Significance
Crop yields are dependent on the number of lateral primordia made by the inflorescence. In maize unbranched mutants, excess lateral primordia are made at the expense of the stem cells located in the center of the meristem. Ultimately, the unbranched mutant meristem lacks enough cells to regenerate and thus, terminates prematurely. This study shows that the duplicate transcription factors unbranched2 and unbranched3 function together to decrease the rate of lateral primordia initiation, thus giving the stem cells of the meristem enough time to regenerate. Variants of the unbranched3 gene affect different aspects of lateral primordia initiation that control crop yield.
Keywords: maize, meristem, yield, inflorescence, QTL
Abstract
The separation of male and female flowers in maize provides the potential for independent regulation of traits that affect crop productivity. For example, tassel branch number controls pollen abundance and length of shedding time, whereas ear row number directly affects kernel yield. Mutations in duplicate SBP-box transcription factor genes unbranched2 (ub2) and ub3 affect both of these yield traits. Double mutants display a decrease in tassel branch number and an increase in ear row number, both of which are enhanced by loss of a related gene called tasselsheath4 (tsh4). Furthermore, triple mutants have more tillers and leaves—phenotypes seen in Corngrass1 mutants that result from widespread repression of SBP-box genes. Immunolocalization of UB2 and UB3 proteins revealed accumulation throughout the meristem but absence from the central domain of the meristem where cells regenerate. Thus, ub2, ub3, and tsh4 function as redundant factors that limit the rate of cell differentiation to the lateral domains of meristems. When these genes are mutated, cells are allocated to lateral primordia at a higher rate, causing a net loss of cells from the central domain and premature termination of the inflorescence. The ub3 locus is tightly linked to quantitative trait loci (QTL) for ear row number and tassel branch number in both the nested association mapping (NAM) and intermated B73 by Mo17 (IBM) populations of maize recombinant inbreds, indicating that this gene may be agronomically important. Analysis of ear and tassel QTL across biparental families suggests that multiple mutations in ub3 independently regulate male and female inflorescence development.
Meristems are groups of totipotent cells responsible for forming all of the tissues and organs of plants throughout their lifecycle, and as such, they have a direct effect on crop yield. Unlike animal systems, where growth is determinate, plants display indeterminate growth and constantly regenerate cells to maintain their apical meristems. If the rate of lateral primordia initiation is unregulated, too many primordia initiate at the expense of the meristem. Hence, coordination is required between pathways that renew the apical meristem and deplete it through initiation of lateral primordia.
The rate of lateral organ initiation from the meristem is characterized by the plastochron index (1). A plastochron is defined as the amount of time between successive lateral organ initiation events. Several genes have been described that regulate plastochron in grasses. The first gene described in maize was terminal ear1 (te1), which encodes an MEI-2–like RNA-binding protein (2) as well as its ortholog in rice PLASTOCHRON2 (PLA2) (3). Both mutants initiate several extra leaves, even into the reproductive phase when leaf initiation is normally suppressed. The pla1 mutant in rice displays a similar phenotype. PLA1 encodes a cytochrome (P450) and is expressed in leaf bases and internodes but not meristems (4), similar to the expression of te1 in maize (2). The fact that these plastochron regulators affect meristem function and yet, are not expressed in the meristem indicates that they may function noncell-autonomously. This idea is supported by work in Arabidopsis, where it was shown that the SQUAMOSA PROMOTER BINDING (SBP)-box transcription factors SPL9 and SPL15 function noncell-autonomously to regulate plastochron index independent of the PLA1 pathway (5). This finding raises the intriguing possibility that SPB-box proteins or their downstream targets may act as mobile signals that travel to meristems to act as inhibitory factors and thereby, regulate the timing of leaf initiation.
Positional cloning of Corngrass1, a dominant phase-change mutant in maize, revealed that it overexpresses miR156, which represses SBP-box genes (6). In Arabidopsis, SBP-box genes have been shown to play a role in regulating the rate of leaf initiation (5), flowering time, and developmental timing (7). In maize, the SBP-box mutant tasselsheath4 (tsh4) showed that these transcription factors also play an important role in suppression of leaf development during the floral phase (8). Interestingly, derepression of floral leaves was linked to reduction of tassel branches in tsh4, raising the possibility that SBP-box genes control partitioning of cells between lateral organs vs. meristems. This hypothesis was supported by studies showing that branch meristem (BM) markers, such as ramosa2 (ra2) (9), are ectopically expressed in the derepressed leaves of tsh4 (10). Such results are consistent with the phytomer concept for plant development, in which leaf, axillary meristem, and internode all form from a common group of progenitor cells that must be directed to specific compartments (11). Thus, if plastochron regulators, such as SBP-box, factors are missing, cells are inappropriately directed to the wrong compartment at the expense of other phytomer components.
A reverse genetic analysis of genes similar to tsh4 revealed a pair of redundant genes [unbranched2 (ub2) and ub3] that functions in the specification of lateral primordia, such as leaves, tassel branches, and kernels. Immunolocalization using anti-UB3 serum showed expression throughout lateral domains of meristems but not in the central domain, where meristem renewal occurs. These results indicate that ub2 and ub3 function together with tsh4 to restrict initiation of lateral primordia, thus allowing cell renewal within the central domain of the meristem. The ub2 and ub3 mutant phenotypes affect several important agronomic properties, and genome-wide association study analysis implicates two independent natural variants at the ub3 locus with increased tassel branch number (TBN) and increased ear row number (ERN), respectively.
Results
ub2 And ub3 Regulate Primordia Initiation.
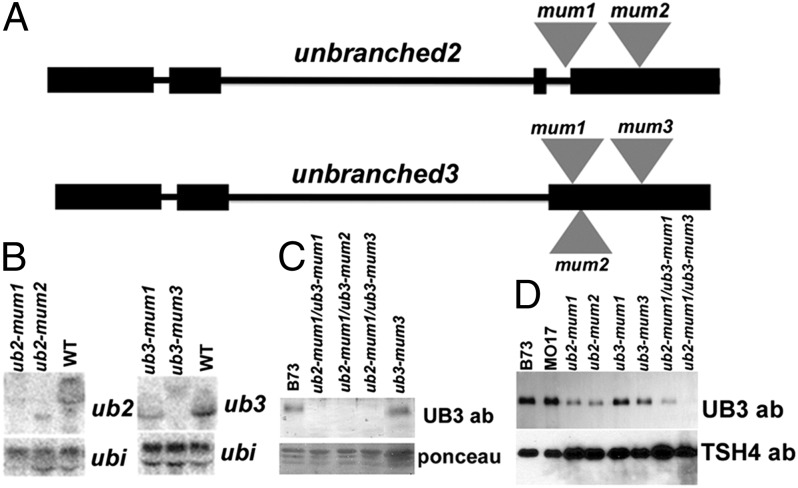
A reverse genetic analysis was performed for duplicate SBP-box genes that group with tsh4, a gene responsible for repression of bract primordia in maize inflorescences (8). These genes, formerly known as TC305612 and TC282500 (6), were renamed ub2 and ub3 based on their mutant phenotypes. UB2 and UB3 share 79% overall amino acid identity and 86.1% identity within the DNA-binding domains (Fig. S1). Phylogenetic analysis showed that ub2 and ub3 are duplicated loci in maize, with single-copy orthologs present in sorghum, rice, and Brachypodium (Fig. S2). The closest maize homolog to ub2 and ub3 is tsh4. Two independent Mutator (Mu) transposon insertions (12) into ub2 were identified: one in a 99-bp intron near the exon border and one into an exon. In addition, three ub3 insertions into the last exon of ub3 were also identified (Fig. 1A). From RNA gel blots, both ub2 alleles completely disrupt transcription, whereas the ub3-mum1 allele only partially reduces transcription (Fig. 1B).
Fig. 1.
Gene structure and expression of ub2 and ub3. (A) Positions of Mu transposon insertions in ub2 and ub3 genes. (B) RNA gel blot analysis of ub2 and ub3 alleles from 3.5-wk-old dissected shoot apices. The ubiquitin (ubi) gene was used as a loading control. (C) Western blot analysis using 3.5-wk-old nuclear extracts from inbred B73, ub2/ub3 double mutants, and ub3-mum3 single mutants. Ponceau protein staining was done as a loading control. (D) Western blot of nuclear extracts from 0.5-cm ear primordia of B73, Mo17, and ub2 and ub3 single and double mutants. The TSH4 antibody was used as a protein loading control.
To test whether these alleles make functional protein, an antibody was raised to full-length UB3 protein. Given the similarity of UB2 and UB3, the serum was predicted to detect both proteins. This result was confirmed by Western blots using nuclei from 3.5-wk-old shoots or 0.5-cm ear primordia, where accumulation was seen in WTs and single mutants but not several double-mutant combinations (Fig. 1 C and D). For example, a band was detected in ub3-mum3 single-mutant shoots and ears, although it lacks functional transcript. This band is likely to correspond to UB2 protein, because ub2-mum1/ub3-mum3 double mutants show no detectable protein (Fig. 1C). In ear tissue, however, the ub2-mum1/ub3-mum1 double mutant had a small amount of protein, indicating that ub3-mum1 is only a partial loss of function (Fig. 1D). Thus, the anti-UB3 antibody recognizes both UB2 and UB3 proteins, and the lack of any protein in ub2-mum1/ub3-mum2 and ub2-mum1/ub3-mum3 double-mutant combinations indicates that these alleles are likely loss-of-function null mutants.
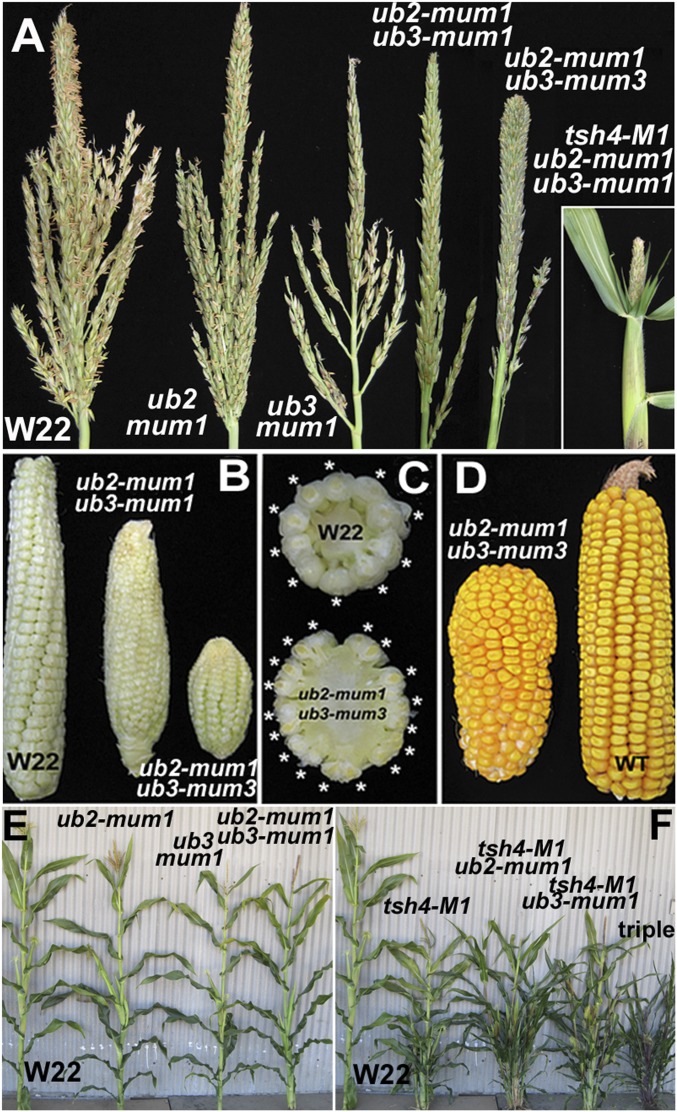
The ub2-mum1 and ub3-mum1 alleles were introgressed into the WT W22 background and observed for phenotypic differences relative to W22. Both ub2-mum1 and ub3-mum1 single mutants showed a modest but significant reduction in TBN, with the ub3-mum1 allele being slightly more severe (Fig. 2A and Fig. S3A). This decrease in TBN was greatly enhanced in three different double-mutant combinations (Fig. 2A and Fig. S3A). In addition, thickening appeared near the tip of the central spike of the tassel of the ub2-mum1/ub3-mum3 double mutant (Fig. 2A), indicating the presence of excess spikelets. A similar reduction in TBN was observed in single mutants of tsh4 (8). To determine whether tsh4 may function redundantly with ub2 and ub3, double- and triple-mutant combinations were made. This putative redundancy was confirmed by a reduction in TBN in two different double-mutant combinations. In addition, two different triple-mutant combinations completely lacked tassel branches (Fig. 2A and Fig. S3A).
Fig. 2.
Floral phenotypes of ub2 and ub3 mutants. (A) Tassel phenotypes of ub2 and ub3 mutant alleles; ub2-mum1, ub3-mum1, and the double mutant are in the W22 inbred background. Triple-mutant tassel in the W22 background is shown in Inset. (B) Ear phenotypes of W22 and double mutants. (C) Razor blade hand sections of the midpoint of the ears in C. Asterisks indicate kernel rows. (D) Fertilized ears of the double mutant compared with WT A619. (E) Mature field-grown plants of W22, ub2, ub3, and the double mutant. (F) Mature field-grown plants of tsh4 and double- and triple-mutant combinations in W22 background showing an increase in tillering.
Defects in ear length and diameter were observed in single and double mutants (Fig. 2 B–D). The most severe length defect was observed in the triple mutant that was approximately one-half that of WT (Fig. S3B), leading to greatly reduced fertility. Significant increases in ear diameter were seen in ub3 single and ub2/ub3 double mutants (Fig. S3C) that also displayed fasciated tips (Fig. 2 B and D). Sections through the midpoint of the double-mutant ears showed nearly two times the number of kernel rows in the double mutants (Fig. 2C). When fertilized, the tips of these ears were disorganized, and the kernels crowded together (Fig. 2D). Taken together, these phenotypes indicate that meristem function during the floral phase of male and female inflorescences is altered by the simultaneous loss of ub2 and ub3.
ub2 And ub3 Repress Tillering and Leaf Initiation Redundantly with tsh4.
Vegetative phenotypes of ub2 and ub3 single and double mutants were observed in the field. Although ub2/ub3 double mutants do not affect tiller number, tillering was significantly increased in all double-mutant combinations with tsh4 (Fig. 2 E and F), with the strongest increase observed in two different triple-mutant combinations (Fig. S4A).
Leaf number was also altered in the double and triple mutants. Because juvenile leaf number is difficult to assay in the field because of senescence and predation, leaf number above the upper most ear node was counted instead. Apart from ub2-mum1/ub3-mum1, which may not be a complete loss of function, double mutants made significantly more leaves, whereas triple mutants made up to two times as many upper leaves (Fig. S4B), mostly found at the base of the tassel (Fig. 2A). Thus, ub2/ub3/tsh4 functions together redundantly to initiate tassel branches and repress excess tiller and leaf initiation. Despite the increase in leaf number, the double and triple mutants flowered at the same time as the WT, suggesting that plastochron is, in fact, shortened in SBP-box mutants.
Lateral Primordia Form at the Expense of the Apical Meristem in Double Mutants.
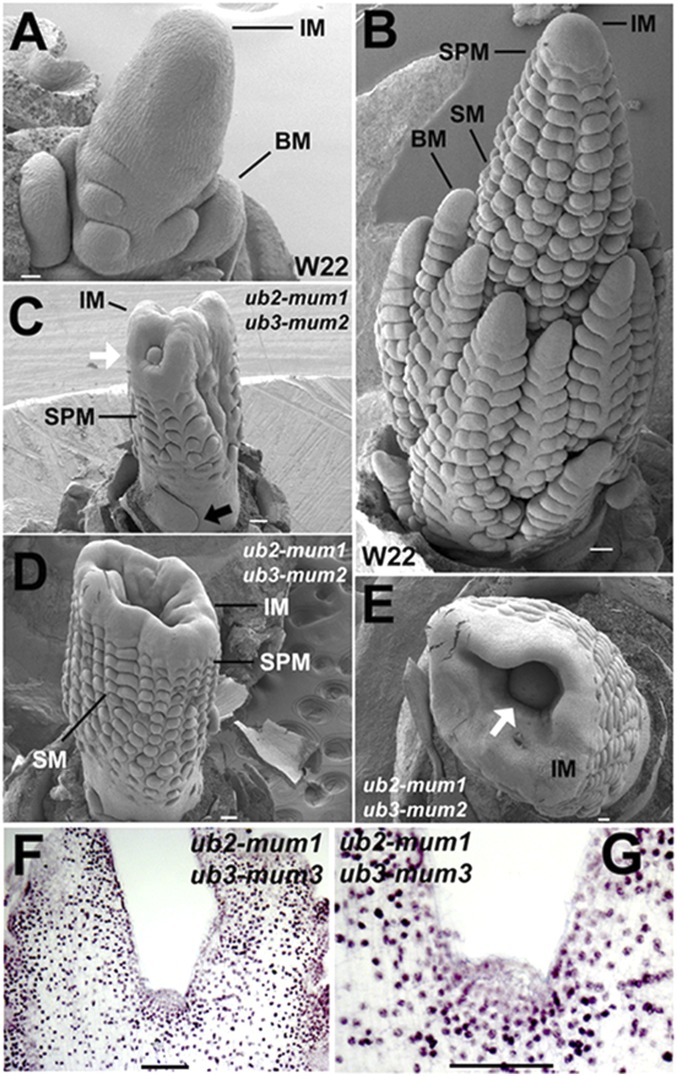
SEM was performed on the double mutants to determine the origin of the extra primordia in the inflorescence. In normal maize tassels, the main inflorescence meristem (IM) first initiates a limited number of branch meristems (BMs) that form the long branches of the tassel (Fig. 3A). Later, the IM initiates several rows of spikelet pair meristems (SPMs), each of which forms two spikelet meristems that ultimately form the flowers and kernels (Fig. 3B). The ub2/ub3 double mutants rarely initiate BMs and often have extra leaves at the base of the tassel instead (Fig. 3C). In addition, widening and fractionation of the IM are often seen at the tip (Fig. 3C). At later stages, the IM continues to widen, and many extra rows of SPMs initiate (Fig. 3D), often at the expense of the main IM that is either left as a sunken residual primordium (Fig. 3E) or completely consumed. Immunolocalization using antibodies to early BM markers, such as KNOTTED1 (KN1) (13–15) and RA2 (10), revealed no expression at the base of the tassel in the double mutant, supporting the observation that BMs do not initiate (Fig. S5 A–C).
Fig. 3.
SEMs and immunolocalization of KN1 in the ub2/ub3 double mutant. (A) Early W22 tassel showing IMs and BMs. (B) Older W22 tassel showing rows of SPMs and spikelet meristems (SMs). (C) Early ub2/ub3 double mutant tassel showing absence of BM and displaced residual IM (white arrow). Black arrow points to extra leaf primordium. (D) Older double mutant showing increased rows of SPM and widened IM. (E) Top view of same double mutant showing sunken residual IM in the center (arrow). (F) KN1 immunolocalization in residual IM of double-mutant tassel. (G) Close-up view of residual IM showing absence of KN1 and vacuolated cells. (Scale bars: A, 30 μm; B–E, 100 μm; F and G, 500 μm.)
We also used the KN1 antibody to follow the fate of the residual IM in the double mutant and determine whether it retained meristem identity. Recessive kn1 mutants are shootless in some backgrounds (16) or have smaller inflorescences with reduced numbers of lateral primordia in others (17). KN1 is found in all meristems, and it is down-regulated in lateral organs (13–15). KN1 immunolocalization experiments showed that the residual meristem of the double mutant has no KN1 expression (Fig. 3 F and G). Moreover, the central cells of the residual meristem appear vacuolated rather than densely cytoplasmic, further indicating that they have lost meristem identity and terminally differentiated (Fig. 3G). These results indicate that meristem maintenance and renewal are compromised in the ub2/ub3 double mutants.
Localization of UB2 and UB3 Proteins.
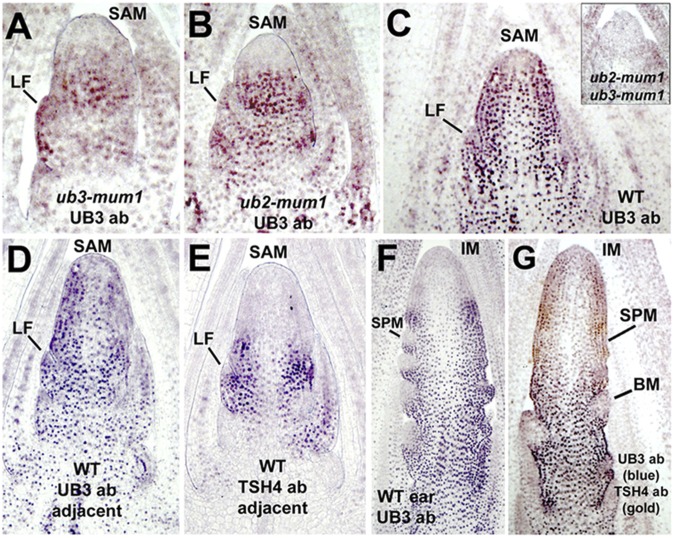
To determine where ub2 and ub3 are expressed, anti-UB3 serum was used for immunolocalization experiments in shoot and floral apices. Because the serum recognizes both UB2 and UB3 proteins, immunolocalization experiments were performed on ub3 and ub2 single mutants to determine the tissue-specific localization of UB2 and UB3 proteins, respectively (Fig. 4 A and B). In the shoot apex, UB2 protein was found in initiating leaf primordia and the base of the shoot apical meristem (SAM) but not in the meristem tip (Fig. 4A). UB3 protein localized in a similar pattern in SAMs of the same age (Fig. 4B). In WT shoot apices, both proteins were found everywhere in the SAM, the stem, and young leaf primordia but absent from the center of the meristem tip, where cell renewal normally occurs (Fig. 4C).
Fig. 4.
Immunolocalization of UB2 and UB3 proteins. (A) UB2 expression in 3-wk-old ub3-mum1 mutant shoot showing expression in leaves (LFs) and the stem. (B) UB3 expression in 3-wk-old ub2-mum1 mutant shoot showing a similar expression pattern. (C) UB2 and UB3 localization in 3.5-wk-old WT shoots showing absence of expression near the tip. Double-mutant negative control showing no localization is shown in Inset. (D) Four-week-old WT transition-stage shoot probed with anti-UB3 serum. (E) Section adjacent to D probed with anti-TSH4 serum. (F) UB2 and UB3 in ear primordia with initiating SPM. (G) UB2 and UB3 (blue) in young tassel initiating BM. The same tassel was also labeled with TSH4 protein (gold) to show complementary expression in SPM above the BZ.
To determine if this meristem expression pattern changes on flowering, florally determined transition-stage meristems were isolated and analyzed (Fig. 4D). Adjacent sections were probed with an anti-TSH4 antibody (8) for comparison (Fig. 4E). UB2 and UB3 proteins continue to be expressed throughout the transition-stage meristem, young leaves, and stem (Fig. 4D), although meristem tip expression is still restricted. In the adjacent section, TSH4 protein was found in a limited domain (Fig. 4E) in leaf primordia and older leaves but not in axillary buds or stems. The expression of TSH4 overlaps completely with UB2 and UB3, consistent with genetic redundancy between all three genes. In female inflorescence ears, UB2 and UB3 were excluded from the tip of the IM as well as the SPMs (Fig. 4F), although high expression was found in bracts subtending the SPM. In early-stage tassels, high UB2 and UB3 expression was found at the base of the tassel, where BMs form, but not in BMs themselves or the inflorescence tips (Fig. 4G). Double labeling with TSH4-specific antibodies showed localization in young bracts subtending the SPM primordia in the upper part of the tassel. At this stage, UB2 and UB3 were expressed in the portion of the tassel that defines the branch zone (BZ) but not the zone that makes SPMs. This complementary expression pattern was confirmed in double-labeling experiments with UB3 and TSH4 or RA2 in radial sections of tassels (Fig. S5 D and E). Thus, UB2 and UB3 are not expressed in the meristems that are affected in the double mutant, including IMs, SPMs, and BMs.
UB3 Maps to Quantitative Trait Loci for TBN and ERN.
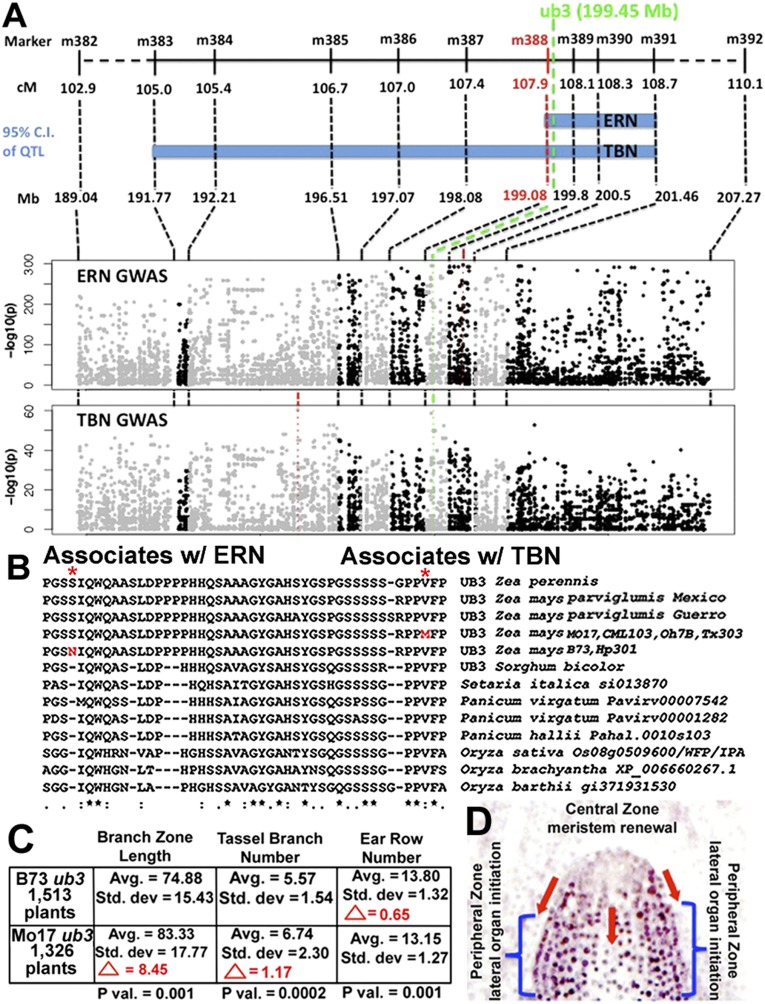
Quantitative trait loci (QTL) analysis of inflorescence traits was done using the nested association mapping (NAM) population. This population was made by crossing B73 with 25 different inbreds and then selfing to create recombinant inbred lines (18), and detected QTL for TBN, ERN, and BZ at the marker most closely linked to ub3 (19). The ub3-linked ERN QTL has the largest effect of any inflorescence QTL in the NAM population and explains 12% of the variance in ERN across the entire NAM population (Figs. S6 and S7). A larger set of HapMap2 SNPs has now become available for the parents of the NAM population (20). From an ∼18-Mb region encompassing the 95% confidence intervals of the ub3-linked ERN and TBN QTL on chromosome 4, we extracted a set of 8,851 polymorphic HapMap2 SNPs that contained no missing data and no heterozygous genotypes among 27 NAM founder lines, including Mo17, and tested each SNP for association with ERN and TBN (Materials and Methods and Fig. 5A). For each trait, we found separate, highly significant nonsynonymous substitutions within the third exon of ub3 downstream of the SBP domain. One variant (Val260Met; 199,457,430 bp) is found in four NAM founder lines (CML103, Mo17, Oh7B, and Tx303) and associated with increased TBN. Another variant (Ser220Asn; 199,457,549 bp) is found in two NAM founders (B73 and Hp301) and associated with increased ERN (Fig. 5B). For both TBN and ERN, genome-wide associations slightly more significant than the nonsynonymous substitution in ub3 were found within the QTL confidence interval (Fig. 5A). These SNPs could represent synthetic associations (21) resulting from either additional alleles at ub3 or closely linked QTL. No correlation was found between QTL effects for TBN and ERN in the ub3 region (Table S1), suggesting that independent mutations, rather than a single pleiotropic mutation, are responsible for the TBN and ERN QTL colocalization.
Fig. 5.
Allelic diversity in ub3 and its association with inflorescence variation with a model for function. (A) Physical and genetic positions of ub3 relative to marker–trait associations for ERN and TBN in the NAM population. The 95% confidence intervals (95% CIs) for ub3-linked ERN and TBN joint linkage QTL are shown in blue. The positions of the most significant markers/SNPs for joint linkage and genome-wide association study (GWAS) analyses of ERN and TBN are shown as red dots. The position of ub3 on each map is shown as green dots. The 95% CI for the ub3-linked ERN QTL extends 2.386 Mb and contains 44 predicted genes, whereas the 95% CI for the ub3-linked TBN QTL extends 9.695 Mb and contains 191 predicted genes. All physical positions refer to AGPv2.1. (B) Clustal alignments of UB3 from teosinte, maize, and other panicoid grasses. Polymorphisms are indicated by asterisks, and amino acid differences are in red. (C) Variation in average BZ length, branch number, and ERN in B73 vs. Mo17 populations of IBM lines. Differences between the two populations are highlighted in red. (D) Model for UB2/UB3 function. UB2/UB3 is not expressed in undifferentiated cells at the tip of the meristem, where renewal occurs. Red arrows signify UB2/UB3 control of cell movement out of the central zone into the peripheral zone, where cell differentiation and lateral organ initiation occur.
Another high-resolution mapping resource, intermated B73 by Mo17 (IBM), was made by crossing B73 with Mo17 and intermating the F1 progeny four times before selfing (22). Using a cleaved amplified polymorphic sequence (CAPS) marker that distinguishes the two polymorphisms in ub3, 167 families derived from the IBM population were genotyped as possessing either the B73 Asn220 allele or the Mo17 Met260 allele. These data were correlated with BZ length, branch number, and ERN scores (19) from 2,839 progeny of 167 genotyped families. Those families that received the Mo17 ub3 allele were found to have, on average, a BZ that was 8.45 mm longer with 1.17 more tassel branches but 0.65 fewer ear rows (Fig. 5C). Thus, the two polymorphisms seem to affect the male and female inflorescences differently: the B73 allele correlates with robust ear growth, whereas the Mo17 allele correlates with robust tassel growth. These results are consistent with the NAM associations. Neither polymorphism is present in teosinte, the progenitor of maize (Fig. 5B). Furthermore, the ub3 locus is not linked to any of the major QTL responsible for differences in inflorescence architecture between maize and teosinte (23). This finding suggests that natural variants in ub3 arose during the process of crop improvement rather than crop domestication.
Discussion
The number of lateral primordia made by the male and female inflorescences of maize is a major determinant of yield. In tassels, such primordia include tassel branches, spikelet pairs, and spikelets, all of which control the length of shedding time and pollen quantity. In ears, the number of spikelet pairs affects kernel yield. Because all lateral primordia are products of meristems, increasing the number of these primordia to improve yield requires alteration of meristem activity. We hypothesize that ub2 and ub3 control the rate at which cells leave the undifferentiated central zone of the meristem and enter the peripheral zone where lateral primordia initiate (Fig. 5D). In ub2 and ub3 double mutants, extra lateral primordia are made too quickly at the expense of the apical meristem, which lacks enough cells to regenerate (Fig. 3 D and E), loses expression of meristem markers, such as kn1 (Fig. 3 F and G), and terminates prematurely. The roles of these genes in regulating the rate of lateral primordia initiation are similar to the functions of SPL9 and SPL15 in vegetative shoots of Arabidopsis (24, 25), although tsh4, ub2, and ub3 primarily affect the floral phase of development.
UB2 and UB3 accumulate throughout the base of the meristem but are excluded from the central tip of the SAM, where cell renewal occurs. Because these SBP-box transcription factors are targets of miR156 (6), the lack of expression in the central tip of the meristem may be a consequence of microRNA repression. In support of this idea, tsh4, which is also targeted by miR156, is expressed in a pattern complementary to miR156 in the IMs (8). Recent reports show repression of miR156 by sugar (26, 27), which itself is a noncell-autonomous mobile signal. Because leaves are carbohydrate sources, perhaps the sugar that they produce travels to the peripheral zone of the meristem to repress microRNA expression and thus, allow ub2 and ub3 expression. It is possible that these sugars either do not travel far enough to the central tip of the SAM or are actively excluded from the central tip of the SAM, allowing miR156 to remain active and repress ub2 and ub3.
It is unclear how SBP-box transcription factors affect the rate of organ initiation without being expressed in the central zone. Classic experiments performed in potato showed that a diffusible substance travels between leaves to control the rate of leaf growth and initiation (28). More recently, it has been shown that leaf-specific promoters driving SPL9 expression in Arabidopsis were able to affect the rate of leaf initiation in the SAM (5). Such results are consistent with the hypothesis that SBP-box transcription factors or their target genes may function as nonautonomous signals that move through the shoot apex to regulate plastochron. Because we observed no sign of UB2 or UB3 protein throughout the IM, SPM, and spikelet meristem or in the apical tips of the vegetative meristem, it is more likely that a downstream factor in the SBP-box gene pathway is the noncell-autonomous signal rather than the SBP-box proteins themselves.
Loss-of-function mutations in ub2/ub3 doubles display seemingly contradictory phenotypes in male vs. female inflorescences with regard to lateral meristems. For example, double mutants initiate several extra rows of SPMs in ears and tassels. In tassels, however, a different type of lateral meristem, the BM, is not made in excess, and the double mutant is nearly unbranched. This difference can be explained by the fact that double mutants make several extra leaves (Fig. S4B). These extra leaves, however, do not simply form in place of tassel branches, because the number of extra leaves does not equal the number of missing tassel branches in the mutants. It is more likely that BM can only form when vegetative growth is suppressed, allowing the tassel to initiate floral structures exclusively. Because both ub2 and ub3 are expressed exclusively in the basal zone of tassels, where BMs are made (Fig. 4G), it is possible that they function to suppress vegetative leaf initiation in that region. These observations suggest that highly branched inflorescences characteristic of panicoid grasses can only be obtained if vegetative leaf initiation is suppressed during the floral phase by SBP-box genes.
Because ub2 and ub3 affect TBN and ERN, two traits that directly affect yield, polymorphisms in these genes were tested for association with agronomically important inflorescence traits. A previous study using the NAM population showed the presence of ub3-linked QTL for TBN and ERN (19). In addition, a previous QTL study in a B73-Mo17 biparental family identified a TBN QTL that mapped close to ub3 (29). A more detailed analysis revealed the existence of two derived polymorphisms in the 3′-end of UB3 that are not present in teosinte, the progenitor of maize, which possesses two-rowed ears or several other panicoid grass orthologs (Fig. 5B). These two polymorphisms in the ub3 gene are associated with distinct effects on male and female inflorescences: the Val260Met mutation associates with higher TBN in inbred lines, such as Mo17, whereas the Ser220Asn mutation associates with higher ERN in inbreds, such as B73 (Fig. 5C). Because both TBN and ERN are yield-related traits in maize, the physical proximity of the candidate SNPs for these traits in ub3 (119 bp apart) could give rise to pseudooverdominance in hybrids that combine two advantageous alleles (e.g., B73 and Mo17). It is unclear at this time how these B73 and Mo17 polymorphisms might affect UB3 function. Western blots comparing B73 and Mo17 ear nuclear extracts appeared similar (Fig. 1D), indicating that the polymorphisms may affect protein function as opposed to protein amount or stability.
The potential for ub3 to be used as a tool to improve the agronomic properties of crop plants is supported by the function of its ortholog in rice, WEALTHY FARMER’S PANICLE (WFP)/IDEAL PLANT ARCHITECTURE (IPA) (Fig. S2). Both WFP and IPA are dominant gain-of-function alleles that improve grain yield in rice by reducing tillering and increasing panicle branching (30, 31), phenotypes that are the opposite of the ub2/ub3/tsh4 loss-of-function phenotype. IPA causes overexpression of OsSPL14 through a mutation in the miR156-binding site that releases the gene from negative regulation by the microRNA (30). Extrapolating from the loss-of-function phenotypes of ub2 and ub3, it is possible that overexpression of these SBP-box genes may delay lateral primordia initiation and differentiation, leaving more time for the meristem to renew and regenerate. After the floral transition, more meristem cells would then be available to produce a more complex inflorescence, thus leading to higher yield. Consequently, these SBP-box genes could be important tools with which to alter plant architecture and theoretically, increase yields in a variety of crop plants.
Materials and Methods
Isolation of ub2 and ub3 and Genetic Analysis.
All ub2 and ub3 alleles were isolated from pools of Mu transposon-mutagenized populations and screened by PCR with Mu-end oligos and either ub2 or ub3 oligos. The ub2-mum1 and ub3-mum1 alleles were introgressed into the W22 background by backcrossing at least two times and then selfing; ub2 and ub3 were followed by PCR using the Mu-end primer Mu9242, the 5F1 primer (TAAAGAAGAGCTGCCGCAAACGCCT) for ub2, and the 7F1 primer (TGCTGGATTTCTCATACCCAAGG) for ub3. Triple mutants were made by crossing the single mutants to tsh4-M1 in the W22 background, selfing, and scoring by PCR. All tassel, leaf, and tillering phenotypes were scored in the field.
Generation of a UB3 Antibody and Immunolocalization.
Full-length UB3 protein coupled to an histidine tag was made into the pET21d expression vector (Novagen). Recombinant protein was isolated under denaturing conditions, dialyzed, and injected into two guinea pigs (Cocalico). The serum was affinity-purified using full-length UB3 protein coupled to a GST tag (Novagen). Immunolocalization was performed as previously described (8).
SEM.
Samples were fixed in formaldehyde acetic acid overnight, critical point-dried, sputter-coated, and examined as previously described (8).
Association of ub3 SNPs with ERN and TBN in the NAM and IBM Populations.
Association analysis using NAM and IBM populations is described in detail in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Katsutoshi Tsuda, Michael Lewis, and China Lunde for critical reading of the manuscript. This work was supported by National Science Foundation Grants PGRP- 1339332 (to G.S.C.) and PGR-1238030 (to P.J.B.) and Binational Agricultural Research and Development Grant Bard IS-4536-12C (to S.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1407401112/-/DCSupplemental.
References
- 1.Steeves TA, Sussex IM. Patterns in Plant Development. 2nd Ed Cambridge Univ Press; Cambridge, United Kingdom: 1989. [Google Scholar]
- 2.Veit B, Briggs SP, Schmidt RJ, Yanofsky MF, Hake S. Regulation of leaf initiation by the terminal ear 1 gene of maize. Nature. 1998;393(6681):166–168. doi: 10.1038/30239. [DOI] [PubMed] [Google Scholar]
- 3.Kawakatsu T, et al. PLASTOCHRON2 regulates leaf initiation and maturation in rice. Plant Cell. 2006;18(3):612–625. doi: 10.1105/tpc.105.037622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miyoshi K, et al. PLASTOCHRON1, a timekeeper of leaf initiation in rice, encodes cytochrome P450. Proc Natl Acad Sci USA. 2004;101(3):875–880. doi: 10.1073/pnas.2636936100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang JW, Schwab R, Czech B, Mica E, Weigel D. Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell. 2008;20(5):1231–1243. doi: 10.1105/tpc.108.058180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chuck G, Cigan AM, Saeteurn K, Hake S. The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat Genet. 2007;39(4):544–549. doi: 10.1038/ng2001. [DOI] [PubMed] [Google Scholar]
- 7.Wu G, Poethig RS. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development. 2006;133(18):3539–3547. doi: 10.1242/dev.02521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chuck G, Whipple C, Jackson D, Hake S. The maize SBP-box transcription factor encoded by tasselsheath4 regulates bract development and the establishment of meristem boundaries. Development. 2010;137(8):1243–1250. doi: 10.1242/dev.048348. [DOI] [PubMed] [Google Scholar]
- 9.Bortiri E, et al. ramosa2 encodes a LATERAL ORGAN BOUNDARY domain protein that determines the fate of stem cells in branch meristems of maize. Plant Cell. 2006;18(3):574–585. doi: 10.1105/tpc.105.039032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chuck G, Bortiri E. The unique relationship between tsh4 and ra2 in patterning floral phytomers. Plant Signal Behav. 2010;5(8):979–981. doi: 10.4161/psb.5.8.12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bossinger G, Rohde W, Lundquist U, Salamini F. Genetics of barley development: Mutant phenotypes and molecular aspects. In: Shewry PR, editor. Barley: Genetics, Biochemistry, Molecular Biology and Biotechnology. Alden Press Ltd; Oxford: 1992. pp. 231–264. [Google Scholar]
- 12.Bensen RJ, et al. Cloning and characterization of the maize An1 gene. Plant Cell. 1995;7(1):75–84. doi: 10.1105/tpc.7.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith LG, Jackson D, Hake S. The expression of Knotted1 marks shoot meristem formation during maize embryogenesis. Dev Genet. 1995;16:344–348. [Google Scholar]
- 14.Smith LG, Greene B, Veit B, Hake S. A dominant mutation in the maize homeobox gene, Knotted-1, causes its ectopic expression in leaf cells with altered fates. Development. 1992;116(1):21–30. doi: 10.1242/dev.116.1.21. [DOI] [PubMed] [Google Scholar]
- 15.Jackson D, Veit B, Hake S. Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development. 1994;120(2):405–413. [Google Scholar]
- 16.Vollbrecht E, Reiser L, Hake S. Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene, knotted1. Development. 2000;127(14):3161–3172. doi: 10.1242/dev.127.14.3161. [DOI] [PubMed] [Google Scholar]
- 17.Kerstetter RA, Laudencia-Chingcuanco D, Smith LG, Hake S. Loss-of-function mutations in the maize homeobox gene, knotted1, are defective in shoot meristem maintenance. Development. 1997;124(16):3045–3054. doi: 10.1242/dev.124.16.3045. [DOI] [PubMed] [Google Scholar]
- 18.McMullen MD, et al. Genetic properties of the maize nested association mapping population. Science. 2009;325(5941):737–740. doi: 10.1126/science.1174320. [DOI] [PubMed] [Google Scholar]
- 19.Brown PJ, et al. Distinct genetic architectures for male and female inflorescence traits of maize. PLoS Genet. 2011;7(11):e1002383. doi: 10.1371/journal.pgen.1002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chia JM, et al. Maize HapMap2 identifies extant variation from a genome in flux. Nat Genet. 2012;44(7):803–807. doi: 10.1038/ng.2313. [DOI] [PubMed] [Google Scholar]
- 21.Brachi B, Morris GP, Borevitz JO. Genome-wide association studies in plants: The missing heritability is in the field. Genome Biol. 2011;12(10):232. doi: 10.1186/gb-2011-12-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee M, et al. Expanding the genetic map of maize with the intermated B73 x Mo17 (IBM) population. Plant Mol Biol. 2002;48(5-6):453–461. doi: 10.1023/a:1014893521186. [DOI] [PubMed] [Google Scholar]
- 23.Wills DM, et al. From many, one: Genetic control of prolificacy during maize domestication. PLoS Genet. 2013;9(6):e1003604. doi: 10.1371/journal.pgen.1003604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang JW, Czech B, Weigel D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell. 2009;138(4):738–749. doi: 10.1016/j.cell.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 25.Schwarz S, Grande AV, Bujdoso N, Saedler H, Huijser P. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol Biol. 2008;67(1-2):183–195. doi: 10.1007/s11103-008-9310-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang L, Xu M, Koo Y, He J, Poethig RS. Sugar promotes vegetative phase change in Arabidopsis thaliana by repressing the expression of MIR156A and MIR156C. eLife. 2013;2:e00260. doi: 10.7554/eLife.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu S, et al. Sugar is an endogenous cue for juvenile-to-adult phase transition in plants. eLife. 2013;2:e00269. doi: 10.7554/eLife.00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Snow R. The young leaf as the inhibiting organ. New Phytol. 1929;28:345–348. [Google Scholar]
- 29.Mickelson S, Stuber CW, Senior L, Kaeppler SM. Quantitative trait loci controlling leaf and tassel traits in a B73 x MO17 population of maize. Crop Sci. 2002;42:1902–1909. [Google Scholar]
- 30.Jiao Y, et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet. 2010;42(6):541–544. doi: 10.1038/ng.591. [DOI] [PubMed] [Google Scholar]
- 31.Miura K, et al. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet. 2010;42(6):545–549. doi: 10.1038/ng.592. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.