Significance
The plague bacillus Yersinia pestis is transmitted by fleas but evolved very recently from Yersinia pseudotuberculosis, a closely related food- and water-borne enteric pathogen. Identifying the specific genetic changes underlying the switch to arthropod-borne transmission is essential to understanding the evolution of Y. pestis and the emergence of plague. We have identified an important early step in this process. The urease enzyme, beneficial for an enteric bacterium, causes acute oral toxicity and significant mortality to fleas. Y. pestis is not toxic to fleas because of a loss-of-function mutation in one of the genes essential for urease activity. Epidemiologic modeling suggests that this mutation was positively selected during evolution because it resulted in a significantly higher probability of Y. pestis transmission.
Keywords: Yersinia urease, plague, evolution, arthropod-borne transmission, pseudogene
Abstract
The arthropod-borne transmission route of Yersinia pestis, the bacterial agent of plague, is a recent evolutionary adaptation. Yersinia pseudotuberculosis, the closely related food-and water-borne enteric species from which Y. pestis diverged less than 6,400 y ago, exhibits significant oral toxicity to the flea vectors of plague, whereas Y. pestis does not. In this study, we identify the Yersinia urease enzyme as the responsible oral toxin. All Y. pestis strains, including those phylogenetically closest to the Y. pseudotuberculosis progenitor, contain a mutated ureD allele that eliminated urease activity. Restoration of a functional ureD was sufficient to make Y. pestis orally toxic to fleas. Conversely, deletion of the urease operon in Y. pseudotuberculosis rendered it nontoxic. Enzymatic activity was required for toxicity. Because urease-related mortality eliminates 30–40% of infective flea vectors, ureD mutation early in the evolution of Y. pestis was likely subject to strong positive selection because it significantly increased transmission potential.
Among the 18 species that compose the genus Yersinia three are pathogenic for humans: Yersinia pestis, the causative agent of plague, and the enteric pathogens Yersinia pseudotuberculosis and Yersinia enterocolitica. Whereas Y. enterocolitica and Y. pseudotuberculosis diverged between 41 and 186 Ma, comparative genomics and population genetic studies indicate that Y. pestis has evolved from Y. pseudotuberculosis only within the last 1,500–6,400 y (1–4). Thus, the two species share most of their genome repertoire but despite this close genetic relationship they cause very different diseases and have very distinct transmission routes. Y. pestis causes acute and often fatal bubonic, septicemic, and pneumonic forms of plague and is transmitted primarily by flea bites (5). In contrast, Y. pseudotuberculosis causes relatively mild food- and water-borne diseases and is transmitted by the fecal–oral route (6).
The recent evolution of flea-borne transmission of Y. pestis has involved sequential gene loss and acquisition steps (7). Y. pestis has acquired two unique plasmids that are not present in Y. pseudotuberculosis: pMT1, required for colonization and bacterial survival in the flea midgut (8), and pPCP1, which is irrelevant to the bacteria–flea interaction but which facilitates bacterial dissemination from the flea bite site (9–11). The major transmission mechanism of Y. pestis depends on its ability to grow as a biofilm in the flea proventriculus, a muscular valve in the flea foregut that connects the esophagus to the midgut (12). The blockage impedes fleas from taking a full blood meal, resulting in repeated feeding attempts, during which some of the bacteria are dislodged from the biofilm and transmitted into the host (12, 13). Selective loss of gene function has been important to the development and stability of Y. pestis biofilm in the flea required for full transmissibility (7, 14, 15).
Studies on the interaction of Y. pseudotuberculosis with fleas revealed another important adaptive change in the evolution of Y. pestis into a flea-borne pathogen. Y. pseudotuberculosis is orally toxic to Xenopsylla cheopis fleas, an important plague vector species. Shortly after taking a blood meal containing Y. pseudotuberculosis, about one-third to one-half of fleas show signs of acute toxicity, including diarrhea and immobility, leading to the death of up to 40% of the infected fleas (16). However, very little is known about the Y. pseudotuberculosis factor(s) responsible for toxicity to fleas; it has been characterized only as a nonsecreted protein specifically active in the flea digestive tract, the production of which is not regulated by temperature—toxicity occurs whether the bacteria are grown at 21 °C or 37 °C (16). The Yersinia toxin complex (Tc) insecticidal-like proteins have been ruled out as the cause of oral toxicity to fleas (16).
In this study, we used comparative proteomics, functional genetics, and biochemical approaches in conjunction with a flea infection model to identify the Yersinia urease enzyme as the flea-toxic factor. Phylogenetics analyses indicate that functional urease activity was lost early in the divergence of Y. pestis and epidemiologic modeling suggests that this gene loss was subject to strong positive selection because it significantly increased transmission probability.
Results
Flea-Toxic Proteins Are Associated with the Y. pseudotuberculosis Cell Membrane.
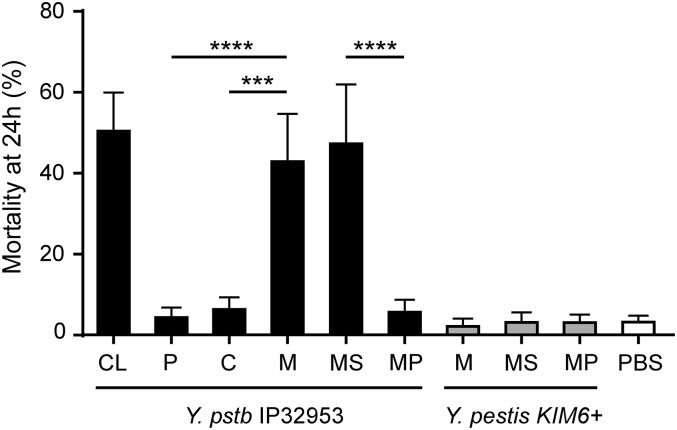
We used a comparative proteomics strategy to identify the Y. pseudotuberculosis flea-toxic factor that has been lost in Y. pestis. X. cheopis fleas were fed on blood containing Y. pseudotuberculosis cellular fractions enriched for periplasmic, cytoplasmic, and membrane proteins. Blood meals containing either the periplasmic or the cytoplasmic fraction were not toxic to fleas, resulting in a mortality no higher than that of control fleas that fed on uninfected blood (Fig. 1). In contrast, fleas that fed on blood containing the membrane fraction were moribund and showed signs of acute toxicity, including diarrhea, immediately after the 1-h feeding period and had a mortality rate at 24 h that was equivalent to that of fleas that received the Y. pseudotuberculosis whole-cell lysate (43% and 50.4%, respectively) (Fig. 1). These results indicate that the Y. pseudotuberculosis protein(s) that are orally toxic to X. cheopis fleas localize to the bacterial membrane.
Fig. 1.
The Y. pseudotuberculosis flea toxin is associated with the bacterial membrane. Mortality of fleas 24 h after feeding on blood containing 8.33 mg/mL of a Y. pseudotuberculosis IP32953 whole-cell lysate (CL) or 2.67–5.67 mg/mL proteins of Y. pseudotuberculosis or Y. pestis periplasmic (P), cytoplasmic (C), membrane (M), membrane supernatant (MS), or membrane pellet (MP) fractions. Data are the mean and range from a minimum of three independent experiments. ***P < 0.001, ****P < 0.0001.
Our attempts to separate the inner and outer membrane proteins with a detergent not toxic for X. cheopis fleas or to prevent excessive loss of protein while removing detergent were unsuccessful. However, some of the proteins in the crude membrane fraction were partially soluble in PBS and were separable by low-speed centrifugation, resulting in membrane supernatant (MS) and membrane pellet (MP) subfractions. The MS and MP fractions were tested for toxicity to fleas. Blood meals containing the MS fraction were acutely toxic to fleas, with ∼50% of fed fleas dead after 24 h, whereas blood meals containing the MP fraction were not toxic (Fig. 1). As expected, the Y. pestis membrane fractions were not toxic to fleas (Fig. 1). Thus, the toxic proteins of Y. pseudotuberculosis exhibiting insecticidal activity against X. cheopis fleas are enriched in the supernatant derived from the membrane fraction.
Differential Proteomic Profiles of Bacterial Membrane Subfractions Identify Candidate Flea Toxins of Y. pseudotuberculosis.
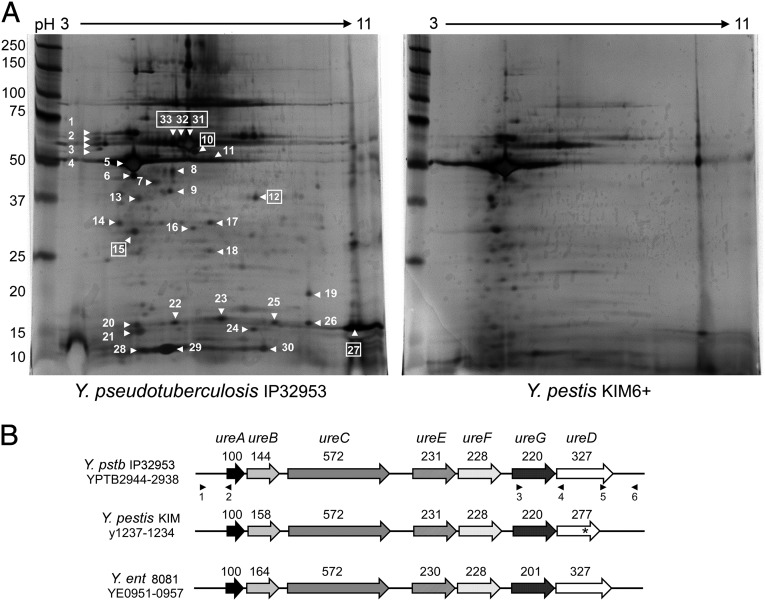
To identify candidate proteins responsible for flea toxicity, protein profiles of the MS subfractions from Y. pseudotuberculosis and Y. pestis were differentiated by 2D gel electrophoresis (2-DE). Thirty-three proteins present in the toxic Y. pseudotuberculosis MS fraction were either absent, differentially produced, or exhibited a slightly different pI compared with the nontoxic Y. pestis MS fraction (Fig. 2A). Mass spectrometry analysis of these identified 19 different proteins is given in Table S1.
Fig. 2.
Urease proteins are present in the toxic subfraction of Y. pseudotuberculosis. (A) Silver-stained 2-DE gels of the membrane supernatant subfraction of Y. pseudotuberculosis and Y. pestis. The numbered spots indicate proteins that were identified as being absent, differentially produced, or with a different pI in Y. pestis compared with Y. pseudotuberculosis profile; boxed numbers indicate urease subunits (Table S1). Molecular mass standards are shown on the left. (B) Organization of the urease cluster in Y. pseudotuberculosis (Y. pstb) IP32953, Y. pestis KIM, and Y. enterocolitica (Y. ent) 8081. The urease locus has a conserved organization in the three species and contains all seven genes that encode the structural (UreABC) and accessory (UreEFGD) proteins of the multimeric urease enzyme. The ureD gene in Y. pestis is a pseudogene (*). The predicted number of amino acids for each protein is noted above the corresponding gene. The positions of primers used for mutagenesis and complementation are indicated by numbered black triangles (identified in Table S2).
Each of the 19 differentially produced Y. pseudotuberculosis proteins had a highly similar homolog (99–100% identity) in Y. pestis. Notably, 4 of the 19 proteins (UreB, UreC, UreD, and UreG) are structural and accessory components of the multimeric urease enzyme encoded by the ureABCEFGD operon (Table S1 and Fig. 2B). The ureD homolog in Y. pestis is a pseudogene owing to a frameshift mutation that introduces a premature stop codon (17). Because of this mutation, Y. pestis strains are phenotypically urease-negative, whereas Y. pseudotuberculosis is urease-positive and able to degrade urea (18). Altogether these data suggest that the intact, functional ureD gene, and by extension the ureolytic phenotype of Y. pseudotuberculosis, might be involved in toxicity to fleas.
The Y. pseudotuberculosis Urease Operon Is Required for Toxicity to Fleas.
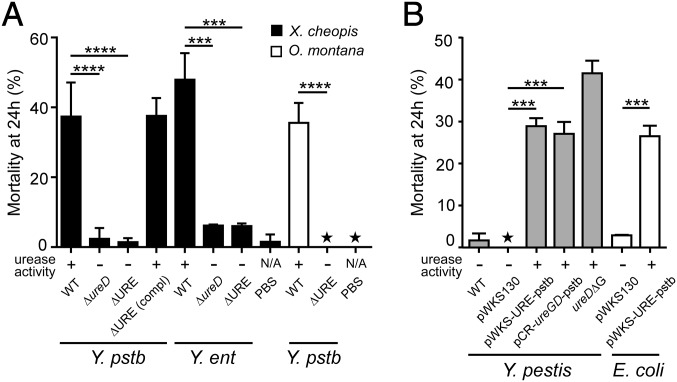
To examine the contribution of the Y. pseudotuberculosis urease in toxicity to fleas, Y. pseudotuberculosis IP32953 strains deleted of the ureD gene (IPΔureD) or of the entire urease cluster (IPΔURE) were constructed. Both mutations completely eliminated the ability of Y. pseudotuberculosis to hydrolyze urea (Fig. 3A). Fleas fed on blood containing the mutant strains did not exhibit any sign of toxicity immediately after feeding or increased mortality after 24 h (Fig. 3A). Identical results were seen for infections of X. cheopis and of Oropsylla montana, an unrelated North American ground squirrel flea species also susceptible to Y. pseudotuberculosis toxicity (Fig. 3A). Y. enterocolitica, which like Y. pseudotuberculosis is orally toxic to fleas (16), was also rendered nontoxic by deletion of the URE operon or of ureD (Fig. 3A). When the Y. pseudotuberculosis URE deletion mutant was complemented with pWKS-UREpstb, a low-copy plasmid containing a wild-type copy of the urease locus and native promoter region, toxicity to fleas was restored, with a mortality rate equivalent to that obtained with the wild-type strain (Fig. 3A). These results show that Y. pseudotuberculosis urease is responsible for the acute oral toxicity and high lethality to fleas.
Fig. 3.
Yersinia urease is responsible for toxicity to X. cheopis and O. montana fleas. (A) Y. pseudotuberculosis and Y. enterocolitica urease mutants are not toxic to fleas. Mortality of fleas 24 h after feeding on blood containing wild-type, ΔureD, ΔURE, or complemented ΔURE (pWKS-UREpstb) strains of Y. pseudotuberuclosis IP32953 (Y. pstb) or Y. enterocolitica 8081 (Y. ent). (B) Reactivation of the silenced urease activity in Y. pestis and expression of Y. pseudotuberculosis urease in Y. pestis and E. coli lead to flea toxicity. Mortality of fleas 24 h after feeding on blood containing wild-type Y. pestis KIM6+ with or without the empty cloning vector (WT, pWKS130), KIM6+ expressing the Y. pstb urease cluster (pWKS-UREpstb), or the Y. pstb ureG-D genes (pCR-ureGDpstb); KIM6+ in which the ureD pseudogene was repaired (ureDΔG); or E. coli containing the empty cloning vector (pWKS130) or expressing the Y. pstb IP32953 urease operon (pWKS-UREpstb). Data are the mean ± SD from a minimum of two experiments except for the strain E. coli (pWKS130) for which only one experiment was realized. ***P < 0.001, ****P < 0.0001, ★the value of both replicates was zero. The result of the in vitro urease test (+ or – phenotype after growth on urea agar) for each bacterial strain is indicated. N/A, not applicable.
Y. pestis Becomes Toxic to Fleas When Urease Activity Is Restored.
Y. pestis and most Escherichia coli strains are nonureolytic and are not orally toxic to X. cheopis fleas (16). To investigate the effect of gain of urease function in these urease-negative strains on fleas, we introduced the pWKS-UREpstb plasmid into Y. pestis KIM6+ and E. coli TOP10. Transformants were able to hydrolyze urea, demonstrating that the Y. pseudotuberculosis URE operon was expressed in E. coli and Y. pestis (Fig. 3B). Because the lack of urease activity in Y. pestis is due to a nonfunctional ureD gene (17), we also introduced the plasmid pCR-ureGDpstb (containing the ureG and ureD genes of Y. pseudotuberculosis) into Y. pestis KIM6+. KIM6+(pCR-ureGDpstb) transformants were ureolytic, demonstrating that the introduction of a functional ureD gene in Y. pestis is sufficient to restore ureolytic activity. The frameshift mutation in Y. pestis that results in a truncated, nonfunctional UreD is due to the insertion of a single guanine residue in a 6-bp poly(G) tract in ureD (17). As expected, site-specific mutation of wild-type Y. pestis that deleted one G residue from the ureD pseudogene, generating the Y. pestis ureDΔG strain, restored the ability to hydrolyze urea (Fig. 3B). Fleas fed on blood containing the urease-positive Y. pestis KIM6+(pWKS-UREpstb), KIM6+(pCR-ureGDpstb), KIM6+ureDΔG, or E. coli (pWKS-UREpstb) strains all experienced excess mortality (27–41%) compared with their urease-negative parent strains (0–1.7%) (Fig. 3B). These results demonstrate that the reactivation of urease activity in Y. pestis restores its ancestral toxicity to fleas and that expression of the URE cluster of Y. pseudotuberculosis in heterologous strains also results in toxicity.
Urease Enzymatic Activity Is Required for Oral Toxicity to Fleas.
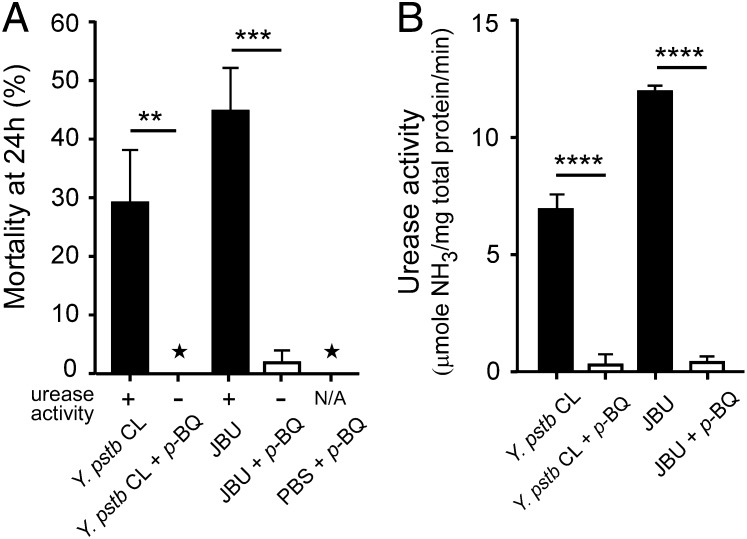
To evaluate the toxicity of urease itself, we fed fleas on a blood meal containing purified jack bean urease (JBU). Immediately after the 1-h feeding period ∼60% of the fleas were moribund and exhibited signs of acute toxicity including diarrhea, as observed after infection with Y. pseudotuberculosis, and the corresponding flea mortality at 24 h was comparable to that observed after an infection with Y. pseudotuberculosis (∼45%, Fig. 4A).
Fig. 4.
Toxicity to fleas is dependent on urease enzymatic activity. (A) Mortality of fleas that fed on blood containing Y. pseudotuberculosis cell lysate (CL) or JBU with or without the urease inhibitor p-Bq. Control fleas were fed on blood containing PBS buffer and p-Bq. Data are means ± SD of two independent experiments. (B) Inhibition of urease enzymatic activity by p-Bq in the samples added to the flea blood meals was verified by using a quantitative urease assay. Data are means ± SD of three independent experiments. **P < 0.01. ***P < 0.001, ****P < 0.0001, ★the value of both replicates was zero.
To show that the enzymatic activity of the urease proteins is required for their insecticidal properties, a Y. pseudotuberculosis cell lysate and a suspension of JBU were treated with the urease inhibitor para-benzoquinone (p-Bq) and then bioassayed in the flea model. Interestingly, in both p-Bq–treated samples the toxic effect was abolished along with the ureolytic activity (Fig. 4 A and B). This result clearly indicates that the toxic activity of Y. pseudotuberculosis and JBU toward X. cheopis is dependent on their ureolytic activity.
Restoration of Urease Activity to Y. pestis Does Not Affect the Flea Infection Phenotype.
Because oral toxicity eliminates many otherwise competent vectors, loss of urease was presumably advantageous to the evolution of the flea-borne transmission. To determine whether urease activity also positively or negatively affects survival within the flea or the ability to produce a transmissible infection, we fed X. cheopis fleas on blood containing KIM6+ (∼2.2 × 108 cfu/mL) or urease-positive KIM6+ ureDΔG (∼5 × 108 cfu/mL). Fed fleas that survived after 24 h were then monitored for mortality, infection rate, and proventricular blockage for 4 wk. Blockage was observed in ∼36% of the fleas infected with wild-type KIM6+ (Fig. 5A), consistent with data previously reported (19). Fleas infected with KIM6+ureDΔG had a slightly lower blockage rate than fleas infected with KIM6+, but the difference was not significant (Fig. 5A). The average infection rate and bacterial load achieved in the fleas during the 4-wk period after infection was similar for both strains (Fig. 5 B and C). Thus, loss of urease activity in Y. pestis did not interfere with the establishment of a transmissible infection in the plague vector.
Fig. 5.
Restoration of urease activity in Y. pestis does not affect infection and blockage rates in fleas that survive the initial acute toxicity. (A) Cumulative blockage rate 4 wk after infection with wild-type Y. pestis KIM6+ or the urease-positive mutant Y. pestis ureDΔG. At least 110 fleas were analyzed per sample. (B) Percentage of fleas infected initially and after 4 wk. (C) Mean bacterial load (cfu) in infected fleas at different times after the infectious blood meal (15–20 fleas per strain and per trial). Data are the mean ± SEM of two trials. NS, nonsignificant difference.
Discussion
Y. pseudotuberculosis, unlike Y. pestis, causes acute oral toxicity to X. cheopis, and previous work determined the toxin to be a protein that is cell-associated rather than secreted (16). Therefore, we used subcellular fractionation and comparative proteomics to identify it. Toxic activity was present mainly in the membrane fraction of Y. pseudotuberculosis lysates, which were found to contain four urease proteins absent or less abundant in the Y. pestis membrane fraction. Urease is a metalloenzyme that catalyzes the hydrolysis of urea to CO2 and two molecules of ammonia, a reaction dependent on the Ni2+-containing active site of the holoenzyme (20). In Yersiniae, the urease cluster is composed of seven genes (21). The ureA, ureB, and ureC genes encode structural subunits and ureE, ureF, ureG, and ureD encode accessory proteins required for the incorporation of nickel ions into the catalytic site. The presence of the urease subunits in the membrane fraction was somewhat surprising because bacterial urease subunits do not contain a signal peptide sequence and are predicted to be cytoplasmic proteins. However, they were easily dissociated into a membrane supernatant subfraction, indicating that they were not integral membrane proteins. Peripheral association of cytoplasmic proteins, including urease, with the membrane is not infrequent in bacteria (22).
It has long been known that all pathogenic Yersinia species except Y. pestis are able to hydrolyze urea (18). The absence of ureolytic activity in Y. pestis is due to a frameshift mutation in ureD resulting from the insertion of an additional G residue in a poly(G) stretch, which introduces a premature stop codon predicted to result in a C-terminal truncation of 54 aa (17). In Klebsiella aerogenes and Heliocbacter pylori, the UreD ortholog is required for recruitment of the Ni2+-carrying UreG and subsequent urease activation, and K. aerogenes ureD deletion mutants, including one that results in a C-terminal truncation, fail to generate active urease (23, 24). Thus, the UreD truncation in Y. pestis likely precludes assembly of the urease activation complex and enzyme activation. Normal localization to the membrane is presumably also affected, accounting for the underrepresentation of all of the urease subunits in 2D gels of the membrane fraction of Y. pestis (Fig. 2A). These phenotypic differences and the correlation between the presence of urease proteins and flea toxicity prompted us to evaluate the role of the urease genes. Our results show that the oral toxicity of Y. pseudotuberculosis and Y. enterocolitica is completely attributable to urease enzymatic activity. Mutational loss of urease activity eliminated oxicity of these two species to fleas; conversely, site-specific repair of the Y. pestis ureD pseudogene and introduction of the Y. pseudotuberculosis urease genes into E. coli was sufficient to make them urease-positive and toxic to fleas (Fig. 3). Toxicity of Y. pseudotuberculosis urease to fleas was not restricted to the rat flea X. cheopis. The bacteria were equivalently toxic to O. montana fleas, a primary vector of plague in North America. Thus, urease-based toxicity pertains to fleas in general and is not something peculiar to X. cheopis.
Urease has been isolated from bacteria, plants, and fungi (25). We found that JBU was also orally toxic to fleas, and that specific inhibition of catalytic activity by the urease inhibitor p-Bq abolished toxicity of both Y. pseudotuberculosis and JBUs. These results show that it is urease enzymatic activity per se, and not some cryptic activity of any of the seven urease proteins, that is responsible for toxicity. Presumably, ammonia released from hydrolysis of urea present in the blood meal reaches cytotoxic levels and is responsible for flea morbidity and mortality. Mice and rats have high blood urea nitrogen levels (2–3 mg/mL). Interestingly, jack bean and soybean ureases have been shown previously to have toxic effects when fed to insects (26, 27). However, insect toxicity of these plant ureases has nothing to do with enzymatic activity, but is due to a 10-kDa toxic peptide fragment released from the urease protein by insect cathepsin-like proteases. A functional urease catalytic site is not required, and only insects that rely on cathepsins B and D as their main digestive enzymes are susceptible (27, 28). In contrast, insects with trypsin-based digestion are not susceptible to plant urease toxicity (27). Fleas probably rely on trypsin-like enzymes to digest their blood meal, because they are the most abundant digestive enzymes of blood-sucking insects (29). Furthermore, bacterial ureases do not contain the 10-kDa entomotoxic peptide sequence responsible for the insecticidal properties of plant ureases (27). The urease of Bacillus thuringiensis has been implicated in an insect larva infection cycle (30). However, in contract to Yersinia urease and fleas, the Bacillus urease is not toxic to these larvae and actually enhances the B. thuringiensis-larva transmission cycle.
Bacterial ureases have been implicated in virulence because their activity protects the bacteria from acidic environments encountered in the host, such as during passage through the stomach (31). For example, urease is required for virulence of Y. enterocolitica (but not Y. pseudotuberculosis) by oral transmission (32, 33). Urease activity was lost very early in the evolutionary divergence from the fecal–oral to the arthropod-borne transmission route. Comparison of ureD in all Y. pseudotuberculosis and Y. pestis strains for which sequence is available shows that none of the Y. pseudotuberculosis genes, but all of the Y. pestis genes, even those of the phylogenetically most ancient Pestoides group, have the additional G in the (G)6 tract between nucleotides 799–805 (4). All Y. pestis ureD sequences also contain six additional identical single nucleotide changes from the Y. pseudotuberculosis IP32953 ureD. Finding the identical ureD pseudogene allele in every isolate from all major phylogenetic branches indicates that the mutation arose and was fixed in the ancestral Y. pestis strain, rather than by homoplasy and convergent evolution (repeated identical mutation in independent phylogenetic branches). The evolutionary loss of urease activity in Y. pestis did not affect virulence or pathogenesis of bubonic plague (17) or the ability to infect or block fleas (Fig. 4). Thus, urease is beneficial for an enteric pathogen and neutral for Y. pestis in the mammal but represented an impediment to the change to the new flea-borne route of transmission because it causes high mortality to the vector upon which it depends. We propose that loss of urease activity was an important step in the evolution of Y. pestis because of its positive effect on vector viability and the probability of transmission from an infected to an uninfected host. This step in the transition to flea-borne transmission is an example of how loss-of-function mutations can be as important to the evolutionary process as gain of function (34).
Fleas can transmit Y. pestis the very next time they feed (early-phase transmission), or after the bacteria multiply and form sufficient biofilm in the flea proventriculus to disrupt blood flow during feeding. Transmission by either mechanism is not very efficient; accordingly, the flea density per host (flea index) required to sustain stable enzootic transmission cycles is relatively high (35, 36). For enzootic maintenance, Y. pestis must be transmitted from an infected host to at least one new host (R0 = 1), and this probability is directly proportional to the number of infective fleas that survive long enough after the infectious blood meal to feed on a naïve host (35). Because urease-related toxicity reduces this number by 30–40%, epidemiologic modeling of plague transmission dynamic (35) indicates that the flea burden required to sustain the transmission cycle would double for a urease-positive clone (Supporting Information and Table S3). This value is greater than the flea densities usually associated with rodents, although the flea index fluctuates seasonally and is highly variable for different flea species and their hosts (35, 37, 38). Thus, transmission probability of a urease-positive Y. pestis would be less likely to reach the prerequisite R0 ≥1 threshold that is essential for enzootic maintenance.
Because it significantly enhanced transmissibility, loss of urease activity was likely subject to strong Darwinian (positive) selection during the early adaptation of Y. pestis to the new transmission route. The selective pressure apparently continues to this day: The mutated ureD allele is present in all Y. pestis isolates worldwide, representing all major phylogenetic branches, despite the fact that this type of mutation (the insertion or deletion of one nucleotide in a homopolymeric run via slipped-strand mispairing) has an inherently high reversion rate to wild type (17, 39). The population genetics evidence and our finding that urease is counterproductive to flea-borne transmission suggests that loss of ureD was not neutral, and that any urease-positive revertants are at such a fitness disadvantage that they are eliminated from the population.
Materials and Methods
Bacterial Strains, Plasmids, and Culture Conditions.
Bacterial strains and plasmids used in this study are listed in Table S4. Mutant strains were generated by standard molecular biology methods that are detailed in Supporting Information. All bacterial strains were routinely cultivated in brain heart infusion (BHI) broth or agar plates at 28 °C (Y. pestis) or 37 °C (Y. pseudotuberculosis and E. coli) unless otherwise stated. When necessary, the media were supplemented with 50 μg/mL kanamycin (Km), 100 μg/mL carbenicillin, or 25 μg/mL chloramphenicol.
Bacterial Cell Lysate and Bacterial Subfractionation.
Yersinia cell lysates were prepared from 200 to 500 mL of overnight 37 °C BHI cultures. The centrifuged cell pellet was washed in PBS and resuspended in 4 mL of PBS and the cells were lysed by French press. Undisrupted bacteria were pelleted by centrifugation at 5,000 × g for 5 min. Periplasmic, cytoplasmic, and membrane fractions were collected as previously described (40). The periplasmic fraction was concentrated to a final volume of ∼1 mL using Amicon ultracentrifugal filters with a 3-kDa cutoff (Millipore). The membrane fraction pellet was washed in PBS, vigorously resuspended in 1 mL PBS, and further fractionated by three rounds of centrifugation at 4,000 × g for 5 min at 4 °C. The resulting supernatant and the final resuspended pellet were referred to as the MS and MP fractions, respectively. Protein concentration was determined with the Quant-iT Protein Assay Kit (Life Technologies).
2-DE.
The first-dimension electrophoresis was performed as follows: 13-cm immobilized 3-11 nonlinear pH gradient strips (GE Healthcare Biosciences) were rehydrated in 250 μL rehydration buffer [7 M Urea, 2 M thiourea, 4% (wt/vol) CHAPS, 1% Triton X-100, 100 mM DTT, 0.5–1% carrier ampholytes (IPGphor Buffer 3-11), and 0.0002% bromophenol blue] containing 75 μg protein of the membrane supernatant fraction from Y. pseudotuberculosis IP32953 or Y. pestis KIM6+ for 12 h at 20 V in an Ethan IPGphor apparatus (GE Healthcare Biosciences). Isoelectric focusing (IEF) was performed using the following parameters: 100 V for 2 h, 500 V for 1 h, 1,000 V for 1 h, 2,000 V for 2 h, 4,000 V for 2 h, 6,000 V for 2 h, and 8,000 V for 8 h. After IEF, reduction and alkylation of thiol groups was performed by immersing strips for 15 min in equilibration buffer [50 mM Tris⋅HCl (pH 8.0), 6 M urea, 30% (vol/vol) glycerol, and 0.002% bromophenol blue] containing 10 mg/mL DTT and 27 mg/mL of iodoacetamide (Sigma-Aldrich). Equilibrated strips were placed horizontally on top of 8–16% Tris-glycine gradient gels (Jules, Inc.). The second-dimension electrophoresis was performed at 200 V for ∼5.5 h. Gels were fixed for 1 h in 40% (vol/vol) methanol and 10% (vol/vol) acetic acid and stained (SilverQuest silverstain kit; Life Technologies). After staining, the gels were conserved at 4 °C in ultrapure water until analysis. Protein spots of interest were excised by hand and sent to the Protein Chemistry section of the Research Technology Branch, National Institute of Allergy and Infectious Diseases, National Institutes of Health for protein identification.
Mass Spectrometry and Protein Identification.
Identification of 2-DE–separated proteins was performed on reduced and alkylated, trypsin-digested samples prepared by standard mass spectrometry protocols. The supernatant and two washes [5% (vol/vol) formic acid in 50% (vol/vol) acetonitrile] of the gel digests were pooled and concentrated by speed vac (Labconco) to dryness in 200-μL polypropylene auto-sampler vials (Sun Sri). The recovered peptides were resuspended in 5 μL of solvent A [0.1% formic acid, 2% (vol/vol) acetonitrile, and 97.9% (vol/vol) water] and analyzed as previously described (41).
Amino acid sequence data were searched against the Uniprot KB/TrEMBl database (May 2012) with an “other bacteria taxonomy filter” and an automatic decoy database search. Proteins were identified using a 1% false discovery rate cutoff and two peptide per protein minimums. For final protein identification the preferred taxonomy was set to Y. pseudotuberculosis strain IP32953 (NCBI NC_006155.1).
Flea Infections.
Approximately three hundred Xenopsylla cheopis fleas were fed for 1 h on heparinized mouse blood or defibrinated rat blood (Bioreclamation) containing live bacteria (∼2 × 108 to ∼2 × 109 cfu/mL), bacterial whole-cell lysates (8.33 mg/mL of total protein), bacterial cell fractions (2.67–5.67 mg/mL of each fraction), or 0.83 mg/mL of JBU type C3 (Sigma-Aldrich), by using an artificial feeding system (19). When the cell lysates and JBU were pretreated with urease inhibitor, the samples were incubated with 250 μM of p-Bq (Sigma-Aldrich), a urease reversible inhibitor (42), for 16–24 h at 4 °C prior to the flea feeding. Control fleas were fed on blood alone or blood containing PBS supplemented with 250 μM of p-Bq. Fleas that took a blood meal were kept at 75% relative humidity and 21 °C. Morbidity (fleas experiencing diarrhea, lying on their sides, and barely moving) was recorded immediately after feeding and mortality was recorded 24 h after the feeding period. Significant differences in mortality were determined by one-way analysis of variance followed by the post hoc Bonferroni test for multiple comparisons.
Blockage of the flea proventriculus was assessed as previously described (12, 19). The data were analyzed by using two-tailed Fisher’s exact test. A sample of 20 fed fleas was collected immediately after infection and at 1 and 28 d after infection and subsequently used for cfu plate count to determine the infection rate and average infectious dose per flea (19). Data were compared by Student t test.
Urease Tests.
Qualitative urease activity was assessed by plating bacteria on Christensen’s urea agar (43) or by mixing 200 μL of an overnight bacterial culture with 2 mL of Stuart’s urea broth (44) and observing the tubes and plates for a color reaction after an overnight incubation at 37 °C.
Urease activity was determined quantitatively by measuring the amount of ammonia released from the urea in the phenol-hypochlorite urease assay (45) adapted for a reaction volume of 200 μL. One microgram of protein from bacterial cell lysates or JBU type C3 (Sigma-Aldrich) in PBS was mixed with 50 mM urea. In some experiments, the cell lysates and JBU were pretreated with 250 μM of p-Bq for 16–24 h at 4 °C. The mixture was incubated for 15 min in a 96-well plate at 21 °C (flea temperature). After the incubation period, 2.30 min after addition of 80 μL phenol nitroprusside and 80 μL alkaline hypochlorite the OD595 was measured in a Synergy 2 multidetection microplate reader using the Gen5 version 1.10 software (Biotek Instruments, Inc.). Urease activity was determined from an NH4Cl standard curve (50–1,000 μM) prepared simultaneously with each assay. Urease activity was expressed as micromoles of NH3 per microgram of protein in cell lysate per minute.
Assays were performed at least three times to determine the rate of ammonia released by minute. Data were analyzed using one-way ANOVA followed by the post hoc Bonferroni test for multiple comparisons.
Supplementary Material
Acknowledgments
We thank Lisa Olano, Raynaldo Martin, and Carl Hammer [Protein Chemistry Section, Research Technologies Branch, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health] for mass spectrometry analysis and protein characterization. We thank Clayton Jarrett for his assistance with X. cheopis and O. montana fleas. We thank Christopher Bosio, James Carroll, Clayton Jarrett, Jeffrey Shannon, Justin Spinner, and Philip Stewart for critical review of the manuscript. This research was supported by the Intramural Research Program of the NIAID, National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 18409.
This article is a PNAS Direct Submission. M.A. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1413209111/-/DCSupplemental.
References
- 1.Achtman M, et al. Microevolution and history of the plague bacillus, Yersinia pestis. Proc Natl Acad Sci USA. 2004;101(51):17837–17842. doi: 10.1073/pnas.0408026101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Achtman M, et al. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc Natl Acad Sci USA. 1999;96(24):14043–14048. doi: 10.1073/pnas.96.24.14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chain PS, et al. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc Natl Acad Sci USA. 2004;101(38):13826–13831. doi: 10.1073/pnas.0404012101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui Y, et al. Historical variations in mutation rate in an epidemic pathogen, Yersinia pestis. Proc Natl Acad Sci USA. 2013;110(2):577–582. doi: 10.1073/pnas.1205750110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perry RD, Fetherston JD. Yersinia pestis—etiologic agent of plague. Clin Microbiol Rev. 1997;10(1):35–66. doi: 10.1128/cmr.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smego RA, Frean J, Koornhof HJ. Yersiniosis I: Microbiological and clinicoepidemiological aspects of plague and non-plague Yersinia infections. Eur J Clin Microbiol Infect Dis. 1999;18(1):1–15. doi: 10.1007/s100960050219. [DOI] [PubMed] [Google Scholar]
- 7.Sun YC, Jarrett CO, Bosio CF, Hinnebusch BJ. Retracing the evolutionary path that led to flea-borne transmission of Yersinia pestis. Cell Host Microbe. 2014;15(5):578–586. doi: 10.1016/j.chom.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hinnebusch BJ, et al. Role of Yersinia murine toxin in survival of Yersinia pestis in the midgut of the flea vector. Science. 2002;296(5568):733–735. doi: 10.1126/science.1069972. [DOI] [PubMed] [Google Scholar]
- 9.Hinnebusch BJ, Fischer ER, Schwan TG. Evaluation of the role of the Yersinia pestis plasminogen activator and other plasmid-encoded factors in temperature-dependent blockage of the flea. J Infect Dis. 1998;178(5):1406–1415. doi: 10.1086/314456. [DOI] [PubMed] [Google Scholar]
- 10.Sebbane F, Jarrett CO, Gardner D, Long D, Hinnebusch BJ. Role of the Yersinia pestis plasminogen activator in the incidence of distinct septicemic and bubonic forms of flea-borne plague. Proc Natl Acad Sci USA. 2006;103(14):5526–5530. doi: 10.1073/pnas.0509544103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sodeinde OA, Sample AK, Brubaker RR, Goguen JD. Plasminogen activator/coagulase gene of Yersinia pestis is responsible for degradation of plasmid-encoded outer membrane proteins. Infect Immun. 1988;56(10):2749–2752. doi: 10.1128/iai.56.10.2749-2752.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jarrett CO, et al. Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J Infect Dis. 2004;190(4):783–792. doi: 10.1086/422695. [DOI] [PubMed] [Google Scholar]
- 13.Bacot AW, Martin CJ. Observations on the mechanism of the tranmission of plague by fleas. J Hygiene. 1914;13(Plague Suppl 3):423–439. [PMC free article] [PubMed] [Google Scholar]
- 14.Erickson DL, Jarrett CO, Callison JA, Fischer ER, Hinnebusch BJ. Loss of a biofilm-inhibiting glycosyl hydrolase during the emergence of Yersinia pestis. J Bacteriol. 2008;190(24):8163–8170. doi: 10.1128/JB.01181-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun YC, Hinnebusch BJ, Darby C. Experimental evidence for negative selection in the evolution of a Yersinia pestis pseudogene. Proc Natl Acad Sci USA. 2008;105(23):8097–8101. doi: 10.1073/pnas.0803525105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erickson DL, et al. Acute oral toxicity of Yersinia pseudotuberculosis to fleas: Implications for the evolution of vector-borne transmission of plague. Cell Microbiol. 2007;9(11):2658–2666. doi: 10.1111/j.1462-5822.2007.00986.x. [DOI] [PubMed] [Google Scholar]
- 17.Sebbane F, Devalckenaere A, Foulon J, Carniel E, Simonet M. Silencing and reactivation of urease in Yersinia pestis is determined by one G residue at a specific position in the ureD gene. Infect Immun. 2001;69(1):170–176. doi: 10.1128/IAI.69.1.170-176.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mollaret HH, Nguyenvan BA, Vandekerkove M, Karimi Y, Eftekhari M. 1964. [ON THE UREASE OF YERSIN’S BACILLUS]. Ann Inst Pasteur (Paris) 107:424–429. French. [PubMed]
- 19.Hinnebusch BJ, Perry RD, Schwan TG. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science. 1996;273(5273):367–370. doi: 10.1126/science.273.5273.367. [DOI] [PubMed] [Google Scholar]
- 20.Carter EL, Flugga N, Boer JL, Mulrooney SB, Hausinger RP. Interplay of metal ions and urease. Metallomics. 2009;1(3):207–221. doi: 10.1039/b903311d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Koning-Ward TF, Robins-Browne RM. Analysis of the urease gene complex of members of the genus Yersinia. Gene. 1996;182(1–2):225–228. doi: 10.1016/s0378-1119(96)00556-2. [DOI] [PubMed] [Google Scholar]
- 22.Bode G, Malfertheiner P, Lehnhardt G, Nilius M, Ditschuneit H. Ultrastructural localization of urease of Helicobacter pylori. Med Microbiol Immunol (Berl) 1993;182(5):233–242. doi: 10.1007/BF00579622. [DOI] [PubMed] [Google Scholar]
- 23.Farrugia MA, Macomber L, Hausinger RP. Biosynthesis of the urease metallocenter. J Biol Chem. 2013;288(19):13178–13185. doi: 10.1074/jbc.R112.446526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee MH, Mulrooney SB, Renner MJ, Markowicz Y, Hausinger RP. Klebsiella aerogenes urease gene cluster: Sequence of ureD and demonstration that four accessory genes (ureD, ureE, ureF, and ureG) are involved in nickel metallocenter biosynthesis. J Bacteriol. 1992;174(13):4324–4330. doi: 10.1128/jb.174.13.4324-4330.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Follmer C. Insights into the role and structure of plant ureases. Phytochemistry. 2008;69(1):18–28. doi: 10.1016/j.phytochem.2007.06.034. [DOI] [PubMed] [Google Scholar]
- 26.Carlini CR, Oliveira AE, Azambuja P, Xavier-Filho J, Wells MA. Biological effects of canatoxin in different insect models: Evidence for a proteolytic activation of the toxin by insect cathepsinlike enzymes. J Econ Entomol. 1997;90(2):340–348. doi: 10.1093/jee/90.2.340. [DOI] [PubMed] [Google Scholar]
- 27.Follmer C, Real-Guerra R, Wasserman GE, Olivera-Severo D, Carlini CR. Jackbean, soybean and Bacillus pasteurii ureases: Biological effects unrelated to ureolytic activity. Eur J Biochem. 2004;271(7):1357–1363. doi: 10.1111/j.1432-1033.2004.04046.x. [DOI] [PubMed] [Google Scholar]
- 28.Ferreira-DaSilva CT, Gombarovits ME, Masuda H, Oliveira CM, Carlini CR. Proteolytic activation of canatoxin, a plant toxic protein, by insect cathepsin-like enzymes. Arch Insect Biochem Physiol. 2000;44(4):162–171. doi: 10.1002/1520-6327(200008)44:4<162::AID-ARCH3>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 29.Lehane M. The Biology of Blood-Sucking in Insects. Cambridge Univ Press; Cambridge, UK: 2005. [Google Scholar]
- 30.Martin PAW, Farrar RR, Jr, Blackburn MB. Survival of diverse Bacillus thuringiensis strains in gypsy moth (Lepidoptera: Lymantriidae) is correlated with urease production. Biol Control. 2009;51(1):147–151. [Google Scholar]
- 31.Ferrero RL, Lee A. The importance of urease in acid protection for the gastric-colonising bacteria Helicobacter pylori and Helicobacter felis sp. nov. Microb Ecol Health Dis. 1991;4(3):121–134. [Google Scholar]
- 32.De Koning-Ward TF, Robins-Browne RM. Contribution of urease to acid tolerance in Yersinia enterocolitica. Infect Immun. 1995;63(10):3790–3795. doi: 10.1128/iai.63.10.3790-3795.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riot B, Berche P, Simonet M. Urease is not involved in the virulence of Yersinia pseudotuberculosis in mice. Infect Immun. 1997;65(5):1985–1990. doi: 10.1128/iai.65.5.1985-1990.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bliven KA, Fisher DJ, Maurelli AT. Characterization of the activity and expression of arginine decarboxylase in human and animal Chlamydia pathogens. FEMS Microbiol Lett. 2012;337(2):140–146. doi: 10.1111/1574-6968.12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lorange EA, Race BL, Sebbane F, Hinnebusch BJ. Poor vector competence of fleas and the evolution of hypervirulence in Yersinia pestis. J Infect Dis. 2005;191(11):1907–1912. doi: 10.1086/429931. [DOI] [PubMed] [Google Scholar]
- 36.Webb CT, Brooks CP, Gage KL, Antolin MF. Classic flea-borne transmission does not drive plague epizootics in prairie dogs. Proc Natl Acad Sci USA. 2006;103(16):6236–6241. doi: 10.1073/pnas.0510090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laudisoit A, Leirs H, Makundi R, Krasnov BR. Seasonal and habitat dependence of fleas parasitic on small mammals in Tanzania. Integr Zool. 2009;4(2):196–212. doi: 10.1111/j.1749-4877.2009.00150.x. [DOI] [PubMed] [Google Scholar]
- 38.Eisen RJ, et al. Flea diversity as an element for persistence of plague bacteria in an East African plague focus. PLoS ONE. 2012;7(4):e35598. doi: 10.1371/journal.pone.0035598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gur-Arie R, et al. Simple sequence repeats in Escherichia coli: Abundance, distribution, composition, and polymorphism. Genome Res. 2000;10(1):62–71. [PMC free article] [PubMed] [Google Scholar]
- 40.Thein M, Sauer G, Paramasivam N, Grin I, Linke D. Efficient subfractionation of gram-negative bacteria for proteomics studies. J Proteome Res. 2010;9(12):6135–6147. doi: 10.1021/pr1002438. [DOI] [PubMed] [Google Scholar]
- 41.Stewart PE, et al. Characterization of the Bat proteins in the oxidative stress response of Leptospira biflexa. BMC Microbiol. 2012;12:290. doi: 10.1186/1471-2180-12-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kot M. Inhibition of jack bean urease by p-benzoquinone: Elucidation of the role of thiols and reversibility of the process. J Enzyme Inhib Med Chem. 2006;21(6):697–701. doi: 10.1080/14756360600889674. [DOI] [PubMed] [Google Scholar]
- 43.Christensen WB. Urea decomposition as a means of differentiating Proteus and paracolon cultures from each other and from Salmonella and Shigella types. J Bacteriol. 1946;52(4):461–466. doi: 10.1128/jb.52.4.461-466.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stuart CA, Van Stratum E, Rustigian R. Further studies on urease production by Proteus and related organisms. J Bacteriol. 1945;49(5):437–444. doi: 10.1128/jb.49.5.437-444.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sangari FJ, Seoane A, Rodríguez MC, Agüero J, García Lobo JM. Characterization of the urease operon of Brucella abortus and assessment of its role in virulence of the bacterium. Infect Immun. 2007;75(2):774–780. doi: 10.1128/IAI.01244-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.