecture was maintained (Fig. 3A), and no significant increase in serum ALT levels was observed (Fig. 3B).
Fig. 3.
Lack of acetaminophen toxicity in Bmal1fx/fxCreAlb mice. (A) H&E-stained histological sections of livers from WT and Bmal1fx/fxCreAlb mice administered APAP at ZT0 and ZT12 and killed 6 h postdose. Note the presence/absence of centrilobular necrosis. (B) Serum ALT levels in WT and Bmal1fx/fxCreAlb mice 6 h after APAP administration at ZT0 and ZT12. Serum ALT levels were markedly elevated in Bmal1fx/fxmice as compared with APAP-treated Bmal1fx/fxCreAlb mice and PBS-treated controls. Data are shown as means ± SEM; *P < 0.05 between treatment and control; #P < 0.05 between WT and Bmal1fx/fxCreAlb. (C) Western blot of APAP adducts in liver. Mice were treated at ZT11 with 300 mg/kg APAP or vehicle control and were killed 1 h after treatment. Liver protein (100 ug) from single animals was loaded into each lane and labeled with anti-APAP antibody. Nonspecific (N.S.) bands are indicated on the right, and APAP adducts are shown on the left. Gel lanes were reordered for presentation but otherwise are unaltered. Data represent means ± SEM. Also see Fig. S2.
Decreased APAP Protein Adducts in Bmal1fx/fxCreAlb Liver at ZT12.
The metabolite NAPQI is an important electrophilic intermediate in APAP toxicity. To determine if NAPQI formation and its electrophilic attack on cellular macromolecules correlated with differential toxicity, we measured APAP protein adducts by Western blot using an antibody raised against APAP-protein conjugates (20). As qualitative evidence for circadian times when NAPQI formation overwhelmed cellular glutathione stores and was free to alkylate cellular proteins, we show extensive APAP protein adduct formation at ZT12 but not at ZT0 in WT mice. In contrast, Bmal1fx/fxCreAlb mice showed no significant protein alkylation at either ZT0 or ZT12 (Fig. 3C and Fig. S2).
Differential Expression of Genes Involved in APAP Metabolism.
To identify candidate enzymes that could be responsible for differential toxicity at ZT12, we analyzed our microarray data to examine the expression of genes with potential relevance to APAP metabolism in mice. Of these, Por, Ugt1a9, Sult1a1, and Abcb1b exhibited consistent down-regulation in Bmal1fx/fxCreAlb liver microarrays compared with WT liver microarrays. No genes were consistently up-regulated (Fig. S3). Quantitative PCR analysis supported the array data for Por, Ugt1a9, and Abcb1b, but Sulf1a1 expression showed no difference in livers of Bmal1fx/fxCreAlb and WT mice (Fig. S4).
Differential Glutathione Depletion in WT and Bmal1fx/fxCreAlb Mice at ZT0 and ZT12.
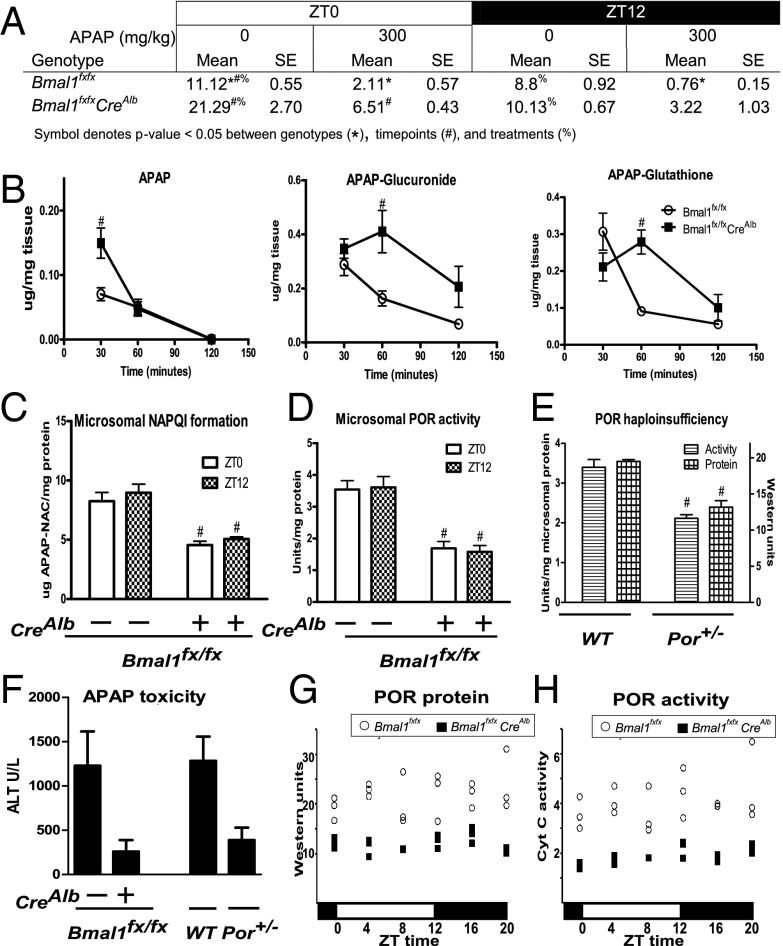
We next evaluated the effect of APAP treatment on total glutathione [oxidized glutathione + glutathione (GSSG + GSH)] content in WT and Bmal1fx/fxCreAlb liver at ZT0 and ZT12. We found higher glutathione levels in control (untreated) liver at ZT0 than at ZT12 in both WT and Bmal1fx/fxCreAlb animals, as is consistent with reports that these levels increase in response to nocturnal feeding (P < 0.05, one-tailed Student t test) (Fig. 4A) (21). Glutathione levels were similar in untreated WT and Bmal1fx/fxCreAlb animals, and APAP treatment decreased liver glutathione at both time points in both WT and Bmal1fx/fxCreAlb mice (P < 0.05, Student one-tailed t test) (Fig. 4A) (6). At ZT0, APAP treatment decreased WT and Bmal1fx/fxCreAlb liver glutathione levels to 19% and 30%, respectively, of the levels in controls (P < 0.05). At ZT12 glutathione in APAP-treated WT animals was depleted to 9% of control levels, consistent with increased toxicity at this time point. In contrast, glutathione in APAP-treated Bmal1fx/fxCreAlb animals was depleted to 32% of control levels (P < 0.05) (Fig. 4A).
Fig. 4.
Differential APAP bioactivation in WT and Bmal1fx/fxCreAlb mice at ZT12. (A) Total liver glutathione (GSH + GSSG) was decreased in WT and Bmal1fx/fxCreAlb mice treated with 300 mg/kg APAP compared with controls at the given zeitgeber times. Post-APAP glutathione levels were lower in WT mice than in Bmal1fx/fxCreAlb mice at both time points Values are from three separate experiments (n ≥ 5). (B) Liver concentrations of APAP and the major metabolites APAP-Gluc and APAP-GS after dosing at ZT11 indicate slower APAP metabolism in Bmal1fx/fxCreAlb mice (n ≥ 4). (C) Liver microsomal metabolism of APAP in Bmal1fx/fxCreAlb mice shows decreased NAPQI (captured as APAP-NAC) formation at ZT0 and ZT12 (n ≥ 4). (D) Bmal1fx/fxCreAlb mice exhibit lower microsomal POR cytochrome c reductase activity than WT mice (n ≥ 5). (E) Por+/− mice exhibit decreased POR activity and protein relative to treated WT mice (n = 3). (F) Por+/− mice exhibit a decreased trend in liver toxicity relative to treated WT mice. (G and H) Bmal1fx/fxCreAlb mice exhibit consistently lower liver POR protein (G) and activity (H) than WT mice. All data represent means ± SEM; for figures B through E #P < 0.05 between WT control and mutant genotype. Also see Figs. S4 and S5.
Altered Kinetics of APAP Metabolism in Bmal1fx/fxCreAlb Mice.
To determine why liver glutathione levels after ZT12 APAP treatment were higher in Bmal1fx/fxCreAlb mice than in WT mice, we monitored levels of APAP and its major metabolites at 30, 60, and 120 min after dosing at ZT11 (Fig. 4B). At 30 min postdose, APAP levels were significantly higher in Bmal1fx/fxCreAlb than in WT liver (P < 0.05), but APAP-glutathione (APAP-GS) and APAP-glucuronide (APAP-Gluc) conjugates showed no significant difference (Fig. 4B). In contrast, at 60 min postdose APAP levels were similar in the two groups, but APAP-GS and APAP-Gluc levels were significantly higher in Bmal1fx/fxCreAlb mice (P < 0.05 for each) (Fig. 4B). At 120 min postdose, APAP was below the limit of detection in four of four WT mice and in three of four Bmal1fx/fxCreAlb mice. APAP-GS and APAP-Gluc were detectable in all animals and showed no difference between genotypes at this time point (Fig. 4B). Cumulative APAP-sulfate levels from the three time points were 18 and 12 ng/mg in the livers of WT and Bmal1fx/fxCreAlb mice, respectively. This minor metabolite made up only ∼1% of total APAP metabolism and was not significantly different between genotypes (Fig. S5). The degradation products APAP-N-acetyl cysteine (APAP-NAC) and APAP-cysteine/APAP-cysteine-glycine (APAP-Cys) also were detected at low levels and reflected levels of the parent compound, APAP-GS (Fig. S5).
Decreased APAP Bioactivation in Bmal1fx/fxCreAlb Microsomes.
We performed in vitro studies to assess microsomal metabolism from WT and Bmal1fxfxCreAlb mouse livers harvested at ZT0 and ZT12 (Fig. 4C). Formation of APAP-NAC, a marker of APAP bioactivation to NAPQI, was measured. Microsomes harvested from WT mice at ZT0 and ZT12 showed no significant difference in APAP-NAC formation (P > 0.05). Bmal1fxfxCreAlb microsomes exhibited lower activity regardless of the time of day. Formation of APAP-NAC in ZT0 and ZT12 microsomes was 45% and 43% of that in WT microsomes at ZT12 (P < 0.05 for each).
To determine if specific APAP-metabolizing cytochromes P450 could account for the decreased APAP-NAC formation in ZT12 microsomes, we performed enzymatic assays for Cyp2e1 and Cyp1a2, because mice lacking these isoforms are protected from APAP hepatotoxicity (22). The activity of Cyp2e1 was 71% of WT (P < 0.05), whereas Cyp1a activity was not significantly different from WT (P = 0.12). Activity of the non-APAP metabolizing Cyp2b was decreased to 76% of control (P < 0.05). Despite these decreases in P450-dependent activities, total P450 content in Bmal1fx/fxCreAlb microsomes was not decreased but instead showed an increase (which was not statistically significant) to ∼150% of control (P = 0.08). (Fig. S6).
Correlation Between POR Activity and APAP Toxicity in Bmal1fx/fxCreAlb and Por+/− Mice.
Our expression data next led us to ask if the basis for decreased P450-dependent bioactivation lies in decreased activity of the P450 electron donor, NADPH-cytochrome P450 oxidoreductase (POR). We observed that Por mRNA displayed circadian oscillation in WT liver and was constitutively decreased in Bmal1fx/fxCreAlb liver (Fig. S3). We found microsomal POR activity was decreased in Bmal1fx/fxCreAlb liver microsomes, being 53% and 55% of WT microsomal POR activity at ZT0 and ZT12, respectively (Fig. 4D). In support of previous findings, reduced POR-dependent cytochrome c reductase activity and protein levels at ZT12 (62% and 67% of control, respectively) were found in liver microsomes generated from haploinsufficient mice heterozygous for Por (Por+/− mice) (Fig. 4E) (23). To determine if this decrease in POR activity alone could protect against APAP toxicity at ZT12, we dosed Bmal1fx/fxCreAlb mice, Por+/− mice, and respective controls at ZT12 and measured serum alanine aminotransferase (ALT) levels 6 h postdose. We found that Bmal1fx/fxCreAlb mice and Por+/− mice were protected similarly from toxicity, showing fivefold and 4.3-fold lower serum ALT values, respectively, relative to controls (P < 0.05 for each).
POR Protein and Activity Are Consistently Low in Bmal1fx/fxCreAlb Mice.
To characterize the circadian oscillation of POR protein and activity, we killed WT and Bmal1fx/fxCreAlb mice every 4 h and performed Western blots and activity assays for POR. We did not observe diurnal oscillations in POR protein or activity in WT mice (Fig. 4 F and G; data are representative of two separate experiments); however POR protein and activity were decreased at all time points in Bmal1fx/fxCreAlb microsomes (Fig. 4 G and H).
Discussion
We set out to define the relative contributions of the central clock and the hepatocyte clock in the chronotoxicity of APAP. To this end, we used a Bmal1fx/fxCreAlb mouse model in which the transcriptional circadian clock within hepatocytes is deleted, whereas peripheral clocks in other cell types, including the central clock within the SCN, are maintained. The power of this model lies in the fact that circadian rhythms of behavioral activity and, importantly, food intake, driven by the central clock can oscillate normally, whereas the hepatocyte clock and its associated biology can be disrupted. The explicit hypothesis tested in this study was that APAP chronotoxicity is the result not only of circadian feeding/fasting-associated metabolism controlled by the central clock but also of transcriptional oscillations of APAP-metabolizing genes driven by the hepatocyte clock.
To validate our mouse model, we first demonstrated that Bmal1fx/fxCreAlb (hepatocyte-null only) mice display normal extrahepatic circadian rhythmicity as demonstrated by locomotor activity, feeding patterns, and circadian gene expression in the SCN and kidney. These data are consistent with data from another conditional Bmal1 model and are in contrast to the global Bmal1-null model (13, 24). We also demonstrated the functional disruption of the hepatocyte clock by describing the marked decrease in the number and amplitude of oscillating transcripts in the liver. This result is in line with that of a different mouse model of hepatocyte circadian disruption in which the hepatocyte clock is arrested by the overexpression of REV-ERB alpha, a repressor of BMAL1 (25). We propose that the small percentage of oscillating liver transcripts not disrupted by hepatocyte deletion of Bmal1 represent transcripts from other cell types in the liver where the albumin promoter is not active (e.g., endothelial, Kupffer, and stellate cells, among others) or represent transcripts that are entrained by the central clock through factors such as feeding or hormones. These results demonstrate that the great majority of transcriptional oscillations in the liver are controlled by the BMAL1-dependent circadian clock in the hepatocyte.
We present data indicating that, although food-induced oscillations in glutathione levels driven by the central clock are an important contributor to APAP chronotoxicity, there also is a significant contribution from bioactivation events driven by the hepatocyte clock. Two observations support this conclusion. First, we report that, although WT mice exhibited APAP chronotoxicity (i.e., toxicity at ZT12 but not at ZT0), Bmal1fx/fxCreAlb mice were consistently resistant. This lack of chronotoxicity was evidenced even though feeding rhythms of WT and Bmal1fx/fxCreAlb mice were similar at both time points. Second, we report that the APAP bioactivation product NAPQI was formed in greater amounts, liver glutathione depletion was greater, and formation of NAPQI-adduction to cellular protein was greater in WT mice than in Bmal1fx/fxCreAlb mice at ZT12. These experiments led us to conclude that, although hepatic glutathione oscillates in response to feeding and is an important nucleophilic sink for NAPQI, the hepatocyte molecular clock driven by Bmal1 plays an important role in APAP toxicity via control of bioactivation to NAPQI.
In support of the idea that APAP bioactivation to NAPQI is under the control of the hepatocyte clock, we offer the following observations. First, we observed slower rates of APAP metabolic clearance in Bmal1fx/fxCreAlb mice at ZT12 than in WT mice (as shown by a decreased rate of disappearance of parent APAP and slower rates of production for both APAP-Gluc and APAP-GS conjugates). Second, depletion of glutathione and formation of APAP-protein adducts also was greater in WT than in Bmal1fx/fxCreAlb animals around ZT12, as is consistent with decreased NAPQI production. Although we observed decreased APAP-GS levels in WT mice at 60 and 120 min postdose, we argue that these lower levels reflect depleted glutathione stores resulting from increased NAPQI production, allowing unconjugated NAPQI to spill over to secondary sites within cellular proteins. This increased electrophilic attack on cellular protein is supported by Western blots showing significant quantities of APAP adducts on cytosolic proteins in WT liver but not in Bmal1fx/fxCreAlb liver 1 h postdose. Third, we directly observed decreased microsomal NAPQI formation in Bmal1fx/fxCreAlb liver, as well as decreased enzyme activities of several individual cytochrome P450-dependent monooxygenases in Bmal1fx/fxCreAlb microsomal assays, as compared with WT. These observations led us to conclude that the rate of APAP bioactivation is decreased at ZT12 in Bmal1fx/fxCreAlb as compared with WT livers. This decreased conversion of APAP to NAPQI allows prolonged clearance of APAP via the glucuronidation pathway and explains much of the protection from chronotoxicity seen in Bmal1fx/fxCreAlb mice at ZT12, a time when glutathione levels are insufficient to prevent toxicity. We interpret these results as suggesting that hepatocyte clock-driven mediators of APAP bioactivation play a significant role in APAP toxicity.
To determine the underlying cause of decreased APAP bioactivation at ZT12, POR was evaluated because of the circadian oscillation of POR mRNA in our own and other studies (26, 27) and its position as the source of electrons for cytochrome P450-dependent metabolism. In addition, our observation of decreased Por expression in Bmal1fx/fxCreAlb was the only candidate gene identified in our gene-expression studies that could explain decreased toxicity (Fig. S3). In keeping with this idea, we observed that enzymatic activity of POR in Bmal1fx/fxCreAlb liver microsomes also was decreased compared with WT and correlated with the decrease in APAP bioactivation seen in Bmal1fx/fxCreAlb microsomes suggesting that this electron donor may be rate limiting for the P450 activities responsible for NAPQI generation. The activity of POR in vitro has been shown to be limiting with respect to electron donation for several different P450 isoforms and substrates (28, 29), and human POR polymorphisms have been reported to contribute to variations in drug metabolism (30). The decrease in APAP-induced hepatotoxicity in Por+/− mice at ZT12 indicates that POR is an important modulator of APAP hepatotoxicity in vivo. In further support of this model, the trend seen in our data of decreased microsomal APAP bioactivation despite increased total P450 content also is found in POR hypomorphic mice (29). These data led us to conclude that the loss of induction of POR transcription/activity by ablation of the hepatocyte clock is a major contributor to the decreased bioactivation and protection from toxicity seen in Bmal1fx/fxCreAlb mice in vivo.
Given POR’s position as the electron donor for cytochromes P450, it became a candidate to explain the circadian control over the metabolic bioactivation of APAP by the circadian clock. Similar to data from other laboratories, our transcriptional analysis showed robust oscillations in Por mRNA in WT mice (14, 26, 27). Moreover, at ZT0 and ZT12, Bmal1fx/fxCreAlb mice showed consistently decreased levels of POR protein and activity under ad libitum feeding and LD 12:12 conditions. Despite clear mRNA rhythms, in two different experiments we were not able to identify circadian oscillations in POR protein or activity under these conditions in WT mice. This result is in contrast to three independent reports that POR protein does display a circadian oscillation in the liver (26, 31, 32). Interestingly, the reported peak phases of POR protein oscillations vary significantly in these prior reports (approximately ZT8, ZT19, and ZT0/24). The basis for these disparate findings is unknown at the present time, but some component, such as ad libitum feeding, which previously has been shown to disrupt circadian amplitude, may be a contributing factor.
Taken together, these data support a scenario in which APAP chronotoxicity is caused by an imbalance between APAP detoxification and bioactivation which is influenced in different ways by two different circadian oscillations. In one, APAP detoxification is regulated by the central circadian clock that regulates feeding schedules which directly drive glutathione levels and indirectly drive some P450 activities (Cyp2e1) in the liver (11, 12). In the other, the hepatocyte circadian clock regulates the transcription of genes involved in APAP oxidation. Our demonstration that Por+/− mice mimic the decreases in Por mRNA, protein, and APAP-induced toxicity seen in Bmal1fx/fxCreAlb mice supports the idea that chronotoxicity of APAP occurs, in part, through the mRNA oscillations of this P450 electron donor.
The regulation of POR by the hepatocyte circadian clock could have far-reaching consequences on drug metabolism beyond that of APAP, given it is the electron donor for all cytochromes P450. Further studies will be needed to address the physiological and toxicological consequences of such regulation under more clinically relevant conditions. Although our animal model is able to separate the effects of two distinct circadian oscillators, it is difficult to predict how natural disruptions in these oscillators affect POR expression and activity. It is known that circadian patterns of feeding and activity can be reset in the SCN quickly after long-distance travel or shift work, but the molecular clock at certain peripheral sites, such as the liver, can take up to 8 d to reset (18). Thus, circadian oscillations in food-induced levels of GSH and transcriptional output genes easily could become asynchronous, and one can assume that the livers of rotating shift workers or frequent intercontinental travelers are rarely entrained. Although the APAP dose used in this study represents extreme acute hepatotoxicity and is well above suggested therapeutic doses, it provides a proof of concept that these populations may be at differential risk from exposure to hepatotoxicants such as APAP. Based upon a more thorough mechanistic understanding of this biology, we may be able to identify such individuals and reduce exposures or alter shift-working paradigms to those more conducive to human health. Potentially of greater importance is that the application of this knowledge to other important toxicants, especially chemotherapeutics, may provide clues that allow better control of dosing paradigms used to treat a variety of important human diseases.
Experimental Procedures
Analysis of Circadian Locomotor Activity and Feeding Rhythms.
Animal procedures were approved by the Animal Care and Use Committee of the School of Medicine and Public Health, University of Wisconsin-Madison. A detailed description of the generation of WT mice and subsequent crosses is included in SI Experimental Procedures. Locomotor activity and wheel-running activity was measured as described previously using Clocklab software (33, 34). Feeding activity was determined by maintaining solitary caged animals and weighing food each day at ZT0 and ZT12 under normal LD 12:12 conditions.
Gene Expression.
In situ hybridization was performed essentially as described (27), using 36-mer oligonucleotide probes (IDT) radiolabeled at the 3′ ends with 33P via terminal I deoxynucleotidyl transferase (Gibco/Invitrogen) (Table S1). SCN signal density was quantified using ImageJ 1.34s software (National Institutes of Health) and normalized to radioactive standards. Total liver or kidney RNA was prepared by using RNeasy Protect (Qiagen). Microarray (Agilent) experimental details and data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus and are accessible through GEO Series accession no. GSE34444 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE34444). Microarray expression profiles were verified by quantitative multiplex-PCR using the Beckman Coulter GeXP system and were analyzed with Beckman Expression Profiler or Taqman RT-PCR. Multiplex PCR primers are listed in Table S1.
Toxicology and Metabolic Studies.
Mice were housed in a selective pathogen-free facility on corncob bedding with food and water ad libitum. In toxicology studies, 9- to 20-wk-old male mice were given i.p. injections of 300 mg/kg APAP (Sigma) prepared in PBS. For ALT measurements and histological analysis animals were killed by CO2 inhalation 6 h after being treated at ZT0 or ZT12. Serum was analyzed by the Clinical Pathology Laboratory (University of Wisconsin, School of Veterinary Medicine). Sections from the left lobe of the liver were fixed in 10% (vol/vol) formalin and embedded in paraffin for staining with H&E and were examined by a blinded observer. For glutathione analysis, Western blots, and APAP metabolite analysis animals were treated with APAP or PBS at ZT11 and were killed 30, 60, or 90 min postdose. Glutathione levels were not affected by PBS treatment and were combined with data from untreated animals for analysis. Total glutathione (GSSG + GSH) was analyzed by HPLC of dinitrophenol derivatives (by B.P.J. using reagents and equipment graciously shared by Jeff Johnson's laboratory, University of Wisconsin, Madison, WI) (35). Western blots were performed on S9 liver homogenates from WT and Bmal1fx/fxCreAlb liver harvested at ZT12. Blots were stained with an antibody raised against APAP protein adducts (a generous gift from Jack Hinson, University of Arkansas for Medical Sciences, Little Rock, AR) (20). APAP metabolites were isolated by extraction in methanol as described by Dai et al. (36) with some modifications as detailed in SI Experimental Procedures. Microsomal APAP bioactivation was determined by measuring conjugation of APAP to N-acetyl-cysteine, with modifications detailed in SI Experimental Procedures (29). Microsomal POR-dependent cytochrome c reductase activity was performed as previously described (37).
Statistical Analysis.
A Student t test, one-tailed to repeat previously reported findings as indicated or two-tailed for new comparisons, was used for single comparisons. For multiple comparisons, a general linear models ANOVA followed by Tukey’s test was performed. A paired t test was used to test disruption between genotypes over matched time points in microarray data. COSOPT was used to identify cycling genes with a periodicity between 20 and 28 h.
Supplementary Material
Acknowledgments
We thank Ruth Sullivan for help with liver histopathology and Jeff Johnson for help with GSH measurements. This work was supported by National Institutes of Health Grants R37-ES005703 and P30-CA014520 (to C.A.B.), T32-CA009135 (to B.E.M.), and T32-ES007015 (to B.P.J.). E.L.M. and A.C.S. were members of, and J.S.T. is an Investigator of The Howard Hughes Medical Institute. J.A.W. is the recipient of a postdoctoral fellowship from The Natural Sciences and Engineering Research Council of Canada.
Footnotes
Conflict of interest statement: J.S.T. is a cofounder of ReSet Therapeutics, Inc., and is a member of its scientific advisory board.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE34444).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1421708111/-/DCSupplemental.
References
- 1.Hinson J, Roberts D, James L. Mechanisms of acetaminophen-induced liver necrosis. Handb Exp Pharmacol. 2010:369–405. doi: 10.1007/978-3-642-00663-0_12. 196: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chun LJ, Tong MJ, Busuttil RW, Hiatt JR. Acetaminophen hepatotoxicity and acute liver failure. J Clin Gastroenterol. 2009;43(4):342–349. doi: 10.1097/MCG.0b013e31818a3854. [DOI] [PubMed] [Google Scholar]
- 3.Myers RP, Li B, Fong A, Shaheen AA, Quan H. Hospitalizations for acetaminophen overdose: A Canadian population-based study from 1995 to 2004. BMC Public Health. 2007;7:143. doi: 10.1186/1471-2458-7-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larson AM, et al. Acute Liver Failure Study Group Acetaminophen-induced acute liver failure: Results of a United States multicenter, prospective study. Hepatology. 2005;42(6):1364–1372. doi: 10.1002/hep.20948. [DOI] [PubMed] [Google Scholar]
- 5.Levi F, Schibler U. Circadian rhythms: Mechanisms and therapeutic implications. Annu Rev Pharmacol Toxicol. 2007;47:593–628. doi: 10.1146/annurev.pharmtox.47.120505.105208. [DOI] [PubMed] [Google Scholar]
- 6.Schnell RC, Bozigian HP, Davies MH, Merrick BA, Johnson KL. Circadian rhythm in acetaminophen toxicity: Role of nonprotein sulfhydryls. Toxicol Appl Pharmacol. 1983;71(3):353–361. doi: 10.1016/0041-008x(83)90022-4. [DOI] [PubMed] [Google Scholar]
- 7.Schnell RC, et al. Factors influencing circadian rhythms in acetaminophen lethality. Pharmacology. 1984;29(3):149–157. doi: 10.1159/000138005. [DOI] [PubMed] [Google Scholar]
- 8.Howell SR, Klaassen C. Circadian variation of hepatic UDP-glucuronic acid and the glucuronidation of xenobiotics in mice. Toxicol Lett. 1991;57(1):73–79. doi: 10.1016/0378-4274(91)90121-l. [DOI] [PubMed] [Google Scholar]
- 9.Matsunaga N, et al. Influence of feeding schedule on 24-h rhythm of hepatotoxicity induced by acetaminophen in mice. J Pharmacol Exp Ther. 2004;311(2):594–600. doi: 10.1124/jpet.104.069062. [DOI] [PubMed] [Google Scholar]
- 10.Kim YC, Lee SJ. Temporal variation in hepatotoxicity and metabolism of acetaminophen in mice. Toxicology. 1998;128(1):53–61. doi: 10.1016/s0300-483x(98)00046-8. [DOI] [PubMed] [Google Scholar]
- 11.Bruckner JV, Ramanathan R, Lee KM, Muralidhara S. Mechanisms of circadian rhythmicity of carbon tetrachloride hepatotoxicity. J Pharmacol Exp Ther. 2002;300(1):273–281. doi: 10.1124/jpet.300.1.273. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez FJ. The 2006 Bernard B. Brodie Award Lecture. Cyp2e1. Drug Metab Dispos. 2007;35(1):1–8. doi: 10.1124/dmd.106.012492. [DOI] [PubMed] [Google Scholar]
- 13.Bunger MK, et al. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103(7):1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes ME, et al. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009;5(4):e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5(2):e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hogenesch JB, Ueda HR. Understanding systems-level properties: Timely stories from the study of clocks. Nat Rev Genet. 2011;12(6):407–416. doi: 10.1038/nrg2972. [DOI] [PubMed] [Google Scholar]
- 17.Okamura H, Doi M, Fustin JM, Yamaguchi Y, Matsuo M. Mammalian circadian clock system: Molecular mechanisms for pharmaceutical and medical sciences. Adv Drug Deliv Rev. 2010;62(9-10):876–884. doi: 10.1016/j.addr.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Damiola F, et al. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14(23):2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Postic C, et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J Biol Chem. 1999;274(1):305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- 20.Matthews AM, Roberts DW, Hinson JA, Pumford NR. Acetaminophen-induced hepatotoxicity. Analysis of total covalent binding vs. specific binding to cysteine. Drug Metab Dispos. 1996;24(11):1192–1196. [PubMed] [Google Scholar]
- 21.Maruyama E, Kojima K, Higashi T, Sakamoto Y. Effect of diet on liver glutathione and glutathione reductase. J Biochem. 1968;63(3):398–399. [PubMed] [Google Scholar]
- 22.Zaher H, et al. Protection against acetaminophen toxicity in CYP1A2 and CYP2E1 double-null mice. Toxicol Appl Pharmacol. 1998;152(1):193–199. doi: 10.1006/taap.1998.8501. [DOI] [PubMed] [Google Scholar]
- 23.Shen AL, O’Leary KA, Kasper CB. Association of multiple developmental defects and embryonic lethality with loss of microsomal NADPH-cytochrome P450 oxidoreductase. J Biol Chem. 2002;277(8):6536–6541. doi: 10.1074/jbc.M111408200. [DOI] [PubMed] [Google Scholar]
- 24.Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci USA. 2008;105(39):15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kornmann B, Schaad O, Reinke H, Saini C, Schibler U. Regulation of circadian gene expression in liver by systemic signals and hepatocyte oscillators. Cold Spring Harb Symp Quant Biol. 2007;72:319–330. doi: 10.1101/sqb.2007.72.041. [DOI] [PubMed] [Google Scholar]
- 26.Gachon F, Olela FF, Schaad O, Descombes P, Schibler U. The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 2006;4(1):25–36. doi: 10.1016/j.cmet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 27.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109(3):307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 28.Waxman DJ, Morrissey JJ, Leblanc GA. Hypophysectomy differentially alters P-450 protein levels and enzyme activities in rat liver: Pituitary control of hepatic NADPH cytochrome P-450 reductase. Mol Pharmacol. 1989;35(4):519–525. [PubMed] [Google Scholar]
- 29.Wu L, et al. Transgenic mice with a hypomorphic NADPH-cytochrome P450 reductase gene: Effects on development, reproduction, and microsomal cytochrome P450. J Pharmacol Exp Ther. 2005;312(1):35–43. doi: 10.1124/jpet.104.073353. [DOI] [PubMed] [Google Scholar]
- 30.Pandey AV, Sproll P. Pharmacogenomics of human P450 oxidoreductase. Front Pharmacol. 2014;5:103. doi: 10.3389/fphar.2014.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mauvoisin D, et al. Circadian clock-dependent and -independent rhythmic proteomes implement distinct diurnal functions in mouse liver. Proc Natl Acad Sci USA. 2014;111(1):167–172. doi: 10.1073/pnas.1314066111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robles MS, Cox J, Mann M. In-vivo quantitative proteomics reveals a key contribution of post-transcriptional mechanisms to the circadian regulation of liver metabolism. PLoS Genet. 2014;10(1):e1004047. doi: 10.1371/journal.pgen.1004047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vitaterna MH, et al. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994;264(5159):719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimomura K, et al. Genome-wide epistatic interaction analysis reveals complex genetic determinants of circadian behavior in mice. Genome Res. 2001;11(6):959–980. doi: 10.1101/gr.171601. [DOI] [PubMed] [Google Scholar]
- 35.Vargas MR, Johnson DA, Johnson JA. Decreased glutathione accelerates neurological deficit and mitochondrial pathology in familial ALS-linked hSOD1(G93A) mice model. Neurobiol Dis. 2011;43(3):543–551. doi: 10.1016/j.nbd.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dai G, He L, Chou N, Wan YJ. Acetaminophen metabolism does not contribute to gender difference in its hepatotoxicity in mouse. Toxicol Sci. 2006;92(1):33–41. doi: 10.1093/toxsci/kfj192. [DOI] [PubMed] [Google Scholar]
- 37.Shen AL, Kasper CB. Differential contributions of NADPH-cytochrome P450 oxidoreductase FAD binding site residues to flavin binding and catalysis. J Biol Chem. 2000;275(52):41087–41091. doi: 10.1074/jbc.M008380200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.