Significance
The blood protein von Willebrand factor (VWF) is required for platelets to stop bleeding at sites of injury, and the metalloprotease ADAMTS13 limits platelet adhesion by cleaving VWF only when flowing blood stretches it, especially within a growing thrombus. This feedback inhibition is essential because ADAMTS13 deficiency causes fatal microvascular thrombosis. How ADAMTS13 recognizes VWF so specifically is not understood. We now find that ADAMTS13 is folded roughly in half so that its distal domains inhibit the metalloprotease domain. VWF relieves this autoinhibition and promotes its own destruction by allosterically activating ADAMTS13. Thus, VWF is both a substrate and a cofactor in this critical regulatory process.
Keywords: hemostasis, metalloproteases, allosteric regulation
Abstract
The metalloprotease ADAMTS13 cleaves von Willebrand factor (VWF) within endovascular platelet aggregates, and ADAMTS13 deficiency causes fatal microvascular thrombosis. The proximal metalloprotease (M), disintegrin-like (D), thrombospondin-1 (T), Cys-rich (C), and spacer (S) domains of ADAMTS13 recognize a cryptic site in VWF that is exposed by tensile force. Another seven T and two complement C1r/C1s, sea urchin epidermal growth factor, and bone morphogenetic protein (CUB) domains of uncertain function are C-terminal to the MDTCS domains. We find that the distal T8-CUB2 domains markedly inhibit substrate cleavage, and binding of VWF or monoclonal antibodies to distal ADAMTS13 domains relieves this autoinhibition. Small angle X-ray scattering data indicate that distal T-CUB domains interact with proximal MDTCS domains. Thus, ADAMTS13 is regulated by substrate-induced allosteric activation, which may optimize VWF cleavage under fluid shear stress in vivo. Distal domains of other ADAMTS proteases may have similar allosteric properties.
After vascular injury, platelets adhere to von Willebrand factor (VWF) multimers bound to endothelial cell surfaces and connective tissue. The force of flowing blood on a growing platelet-rich thrombus stretches the central A2 domain of VWF and exposes a Tyr1605-Met1606 cleavage site for ADAMTS13 (Fig. 1A) (1–5), a metalloprotease that severs VWF and releases adherent platelets. Deficiency of ADAMTS13 disrupts this feedback regulatory mechanism and causes thrombotic thrombocytopenic purpura (TTP), which is characterized by life-threatening microvascular thrombosis (3, 6, 7).
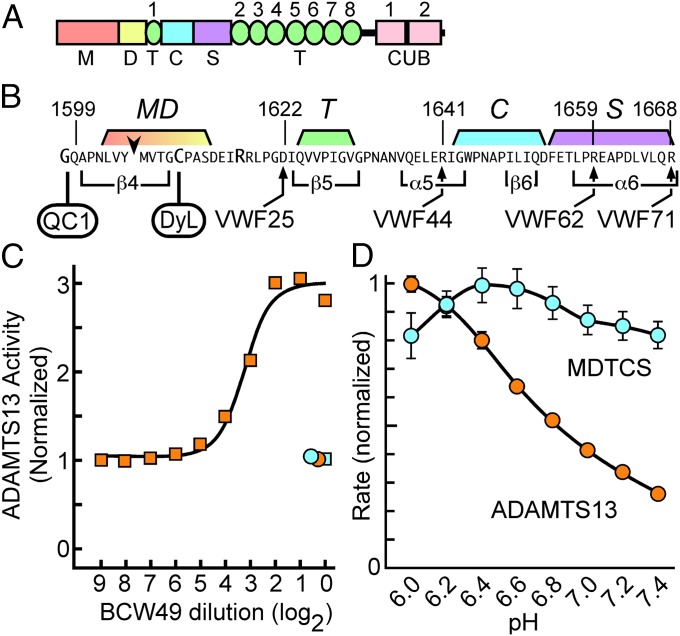
Fig. 1.
Activation of ADAMTS13 by autoantibodies from a patient with TTP or by low pH. (A) Structure of ADAMTS13. (B) Fluorogenic substrates terminate at VWF residues indicated by arrows. Each substrate has Lys1617 replaced with Arg, N-terminal Gly modified with IRDye QC-1 (QC1), and Asn1610 replaced by Cys and modified with DyLight 633 (DyL) (22). The arrowhead indicates the cleaved Tyr-Met bond. Secondary structure elements of the VWF A2 domain (11) are indicated below and segments that interact with specific ADAMTS13 domains (13) are indicated above the sequence. (C) BCW49 plasma activated ADAMTS13 with a titer of 9.6 U at pH 7.4 (orange squares), but not at pH 6.0 (orange circle). BCW49 plasma did not activate MDTCS at pH 6 (blue circle) or pH 7.4 (blue circle). (D) Rates of VWF71 cleavage were determined as a function of pH for ADAMTS13 (orange circles) and MDTCS (blue circles). Error bars indicate 95% confidence intervals and if not shown are smaller than the symbols.
The recognition and cleavage of VWF is a formidable challenge. VWF and ADAMTS13 occur at ∼10 µg/mL and ∼1 µg/mL, respectively, compared with total plasma protein of ∼80,000 µg/mL. ADAMTS13 is constitutively active and has no known inhibitors in vivo. Nevertheless, VWF is the only identified ADAMTS13 substrate, and VWF is resistant to cleavage until subjected to fluid shear stress (8), adsorbed on a surface (9), or treated with denaturants (8, 10). This specificity depends on structural features of both ADAMTS13 and VWF that have not been characterized fully.
The proximal metalloprotease (M), disintegrin-like (D), thrombospondin-1 (T), Cys-rich (C), and spacer (S) domains domains of ADAMTS13 bind to cryptic sites that are uncovered by unfolding VWF domain A2 (11-15) (Fig. 1B), and these interactions are required for efficient cleavage of VWF or peptide substrates. More distal ADAMTS13 domains bind to sites in or near VWF domain D4 that are always available (16–18). Deletion of distal ADAMTS13 domains impairs the cleavage of VWF multimers in vitro (16, 19) and increases VWF-dependent microvascular thrombosis in vivo (20) but accelerates the cleavage of peptide substrates (12, 13). In addition, ADAMTS13 cleaves guanidine hydrochloride-treated VWF multimers with an apparent Km of ∼15 nM (21), which is 100-fold lower than the Km of ∼1.6–1.7 µM for peptide substrates that are based on the sequence of VWF domain A2 (12, 14). These striking differences suggest that distal T or complement c1r/c1s, sea urchin epidermal growth factor, and bone morphogenetic protein (CUB) domains regulate ADAMTS13 activity. We have now shown that these distal domains inhibit ADAMTS13, and binding to VWF relieves this autoinhibition.
Results
Activation of ADAMTS13 by Antibodies and Low pH.
Evidence for allosteric regulation was obtained unexpectedly in the course of analyzing plasma samples from patients with TTP. The majority of adult patients with acquired TTP have autoantibodies that inhibit ADAMTS13 and reduce its activity in plasma to <5% of normal, but one patient proved to be a remarkable exception. When assayed with a fluorogenic ADAMTS13 substrate, VWF71 (Fig. 1B) (22), patient BCW49 had high-titer autoantibodies that paradoxically activated exogenous ADAMTS13 ∼threefold (Fig. 1C). Activation occurred at pH 7.4, which is characteristic of blood, but not at pH 6, which is used routinely for clinical ADAMTS13 assays (23). Furthermore, BCW49 plasma had no effect on the activity of MDTCS at either pH 7.4 or pH 6.
The loss of pH dependence for MDTCS suggested a regulatory function for the distal domains that are missing from this truncated ADAMTS13 construct. We (22) and others (23, 24) have observed that full-length ADAMTS13 is most active at pH 6, with markedly decreased activity at pH 7.4 (Fig. 1D and Fig. S1). This phenomenon has been attributed to ionization of a Zn2+-bound water molecule in the active site (24). However, MDTCS had approximately the same activity at both pH values, which excludes titration of the active site or the substrate. Instead, distal domains mediate the autoinhibition of ADAMTS13 at physiological pH. The protonation of specific residues at pH 6 alters critical interactions between distal and proximal ADAMTS13 domains and relieves this autoinhibition (Fig. 1D). The autoantibodies of patient BCW49 also can induce an activated conformation of ADAMTS13 even at physiological pH (Fig. 1C).
Allosteric Activation of ADAMTS13 by VWF.
We screened anti-ADAMTS13 monoclonal antibodies for properties similar to those of BCW49 plasma and identified several that increased the rate of VWF71 cleavage (Fig. 2A). When combined at saturating concentrations (Fig. S2), certain antibodies that recognized T67, T8, and CUB domains had additive effects and activated ADAMTS13 up to ∼4.2-fold at pH 7.4 (Fig. 2A), which is comparable to the magnitude of ADAMTS13 activation achieved by changing to pH 6 (Fig. 1D). These antibodies had markedly decreased effects on ADAMTS13 activity at pH 6 (Fig. S2). Autoantibodies in BCW49 plasma also bound to the CUB domains (Fig. 2B). These data suggest that specific distal T and/or CUB domains contribute to autoinhibition of ADAMTS13 at physiological pH.
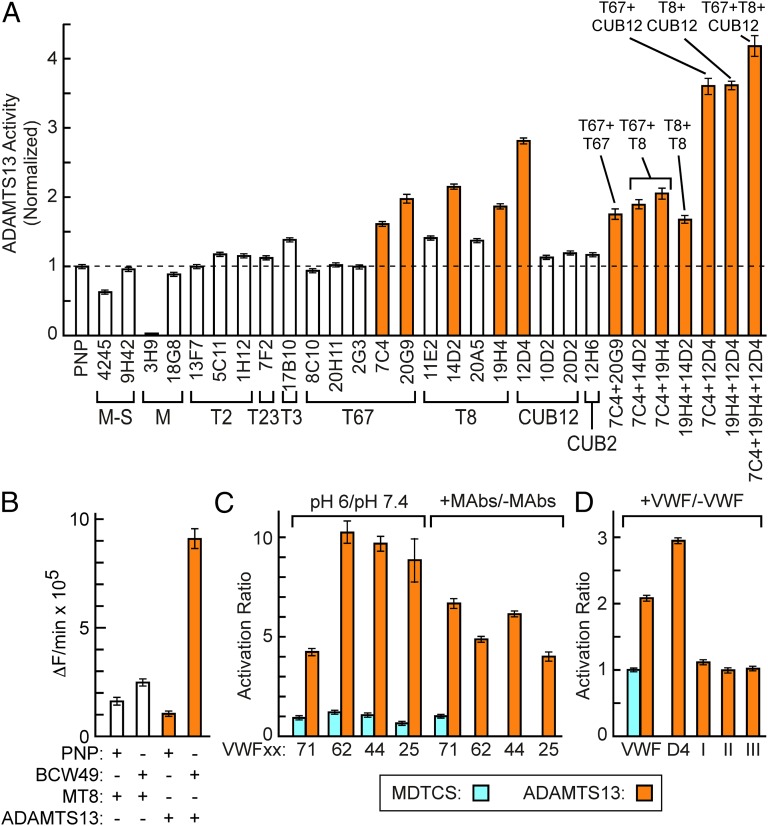
Fig. 2.
Allosteric activation of ADAMTS13. (A) Monoclonal anti-ADAMTS13 antibodies were added to ADAMTS13 assays with pooled normal plasma (PNP). Reaction rates were normalized to the rate with PNP only. The location of epitopes is indicated. Activation values >1.5× control (orange bars) are significantly different from the PNP reference condition (P < 0.05). (B) Recombinant ADAMTS13 and MT8 were immunoprecipitated with antibodies in PNP or BCW49 plasma, and the recovered enzyme was assayed with substrate VWF71. (C and D) MDTCS (blue bars) and ADAMTS13 (orange bars) were assayed with the indicated fluorogenic substrates (Fig. 1B). (C) Bars indicate the ratio of reaction rates at pH 6 and pH 7.4 (Left), or the ratio of reaction rates with and without activating monoclonal antibodies 7C4, 19H4, and 12D4 (Right). (D) Plasma VWF multimers (0.45 µM) or VWF D4 monomers (40 µM) increased the activity of ADAMTS13 toward VWF71. SPI (2 µM), SPII (2 µM), and SPIII (0.54 µM) fragments of VWF, which are cleaved within the D4 domain by V8 protease, had little or no effect on ADAMTS13 activity. Error bars indicate 95% confidence intervals.
Only some monoclonal antibodies that recognize a particular distal domain can activate ADAMTS13 (Fig. 2A), and these differences correlate with epitope specificity. For example, anti-T67 antibody 7C4 activates ADAMTS13 but 8C10 does not, and these antibodies bind distinct sites on ADAMTS13. Activating anti-T8 antibodies 14D2 and 19H4 bind competitively to ADAMTS13 at a site distinct from that recognized by nonactivating anti-T8 antibodies 11E2 and 20A5. Finally, anti-CUB antibodies 12D4 and 12H6 recognize different epitopes, but 12D4 activates ADAMTS13, whereas 12H6 does not (Fig. 2A). These results suggest that activating antibodies bind and stabilize an activated ADAMTS conformation, whereas nonactivating antibodies show no preference for such a conformation.
Several proximal ADAMTS13 domains participate in recognition of the extended VWF A2 domain that contains the scissile Tyr1605-Met1606 bond (12–15) (Fig. 1B). We prepared a series of fluorogenic substrates to determine whether activation involved specific proximal ADAMTS13 domains that engage distinct segments of VWF (Fig. 1B). As expected (13), the cleavage rate decreased as the substrate length decreased. However, the magnitude of activation by low pH (Fig. 2C) increased to ∼ninefold even for the shortest substrate VWF25, even though VWF25 interacts only with the MD domains of ADAMTS13 (13). The activation ratios, which correspond to the change in free energy of transition state stabilization (∆∆GT) (25), do not depend strongly on substrate length. In addition, antibodies that increased the cleavage of fluorogenic substrates by ADAMTS13 had no effect on MDTCS (Fig. 2C).
More importantly, adding VWF or recombinant VWF D4 markedly increased fluorogenic substrate cleavage (Fig. 2D). Staphylococcal V8 protease cleaves VWF after Glu2129 between the VWD4 and C8-4 subdomains of D4 and more slowly after Glu1673 between the A2 and A3 domains (26) (Fig. S3). These proteolytic fragments—SPI, SPII, and SPIII—had little effect on ADAMTS13 (Fig. 2D), suggesting that a functional ADAMTS13 binding site requires cooperation between motifs on both sides of the V8 cleavage site in the D4 domain.
These results indicate that the VWF D4 domain binds distal ADAMTS13 domains and allosterically activates the N-terminal M domain. The interaction of these distal domains with proximal MDTCS domains might be expected to reduce the affinity of ADAMTS13 binding to VWF A2 domain sequences that contain the scissile Tyr-Met bond, and this prediction was confirmed by using ADAMTS13(E225Q) variants that can bind substrates normally but cannot cleave them (17) (Fig. S4). MDTCS bound to a chimeric GST-VWF73 construct with a Kd of 210 ± 30 nM (SE), whereas ADAMTS13 bound slightly less tightly with a Kd of 440 ± 50 nM (SE).
The mutation G1505E, which was identified in patients with von Willebrand disease type 2A, prevents complete folding of VWF domain A2, exposes the scissile bond, and allows ADAMTS13 to cleave it rapidly and completely in the absence of shear stress (27). MDTCS bound to VWF construct A1-CK(G1505E) but did not bind to wild-type A1-CK (with a folded A2 domain) or to construct D4-CK (with no A2 domain). In contrast, ADAMTS13 did bind to VWF constructs D4-CK and A1-CK, presumably at the D4 site. ADAMTS13 also bound to A1-CK(G1505E) with increased affinity, which is consistent with binding to both A2 and D4 sites (Fig. S4).
ADAMTS13 Distal Domains Facilitate VWF Cleavage Under Shear Stress.
The natural substrate of ADAMTS13, multimeric plasma VWF, is resistant to cleavage until subjected to fluid shear stress that exposes the cleavage site in the A2 domain. The time course of this reaction can be analyzed by vortexing plasma VWF with ADAMTS13 variants and detecting specific cleavage products by Western blotting (16, 17). Under these conditions, full-length ADAMTS13 cleaved VWF ∼1.7-fold faster than MDTCS (Fig. 3 A and C), even though ADAMTS13 is ∼fourfold less active toward small peptide substrates (Fig. 1). The preferential cleavage of VWF by ADAMTS13 is not explained by an effect of shear stress on the enzyme, because vortexing ADAMTS13 with VWF71 for 10 min increased product generation only 1.15 ± 0.07-fold (SD); this small effect of vortexing may be due to frictional heating of the reaction.
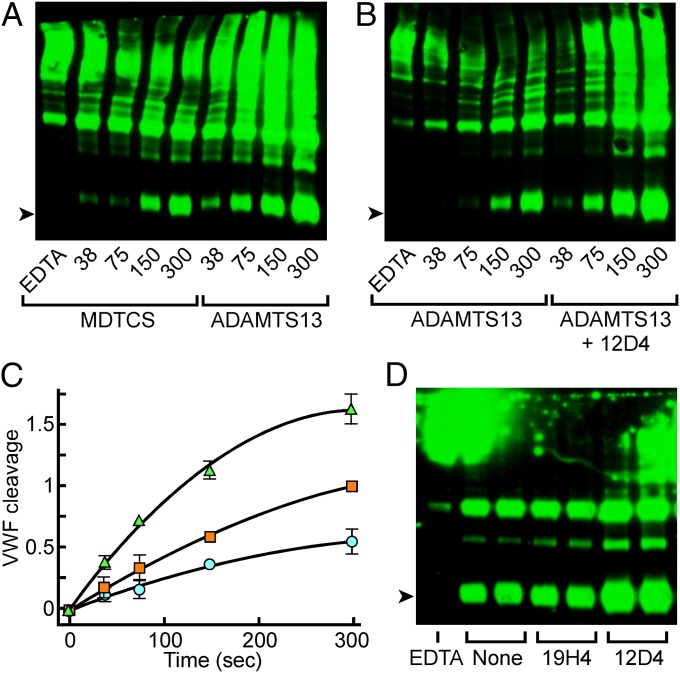
Fig. 3.
Cleavage of VWF by ADAMTS13 under fluid shear stress. VWF cleavage products were detected by SDS/PAGE and immunoblotting with anti-VWF. The arrowheads indicate the 350-kDa VWF fragment produced by ADAMTS13. Plasma VWF (40 nM) was sheared for the indicated times (seconds) with (A) MDTCS or ADAMTS13 (50 nM), or (B) with ADAMTS13 (50 nM) without or with monoclonal antibody 12D4 (40 µg/mL). (C) Densitometric analysis to compare the rate of VWF cleavage by ADAMTS13 without (orange squares) or with (green triangles) monoclonal antibody 12D4, or by MDTCS (blue circles). Values are normalized to cleavage at 300 s by ADAMTS13 alone and plotted as the mean ± SE for two experiments. (D) Duplicate reactions with ADAMTS13 were sheared for 300 s without or with monoclonal antibody 19H4 (100 µg/mL) or 12D4 (40 µg/mL). Control reactions were sheared for 300 s in the presence of 10 mM EDTA.
Anti-CUB antibody 12D4 increased the rate of VWF cleavage ∼twofold (Fig. 3 B and C). Thus, distal ADAMTS13 domains that bind VWF domain D4 confer an advantage for cleaving VWF that can be increased further by allosteric activation. Although anti-T8 antibody 19H4 also accelerates the cleavage of peptide substrates (Fig. 2A), it had no effect on the cleavage of VWF (Fig. 3D).
ADAMTS13 Distal Domains Are Adjacent to Its Proximal Domains.
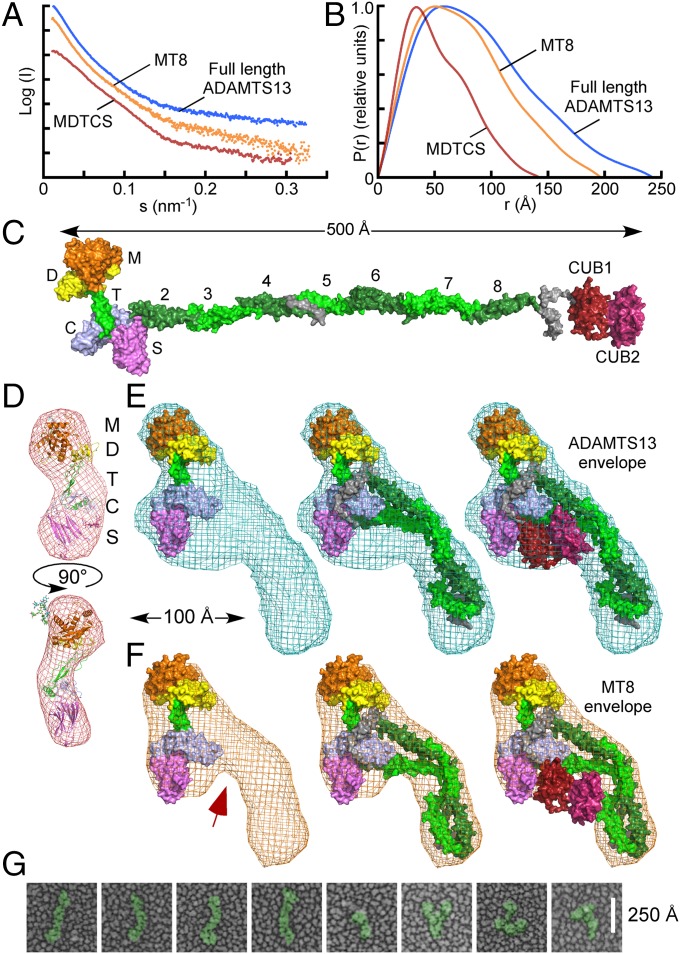
The regulation of ADAMTS13 metalloprotease activity by its distal T-CUB domains implies that proximal and distal domains are close enough to make contact, and small angle X-ray scattering (SAXS) data (Fig. 4A) indicate that ADAMTS13 does have such a structure. Pair distance distributions (Fig. 4B) show that MDTCS, MT8, and ADAMTS13 have maximum dimensions of ∼140 Å, ∼200 Å, and ∼230 Å, respectively. For comparison, a molecular model of ADAMTS13 has a maximum extended length of ∼500 Å (Fig. 4C). Agreement was excellent between an atomic model for MDTCS and experimental scattering profiles (χ = 1.86) (28) (Fig. 4A) or ab initio envelopes (Fig. 4D). The envelopes for full-length ADAMTS13 (Fig. 4E) are too compact for fully extended ADAMTS13 but readily accommodate a folded or condensed conformation. Specific flexible segments have not been identified, but the extended linkers after T4 and T8 are susceptible to proteolysis by thrombin, plasmin, or elastase (29, 30) and may act as hinges. Comparisons of SAXS envelopes for ADAMTS13 and construct MT8, which lacks both CUB domains, indicate that the CUB domains are adjacent to proximal ADAMTS13 domains and occupy a bulge in the envelope for ADAMTS13 (Fig. 4E) that is missing from the envelope for MT8, which cannot accommodate the CUB domains in this location (Fig. 4F). This folded conformation would enable distal T7-CUB domains to allosterically regulate ADAMTS13 by variably occluding the active site and substrate binding exosites. The occurrence of condensed ADAMTS13 is supported by quick-freeze deep-etch EM, which shows both elongated and folded conformations for ADAMTS13 (Fig. 4G).
Fig. 4.
ADAMTS13 structure by EM and SAXS. Scattering profiles (A) and pair distribution functions (B) for MDTCS, (red) MT8 (orange), and ADAMTS13 (blue). (C) Molecular model of ADAMTS13. (D–F) Ab initio envelopes calculated from scattering profiles for (D) MDTCS (red mesh), (E) ADAMTS13 (blue mesh), or (F) MT8 (orange mesh) superimposed on an atomic model of MDTCS (D, cartoon; E and F, surface), MT8, or ADAMTS13 (E and F, surface). The arrow indicates a difference between MT8 and ADAMTS13 maps that corresponds to the proposed location of CUB domains. (G) Quick-freeze deep-etch EM of ADAMTS13.
Discussion
Our results indicate that VWF promotes its own destruction by allosterically activating ADAMTS13, and this mechanism could have evolved to balance the competing needs to promote VWF-dependent hemostasis and prevent VWF-dependent thrombosis. In addition, the finding that VWF activates ADAMTS13 raises the possibility that other ADAMTS13 cofactors occur on cell surfaces, in blood, or in extracellular matrix.
For human ADAMTS13, binding to VWF D4 allosterically activates ADAMTS13, increasing its catalytic activity, but binding to VWF D4 also positions ADAMTS13 on the substrate where it can act rapidly when shear stress exposes the scissile bond in the adjacent A2 domain. These two mechanisms work together to focus proteolytic activity on VWF multimers. However, mouse ADAMTS13 may rely mainly on substrate binding rather than allostery.
Some mouse strains express a “long” Adamts13 that is similar to full-length human ADAMTS13. Other strains have an intracisternal A-particle (IAP) retrotransposon inserted in the Adamts13 gene and express a “short” Adamts13 that is truncated after the T6 (31). Long Adamts13 cleaves VWF multimers ninefold faster than short Adamts13, which is consistent with the role we propose for binding of distal domains to VWF D4 (19). However, long and short Adamts13 reportedly have similar activity toward small substrates that are similar to VWF71 (19, 31). Therefore, in contrast to human ADAMTS13, deletion of the distal T7-CUB2 domains may not activate mouse Adamts13, which suggests that allosteric activation is less significant than for human ADAMTS13 or depends on domains other than T7-CUB2. Further study is required to determine the relative importance of substrate binding and allosteric activation for ADAMTS13 in humans, mice, and other vertebrates.
The allosteric properties of ADAMTS13 are likely to have clinical relevance. Relatively modest decreases in ADAMTS13 activity seem to increase the risk of ischemic stroke and myocardial infarction (32–34). In animal models, ADAMTS13 deficiency promotes vascular inflammation and atherosclerosis (35). Therefore, inherited or acquired defects in the allosteric regulation of ADAMTS13 could inhibit VWF cleavage and contribute to thrombosis in these conditions.
Conversely, agents that block allosteric activation could be useful to treat bleeding caused by excessive proteolysis of VWF, which occurs mainly in two settings. Von Willebrand disease type 2A frequently is due to mutations like G1505E (27) that impair the folding of the VWF A2 domain and allow cleavage by ADAMTS13 even in the absence of fluid shear stress. As a result, the VWF multimers that circulate in the blood are too small for effective hemostasis (36, 37). Acquired von Willebrand syndrome also is characterized by deficiency of the largest VWF multimers, often as a result of pathologically increased fluid shear stress that promotes cleavage by ADAMTS13. For example, this type of acquired von Willebrand syndrome is very common in patients with implanted left ventricular assist devices, who experience recurrent major gastrointestinal bleeding as a result (38). Because ADAMTS13 can be allosteric activated by ∼fourfold to ∼10-fold, an inhibitor of allosteric activation could reduce ADAMTS13 activity to no lower than ∼10%, which is not low enough to cause TTP (6, 7) but may be useful to treat bleeding. The feasibility of this approach is supported by a study demonstrating that a monoclonal antibody against the VWF D4 domain inhibits ADAMTS13 binding and markedly reduces the excessive degradation of VWF multimers in an ex vivo model using a HeartMate II left ventricular assist device (39).
Like ADAMTS13, other ADAMTS proteases and related ADAMTSL proteins also have complex C-terminal structures with up to 15 T repeats and variable numbers of proteoglycan, a protease and lacunin (PLAC) domain, and Gon-1 modules that are likely to mediate regulatory protein–protein interactions (40, 41). For example, N-terminal procollagen processing depends on ADAMTS2, and defects in ADAMTS2 cause a variant of Ehlers-Danlos syndrome (41). Microfibrils regulate the activation of TGF-β, which is force-dependent (42), as is the processing of VWF by ADAMTS13 (8, 11). The biogenesis of microfibrils involves several ADAMTS and ADAMTSL proteins, and mutations in them cause developmental disorders such as Weill-Marchesani syndrome (ADAMTS10, ADAMTS17), geleophysic dysplasia (ADAMTSL2), and ectopia lentis (ADAMTS17, ADAMTSL4) (41, 43). The distal domains of these homologous proteins share many structural features with ADAMTS13 and may have similar allosteric properties that are essential for their biological functions.
Materials and Methods
Antibodies.
Mouse monoclonal antibodies to ADAMTS13 were made by DNA immunization with a plasmid encoding human ADAMTS13, i.p. injection of recombinant human ADAMTS13, and isolation of hybridomas by standard methods (44). Epitopes were localized and binding affinities determined by ELISAs (44) and immunoprecipitation (45) using a panel of ADAMTS13 constructs with C-terminal truncations or internal domain deletions.
Plasma samples from patients with TTP or healthy donors were obtained with informed consent under protocols approved by the institutional review board of Washington University or BloodCenter of Wisconsin.
ADAMTS13 Assays.
Fluorogenic peptide substrate FRETS-rVWF71 (VWF71) has an N-terminal Gly followed by VWF Gln1599-Arg1668 with mutations N1610C and K1617R. The N terminus is modified with IRDye QC-1 N-hydroxysuccinimide ester (Li-COR), and Cys1610 is modified with DyLight 633 maleimide (Thermo Scientific) (22). FRETS-rVWF25, FRETS-rVWF44, and FRETS-rVWF62 were prepared similarly except with C-terminal residues Asp1622, Arg1641, and Arg1659, respectively.
ADAMTS13 was assayed with fluorogenic substrates as previously described (22). Standard assays included 50 mM Bis-Tris (pH 6) or Hepes (pH 7.4), 150 mM NaCl, 10 mM CaCl2, 0.05% Tween-20, 1 mg/mL BSA, 1 µM fluorogenic substrate, and PNP or recombinant ADAMTS13 variants in a total volume of 200 µL. Reaction was initiated by adding buffer containing substrate to enzyme in 96-well white microplates (Optiplate-96, PerkinElmer) at 30 °C. Product generation was monitored as an increase in fluorescence using a Victor2V Multilabel Counter (PerkinElmer) or Synergy H1 Hybrid Multi-Mode Microplate Reader (Biotek) with 635 ± 10-nm excitation and 660 ± 10-nm emission filters. Assays by this method have an interassay coefficient of variation of <2% (22).
ADAMTS13 activity as a function of pH was assayed similarly, except the buffer included 20 mM Bis-Tris, 20 mM Hepes, and 20 mM Tris⋅HCl at pH values between pH 6 and pH 7.4 in increments of 0.2 pH unit.
Shear-induced cleavage of VWF was assayed as described previously (16, 17), with modifications. Reactions (30 µL total volume) were performed in 50 mM Hepes (pH 7.4), 150 mM NaCl, 5 mM CaCl2, and 1 mg/mL BSA at room temperature with plasma VWF (40 nM) and ADAMTS13 variants (50 nM) without or with 10 mM EDTA, without or with fluid shear stress, and with or without antibody 12D4 or 19H4. To be sure that ADAMTS13 was saturated with antibodies, the final concentrations of 19H4 (100 µg/mL) and 12D4 (40 µg/mL) were ∼20-fold higher than the EC50 for activating ADAMTS13 toward VWF71 (Fig. S2). Reactions were stopped by adding EDTA to 10 mM. Samples (1–5 µL) were mixed with 2× Tris-glycine SDS sample buffer, degassed, heated 10 min at 70 °C, and analyzed by electrophoresis on Novex 4% Tris-glycine SDS gels. Gels were immersed in PBS containing 1 mM β-mercaptoethanol for 10 min with gentle shaking, washed twice with PBS, and rinsed with water. Proteins were transferred to a PVDF membrane for 10 min at 23 V using an iBlot device (Thermo Scientific Life Technologies). The membrane was washed in PBS for 5 min, blocked in casein blocking buffer for 1 h, and incubated overnight in blocking buffer containing 1:2,500 HRP-labeled rabbit anti-human VWF antibody (P226, Dako). Labeled proteins were detected by chemiluminescence (WesternBright ECL HRP, Advansta) and analyzed with Image Studio 4.0 (LI-COR).
ADAMTS13 and VWF Proteins.
SI Materials and Methods includes detailed procedures for the further purification of human plasma VWF (Laboratoire Français du Fractionnement et des Biotechnologies, Lille, France), preparation of staphylococcal V8 protease fragments SPI, SPII, and SPIII, and the production of recombinant ADAMTS13 and VWF variants for enzymatic characterization and antibody epitope localization. The concentration of proteins was determined in BCA protein assays (Thermo Scientific Pierce) standardized with BSA.
Activation Assays.
Pooled normal Li+-heparin plasma (PNP) was prepared from ≥35 healthy donors. Plasma samples from patients with TTP were screened for activation of ADAMTS13 in assays (200 µL) with FRETS-rVWF71, 50 µL PNP, and 50 µL of plasma. Monoclonal antibodies were screened with 25 µL PNP and ∼2 µg of antibody per reaction. VWF and VWF fragments were added at varying concentrations. EC50 values for activators were determined from assays of serial dilutions and nonlinear regression (22). Differences between mean values were assessed with the unpaired Student t test.
Binding Assays.
Dissociation constants (Kd) were determined by biolayer interferometry (BLI) using an Octet RED96 (ForteBio). GST-VWF73 (12) was captured by biotinylated anti-GST antibody bound to streptavidin-biosensors. D4-CK, A1-CK, and A1-CK(G1505E) were captured on Ni-NTA biosensors. Binding to ADAMTS13 proteins was performed in 20 mM Hepes (pH 7.4), 150 mM NaCl, 2 mg/mL BSA, and 0.02% Tween 20. A buffer-only reference was subtracted from all curves. Affinities were determined from global kinetic analysis for a 1:1 binding model using Octet RED software, version 5.2.
Molecular Modeling.
An atomic model of MDTCS was constructed from the crystallographic structure of DTCS domains (15) and a homology model of MD domains (13) that was based on ADAMTS4 (46).
Full-length ADAMTS13 was built using HHpred (47) to model distal T domains, CUB domains, and linkers after T4 and T8. Templates for T domains included 1w0r, 3t5o, 3ghm, 3r6b, 1lsl, and 1vex. Templates for CUB domains included 2qqk, 3kq4, 1spp, 2qqm, and 2wno.
Electron Microscopy.
Deep-etch replicas were prepared by adsorbing ADAMTS13 on acid-cleaned glass chips, fixing with 2% glutaraldehyde, and rinsing with water before quick-freezing, freeze-drying, platinum-replication, and electron microscopy (48–50).
Small Angle X-ray Scattering.
SAXS data were collected for ADAMTS13 variants on the SIBYLS beamline 12.3.1.2 at the Advanced Light Source (ALS), a national user facility operated by the Lawrence Berkeley National Laboratory and supported by the Director, Office of Science, Office of Basic Energy Sciences, of the US Department of Energy under Contract DE-AC02-05CH11231 (51). All proteins were monodisperse and yielded similar scattering profiles at three or four concentrations between 0.5 mg/mL and 3.0 mg/mL. Pair distance distribution functions were obtained using DATGNOM (52).
The quality of the SAXS data was evaluated by comparison with an atomic model of MDTCS. The M domain was modeled (13) on ADAMTS4 (46), DTCS13 domains were from the corresponding crystal structure (15), and N-linked oligosaccharides were added with GLYPROT (53). This structure was compared with the experimental scattering profiles using CRYSOL (28), and agreement was excellent with χ = 1.86 (28). Models (envelopes) were generated from scattering profiles using DAMMIN (54), averaged (n = 15) using DAMAVER (55), and superimposed on the atomic structure of MDTCS using SUPCOMB (56).
Supplementary Material
Acknowledgments
We thank Bruce Linders (Washington University) for tissue culture to prepare ADAMTS13 variants, and Kevin Dyer and Dr. Gregory Hura (Lawrence Berkeley Laboratory) for assistance with SAXS data collection. This work was supported by National Institutes of Health Grants R01 HL72917, R01 HL89746, U54 HL112303, and T32 HL007088, and by the Fonds voor wetenschappelijk onderzoek Flanders Grant G.0584.11, Belgium.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1413282112/-/DCSupplemental.
References
- 1.Zheng X, et al. Structure of von Willebrand factor-cleaving protease (ADAMTS13), a metalloprotease involved in thrombotic thrombocytopenic purpura. J Biol Chem. 2001;276(44):41059–41063. doi: 10.1074/jbc.C100515200. [DOI] [PubMed] [Google Scholar]
- 2.Soejima K, et al. A novel human metalloprotease synthesized in the liver and secreted into the blood: Possibly, the von Willebrand factor-cleaving protease? J Biochem. 2001;130(4):475–480. doi: 10.1093/oxfordjournals.jbchem.a003009. [DOI] [PubMed] [Google Scholar]
- 3.Levy GG, et al. Mutations in a member of the ADAMTS gene family cause thrombotic thrombocytopenic purpura. Nature. 2001;413(6855):488–494. doi: 10.1038/35097008. [DOI] [PubMed] [Google Scholar]
- 4.Fujikawa K, Suzuki H, McMullen B, Chung D. Purification of human von Willebrand factor-cleaving protease and its identification as a new member of the metalloproteinase family. Blood. 2001;98(6):1662–1666. doi: 10.1182/blood.v98.6.1662. [DOI] [PubMed] [Google Scholar]
- 5.Gerritsen HE, Robles R, Lämmle B, Furlan M. Partial amino acid sequence of purified von Willebrand factor-cleaving protease. Blood. 2001;98(6):1654–1661. doi: 10.1182/blood.v98.6.1654. [DOI] [PubMed] [Google Scholar]
- 6.Furlan M, et al. von Willebrand factor-cleaving protease in thrombotic thrombocytopenic purpura and the hemolytic-uremic syndrome. N Engl J Med. 1998;339(22):1578–1584. doi: 10.1056/NEJM199811263392202. [DOI] [PubMed] [Google Scholar]
- 7.Tsai HM, Lian EC. Antibodies to von Willebrand factor-cleaving protease in acute thrombotic thrombocytopenic purpura. N Engl J Med. 1998;339(22):1585–1594. doi: 10.1056/NEJM199811263392203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsai HM. Physiologic cleavage of von Willebrand factor by a plasma protease is dependent on its conformation and requires calcium ion. Blood. 1996;87(10):4235–4244. [PubMed] [Google Scholar]
- 9.Majerus EM, Anderson PJ, Sadler JE. Binding of ADAMTS13 to von Willebrand factor. J Biol Chem. 2005;280(23):21773–21778. doi: 10.1074/jbc.M502529200. [DOI] [PubMed] [Google Scholar]
- 10.Furlan M, Robles R, Lämmle B. Partial purification and characterization of a protease from human plasma cleaving von Willebrand factor to fragments produced by in vivo proteolysis. Blood. 1996;87(10):4223–4234. [PubMed] [Google Scholar]
- 11.Zhang Q, et al. Structural specializations of A2, a force-sensing domain in the ultralarge vascular protein von Willebrand factor. Proc Natl Acad Sci USA. 2009;106(23):9226–9231. doi: 10.1073/pnas.0903679106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao W, Anderson PJ, Majerus EM, Tuley EA, Sadler JE. Exosite interactions contribute to tension-induced cleavage of von Willebrand factor by the antithrombotic ADAMTS13 metalloprotease. Proc Natl Acad Sci USA. 2006;103(50):19099–19104. doi: 10.1073/pnas.0607264104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao W, Anderson PJ, Sadler JE. Extensive contacts between ADAMTS13 exosites and von Willebrand factor domain A2 contribute to substrate specificity. Blood. 2008;112(5):1713–1719. doi: 10.1182/blood-2008-04-148759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zanardelli S, et al. ADAMTS13 substrate recognition of von Willebrand factor A2 domain. J Biol Chem. 2006;281(3):1555–1563. doi: 10.1074/jbc.M508316200. [DOI] [PubMed] [Google Scholar]
- 15.Akiyama M, Takeda S, Kokame K, Takagi J, Miyata T. Crystal structures of the noncatalytic domains of ADAMTS13 reveal multiple discontinuous exosites for von Willebrand factor. Proc Natl Acad Sci USA. 2009;106(46):19274–19279. doi: 10.1073/pnas.0909755106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang P, Pan W, Rux AH, Sachais BS, Zheng XL. The cooperative activity between the carboxyl-terminal TSP1 repeats and the CUB domains of ADAMTS13 is crucial for recognition of von Willebrand factor under flow. Blood. 2007;110(6):1887–1894. doi: 10.1182/blood-2007-04-083329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feys HB, Anderson PJ, Vanhoorelbeke K, Majerus EM, Sadler JE. Multi-step binding of ADAMTS-13 to von Willebrand factor. J Thromb Haemost. 2009;7(12):2088–2095. doi: 10.1111/j.1538-7836.2009.03620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zanardelli S, et al. A novel binding site for ADAMTS13 constitutively exposed on the surface of globular VWF. Blood. 2009;114(13):2819–2828. doi: 10.1182/blood-2009-05-224915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou W, Bouhassira EE, Tsai HM. An IAP retrotransposon in the mouse ADAMTS13 gene creates ADAMTS13 variant proteins that are less effective in cleaving von Willebrand factor multimers. Blood. 2007;110(3):886–893. doi: 10.1182/blood-2007-01-070953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banno F, et al. The distal carboxyl-terminal domains of ADAMTS13 are required for regulation of in vivo thrombus formation. Blood. 2009;113(21):5323–5329. doi: 10.1182/blood-2008-07-169359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson PJ, Kokame K, Sadler JE. Zinc and calcium ions cooperatively modulate ADAMTS13 activity. J Biol Chem. 2006;281(2):850–857. doi: 10.1074/jbc.M504540200. [DOI] [PubMed] [Google Scholar]
- 22.Muia J, et al. An optimized fluorogenic ADAMTS13 assay with increased sensitivity for the investigation of patients with thrombotic thrombocytopenic purpura. J Thromb Haemost. 2013;11(8):1511–1518. doi: 10.1111/jth.12319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kokame K, Nobe Y, Kokubo Y, Okayama A, Miyata T. FRETS-VWF73, a first fluorogenic substrate for ADAMTS13 assay. Br J Haematol. 2005;129(1):93–100. doi: 10.1111/j.1365-2141.2005.05420.x. [DOI] [PubMed] [Google Scholar]
- 24.Di Stasio E, et al. Mechanistic studies on ADAMTS13 catalysis. Biophys J. 2008;95(5):2450–2461. doi: 10.1529/biophysj.108.131532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wells JA. Additivity of mutational effects in proteins. Biochemistry. 1990;29(37):8509–8517. doi: 10.1021/bi00489a001. [DOI] [PubMed] [Google Scholar]
- 26.Girma JP, Chopek MW, Titani K, Davie EW. Limited proteolysis of human von Willebrand factor by Staphylococcus aureus V-8 protease: Isolation and partial characterization of a platelet-binding domain. Biochemistry. 1986;25(11):3156–3163. doi: 10.1021/bi00359a013. [DOI] [PubMed] [Google Scholar]
- 27.Hassenpflug WA, et al. Impact of mutations in the von Willebrand factor A2 domain on ADAMTS13-dependent proteolysis. Blood. 2006;107(6):2339–2345. doi: 10.1182/blood-2005-04-1758. [DOI] [PubMed] [Google Scholar]
- 28.Svergun DI, Barberato C, Koch MHJ. CRYSOL—a program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Cryst. 1995;28:768–773. [Google Scholar]
- 29.Crawley JT, et al. Proteolytic inactivation of ADAMTS13 by thrombin and plasmin. Blood. 2005;105(3):1085–1093. doi: 10.1182/blood-2004-03-1101. [DOI] [PubMed] [Google Scholar]
- 30.Hiura H, et al. Proteolytic fragmentation and sugar chains of plasma ADAMTS13 purified by a conformation-dependent monoclonal antibody. J Biochem. 2010;148(4):403–411. doi: 10.1093/jb/mvq075. [DOI] [PubMed] [Google Scholar]
- 31.Banno F, Kaminaka K, Soejima K, Kokame K, Miyata T. Identification of strain-specific variants of mouse Adamts13 gene encoding von Willebrand factor-cleaving protease. J Biol Chem. 2004;279(29):30896–30903. doi: 10.1074/jbc.M314184200. [DOI] [PubMed] [Google Scholar]
- 32.Crawley JT, Lane DA, Woodward M, Rumley A, Lowe GD. Evidence that high von Willebrand factor and low ADAMTS-13 levels independently increase the risk of a non-fatal heart attack. J Thromb Haemost. 2008;6(4):583–588. doi: 10.1111/j.1538-7836.2008.02902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bongers TN, et al. Lower levels of ADAMTS13 are associated with cardiovascular disease in young patients. Atherosclerosis. 2009;207(1):250–254. doi: 10.1016/j.atherosclerosis.2009.04.013. [DOI] [PubMed] [Google Scholar]
- 34.Andersson HM, et al. High VWF, low ADAMTS13, and oral contraceptives increase the risk of ischemic stroke and myocardial infarction in young women. Blood. 2012;119(6):1555–1560. doi: 10.1182/blood-2011-09-380618. [DOI] [PubMed] [Google Scholar]
- 35.Gandhi C, Khan MM, Lentz SR, Chauhan AK. ADAMTS13 reduces vascular inflammation and the development of early atherosclerosis in mice. Blood. 2012;119(10):2385–2391. doi: 10.1182/blood-2011-09-376202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ginsburg D, Bowie EJW. Molecular genetics of von Willebrand disease. Blood. 1992;79(10):2507–2519. [PubMed] [Google Scholar]
- 37.Nichols WL, et al. von Willebrand disease (VWD): Evidence-based diagnosis and management guidelines, the National Heart, Lung, and Blood Institute (NHLBI) Expert Panel report (USA) Haemophilia. 2008;14(2):171–232. doi: 10.1111/j.1365-2516.2007.01643.x. [DOI] [PubMed] [Google Scholar]
- 38.Suarez J, et al. Mechanisms of bleeding and approach to patients with axial-flow left ventricular assist devices. Circ Heart Fail. 2011;4(6):779–784. doi: 10.1161/CIRCHEARTFAILURE.111.962613. [DOI] [PubMed] [Google Scholar]
- 39.Rauch A, et al. Antibody-based prevention of von Willebrand factor degradation mediated by circulatory assist devices. Thromb Haemost. 2014;112(5):1014–1023. doi: 10.1160/TH14-02-0148. [DOI] [PubMed] [Google Scholar]
- 40.Apte SS. A disintegrin-like and metalloprotease (reprolysin-type) with thrombospondin type 1 motif (ADAMTS) superfamily: functions and mechanisms. J Biol Chem. 2009;284(46):31493–31497. doi: 10.1074/jbc.R109.052340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Le Goff C, Cormier-Daire V. The ADAMTS(L) family and human genetic disorders. Hum Mol Genet. 2011;20(R2):R163–R167. doi: 10.1093/hmg/ddr361. [DOI] [PubMed] [Google Scholar]
- 42.Shi M, et al. Latent TGF-β structure and activation. Nature. 2011;474(7351):343–349. doi: 10.1038/nature10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hubmacher D, Apte SS. Genetic and functional linkage between ADAMTS superfamily proteins and fibrillin-1: A novel mechanism influencing microfibril assembly and function. Cell Mol Life Sci. 2011;68(19):3137–3148. doi: 10.1007/s00018-011-0780-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feys HB, et al. ADAMTS-13 plasma level determination uncovers antigen absence in acquired thrombotic thrombocytopenic purpura and ethnic differences. J Thromb Haemost. 2006;4(5):955–962. doi: 10.1111/j.1538-7836.2006.01833.x. [DOI] [PubMed] [Google Scholar]
- 45.Feys HB, et al. Thrombotic thrombocytopenic purpura directly linked with ADAMTS13 inhibition in the baboon (Papio ursinus) Blood. 2010;116(12):2005–2010. doi: 10.1182/blood-2010-04-280479. [DOI] [PubMed] [Google Scholar]
- 46.Mosyak L, et al. Crystal structures of the two major aggrecan degrading enzymes, ADAMTS4 and ADAMTS5. Protein Sci. 2008;17(1):16–21. doi: 10.1110/ps.073287008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33(Web Server issue):W244-8. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heuser JE, et al. Synaptic vesicle exocytosis captured by quick freezing and correlated with quantal transmitter release. J Cell Biol. 1979;81(2):275–300. doi: 10.1083/jcb.81.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heuser JE. Procedure for freeze-drying molecules adsorbed to mica flakes. J Mol Biol. 1983;169(1):155–195. doi: 10.1016/s0022-2836(83)80179-x. [DOI] [PubMed] [Google Scholar]
- 50.Heuser J. Protocol for 3-D visualization of molecules on mica via the quick-freeze, deep-etch technique. J Electron Microsc Tech. 1989;13(3):244–263. doi: 10.1002/jemt.1060130310. [DOI] [PubMed] [Google Scholar]
- 51.Hura GL, et al. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nat Methods. 2009;6(8):606–612. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Cryst. 1992;25(4):495–503. [Google Scholar]
- 53.Bohne-Lang A, von der Lieth CW. GlyProt: In silico glycosylation of proteins. Nucleic Acids Res. 2005;33(Web Server issue):W214-9. doi: 10.1093/nar/gki385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J. 1999;76(6):2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J Appl Cryst. 2003;36(3-1):860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kozin MB, Svergun DI. Automated matching of high- and low-resolution structural models. J Appl Cryst. 2001;34(1):33–41. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.